

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0369a.2
(1602 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
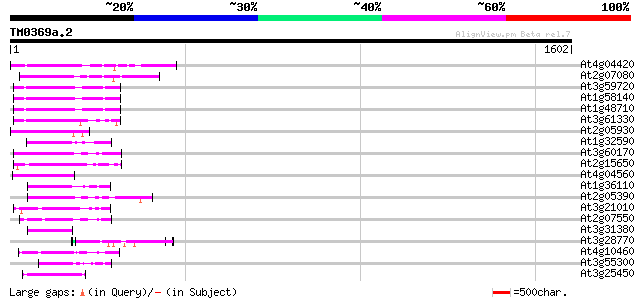
Score E
Sequences producing significant alignments: (bits) Value
At4g04420 putative transposon protein 131 4e-30
At2g07080 putative gag-protease polyprotein 112 2e-24
At3g59720 copia-type reverse transcriptase-like protein 94 8e-19
At1g58140 hypothetical protein 92 2e-18
At1g48710 hypothetical protein 92 3e-18
At3g61330 copia-type polyprotein 91 5e-18
At2g05930 copia-like retroelement pol polyprotein 86 2e-16
At1g32590 hypothetical protein, 5' partial 82 2e-15
At3g60170 putative protein 79 3e-14
At2g15650 putative retroelement pol polyprotein 78 5e-14
At4g04560 putative transposon protein 73 2e-12
At1g36110 hypothetical protein 71 4e-12
At2g05390 putative retroelement pol polyprotein 70 8e-12
At3g21010 unknown protein 68 4e-11
At2g07550 putative retroelement pol polyprotein 61 6e-09
At3g31380 hypothetical protein 60 1e-08
At3g28770 hypothetical protein 58 5e-08
At4g10460 putative retrotransposon 57 9e-08
At3g55300 putative protein 55 3e-07
At3g25450 hypothetical protein 54 7e-07
>At4g04420 putative transposon protein
Length = 1008
Score = 131 bits (329), Expect = 4e-30
Identities = 115/503 (22%), Positives = 228/503 (44%), Gaps = 66/503 (13%)
Query: 1 LESFFLGFDADLWDIIVDGYERPV--DTDGKKIPRSE---MTADQKKLYSQHHKARAILL 55
+ + G + W G++ PV DG+ + +++ A++ K + + +A +++
Sbjct: 28 MRALIRGLGKEAWIATSIGWKAPVIKGEDGEDVLKTKDQWNDAEEAKAKA-NSRALSLIF 86
Query: 56 SAISYEEYQKITDREFAKGIFDSLKMSHEGNKKVKESKALSLIQKYESFIMEPNESIEEM 115
+ ++ ++++I + E AK +D L ++EG VK S+ L ++E+ ME E+IEE
Sbjct: 87 NFVNQNQFKRIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQFENLSMEETENIEEF 146
Query: 116 FSRFQLLVAGIRPLNKSYTTKDHVIRIIRCLPESWMPLVTSIELTRDVENMSLEELISIL 175
+ + + L K Y K V +++RCLP + T++ + D +++ EE++ +L
Sbjct: 147 SGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSLDTDSIDFEEVVGML 206
Query: 176 KCHELKRSEMQDLKKKSIALKSKSEKAKVEKSKALQAEEEESEEASEDSDEDELTLISKR 235
+ +EL+ + + K +AL + ++K ++++ K D +++++K
Sbjct: 207 QAYELEITSGKGGYSKGLALAASAKKNEIQELK------------------DTMSMMAKD 248
Query: 236 LNRIWKHRQSKFKGSGRAKGKYESSGQKKSSIKEVTCFECKESGHYKSDCPKLK-KDKKP 294
+R + + K G + +Y K+ E+ C EC+ GH K++CP LK KD K
Sbjct: 249 FSRAMRRVEKKGFGRNQGTDRYRDRSSKRD---EIQCHECQGYGHIKAECPSLKRKDLKC 305
Query: 295 KK--------------HFKTKKSLMVTFDESESEDVDSDGEVQGLMAIV-----KDKGAE 335
+ K KS + ES+S D DS+ ++G ++ V KD+ ++
Sbjct: 306 SECNGLGHTKFDCVGSKSKPDKSCS-SESESDSNDGDSEDYIKGFVSFVGIIEEKDESSD 364
Query: 336 SKEAVDSDSESEGDPNSDDENEVFASFSTSELKHALSDIMDKYNSLLSTHKKLKKNLSAV 395
S EA D ++ D +SD E +V +I +++ L + L K A
Sbjct: 365 S-EADGEDEDNSADEDSDIEKDV--------------NINEEFRKLYDSWLMLSKEKVAW 409
Query: 396 SKTPSEHEKIISDLKNENHALVNSNSVLKNQIAKLEE---IVACDASDCRNESKYEKSFQ 452
+ + +++ LK E A NS L + + EE ++ + SD R S
Sbjct: 410 LEEKLKVQELTEKLKGELTAANQKNSELIQKCSVAEEKNRELSQELSDTRKNIHMLNSGT 469
Query: 453 RFLAKSVDRSLMASMIYGVSRNG 475
+ L + + +G+ NG
Sbjct: 470 KDLDSILAAGRVGKSNFGLGYNG 492
>At2g07080 putative gag-protease polyprotein
Length = 627
Score = 112 bits (279), Expect = 2e-24
Identities = 98/424 (23%), Positives = 189/424 (44%), Gaps = 47/424 (11%)
Query: 27 DGKKI--PRSEMTADQKKLYSQHHKARAILLSAISYEEYQKITDREFAKGIFDSLKMSHE 84
DG KI P++ TA++K + +A + + + +E++ I + AK +D+L+ SHE
Sbjct: 37 DGFKITKPKANWTAEEKLQSKFNARAMKAIFNGVDEDEFKLIQGCKSAKQAWDTLQKSHE 96
Query: 85 GNKKVKESKALSLIQKYESFIMEPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRIIR 144
G VK ++ + ++E MEP+E I + S+ L + K+Y + V +++R
Sbjct: 97 GTSSVKRTRLDHIATQFEYLKMEPDEKIVKFSSKISALANEAEVMGKTYKDQKLVKKLLR 156
Query: 145 CLPESWMPLVTSIELTRDVENMSLEELISILKCHELKRSEMQDLKKKSIALKSKSEKAKV 204
CLP + + + + + +S +L+ +LK E+K + +K K
Sbjct: 157 CLPPKFAAHKAVMRVAGNTDKISFVDLVGMLKLEEMKADQ---------------DKVKP 201
Query: 205 EKSKALQAEEEESEEASEDSDEDELTLISKRLNRIWKHRQSKFKGSGRAKGKYESSGQKK 264
K+ A A ++ SE+ E +D + L+++ + K + + S + E+ +K
Sbjct: 202 SKNIAFNA-DQGSEQFQE--IKDGMALLARNFGKALKRVEIDGERSRGRFSRSENDDLRK 258
Query: 265 SSIKEVTCFECKESGHYKSDCPKLKKDKK---------------PKKHFKTKKSLMVTFD 309
KE+ C+EC GH K +CP K+ + P K K K+ +++F
Sbjct: 259 K--KEIQCYECGGFGHIKPECPITKRKEMKCLKCKGVGHTKFECPNKS-KLKEKSLISFS 315
Query: 310 ESESEDVDSDGEVQGLMAIVKDKGAESKEAVDSDSESEGDPNSDDENEVFASFSTSELKH 369
+SES+D +GE SK D+DS+ + + N D+ V K
Sbjct: 316 DSESDD---EGEELLNFVAFMASSDSSKFMSDTDSDCDEELNPKDKYRVLYDSWVQLSKD 372
Query: 370 ALSDIMDKYN---SLLSTHKKLKKNLSAVS---KTPSEHEKIISDLKNENHALVNSNSVL 423
L + +K L + + K+ LS ++ + ++K + L+ E H + +L
Sbjct: 373 KLKLVKEKLTLEAKLANVSTEDKQKLSGITVDGNSQDYYQKKLDCLQKECHRERDRTKLL 432
Query: 424 KNQI 427
+ ++
Sbjct: 433 EREL 436
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 93.6 bits (231), Expect = 8e-19
Identities = 72/320 (22%), Positives = 153/320 (47%), Gaps = 36/320 (11%)
Query: 11 DLWDIIVDGYERPVDTDGKKIPRSEMTADQKKLYSQHHKARAILLSAISYEEYQKITDRE 70
D+W+I+ G+ P + + + D +K + KA ++ + + ++K+ +
Sbjct: 33 DVWEIVEKGFIEPENEGSLSQTQKDGLRDSRK---RDKKALCLIYQGLDEDTFEKVVEAT 89
Query: 71 FAKGIFDSLKMSHEGNKKVKESKALSLIQKYESFIMEPNESIEEMFSRFQLLVAGIRPLN 130
AK ++ L+ S++G +VK+ + +L ++E+ M+ E + + FSR + ++
Sbjct: 90 SAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYFSRVLTVTNNLKRNG 149
Query: 131 KSYTTKDHVIRIIRCLPESWMPLVTSIELTRDVENMSLEELISILKCHELKRSEMQDLKK 190
+ + +++R L + +VT IE T+D+E M++E+L+ L+ +E K+ + +D+
Sbjct: 150 EKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSLQAYEEKKKKKEDI-- 207
Query: 191 KSIALKSKSEKAKVEKSKALQAEEEESEEASEDSDEDELTLISK---RLNRIWK----HR 243
VE+ +Q +EE+ ++ + ++ + R W+ +
Sbjct: 208 -------------VEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWRPHEDNT 254
Query: 244 QSKFKGSGRAKGK-YESSGQKKSSIKEVTCFECKESGHYKSDC--PKLKKDKKPKKHFKT 300
+ + S R +GK + S KSS+K C+ C + GHY S+C P KK K+ + +
Sbjct: 255 NQRGENSSRGRGKGHPKSRYDKSSVK---CYNCGKFGHYASECKAPSNKKFKEKANYVEE 311
Query: 301 K-----KSLMVTFDESESED 315
K LM ++ + E E+
Sbjct: 312 KIQEEDMLLMASYKKDEQEE 331
>At1g58140 hypothetical protein
Length = 1320
Score = 92.0 bits (227), Expect = 2e-18
Identities = 71/320 (22%), Positives = 153/320 (47%), Gaps = 36/320 (11%)
Query: 11 DLWDIIVDGYERPVDTDGKKIPRSEMTADQKKLYSQHHKARAILLSAISYEEYQKITDRE 70
D+W+I+ G+ P + + + D +K + KA ++ + + ++K+ +
Sbjct: 33 DVWEIVEKGFIEPENEGSLSQTQKDGLRDSRK---RDKKALCLIYQGLDEDTFEKVVEAT 89
Query: 71 FAKGIFDSLKMSHEGNKKVKESKALSLIQKYESFIMEPNESIEEMFSRFQLLVAGIRPLN 130
AK ++ L+ S++G +VK+ + +L ++E+ M+ E + + FSR + ++
Sbjct: 90 SAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYFSRVLTVTNNLKRNG 149
Query: 131 KSYTTKDHVIRIIRCLPESWMPLVTSIELTRDVENMSLEELISILKCHELKRSEMQDLKK 190
+ + +++R L + +VT IE T+D+E M++E+L+ L+ +E K+ + +D+
Sbjct: 150 EKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSLQAYEEKKKKKEDI-- 207
Query: 191 KSIALKSKSEKAKVEKSKALQAEEEESEEASEDSDEDELTLISK---RLNRIWK----HR 243
VE+ +Q +EE+ ++ + ++ + R W+ +
Sbjct: 208 -------------VEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWRPHEDNT 254
Query: 244 QSKFKGSGRAKGK-YESSGQKKSSIKEVTCFECKESGHYKSDC--PKLKKDKKPKKHFKT 300
+ + S R +GK + S KSS+K C+ C + GHY S+C P KK ++ + +
Sbjct: 255 NQRGENSSRGRGKGHPKSRYDKSSVK---CYNCGKFGHYASECKAPSNKKFEEKANYVEE 311
Query: 301 K-----KSLMVTFDESESED 315
K LM ++ + E E+
Sbjct: 312 KIQEEDMLLMASYKKDEQEE 331
>At1g48710 hypothetical protein
Length = 1352
Score = 91.7 bits (226), Expect = 3e-18
Identities = 70/320 (21%), Positives = 153/320 (46%), Gaps = 36/320 (11%)
Query: 11 DLWDIIVDGYERPVDTDGKKIPRSEMTADQKKLYSQHHKARAILLSAISYEEYQKITDRE 70
D+W+I+ G+ P + + + D +K + KA ++ + + ++K+ +
Sbjct: 33 DVWEIVEKGFIEPENEGSLSQTQKDGLRDSRK---RDKKALCLIYQGLDEDTFEKVVEAT 89
Query: 71 FAKGIFDSLKMSHEGNKKVKESKALSLIQKYESFIMEPNESIEEMFSRFQLLVAGIRPLN 130
AK ++ L+ S++G +VK+ + +L ++E+ M+ E + + FSR + ++
Sbjct: 90 SAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYFSRVLTVTNNLKRNG 149
Query: 131 KSYTTKDHVIRIIRCLPESWMPLVTSIELTRDVENMSLEELISILKCHELKRSEMQDLKK 190
+ + +++R L + +VT IE T+D+E M++E+L+ L+ +E K+ + +D+
Sbjct: 150 EKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSLQAYEEKKKKKEDI-- 207
Query: 191 KSIALKSKSEKAKVEKSKALQAEEEESEEASEDSDEDELTLISK---RLNRIWK----HR 243
+E+ +Q +EE+ ++ + ++ + R W+ +
Sbjct: 208 -------------IEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWRPHEDNT 254
Query: 244 QSKFKGSGRAKGK-YESSGQKKSSIKEVTCFECKESGHYKSDC--PKLKKDKKPKKHFKT 300
+ + S R +GK + S KSS+K C+ C + GHY S+C P KK ++ + +
Sbjct: 255 NQRGENSSRGRGKGHPKSRYDKSSVK---CYNCGKFGHYASECKAPSNKKFEEKANYVEE 311
Query: 301 K-----KSLMVTFDESESED 315
K LM ++ + E E+
Sbjct: 312 KIQEEDMLLMASYKKDEQEE 331
>At3g61330 copia-type polyprotein
Length = 1352
Score = 90.9 bits (224), Expect = 5e-18
Identities = 69/331 (20%), Positives = 151/331 (44%), Gaps = 58/331 (17%)
Query: 11 DLWDIIVDGYERPVDTDGKKIPRSEMTADQKKLYSQHHKARAILLSAISYEEYQKITDRE 70
D+W+I+ G+ P + + + D +K + KA ++ + + ++K+ +
Sbjct: 33 DVWEIVEKGFIEPENEGSLSQTQKDGLRDSRK---RDKKALCLIYQGLDEDTFEKVVEAT 89
Query: 71 FAKGIFDSLKMSHEGNKKVKESKALSLIQKYESFIMEPNESIEEMFSRFQLLVAGIRPLN 130
AK ++ L+ S++G +VK+ + +L ++E+ M+ E + + FSR + ++
Sbjct: 90 SAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYFSRVLTVTNNLKRNG 149
Query: 131 KSYTTKDHVIRIIRCLPESWMPLVTSIELTRDVENMSLEELISILKCHELKRSEMQDLKK 190
+ + +++R L + +VT IE T+D+E M++E+L+ L+ +E K+ + +D+ +
Sbjct: 150 EKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSLQAYEEKKKKKEDIAE 209
Query: 191 KSIALKSKSE------------------KAKVEKSKALQAEEEESEEASEDSDEDELTLI 232
+ + ++ E + + + E+ + + E+S
Sbjct: 210 QVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWRPHEDNTNQRGENS-------- 261
Query: 233 SKRLNRIWKHRQSKFKGSGRAKGKYESSGQKKSSIKEVTCFECKESGHYKSDCPKLKKDK 292
S+ +G G K +Y+ KSS+K C+ C + GHY S+C K +K
Sbjct: 262 ------------SRGRGKGHPKSRYD-----KSSVK---CYNCGKFGHYASEC-KAPSNK 300
Query: 293 K--PKKHFKTKK------SLMVTFDESESED 315
K K H+ +K LM ++ + E ++
Sbjct: 301 KFEEKAHYVEEKIQEEDMLLMASYKKDEQKE 331
>At2g05930 copia-like retroelement pol polyprotein
Length = 916
Score = 85.5 bits (210), Expect = 2e-16
Identities = 58/250 (23%), Positives = 120/250 (47%), Gaps = 23/250 (9%)
Query: 1 LESFFLGFDADLWDIIVDGYERPV--DTDGKKIPRSE---MTADQKKLYSQHHKARAILL 55
+ + G + W G++ PV DG+ + ++E A++ K + + +A +++
Sbjct: 40 MRALIRGLGKEAWIATSIGWKAPVIKGEDGEDVLKTEDQWNDAEEAKA-TANSRALSLIF 98
Query: 56 SAISYEEYQKITDREFAKGIFDSLKMSHEGNKKVKESKALSLIQKYESFIMEPNESIEEM 115
++++ ++++I + E AK +D L ++EG VK S+ L ++E+ ME E+IEE
Sbjct: 99 NSVNQNQFKQIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQFENLTMEETENIEEF 158
Query: 116 FSRFQLLVAGIRPLNKSYTTKDHVIRIIRCLPESWMPLVTSIELTRDVENMSLEELISIL 175
+ + + L K Y K V +++RCLP + T++ + D ++ EE++ +
Sbjct: 159 SGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSLDTNSIDFEEVVGMF 218
Query: 176 KCHELK-----------RSEMQDLKKKSIALKSKSEKAKVE------KSKALQAEEEESE 218
+ +EL+ ++E LK+K + ++ KSK ++ ESE
Sbjct: 219 QAYELEITSGKGGYGHIKAECPSLKRKDLKCSECKGLGHIKFDCVGSKSKPDRSCSSESE 278
Query: 219 EASEDSDEDE 228
S D D ++
Sbjct: 279 SDSNDGDSED 288
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 82.0 bits (201), Expect = 2e-15
Identities = 58/248 (23%), Positives = 114/248 (45%), Gaps = 46/248 (18%)
Query: 48 HKARAILLSAISYEEYQKITDREFAKGIFDSLKMSHEGNKKVKESKALSLIQKYESFIME 107
HK + L ++I + I +E +K +++S+K ++GN +V+ ++ L + +E M+
Sbjct: 26 HKVKNYLFASIDKTILKTILQKETSKDLWESMKRKYQGNDRVQSAQLQRLRRSFEVLEMK 85
Query: 108 PNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRIIRCLPESWMPLVTSIELTRDVENMS 167
E+I FSR + +R L + V +I+R L E + +V +IE + +++ ++
Sbjct: 86 IGETITGYFSRVMEITNDMRNLGEDMPDSKVVEKILRTLVEKFTYVVCAIEESNNIKELT 145
Query: 168 LEELISILKCHELKRSEMQDLKKKSIALKSKSEKAKVEKSKALQAEEEESEEASEDSDED 227
++ L S L HE Q+L + + + + L+AE +
Sbjct: 146 VDGLQSSLMVHE------QNLSRHDV------------EERVLKAETQ------------ 175
Query: 228 ELTLISKRLNRIWKHRQSKFKGS----GRAKGKYESSGQKKSSIKEVTCFECKESGHYKS 283
W+ + +G GR +G Y+ G+ + V CF+C + GHYK+
Sbjct: 176 ------------WRPDGGRGRGGSPSRGRGRGGYQGRGRGYVNRDTVECFKCHKMGHYKA 223
Query: 284 DCPKLKKD 291
+CP +K+
Sbjct: 224 ECPSWEKE 231
>At3g60170 putative protein
Length = 1339
Score = 78.6 bits (192), Expect = 3e-14
Identities = 66/308 (21%), Positives = 133/308 (42%), Gaps = 45/308 (14%)
Query: 11 DLWDIIVDGYERPVDTDGKKIPRSEMTADQKKLYSQHHKARAILLSAISYEEYQKITDRE 70
+LW ++ +G V ++ KL + K + L AI E + I D+
Sbjct: 34 ELWRLVEEGIPAIVVGTTPVSEAQRSAVEEAKL--KDLKVKNFLFQAIDREILETILDKS 91
Query: 71 FAKGIFDSLKMSHEGNKKVKESKALSLIQKYESFIMEPNESIEEMFSRFQLLVAGIRPLN 130
+K I++S+K ++G+ KVK ++ +L +++E M+ E I+ R +V ++
Sbjct: 92 TSKAIWESMKKKYQGSTKVKRAQLQALRKEFELLAMKEGEKIDTFLGRTLTVVNKMKTNG 151
Query: 131 KSYTTKDHVIRIIRCLPESWMPLVTSIELTRDVENMSLEELISILKCHELKRSEMQDLKK 190
+ V +I+R L + +V SIE + D+ +S++EL L HE + +
Sbjct: 152 EVMEQSTIVSKILRSLTPKFNYVVCSIEESNDLSTLSIDELHGSLLVHEQRLN------- 204
Query: 191 KSIALKSKSEKAKVEKSKALQAEEEESEEASEDSDEDELTLISKRLNRIWKHRQSKFKGS 250
V++ +AL+ EE + F+GS
Sbjct: 205 -----------GHVQEEQALKVTHEERPSQGRG--------------------RGVFRGS 233
Query: 251 -GRAKGKYESSGQKKSSIKEVTCFECKESGHYKSDCPKLKKDKKPKKHFKTKKSLMVTFD 309
GR +G+ G+ ++ V C++C GH++ +CP+ +K+ + + ++ L++ +
Sbjct: 234 RGRGRGR----GRSGTNRAIVECYKCHNLGHFQYECPEWEKNANYAELEEEEELLLMAYV 289
Query: 310 ESESEDVD 317
E + D
Sbjct: 290 EQNQANRD 297
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 77.8 bits (190), Expect = 5e-14
Identities = 69/317 (21%), Positives = 139/317 (43%), Gaps = 51/317 (16%)
Query: 12 LWDIIVDGY--------ERPVDTDGKKIPRSEMTADQKKLYSQHHKARAILLSAISYEEY 63
LW ++ +G E P K + +T D L IL +A++ + +
Sbjct: 33 LWSVVEEGVPVEPVQAEETPETARAKTLREEAVTNDTMALQ--------ILQTAVTDQIF 84
Query: 64 QKITDREFAKGIFDSLKMSHEGNKKVKESKALSLIQKYESFIMEPNESIEEMFSRFQLLV 123
+I +K +D LK ++G+ +V+ K SL ++YE+ M N++I+ + +L
Sbjct: 85 SRIAAASSSKEAWDVLKDEYQGSPQVRLVKLQSLRREYENLKMYDNDNIKTFTDKLIVLE 144
Query: 124 AGIRPLNKSYTTKDHVIRIIRCLPESWMPLVTSIELTRDVENMSLEELISILKCHELK-R 182
+ + T + +I+ LP + +V+ +E TRD++ +++ EL+ ILK E +
Sbjct: 145 IQLTYHGEKKTNTQLIQKILISLPAKFDSIVSVLEQTRDLDALTMSELLGILKAQEARVT 204
Query: 183 SEMQDLKKKSIALKSKSEKAKVEKSKALQAEEEESEEASEDSDEDELTLISKRLNRIWKH 242
+ + K+ + ++SK ++ ++ ++ + ++ KH
Sbjct: 205 AREESTKEGAFYVRSKGRESGFKQDNTNNRVNQDKKWCG--------------FHKSSKH 250
Query: 243 RQSKFKGSGRAKGKYESSGQKKSSIKEVTCFECKESGHYKSDCPKLKKDKKPKKHFKTKK 302
+ + R K K + G+ K S + C++C + GHY ++C K K+
Sbjct: 251 TEEEC----REKPKNDDHGKNKRS--NIKCYKCGKIGHYANEC-----------RSKNKE 293
Query: 303 SLMVTFDESESEDVDSD 319
VT +E EDV+ D
Sbjct: 294 RAHVTLEE---EDVNED 307
>At4g04560 putative transposon protein
Length = 590
Score = 72.8 bits (177), Expect = 2e-12
Identities = 37/181 (20%), Positives = 93/181 (50%), Gaps = 4/181 (2%)
Query: 9 DADLWDIIVDGYERPVDTDGKKI----PRSEMTADQKKLYSQHHKARAILLSAISYEEYQ 64
D D W + DG+ P D K+ R+E AD+K + + +A +++ ++ ++
Sbjct: 368 DMDAWFAVEDGWMPPTTKDAKRDIVSKSRTEWIADEKTAANHNSQALSVIFGSLLRNKFT 427
Query: 65 KITDREFAKGIFDSLKMSHEGNKKVKESKALSLIQKYESFIMEPNESIEEMFSRFQLLVA 124
++ AK +++ L++S E VK ++ L ++E+ ME ES+++ + +
Sbjct: 428 QVQGCLSAKEVWEILQVSFECTNNVKRTRLDMLASEFENLTMEAEESVDDFNGKLSSITQ 487
Query: 125 GIRPLNKSYTTKDHVIRIIRCLPESWMPLVTSIELTRDVENMSLEELISILKCHELKRSE 184
L K+Y K V + +R LP+ + ++I+++ + + + ++++ +++ ++ + E
Sbjct: 488 EAVVLGKTYKDKKMVKKFLRSLPDKFQSHKSAIDVSLNSDQLKFDQVVGMMQAYDTDKEE 547
Query: 185 M 185
+
Sbjct: 548 I 548
>At1g36110 hypothetical protein
Length = 745
Score = 71.2 bits (173), Expect = 4e-12
Identities = 52/238 (21%), Positives = 99/238 (40%), Gaps = 36/238 (15%)
Query: 50 ARAILLSAISYEEYQKITDREFAKGIFDSLKMSHEGNKKVKESKALSLIQKYESFIMEPN 109
AR ++ +I ++ AK ++DS+K + G +VKE++ +L+ ++E M+ +
Sbjct: 60 ARGLIFQSIPESLTLQVGTLATAKLVWDSIKTRYVGADRVKEARLQTLMAEFEKMKMKES 119
Query: 110 ESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRIIRCLP-ESWMPLVTSIELTRDVENMSL 168
E I+ R L L + T V + + LP ++ ++ S+E D+ N S
Sbjct: 120 EKIDVFAGRLAELATRSDALGSNIETSKLVKKFLNALPLRKYIHIIASLEQVLDLNNTSF 179
Query: 169 EELISILKCHELKRSEMQDLKKKSIALKSKSEKAKVEKSKALQAEEEESEEASEDSDEDE 228
E+++ +K +E + EE+E D+ +
Sbjct: 180 EDIVGRIKVYE---------------------------ERVWDGEEQED-------DQGK 205
Query: 229 LTLISKRLNRIWKHRQSKFKGSGRAKGKYESSGQKKSSIKEVTCFECKESGHYKSDCP 286
L + W + + +G G G+ G+ +VTC+ C + GHY S+CP
Sbjct: 206 LMYANTDTQDSWYASRGRGQGGG-FNGRGRGRGRGSRDTSKVTCYRCDKLGHYASNCP 262
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 70.5 bits (171), Expect = 8e-12
Identities = 70/375 (18%), Positives = 158/375 (41%), Gaps = 67/375 (17%)
Query: 50 ARAILLSAISYEEYQKITDREFAKGIFDSLKMSHEGNKKVKESKALSLIQKYESFIMEPN 109
ARA+L ++ ++ + +K +++++K + G ++VKE+K +L+ +++ M+ N
Sbjct: 27 ARALLFQSVPESTILQVGKHKTSKAMWEAIKTRNLGAERVKEAKLQTLMAEFDRLNMKDN 86
Query: 110 ESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRIIRCLP-ESWMPLVTSIELTRDVENMSL 168
E+I+E R + L + V + ++ LP + ++ ++ ++E D+
Sbjct: 87 ETIDEFVGRISEISTKSESLGEEIEESKIVKKFLKSLPRKKYIHIIAALEQILDLNTTGF 146
Query: 169 EELISILKCHELKRSEMQDLKKKSIALKSKSEKAKVEKSKALQAEEEESEEASEDSDEDE 228
E+++ +K +E + + D + E+ K + A E S +
Sbjct: 147 EDIVGRMKTYEDRVCDEDD--------------SPEEQGKLMYANSESSYDTRGG----- 187
Query: 229 LTLISKRLNRIWKHRQSKFKGSGRAKGKYESSGQKKSSIKEVTCFECKESGHYKSDCPKL 288
+ + + SGR +G Y G ++ +V C+ C ++GHY S+C
Sbjct: 188 -------------RGRGRGRSSGRGRGGY---GYQQRDKSKVICYRCDKTGHYASEC--- 228
Query: 289 KKDKKPKKHFKTKKSLMVTFDESESEDVDSDGEVQGLM----AIVKDKGAESKEAVDSDS 344
+ L + + + ++ + D E++ LM + ++ + KE ++ S
Sbjct: 229 -----------LDRLLKLIKAQEQQQNNEDDDEIESLMMHEVVYLNERSVKPKE-FEACS 276
Query: 345 ESEGDPNSDDENEVFASFS-TSELKHALS-----------DIMDKYNSLLSTHKKLKKNL 392
++ ++ N + + S+L ++ DI K + +L T ++K L
Sbjct: 277 DNSWYLDNGASNHMTGNLQWFSKLNEMITGKVRFGDDSRIDIKGKGSIVLITKGGIRKTL 336
Query: 393 SAVSKTPSEHEKIIS 407
+ V P IIS
Sbjct: 337 TDVYFIPDLKSNIIS 351
>At3g21010 unknown protein
Length = 438
Score = 68.2 bits (165), Expect = 4e-11
Identities = 67/282 (23%), Positives = 119/282 (41%), Gaps = 47/282 (16%)
Query: 12 LWDIIVDGYERPVDTDGKKIP------RSEMTADQKKLYSQHHKARAILLSAISYEEYQK 65
LWD++ +G V D K+P ++E + + L + KA IL S+++ ++K
Sbjct: 33 LWDVVENG----VPPDPSKVPELAATIQAEEFSKWRDLAEKDMKALQILQSSLTDSAFRK 88
Query: 66 ITDREFAKGIFDSLKMSHEGNKKVKESKALSLIQKYESFIMEPNESIEEMFSRFQLLVAG 125
AK ++DSL+ +GN + ++K L +++E M E I R +V
Sbjct: 89 TLSASSAKDLWDSLE---KGNNE--QAKLRRLEKQFEELSMNEGEPINPYIDRVIEIVEQ 143
Query: 126 IRPLNKSYTTKDHVIRIIRCLPESWMPLVTSIELTRDVENMSLEELISILKCHELKRSEM 185
+R + + + +++ L S+ + + D++NMSL+ + + ++ E
Sbjct: 144 LRRFKIAKSDYQVIEKVLATLSGSYDDVAPVLGDLVDLKNMSLKSFVELFYVYDSITEES 203
Query: 186 QDLKKKSIALKSKSEKAKVEKSKALQAEEEESEEASEDSDEDELTLISKRLNRIWKHRQS 245
L K+ L+S +E E+ L +K H+Q
Sbjct: 204 IHLMLKANRLRSMAE-------------------------EESCVLCNKN-----NHKQE 233
Query: 246 KFKGSGRAKGKYESSGQKKSSIKEVTCFECKESGHYKSDCPK 287
S GK SSG ++S K+ CF+C E GH DC K
Sbjct: 234 DCSSSIPKAGK--SSGARQSKPKKGKCFQCGERGHKAKDCNK 273
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 60.8 bits (146), Expect = 6e-09
Identities = 60/265 (22%), Positives = 108/265 (40%), Gaps = 60/265 (22%)
Query: 29 KKIPRSEMTADQKKLYSQHHKARAILLSAISYEEYQKITDREFAKGIF---DSLKMSHEG 85
+K+ + E ++KK KAR+ ++ +++ +KI A + D L MS
Sbjct: 59 EKLEKFEALEEKKK------KARSAIVLSVTDRVLRKIKKESTAAAMLLALDKLYMSKAL 112
Query: 86 NKKVKESKALSLIQKYESFIMEPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRIIRC 145
++ QK SF M N S+E F ++ + +N + +D I ++
Sbjct: 113 PNRIYPK------QKLYSFKMSENLSVEGNIDEFLQIITDLENMNVIISDEDQAILLLTA 166
Query: 146 LPESWMPLVTSIELTRDVENMSLEELISILKCHELKRSEMQDLKKKSIALKSKSEKAKVE 205
LP+++ L +++ + ++L+E+ + + EL E V+
Sbjct: 167 LPKAFDQLKDTLKYSSGKSILTLDEVAAAIYSKEL-------------------ELGSVK 207
Query: 206 KSKALQAEEEESEEASEDSDEDELTLISKRLNRIWKHRQSKFKGSGRAKGKYESSGQKKS 265
KS +QAE ++ +E+ KG G KGK G+K
Sbjct: 208 KSIKVQAEGLYVKDKNEN------------------------KGKGEQKGK--GKGKKGK 241
Query: 266 SIKEVTCFECKESGHYKSDCPKLKK 290
S K+ C+ C E GH++S CP K
Sbjct: 242 SKKKPGCWTCGEEGHFRSSCPNQNK 266
>At3g31380 hypothetical protein
Length = 262
Score = 59.7 bits (143), Expect = 1e-08
Identities = 33/131 (25%), Positives = 69/131 (52%), Gaps = 1/131 (0%)
Query: 50 ARAILLSAISYEEYQKITDREFAKGIFDSLKMSHEGNKKVKESKALSLIQKYESFIMEPN 109
AR +L+ +I ++ + + AK ++DS+K H G ++VKE++ +L+ +E M+
Sbjct: 2 ARGLLVQSIPEAFTLQVGNLQTAKEVWDSIKTRHVGAERVKEARVQTLMADFEKMKMKEA 61
Query: 110 ESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRIIRCLP-ESWMPLVTSIELTRDVENMSL 168
E I++ R L PL + V + + LP + ++ ++ S+E D+ N +
Sbjct: 62 EKIDDFAGRLSELSTKSAPLGVNIEVPKLVKKFLNSLPRKKYIHIIASLEQVLDLNNTTF 121
Query: 169 EELISILKCHE 179
E+++ +K +E
Sbjct: 122 EDIVGCMKVNE 132
>At3g28770 hypothetical protein
Length = 2081
Score = 57.8 bits (138), Expect = 5e-08
Identities = 69/289 (23%), Positives = 126/289 (42%), Gaps = 26/289 (8%)
Query: 189 KKKSIALKSKSEKAKVEKSKALQAEEEESEEASEDSDEDELTLISKRLNRIWKHRQSKFK 248
KKK + S +K + +K + + E ++ E+ +++ + E + + + + ++S+
Sbjct: 946 KKKKESKNSNMKKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENKDNKEKKESEDS 1005
Query: 249 GS-GRAKGKYESSGQKKSSIKEVTCFECKESGHYK-----SDCPKLKKDKKPKKHFKTKK 302
S R K +YE +KKS KE E K+S K S+ K KK+K+ + K KK
Sbjct: 1006 ASKNREKKEYE---EKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKK 1062
Query: 303 SLMVTFDESESEDVDSDGEVQGLMAIVKDKGAESKEAVDSDS-ESEGDPNSDDENEVFAS 361
T ++ ESE+ S K + KE D+ S + E D ++E S
Sbjct: 1063 KEEETKEKKESENHKS------------KKKEDKKEHEDNKSMKKEEDKKEKKKHEESKS 1110
Query: 362 FSTSELKHALSDIMDKYNSLLSTHKKLKK---NLSAVSKTPSEHEKIISDLKNENHALVN 418
E K + + D+ ++ K KK ++ V K + EK ++ K+E + +
Sbjct: 1111 RKKEEDKKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIES 1170
Query: 419 SNSVLKNQIAKLEEIVACDASDCRNESKYEKSFQRFLAKSVDRSLMASM 467
S S KN++ K E+ + D + + E ++ DR S+
Sbjct: 1171 SKS-QKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSV 1218
Score = 49.7 bits (117), Expect = 1e-05
Identities = 55/269 (20%), Positives = 105/269 (38%), Gaps = 21/269 (7%)
Query: 176 KCHELKRSEMQDLKKKSIALKSKSEKAKVEKSKALQAEEEESEEASEDSDEDELTLISKR 235
K + KR E ++KS K +S K +K + E++ESE ED+
Sbjct: 1032 KSQDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKSKKKEDKK---EHE 1088
Query: 236 LNRIWKHRQSKFKGSGRAKGKYESSGQKKSSIKEVTCFECKESGHYKSDCPKLKKDKKPK 295
N+ K + K + K + S +K+ K++ E + S K D K +KK
Sbjct: 1089 DNKSMKKEEDK---KEKKKHEESKSRKKEEDKKDMEKLEDQNSNKKKED----KNEKKKS 1141
Query: 296 KHFKTKKSLMVTFDESESEDVDSDGEVQGLMAIVKDKGAESKEAVDSDSESEGDPNSDDE 355
+H K K ++ E+E+ E++ ++ E + +S D E
Sbjct: 1142 QHVKLVKKESDKKEKKENEEKSETKEIE-------SSKSQKNEVDKKEKKSSKDQQKKKE 1194
Query: 356 NEVFASFSTSELKHALSDIMDKYNSLLSTHKKLKKNLSAVSKTPSEHEKIISDLKNENHA 415
E+ S E K ++ K + + +KK K+ +K + + + +
Sbjct: 1195 KEMKES---EEKKLKKNEEDRKKQTSVEENKKQKETKKEKNKPKDDKKNTTKQSGGKKES 1251
Query: 416 LVNSNSVLKNQIAKLEEIVACDASDCRNE 444
+ + + +NQ K + D+ + +NE
Sbjct: 1252 MESESKEAENQ-QKSQATTQADSDESKNE 1279
Score = 49.3 bits (116), Expect = 2e-05
Identities = 63/310 (20%), Positives = 122/310 (39%), Gaps = 45/310 (14%)
Query: 176 KCHELKRS--EMQDLKKKSIALKSKSEKAKVEKSKALQAEEEESEEASEDSDEDELTLIS 233
K HE +S + +D K+K +SKS K + +K + E++ S + ED +E + S
Sbjct: 1085 KEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDMEKLEDQNSNKKKEDKNEKKK---S 1141
Query: 234 KRLNRIWKHRQSKFKGSGRAKGKYESSGQKKSSIKEVTCFECKESGHY---------KSD 284
+ + + K K K K + + KS EV E K S +S+
Sbjct: 1142 QHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNEVDKKEKKSSKDQQKKKEKEMKESE 1201
Query: 285 CPKLKKDKKP----------KKHFKTKKSLMVTFDESESEDVDSDGEVQGL--------- 325
KLKK+++ KK +TKK D+ ++ S G+ + +
Sbjct: 1202 EKKLKKNEEDRKKQTSVEENKKQKETKKEKNKPKDDKKNTTKQSGGKKESMESESKEAEN 1261
Query: 326 ----MAIVKDKGAESKEAVDSDSESEGDPNSDDE-------NEVFASFSTSELKHALSDI 374
A + ESK + ++S+ D +SD + NE+ + ++
Sbjct: 1262 QQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESKNEILMQADSQATTQRNNEE 1321
Query: 375 MDKYNSLLSTHKKLKKNLSAVSKTPSEHEKIISDLKNENHALVNSNSVLKNQIAKLEEIV 434
K + ++ +KK K+ +K + + + ++ + + +NQ K +
Sbjct: 1322 DRKKQTSVAENKKQKETKEEKNKPKDDKKNTTKQSGGKKESMESESKEAENQ-QKSQATT 1380
Query: 435 ACDASDCRNE 444
D+ + +NE
Sbjct: 1381 QADSDESKNE 1390
Score = 47.8 bits (112), Expect = 5e-05
Identities = 61/289 (21%), Positives = 114/289 (39%), Gaps = 26/289 (8%)
Query: 179 ELKRSEMQDLKKKSIALKSKSEKAKVEKSKALQAEEEESEEASEDSDEDELTLISKRLNR 238
E K ++ KK+ + K + K + + + E+EES + E+E + N
Sbjct: 1017 EKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENH 1076
Query: 239 IWKHRQSKFKGSGRAKGKYESSGQKKSSIKEVTCFECKESGHYKSDCPKLKKDKKPKKHF 298
K ++ K K ++ KK K+ ++ H +S K ++DKK +
Sbjct: 1077 KSKKKEDK-------KEHEDNKSMKKEEDKK------EKKKHEESKSRKKEEDKKDMEKL 1123
Query: 299 KTKKSLMVTFDESESEDVDSDGEVQGLMAIVKDKGAESKEAVDSDSES-EGDPNSDDENE 357
+ + S + ED + + Q + + K+ + K+ + SE+ E + + +NE
Sbjct: 1124 EDQNS------NKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNE 1177
Query: 358 VFASFSTSELKHALSDIMDKYNSLL-STHKKLKKNLSAVSK-TPSEHEKIISDLKNENHA 415
V E K + K + S KKLKKN K T E K + K E +
Sbjct: 1178 V----DKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKETKKEKNK 1233
Query: 416 LVNSNSVLKNQIAKLEEIVACDASDCRNESKYEKSFQRFLAKSVDRSLM 464
+ Q +E + ++ + N+ K + + Q +S + LM
Sbjct: 1234 PKDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQADSDESKNEILM 1282
Score = 42.0 bits (97), Expect = 0.003
Identities = 57/288 (19%), Positives = 119/288 (40%), Gaps = 28/288 (9%)
Query: 176 KCHELKRSEMQDLKKKSIALKSKSEKAKVEKSKALQAEEEESEEASE--DSDEDELTLIS 233
K E K+ + + K K K + +S++ +AE ++ +A+ DSDE + ++
Sbjct: 1223 KQKETKKEKNKPKDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQADSDESKNEILM 1282
Query: 234 KRLNRIWKHRQSKFKGSGRAKGKYESSGQKKSSI---------KEVTCFECKESGHYKSD 284
+ ++ H S+ S +K + +++ K+ + E K+ K +
Sbjct: 1283 QADSQADSHSDSQ-ADSDESKNEILMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEE 1341
Query: 285 CPKLKKDKK-PKKHFKTKKSLMVTFDESESEDVDSDGEVQGLMAIVKDKGAESKEAV--- 340
K K DKK K KK M ESES++ ++ + Q D ESK +
Sbjct: 1342 KNKPKDDKKNTTKQSGGKKESM----ESESKEAENQQKSQATTQADSD---ESKNEILMQ 1394
Query: 341 ---DSDSESEGDPNSDD-ENEVFASFSTSELKHALSDIMDKYNSLLSTHKKLKKNLSAVS 396
+DS S+ +SD+ +NE+ + ++ K + ++ +KK K+ +
Sbjct: 1395 ADSQADSHSDSQADSDESKNEILMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEKN 1454
Query: 397 KTPSEHEKIISDLKNENHALVNSNSVLKNQIAKLEEIVACDASDCRNE 444
K + + + ++ + + +NQ K + ++ + +NE
Sbjct: 1455 KPKDDKKNTTEQSGGKKESMESESKEAENQ-QKSQATTQGESDESKNE 1501
Score = 40.4 bits (93), Expect = 0.008
Identities = 52/228 (22%), Positives = 94/228 (40%), Gaps = 19/228 (8%)
Query: 190 KKSIALKSKSEKAKVEKSKALQAEEEESEEASEDSDEDELTLISKRLNRIWKHRQSKFKG 249
KK+ K E+ K +++ ++ E++E+ DS +D+ + +I+ +SK
Sbjct: 686 KKNDGSSEKGEEGKENNKDSMEDKKLENKESQTDSKDDKSVDDKQEEAQIYGG-ESKDDK 744
Query: 250 SGRAKGKYESSGQKKSSIKEVTCFECKESGHY--KSDCPKL----KKDKKPKKHFKTKKS 303
S AKGK + S + K + KE K + K+ KK+ K K +TK +
Sbjct: 745 SVEAKGKKKESKENKKTKTNENRVRNKEENVQGNKKESEKVEKGEKKESKDAKSVETKDN 804
Query: 304 LMVTFDESESEDVDSDGEVQGLMAIVKDKGAESKEAVDSDSESEGDPNSDDENEVFASFS 363
++ E+ E + GE K+ ESK+ +++ + + D N V
Sbjct: 805 KKLSSTENRDEAKERSGEDN------KEDKEESKDYQSVEAKEKNENGGVDTN-VGNKED 857
Query: 364 TSELKHALSDIMDKYNSLLSTHKKLKKNLSAVSKTPSEHEKIISDLKN 411
+ +LK S + K N + +KK V + K + D N
Sbjct: 858 SKDLKDDRS-VEVKANK----EESMKKKREEVQRNDKSSTKEVRDFAN 900
Score = 37.7 bits (86), Expect = 0.054
Identities = 67/335 (20%), Positives = 138/335 (41%), Gaps = 33/335 (9%)
Query: 154 VTSIELTRDVENM--SLEE--LISILKCHELKRSEMQDLKKKSIALKSKSEKAK----VE 205
V++I+ ++VE S+E+ + L+ E +SE++ K ++ K E+A+ V
Sbjct: 326 VSTIDNEKEVEGQGESIEDSDIEKNLESKEDVKSEVEAAKNAGSSMTGKLEEAQRNNGVS 385
Query: 206 KSKALQAEEEESEEASED-------SDEDELTLISKRLNRIWKHRQS--KFKGSG-RAKG 255
++ + +E + S E++ D +DED K+ N+ H + KG K
Sbjct: 386 TNETMNSENKGSGESTNDKMVNATTNDEDH-----KKENKEETHENNGESVKGENLENKA 440
Query: 256 KYESSGQKKSSIKEVTCFECKESGHYKSDCPKLKKDKKPKKHFKTKKSLMVTFDESESED 315
E S + ++ +V E K + ++ ++ ++ + + + + ++ E+
Sbjct: 441 GNEESMKGENLENKVGNEELKGNASVEAKTNNESSKEEKREESQRSNEVYMNKETTKGEN 500
Query: 316 VDSDGEVQGLMAIVKDKGAESKEAVDSDSESEGDPNSDDENEVFASFSTSELKHALSDIM 375
V+ GE G KD E+KE D + + D N D N +++ + +S
Sbjct: 501 VNIQGESIG--DSTKDNSLENKE----DVKPKVDANESDGNSTKERHQEAQVNNGVSTED 554
Query: 376 DKYNSLLSTHKKLKKNLSAVSKTPSEH--EKIISDLKNENHALVNSNSVLKNQIAKLEEI 433
+++ + +K V+ +H EK N ++ N N K +L++
Sbjct: 555 KNLDNIGADEQKKNDKSVEVTTNDGDHTKEKREETQGNNGESVKNENLENKEDKKELKDD 614
Query: 434 VACDASDCRNESKYEKSFQRFLAKSVDRSLMASMI 468
+ A S EK Q K D S+ + ++
Sbjct: 615 ESVGAKTNNETSLEEKREQ--TQKGHDNSINSKIV 647
Score = 32.7 bits (73), Expect = 1.7
Identities = 42/165 (25%), Positives = 68/165 (40%), Gaps = 17/165 (10%)
Query: 197 SKSEKAKVEKSKALQAEEEESEEASEDSDEDELTLISKRLNRIWKHRQSKFKGSGRAKGK 256
+KSE +V+K+ + EE +E ++DS ED K+L K Q+ K K
Sbjct: 679 TKSE-VEVKKNDGSSEKGEEGKENNKDSMED------KKLEN--KESQTDSKDDKSVDDK 729
Query: 257 YESSGQKKSSIKEVTCFECKESGHYKSDCPKLKKDKKPKKHFKTKKSLMVTFDESESEDV 316
E + K+ E K K + + KK K + + K+ V ++ ESE V
Sbjct: 730 QEEAQIYGGESKDDKSVEAKGK---KKESKENKKTKTNENRVRNKEE-NVQGNKKESEKV 785
Query: 317 DSDGEVQGLMA----IVKDKGAESKEAVDSDSESEGDPNSDDENE 357
+ + + A +K S E D E G+ N +D+ E
Sbjct: 786 EKGEKKESKDAKSVETKDNKKLSSTENRDEAKERSGEDNKEDKEE 830
Score = 32.3 bits (72), Expect = 2.3
Identities = 85/389 (21%), Positives = 143/389 (35%), Gaps = 50/389 (12%)
Query: 175 LKCHELKRSEMQDLKKKSIALKSKSEKAKVEKSKALQAEEEESEEASEDSD--------E 226
+K E E+ K+I +K + E++K K+ +E +EE S+DS+ E
Sbjct: 1588 VKGKEDNGDEVGKENSKTIEVKGRHEESKDGKTNENGGKEVSTEEGSKDSNIVERNGGKE 1647
Query: 227 DELTLISKRLNRIWKHRQSKFK-GSGRAKGKYESSGQ-KKSSIKEVTCFECKESGHYKSD 284
D + S+ + + + G GK E + K++S KE + + E G
Sbjct: 1648 DSIKEGSEDGKTVEINGGEELSTEEGSKDGKIEEGKEGKENSTKEGSKDDKIEEG----- 1702
Query: 285 CPKLKKDKKPKKHFKTKKSLMVTFD-ESESEDVDSDGEVQGLMAIVKDKGAESKEAVDSD 343
K+ K+ K K + D E+ E+ DG KD + V D
Sbjct: 1703 --MEGKENSTKESSKDGKINEIHGDKEATMEEGSKDGGTNSTGKDSKDSKSVEINGVKDD 1760
Query: 344 SESEGDPNSDDENEVFASFSTSELKHALSDIMDKYNSL------------LSTHKKLKKN 391
S + D + D NE+ + +K +++I NSL L T+K KN
Sbjct: 1761 SLKD-DSKNGDINEI-NNGKEDSVKDNVTEIQGNDNSLTNSTSSEPNGDKLDTNKDSMKN 1818
Query: 392 L-----------SAVSKTPSEHEKIISDLKNENHALVNSN-SVLKNQIAKLEEIVACDAS 439
S +T E +S + N+N V SN + + NQ + + +
Sbjct: 1819 NTMEAQGGSNGDSTNGETEETKESNVS-MNNQNMQDVGSNENSMNNQTTGTGDDIISTTT 1877
Query: 440 DCRNESKYEKSFQRFLAKSVDRSLMASMIYGVSRNGMHGIGYSKPIRNEPSVSKAKSLYE 499
D ES K F++ ++S + + Y P+ + ++S
Sbjct: 1878 D--TESNTSKEVTSFISNLEEKSPGTQEFQSFFQKLKDYMKYLCPVSSTFEAKDSRSYMS 1935
Query: 500 CFVPSGTILPDPVPAKIAKNPLKKGSFSM 528
+ T L D + AK K GS M
Sbjct: 1936 EMISMATKLSDAMAVLQAK---KSGSGQM 1961
Score = 31.2 bits (69), Expect = 5.1
Identities = 50/205 (24%), Positives = 82/205 (39%), Gaps = 38/205 (18%)
Query: 182 RSEMQDLKKKSIALKSKSEKAKVEK------SKALQAEEEESEEASEDSDEDELTLISKR 235
R EM ++ K L + + +EK + +L E++ + E S E +T I K
Sbjct: 120 RKEMFEVMKSLTELHAAIGRVIIEKHIKGDEAMSLSLEQKNAVETSITQWEQTITRIVKI 179
Query: 236 LNRIWKHRQSKFKGSGRAKGKYESSGQKKS----SIKEVTCFECKESGHYKSD------- 284
+ + K K S A + SS + + S T E ES K D
Sbjct: 180 VVEV------KSKSSSEASSEESSSTEHNNVTTGSNMVETNGENSESTQEKGDGVEGSNG 233
Query: 285 ------------CPKLKKDKKPKKHFKTKKSLMVTFDESESEDVDSDGEVQGLMAIVKDK 332
LK+ ++ +TK++ + + ++V+ GE G AI +K
Sbjct: 234 GDVSMENLQGNKVEDLKEGNNVVENGETKENNGENVESNNEKEVEGQGESIGDSAI--EK 291
Query: 333 GAESKEAVDSDSE-SEGDPNSDDEN 356
ESKE V S+ E ++ D +S EN
Sbjct: 292 NLESKEDVKSEVEAAKNDGSSMTEN 316
>At4g10460 putative retrotransposon
Length = 1230
Score = 57.0 bits (136), Expect = 9e-08
Identities = 67/295 (22%), Positives = 122/295 (40%), Gaps = 58/295 (19%)
Query: 25 DTDGKKIPRSEMTADQKKLYSQHHKARAILLSAISYEEYQKITDREFAKGIFDSLKMSHE 84
+ +GK+ SE + + + KAR+ ++ ++S + +K + A + ++L
Sbjct: 50 EEEGKE---SEKGDKEALMEEKRQKARSTIVLSVSDQVLRKSKKEKTAPSMLEALD---- 102
Query: 85 GNKKVKESKALS----LIQKYESFIMEPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVI 140
K+ SKAL L QK S+ M+ N S+E F L+A + N + +D I
Sbjct: 103 ---KLYMSKALPNRIYLKQKLYSYKMQENLSVEGNIDEFLRLIADLENTNVLVSDEDQAI 159
Query: 141 RIIRCLPESWMPLVTSIELTRDVENMSLEELISILKCHELKRSEMQDLKKKSIALKSKSE 200
++ LP+ + L +++ +S++E+++ + EL+ KKSI + ++E
Sbjct: 160 LLLMSLPKQFDQLKDTLKYGSGRTTLSVDEVVAAIYSKELELGS----NKKSI--RGQAE 213
Query: 201 KAKVEKSKALQAEEEESEEASEDSDEDELTLISKRLNRIWKHRQSKFKGSGRAKGKYESS 260
V+ + E+ E+ + KG R++ K
Sbjct: 214 GLYVKDKPETRGMSEQKEKGN--------------------------KGRSRSRSK---- 243
Query: 261 GQKKSSIKEVTCFECKESGHYKSDCP-KLKKDKKPKKHFKTKKSLMVTFDESESE 314
G K C+ C E GH+K+ CP K K+ K K K T + SE
Sbjct: 244 GWK-------GCWICGEEGHFKTSCPNKGKQQNKGKDQASGSKGEAATIKGNTSE 291
>At3g55300 putative protein
Length = 262
Score = 55.5 bits (132), Expect = 3e-07
Identities = 53/209 (25%), Positives = 90/209 (42%), Gaps = 38/209 (18%)
Query: 83 HEGNKKVKE-SKALSLIQKYESFIMEPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIR 141
+EGN + K + + L K+ SF M N+SIEE LVA + L+ + + +D ++
Sbjct: 36 YEGNYQTKTLANMIYLKHKFTSFKMVENKSIEENLDTSLKLVADLASLDINTSDEDQALQ 95
Query: 142 IIRCLPESWMPLVTSIELTRDVENMSLEELISILKCHELKRSEMQDLKKKSIALKSKSEK 201
+ LP + LV +++ + ++L +IS E+ +LK+K + SK +
Sbjct: 96 FMAGLPPQYDSLVDTLKYGSGKDTLTLNAVISSAYSKEI------ELKEKGLLNVSKPD- 148
Query: 202 AKVEKSKALQAEEEESEEASEDSDEDELTLISKRLNRIWKHRQSKFKGSGRAKGKYESSG 261
S+AL + R N K + GS GK++S
Sbjct: 149 -----SEALLGD------------------FKGRSNN--KGNKRPCTGSSSGNGKFQSRP 183
Query: 262 QKKSSIKEVTCFECKESGHYKSDCPKLKK 290
+S CF C + H+K +CP+ KK
Sbjct: 184 SNNTS-----CFICGKEDHWKRECPQRKK 207
>At3g25450 hypothetical protein
Length = 1343
Score = 53.9 bits (128), Expect = 7e-07
Identities = 37/181 (20%), Positives = 94/181 (51%), Gaps = 8/181 (4%)
Query: 37 TADQKKLYSQHHKARAILLSAISYEEYQKITDREFAKGIFDSLKMSHEGNKKVKESKALS 96
+AD++K + ARA+L +I ++ ++ + +++++K + G ++VKE++ +
Sbjct: 54 SADEEK----NDMARALLFQSIPESLILQVGKQKTSSAVWEAIKSRNLGAERVKEARLQT 109
Query: 97 LIQKYESFIMEPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRIIRCLP-ESWMPLVT 155
L+ +++ M+ +E+I++ R + L + V + ++ LP + ++ +V
Sbjct: 110 LMAEFDKLKMKDSETIDDYVGRISEITTKAAALGEDIEESKIVKKFLKSLPRKKYIHIVA 169
Query: 156 SIELTRDVENMSLEELISILKCHELKRSEMQDLKKKSIALKSKSEKAKVEKSKALQAEEE 215
++E D++ + E++ +K +E + + D + L ++ E+ V+ L+ EEE
Sbjct: 170 ALEQVLDLKTTTFEDIAGRIKTYEDRVWDDDDSHEDQGKLMTEVEEEVVDD---LEEEEE 226
Query: 216 E 216
E
Sbjct: 227 E 227
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.355 0.156 0.545
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,663,341
Number of Sequences: 26719
Number of extensions: 1152545
Number of successful extensions: 14509
Number of sequences better than 10.0: 392
Number of HSP's better than 10.0 without gapping: 82
Number of HSP's successfully gapped in prelim test: 327
Number of HSP's that attempted gapping in prelim test: 12461
Number of HSP's gapped (non-prelim): 1470
length of query: 1602
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1489
effective length of database: 8,299,349
effective search space: 12357730661
effective search space used: 12357730661
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.6 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0369a.2


