

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0365.13
(1615 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
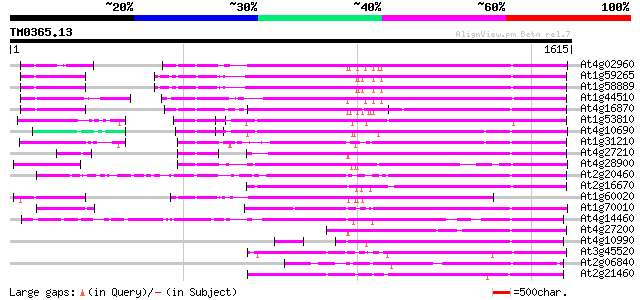
Score E
Sequences producing significant alignments: (bits) Value
At4g02960 putative polyprotein of LTR transposon 833 0.0
At1g59265 polyprotein, putative 832 0.0
At1g58889 polyprotein, putative 831 0.0
At1g44510 polyprotein, putative 813 0.0
At4g16870 retrotransposon like protein 762 0.0
At1g53810 746 0.0
At4g10690 retrotransposon like protein 736 0.0
At1g31210 putative reverse transcriptase 725 0.0
At4g27210 putative protein 707 0.0
At4g28900 putative protein 691 0.0
At2g20460 putative retroelement pol polyprotein 654 0.0
At2g16670 putative retroelement pol polyprotein 626 e-179
At1g60020 hypothetical protein 612 e-175
At1g70010 hypothetical protein 609 e-174
At4g14460 retrovirus-related like polyprotein 593 e-169
At4g27200 putative protein 510 e-144
At4g10990 putative retrotransposon polyprotein 462 e-130
At3g45520 copia-like polyprotein 461 e-129
At2g06840 putative retroelement pol polyprotein 451 e-126
At2g21460 putative retroelement pol polyprotein 435 e-122
>At4g02960 putative polyprotein of LTR transposon
Length = 1456
Score = 833 bits (2153), Expect = 0.0
Identities = 502/1253 (40%), Positives = 684/1253 (54%), Gaps = 160/1253 (12%)
Query: 440 QGQFPAPTAQTPQFPRPPQQQVSTG----MHPQAMLATVPAHHSSPVTWCPDSGATHHVT 495
QG Q QF QQ ST P+A LA ++++ W DSGATHH+T
Sbjct: 264 QGHSAKRCPQLHQFQSTTNQQQSTSPFTPWQPRANLAVNSPYNAN--NWLLDSGATHHIT 321
Query: 496 NNPGIFVDHVPSSSQEQLLVGNGQGLPIQSIGSALLPSSFNPTASLILKKLLLVPSITKN 555
++ H P + + +++ +G +PI GSA LP+S + SL L K+L VP+I KN
Sbjct: 322 SDFNNLSFHQPYTGGDDVMIADGSTIPITHTGSASLPTS---SRSLDLNKVLYVPNIHKN 378
Query: 556 LVSVSRFARDNNVFFSFHANHCSVHHQDTYELLLTGTVGDDGLYYFDGPLTKASSQPQSS 615
L+SV R N V F V +T LL G D+ LY +
Sbjct: 379 LISVYRLCNTNRVSVEFFPASFQVKDLNTGVPLLQGKTKDE-LYEW-------------- 423
Query: 616 SPLSSIQACLASKSAANKASPAIDLQSATVSPIVPSLVPCNNADSVNVSSTVSDLVSCNS 675
P++S QA S +KA+
Sbjct: 424 -PIASSQAVSMFASPCSKAT---------------------------------------- 442
Query: 676 GSGISVYDLWHSRLGHPHHDVLKQTLSLCNISVPSNKSVFSFCKACCLSKLHRLPSSSSN 735
+ WHSRLGHP +L +S ++ V + C C ++K H++P S+S
Sbjct: 443 ------HSSWHSRLGHPSLAILNSVISNHSLPVLNPSHKLLSCSDCFINKSHKVPFSNST 496
Query: 736 TVYNTPFELVFADLWGPASLESSCGFLYFLTCVDACTKYTWVFPLKKKSDTCATFINFQA 795
+ P E +++D+W L S + Y++ VD T+YTW++PLK+KS TFI F++
Sbjct: 497 ITSSKPLEYIYSDVWSSPIL-SIDNYRYYVIFVDHFTRYTWLYPLKQKSQVKDTFIIFKS 555
Query: 796 MVELQFGHKLKSVQTDGGGEFKPLTSHFQKLGIIHRLTCPHTHHQNGSVERKHRHIVETG 855
+VE +F ++ ++ +D GGEF L + + GI H + PHT NG ERKHRHIVE G
Sbjct: 556 LVENRFQTRIGTLYSDNGGEFVVLRDYLSQHGISHFTSPPHTPEHNGLSERKHRHIVEMG 615
Query: 856 LALLSHASMPLQFWDHAFLAATYLINRMSTTTLQGASPYFKLYGQYPDFKSLKVFGSACF 915
L LLSHAS+P +W +AF A YLINR+ T LQ SP+ KL+GQ P+++ LKVFG AC+
Sbjct: 616 LTLLSHASVPKTYWPYAFSVAVYLINRLPTPLLQLQSPFQKLFGQPPNYEKLKVFGCACY 675
Query: 916 PFLRPYNSNKLSLHSKECVFIGYSINHKGYKCLD-QSGRIYISKDVLFHEHRFPY----- 969
P+LRPYN +KL SK+C F+GYS+ Y CL +GR+Y S+ V F E FP+
Sbjct: 676 PWLRPYNRHKLEDKSKQCAFMGYSLTQSAYLCLHIPTGRLYTSRHVQFDERCFPFSTTNF 735
Query: 970 -------------------------PILFPT--------DHSSSSSAEFYPLSTIPIISH 996
P++ P D S + PL T + S
Sbjct: 736 GVSTSQEQRSDSAPNWPSHTTLPTTPLVLPAPPCLGPHLDTSPRPPSSPSPLCTTQVSSS 795
Query: 997 TAP------PSSPVLAASGHSSPGPQDSPQQQQ-----------PPSATLSADDPSTSTP 1039
P PSS A H+ P P P Q Q P + S + P+ ++P
Sbjct: 796 NLPSSSISSPSSSEPTAPSHNGPQPTAQPHQTQNSNSNSPILNNPNPNSPSPNSPNQNSP 855
Query: 1040 AVVSP------PVPSSSAQSADTSASV---------VAAAPPVI-------VNTHPMQTR 1077
SP P PS+S ++ +S V APP+I VNTH M TR
Sbjct: 856 LPQSPISSPHIPTPSTSISEPNSPSSSSTSTPPLPPVLPAPPIIQVNAQAPVNTHSMATR 915
Query: 1078 AKNGIVKPRLQ---PTLLLTHLEPTSVKQAMKDVKWYKAMQEEYTALMNNGTWTLVPLPA 1134
AK+GI KP + T L + EP + QAMKD +W +AM E A + N TW LVP P
Sbjct: 916 AKDGIRKPNQKYSYATSLAANSEPRTAIQAMKDDRWRQAMGSEINAQIGNHTWDLVPPPP 975
Query: 1135 -NRTPVGCKWIYRIKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILS 1193
+ T VGC+WI+ K N DGS+N+YKARLVAKGY+Q G DY+ETFSPV+K ++R++L
Sbjct: 976 PSVTIVGCRWIFTKKFNSDGSLNRYKARLVAKGYNQRPGLDYAETFSPVIKSTSIRIVLG 1035
Query: 1194 LAISRGWPLQQLDVNNAFLNGVLEEEVYMVQPPGFEHKDK-NLVCKLQKALYGLKQAPRA 1252
+A+ R WP++QLDVNNAFL G L +EVYM QPPGF KD+ + VC+L+KA+YGLKQAPRA
Sbjct: 1036 VAVDRSWPIRQLDVNNAFLQGTLTDEVYMSQPPGFVDKDRPDYVCRLRKAIYGLKQAPRA 1095
Query: 1253 WFHRLKEVLIRFGFQASKCDPSLFTYYSSQGCIYMLVYVDDIIITGASMSLIQQLTAQLH 1312
W+ L+ L+ GF S D SLF + IYMLVYVDDI+ITG L++ L
Sbjct: 1096 WYVELRTYLLTVGFVNSISDTSLFVLQRGRSIIYMLVYVDDILITGNDTVLLKHTLDALS 1155
Query: 1313 SIFALKQLGQLDYFLGVQVTHLSNGNLLLNQTKYISDLLTKVNMLDAAAISTPMQCGVKL 1372
F++K+ L YFLG++ + G L L+Q +Y DLL + NML A ++TPM KL
Sbjct: 1156 QRFSVKEHEDLHYFLGIEAKRVPQG-LHLSQRRYTLDLLARTNMLTAKPVATPMATSPKL 1214
Query: 1373 SKNEGSALKDPTEYRSVVGALQYATITRPEISFAVNKVCQFMAQPHEEHWKAVKRILRYL 1432
+ + G+ L DPTEYR +VG+LQY TRP++S+AVN++ Q+M P ++HW A+KR+LRYL
Sbjct: 1215 TLHSGTKLPDPTEYRGIVGSLQYLAFTRPDLSYAVNRLSQYMHMPTDDHWNALKRVLRYL 1274
Query: 1433 KGTLTHGILLQPCSMTRPLPLIAYCDADWGSDPDDRRSTSGSCVFLGYNLISWTAKKQSL 1492
GT HGI L+ L L AY DADW D DD ST+G V+LG++ ISW++KKQ
Sbjct: 1275 AGTPDHGIFLK---KGNTLSLHAYSDADWAGDTDDYVSTNGYIVYLGHHPISWSSKKQKG 1331
Query: 1493 VARSSTEAEYRSLANTTAELLWVQSLLTELKIPFT-IPTVYCDNMSTVILTHNPILHTRT 1551
V RSSTEAEYRS+ANT++EL W+ SLLTEL I + P +YCDN+ L NP+ H+R
Sbjct: 1332 VVRSSTEAEYRSVANTSSELQWICSLLTELGIQLSHPPVIYCDNVGATYLCANPVFHSRM 1391
Query: 1552 KHMEMDIFFVREKVQDKSLLVQHVPSEHQKADIFTKALSPTRFLLLKDKLNVV 1604
KH+ +D F+R +VQ +L V HV + Q AD TK LS F K+ V+
Sbjct: 1392 KHIALDYHFIRNQVQSGALRVVHVSTHDQLADTLTKPLSRVAFQNFSRKIGVI 1444
Score = 73.2 bits (178), Expect = 1e-12
Identities = 48/212 (22%), Positives = 97/212 (45%), Gaps = 23/212 (10%)
Query: 30 KLDDSNFYSWKQQVEGVIRSHKLQKFVQ-DPPMIPEKFLTEEDKITENVNPLFADWEQQD 88
KL +N+ W +QV + ++L F+ PM P T+ VNP + W +QD
Sbjct: 25 KLTSTNYLMWSRQVHALFDGYELAGFLDGSTPMPPATIGTDA---VPRVNPDYTRWRRQD 81
Query: 89 SLLFTWLLSTLSPAILPNVIQCVHSYQVWEEIHSFFNAKSRAQSTQLRNELKNITKGTKT 148
L+++ +L +S ++ P V + + Q+WE + + S TQLR
Sbjct: 82 KLIYSAILGAISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQLR------------ 129
Query: 149 ASEFLQRIKTIVNTLASIGEPVTFRDHLEAIFDGLSEEYSALMTVIYNQTTYFTISEVEA 208
F+ R + LA +G+P+ + +E + + L ++Y ++ I + T +++E+
Sbjct: 130 ---FITRF----DQLALLGKPMDHDEQVERVLENLPDDYKPVIDQIAAKDTPPSLTEIHE 182
Query: 209 MVISHEARFDRMRKKQLADTTPAVFLAQAQNS 240
+I+ E++ + ++ T V + N+
Sbjct: 183 RLINRESKLLALNSAEVVPITANVVTHRNTNT 214
>At1g59265 polyprotein, putative
Length = 1466
Score = 832 bits (2150), Expect = 0.0
Identities = 512/1268 (40%), Positives = 692/1268 (54%), Gaps = 161/1268 (12%)
Query: 416 PYSHATDMQGMQAAGMSFVRPWLPQGQFPAPTAQTPQFPRPPQQQVSTGMHPQAMLATVP 475
PY + G+Q G S R Q + +Q P P P Q P+A LA
Sbjct: 274 PYLGKCQICGVQ--GHSAKRCSQLQHFLSSVNSQQPPSPFTPWQ-------PRANLALGS 324
Query: 476 AHHSSPVTWCPDSGATHHVTNNPGIFVDHVPSSSQEQLLVGNGQGLPIQSIGSALLPSSF 535
+ S+ W DSGATHH+T++ H P + + ++V +G +PI GS L +
Sbjct: 325 PYSSN--NWLLDSGATHHITSDFNNLSLHQPYTGGDDVMVADGSTIPISHTGSTSLSTKS 382
Query: 536 NPTASLILKKLLLVPSITKNLVSVSRFARDNNVFFSFHANHCSVHHQDTYELLLTGTVGD 595
P L L +L VP+I KNL+SV R N V F V +T LL G D
Sbjct: 383 RP---LNLHNILYVPNIHKNLISVYRLCNANGVSVEFFPASFQVKDLNTGVPLLQGKTKD 439
Query: 596 DGLYYFDGPLTKASSQPQSSSPLSSIQACLASKSAANKASPAIDLQSATVSPIVPSLVPC 655
+ ++ P+ SS P+S ASP
Sbjct: 440 E---LYEWPIA-------SSQPVSLF------------ASP------------------- 458
Query: 656 NNADSVNVSSTVSDLVSCNSGSGISVYDLWHSRLGHPHHDVLKQTLSLCNISVPSNKSVF 715
S + + WH+RLGHP +L +S ++SV + F
Sbjct: 459 ---------------------SSKATHSSWHARLGHPAPSILNSVISNYSLSVLNPSHKF 497
Query: 716 SFCKACCLSKLHRLPSSSSNTVYNTPFELVFADLWGPASLESSCGFLYFLTCVDACTKYT 775
C C ++K +++P S S P E +++D+W L S + Y++ VD T+YT
Sbjct: 498 LSCSDCLINKSNKVPFSQSTINSTRPLEYIYSDVWSSPIL-SHDNYRYYVIFVDHFTRYT 556
Query: 776 WVFPLKKKSDTCATFINFQAMVELQFGHKLKSVQTDGGGEFKPLTSHFQKLGIIHRLTCP 835
W++PLK+KS TFI F+ ++E +F ++ + +D GGEF L +F + GI H + P
Sbjct: 557 WLYPLKQKSQVKETFITFKNLLENRFQTRIGTFYSDNGGEFVALWEYFSQHGISHLTSPP 616
Query: 836 HTHHQNGSVERKHRHIVETGLALLSHASMPLQFWDHAFLAATYLINRMSTTTLQGASPYF 895
HT NG ERKHRHIVETGL LLSHAS+P +W +AF A YLINR+ T LQ SP+
Sbjct: 617 HTPEHNGLSERKHRHIVETGLTLLSHASIPKTYWPYAFAVAVYLINRLPTPLLQLESPFQ 676
Query: 896 KLYGQYPDFKSLKVFGSACFPFLRPYNSNKLSLHSKECVFIGYSINHKGYKCLD-QSGRI 954
KL+G P++ L+VFG AC+P+LRPYN +KL S++CVF+GYS+ Y CL Q+ R+
Sbjct: 677 KLFGTSPNYDKLRVFGCACYPWLRPYNQHKLDDKSRQCVFLGYSLTQSAYLCLHLQTSRL 736
Query: 955 YISKDVLFHEHRFPYPILFPT-----DHSSSSSAEFYPLSTIPIIS------------HT 997
YIS+ V F E+ FP+ T + SS + P +T+P + H
Sbjct: 737 YISRHVRFDENCFPFSNYLATLSPVQEQRRESSCVWSPHTTLPTRTPVLPAPSCSDPHHA 796
Query: 998 A-PPSSP----------------VLAASGHSSP--------GPQDSPQQQQPPSATLSAD 1032
A PPSSP ++S SSP GPQ + Q Q + T S+
Sbjct: 797 ATPPSSPSAPFRNSQVSSSNLDSSFSSSFPSSPEPTAPRQNGPQPTTQPTQTQTQTHSSQ 856
Query: 1033 DPSTSTPAVVSP-----------------PVPSSSAQSADTS---ASVVAAAPPVI---- 1068
+ S + P SP P P++SA S+ TS S++ PP +
Sbjct: 857 NTSQNNPTNESPSQLAQSLSTPAQSSSSSPSPTTSASSSSTSPTPPSILIHPPPPLAQIV 916
Query: 1069 -------VNTHPMQTRAKNGIVKPRLQPTL---LLTHLEPTSVKQAMKDVKWYKAMQEEY 1118
+NTH M TRAK GI+KP + +L L EP + QA+KD +W AM E
Sbjct: 917 NNNNQAPLNTHSMGTRAKAGIIKPNPKYSLAVSLAAESEPRTAIQALKDERWRNAMGSEI 976
Query: 1119 TALMNNGTWTLVPLPANR-TPVGCKWIYRIKENPDGSINKYKARLVAKGYSQVQGFDYSE 1177
A + N TW LVP P + T VGC+WI+ K N DGS+N+YKARLVAKGY+Q G DY+E
Sbjct: 977 NAQIGNHTWDLVPPPPSHVTIVGCRWIFTKKYNSDGSLNRYKARLVAKGYNQRPGLDYAE 1036
Query: 1178 TFSPVVKPITVRLILSLAISRGWPLQQLDVNNAFLNGVLEEEVYMVQPPGFEHKDK-NLV 1236
TFSPV+K ++R++L +A+ R WP++QLDVNNAFL G L ++VYM QPPGF KD+ N V
Sbjct: 1037 TFSPVIKSTSIRIVLGVAVDRSWPIRQLDVNNAFLQGTLTDDVYMSQPPGFIDKDRPNYV 1096
Query: 1237 CKLQKALYGLKQAPRAWFHRLKEVLIRFGFQASKCDPSLFTYYSSQGCIYMLVYVDDIII 1296
CKL+KALYGLKQAPRAW+ L+ L+ GF S D SLF + +YMLVYVDDI+I
Sbjct: 1097 CKLRKALYGLKQAPRAWYVELRNYLLTIGFVNSVSDTSLFVLQRGKSIVYMLVYVDDILI 1156
Query: 1297 TGASMSLIQQLTAQLHSIFALKQLGQLDYFLGVQVTHLSNGNLLLNQTKYISDLLTKVNM 1356
TG +L+ L F++K +L YFLG++ + G L L+Q +YI DLL + NM
Sbjct: 1157 TGNDPTLLHNTLDNLSQRFSVKDHEELHYFLGIEAKRVPTG-LHLSQRRYILDLLARTNM 1215
Query: 1357 LDAAAISTPMQCGVKLSKNEGSALKDPTEYRSVVGALQYATITRPEISFAVNKVCQFMAQ 1416
+ A ++TPM KLS G+ L DPTEYR +VG+LQY TRP+IS+AVN++ QFM
Sbjct: 1216 ITAKPVTTPMAPSPKLSLYSGTKLTDPTEYRGIVGSLQYLAFTRPDISYAVNRLSQFMHM 1275
Query: 1417 PHEEHWKAVKRILRYLKGTLTHGILLQPCSMTRPLPLIAYCDADWGSDPDDRRSTSGSCV 1476
P EEH +A+KRILRYL GT HGI L+ L L AY DADW D DD ST+G V
Sbjct: 1276 PTEEHLQALKRILRYLAGTPNHGIFLK---KGNTLSLHAYSDADWAGDKDDYVSTNGYIV 1332
Query: 1477 FLGYNLISWTAKKQSLVARSSTEAEYRSLANTTAELLWVQSLLTELKIPFT-IPTVYCDN 1535
+LG++ ISW++KKQ V RSSTEAEYRS+ANT++E+ W+ SLLTEL I T P +YCDN
Sbjct: 1333 YLGHHPISWSSKKQKGVVRSSTEAEYRSVANTSSEMQWICSLLTELGIRLTRPPVIYCDN 1392
Query: 1536 MSTVILTHNPILHTRTKHMEMDIFFVREKVQDKSLLVQHVPSEHQKADIFTKALSPTRFL 1595
+ L NP+ H+R KH+ +D F+R +VQ +L V HV + Q AD TK LS T F
Sbjct: 1393 VGATYLCANPVFHSRMKHIAIDYHFIRNQVQSGALRVVHVSTHDQLADTLTKPLSRTAFQ 1452
Query: 1596 LLKDKLNV 1603
K+ V
Sbjct: 1453 NFASKIGV 1460
Score = 107 bits (266), Expect = 7e-23
Identities = 53/187 (28%), Positives = 102/187 (54%), Gaps = 2/187 (1%)
Query: 30 KLDDSNFYSWKQQVEGVIRSHKLQKFVQDPPMIPEKFLTEEDKITENVNPLFADWEQQDS 89
KL +N+ W +QV + ++L F+ +P + + VNP + W++QD
Sbjct: 25 KLTSTNYLMWSRQVHALFDGYELAGFLDGSTTMPPATIGTD--AAPRVNPDYTRWKRQDK 82
Query: 90 LLFTWLLSTLSPAILPNVIQCVHSYQVWEEIHSFFNAKSRAQSTQLRNELKNITKGTKTA 149
L+++ +L +S ++ P V + + Q+WE + + S TQLR +LK TKGTKT
Sbjct: 83 LIYSAVLGAISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQLRTQLKQWTKGTKTI 142
Query: 150 SEFLQRIKTIVNTLASIGEPVTFRDHLEAIFDGLSEEYSALMTVIYNQTTYFTISEVEAM 209
+++Q + T + LA +G+P+ + +E + + L EEY ++ I + T T++E+
Sbjct: 143 DDYMQGLVTRFDQLALLGKPMDHDEQVERVLENLPEEYKPVIDQIAAKDTPPTLTEIHER 202
Query: 210 VISHEAR 216
+++HE++
Sbjct: 203 LLNHESK 209
>At1g58889 polyprotein, putative
Length = 1466
Score = 831 bits (2146), Expect = 0.0
Identities = 511/1268 (40%), Positives = 691/1268 (54%), Gaps = 161/1268 (12%)
Query: 416 PYSHATDMQGMQAAGMSFVRPWLPQGQFPAPTAQTPQFPRPPQQQVSTGMHPQAMLATVP 475
PY + G+Q G S R Q + +Q P P P Q P+A LA
Sbjct: 274 PYLGKCQICGVQ--GHSAKRCSQLQHFLSSVNSQQPPSPFTPWQ-------PRANLALGS 324
Query: 476 AHHSSPVTWCPDSGATHHVTNNPGIFVDHVPSSSQEQLLVGNGQGLPIQSIGSALLPSSF 535
+ S+ W DSGATHH+T++ H P + + ++V +G +PI GS L +
Sbjct: 325 PYSSN--NWLLDSGATHHITSDFNNLSLHQPYTGGDDVMVADGSTIPISHTGSTSLSTKS 382
Query: 536 NPTASLILKKLLLVPSITKNLVSVSRFARDNNVFFSFHANHCSVHHQDTYELLLTGTVGD 595
P L L +L VP+I KNL+SV R N V F V +T LL G D
Sbjct: 383 RP---LNLHNILYVPNIHKNLISVYRLCNANGVSVEFFPASFQVKDLNTGVPLLQGKTKD 439
Query: 596 DGLYYFDGPLTKASSQPQSSSPLSSIQACLASKSAANKASPAIDLQSATVSPIVPSLVPC 655
+ ++ P+ SS P+S ASP
Sbjct: 440 E---LYEWPIA-------SSQPVSLF------------ASP------------------- 458
Query: 656 NNADSVNVSSTVSDLVSCNSGSGISVYDLWHSRLGHPHHDVLKQTLSLCNISVPSNKSVF 715
S + + WH+RLGHP +L +S ++SV + F
Sbjct: 459 ---------------------SSKATHSSWHARLGHPAPSILNSVISNYSLSVLNPSHKF 497
Query: 716 SFCKACCLSKLHRLPSSSSNTVYNTPFELVFADLWGPASLESSCGFLYFLTCVDACTKYT 775
C C ++K +++P S S P E +++D+W L S + Y++ VD T+YT
Sbjct: 498 LSCSDCLINKSNKVPFSQSTINSTRPLEYIYSDVWSSPIL-SHDNYRYYVIFVDHFTRYT 556
Query: 776 WVFPLKKKSDTCATFINFQAMVELQFGHKLKSVQTDGGGEFKPLTSHFQKLGIIHRLTCP 835
W++PLK+KS TFI F+ ++E +F ++ + +D GGEF L +F + GI H + P
Sbjct: 557 WLYPLKQKSQVKETFITFKNLLENRFQTRIGTFYSDNGGEFVALWEYFSQHGISHLTSPP 616
Query: 836 HTHHQNGSVERKHRHIVETGLALLSHASMPLQFWDHAFLAATYLINRMSTTTLQGASPYF 895
HT NG ERKHRHIVETGL LLSHAS+P +W +AF A YLINR+ T LQ SP+
Sbjct: 617 HTPEHNGLSERKHRHIVETGLTLLSHASIPKTYWPYAFAVAVYLINRLPTPLLQLESPFQ 676
Query: 896 KLYGQYPDFKSLKVFGSACFPFLRPYNSNKLSLHSKECVFIGYSINHKGYKCLD-QSGRI 954
KL+G P++ L+VFG AC+P+LRPYN +KL S++CVF+GYS+ Y CL Q+ R+
Sbjct: 677 KLFGTSPNYDKLRVFGCACYPWLRPYNQHKLDDKSRQCVFLGYSLTQSAYLCLHLQTSRL 736
Query: 955 YISKDVLFHEHRFPYPILFPT-----DHSSSSSAEFYPLSTIPIIS------------HT 997
YIS+ V F E+ FP+ T + SS + P +T+P + H
Sbjct: 737 YISRHVRFDENCFPFSNYLATLSPVQEQRRESSCVWSPHTTLPTRTPVLPAPSCSDPHHA 796
Query: 998 A-PPSSP----------------VLAASGHSSP--------GPQDSPQQQQPPSATLSAD 1032
A PPSSP ++S SSP GPQ + Q Q + T S+
Sbjct: 797 ATPPSSPSAPFRNSQVSSSNLDSSFSSSFPSSPEPTAPRQNGPQPTTQPTQTQTQTHSSQ 856
Query: 1033 DPSTSTPAVVSP-----------------PVPSSSAQSADTS---ASVVAAAPPVI---- 1068
+ S + P SP P P++SA S+ TS S++ PP +
Sbjct: 857 NTSQNNPTNESPSQLAQSLSTPAQSSSSSPSPTTSASSSSTSPTPPSILIHPPPPLAQIV 916
Query: 1069 -------VNTHPMQTRAKNGIVKPRLQPTL---LLTHLEPTSVKQAMKDVKWYKAMQEEY 1118
+NTH M TRAK GI+KP + +L L EP + QA+KD +W AM E
Sbjct: 917 NNNNQAPLNTHSMGTRAKAGIIKPNPKYSLAVSLAAESEPRTAIQALKDERWRNAMGSEI 976
Query: 1119 TALMNNGTWTLVPLPANR-TPVGCKWIYRIKENPDGSINKYKARLVAKGYSQVQGFDYSE 1177
A + N TW LVP P + T VGC+WI+ K N DGS+N+YKAR VAKGY+Q G DY+E
Sbjct: 977 NAQIGNHTWDLVPPPPSHVTIVGCRWIFTKKYNSDGSLNRYKARFVAKGYNQRPGLDYAE 1036
Query: 1178 TFSPVVKPITVRLILSLAISRGWPLQQLDVNNAFLNGVLEEEVYMVQPPGFEHKDK-NLV 1236
TFSPV+K ++R++L +A+ R WP++QLDVNNAFL G L ++VYM QPPGF KD+ N V
Sbjct: 1037 TFSPVIKSTSIRIVLGVAVDRSWPIRQLDVNNAFLQGTLTDDVYMSQPPGFIDKDRPNYV 1096
Query: 1237 CKLQKALYGLKQAPRAWFHRLKEVLIRFGFQASKCDPSLFTYYSSQGCIYMLVYVDDIII 1296
CKL+KALYGLKQAPRAW+ L+ L+ GF S D SLF + +YMLVYVDDI+I
Sbjct: 1097 CKLRKALYGLKQAPRAWYVELRNYLLTIGFVNSVSDTSLFVLQRGKSIVYMLVYVDDILI 1156
Query: 1297 TGASMSLIQQLTAQLHSIFALKQLGQLDYFLGVQVTHLSNGNLLLNQTKYISDLLTKVNM 1356
TG +L+ L F++K +L YFLG++ + G L L+Q +YI DLL + NM
Sbjct: 1157 TGNDPTLLHNTLDNLSQRFSVKDHEELHYFLGIEAKRVPTG-LHLSQRRYILDLLARTNM 1215
Query: 1357 LDAAAISTPMQCGVKLSKNEGSALKDPTEYRSVVGALQYATITRPEISFAVNKVCQFMAQ 1416
+ A ++TPM KLS G+ L DPTEYR +VG+LQY TRP+IS+AVN++ QFM
Sbjct: 1216 ITAKPVTTPMAPSPKLSLYSGTKLTDPTEYRGIVGSLQYLAFTRPDISYAVNRLSQFMHM 1275
Query: 1417 PHEEHWKAVKRILRYLKGTLTHGILLQPCSMTRPLPLIAYCDADWGSDPDDRRSTSGSCV 1476
P EEH +A+KRILRYL GT HGI L+ L L AY DADW D DD ST+G V
Sbjct: 1276 PTEEHLQALKRILRYLAGTPNHGIFLK---KGNTLSLHAYSDADWAGDKDDYVSTNGYIV 1332
Query: 1477 FLGYNLISWTAKKQSLVARSSTEAEYRSLANTTAELLWVQSLLTELKIPFT-IPTVYCDN 1535
+LG++ ISW++KKQ V RSSTEAEYRS+ANT++E+ W+ SLLTEL I T P +YCDN
Sbjct: 1333 YLGHHPISWSSKKQKGVVRSSTEAEYRSVANTSSEMQWICSLLTELGIRLTRPPVIYCDN 1392
Query: 1536 MSTVILTHNPILHTRTKHMEMDIFFVREKVQDKSLLVQHVPSEHQKADIFTKALSPTRFL 1595
+ L NP+ H+R KH+ +D F+R +VQ +L V HV + Q AD TK LS T F
Sbjct: 1393 VGATYLCANPVFHSRMKHIAIDYHFIRNQVQSGALRVVHVSTHDQLADTLTKPLSRTAFQ 1452
Query: 1596 LLKDKLNV 1603
K+ V
Sbjct: 1453 NFASKIGV 1460
Score = 107 bits (266), Expect = 7e-23
Identities = 53/187 (28%), Positives = 102/187 (54%), Gaps = 2/187 (1%)
Query: 30 KLDDSNFYSWKQQVEGVIRSHKLQKFVQDPPMIPEKFLTEEDKITENVNPLFADWEQQDS 89
KL +N+ W +QV + ++L F+ +P + + VNP + W++QD
Sbjct: 25 KLTSTNYLMWSRQVHALFDGYELAGFLDGSTTMPPATIGTD--AAPRVNPDYTRWKRQDK 82
Query: 90 LLFTWLLSTLSPAILPNVIQCVHSYQVWEEIHSFFNAKSRAQSTQLRNELKNITKGTKTA 149
L+++ +L +S ++ P V + + Q+WE + + S TQLR +LK TKGTKT
Sbjct: 83 LIYSAVLGAISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQLRTQLKQWTKGTKTI 142
Query: 150 SEFLQRIKTIVNTLASIGEPVTFRDHLEAIFDGLSEEYSALMTVIYNQTTYFTISEVEAM 209
+++Q + T + LA +G+P+ + +E + + L EEY ++ I + T T++E+
Sbjct: 143 DDYMQGLVTRFDQLALLGKPMDHDEQVERVLENLPEEYKPVIDQIAAKDTPPTLTEIHER 202
Query: 210 VISHEAR 216
+++HE++
Sbjct: 203 LLNHESK 209
>At1g44510 polyprotein, putative
Length = 1459
Score = 813 bits (2099), Expect = 0.0
Identities = 497/1238 (40%), Positives = 679/1238 (54%), Gaps = 154/1238 (12%)
Query: 436 PWLPQGQFPAPTAQTPQFPRPPQQQVSTGMHPQAMLATVPAHHSSPVTWCPDSGATHHVT 495
P L Q PA ++ F T P+A LA + ++P W DSGATHH+T
Sbjct: 300 PQLQAMQLPASSSAHSPF---------TPWQPRANLAIGSPYAANP--WLLDSGATHHIT 348
Query: 496 NNPGIFVDHVPSSSQEQLLVGNGQGLPIQSIGSALLPSSFNPTASLILKKLLLVPSITKN 555
++ H P + E +++ +G GL I+ GS LPS L L K+L VP I KN
Sbjct: 349 SDLNALSLHQPYNGGEYVMIADGTGLTIKQTGSTFLPSQ---NRDLALHKVLYVPDIRKN 405
Query: 556 LVSVSRFARDNNVFFSFHANHCSVHHQDTYELLLTGTVGDDGLYYF---DGPLTKASSQP 612
L+SV R N V F V +T LLL G DD LY + + P T + P
Sbjct: 406 LISVYRLCNTNQVSVEFFPASFQVKDLNTGTLLLQGRTKDD-LYEWPVTNPPATALFTSP 464
Query: 613 QSSSPLSSIQACLASKSAANKASPAIDLQSATVSPIVPSLVPCNNADSVNVSSTVSDLVS 672
+ LSS + L SA+ I+ +L+ + S+ VS S+ S
Sbjct: 465 SPKTTLSSWHSRLGHPSAS----------------ILNTLL---SKFSLPVSVASSNKTS 505
Query: 673 CNSGSGISVYDLWHSRLGHPHHDVLKQTLSLCNISVPSNKSVFSFCKACCLSKLHRLPSS 732
C+ C I NKS H+LP +
Sbjct: 506 CSD----------------------------CLI----NKS-------------HKLPFA 520
Query: 733 SSNTVYNTPFELVFADLWGPASLESSCGFLYFLTCVDACTKYTWVFPLKKKSDTCATFIN 792
+S+ ++P E +F D+W + + S + Y+L VD T+YTW++PL++KS ATFI
Sbjct: 521 TSSIHSSSPLEYIFTDVW-TSPIISHDNYKYYLVLVDHYTRYTWLYPLQQKSQVKATFIA 579
Query: 793 FQAMVELQFGHKLKSVQTDGGGEFKPLTSHFQKLGIIHRLTCPHTHHQNGSVERKHRHIV 852
F+A+VE +F K++++ +D GGEF L GI H + PHT NG ERKHRHIV
Sbjct: 580 FKALVENRFQAKIRTLYSDNGGEFIALRDFLVSNGISHLTSPPHTPEHNGLSERKHRHIV 639
Query: 853 ETGLALLSHASMPLQFWDHAFLAATYLINRMSTTTLQGASPYFKLYGQYPDFKSLKVFGS 912
ETGL LL+ AS+P ++W +AF A YLINRM T L SP+ KL+G P+++ L+VFG
Sbjct: 640 ETGLTLLTQASVPREYWTYAFATAVYLINRMPTPVLCLQSPFQKLFGSSPNYQRLRVFGC 699
Query: 913 ACFPFLRPYNSNKLSLHSKECVFIGYSINHKGYKCLD-QSGRIYISKDVLFHEHRFPY-- 969
CFP+LRPY NKL SK CVF+GYS+ Y CLD + R+Y S+ V+F E +P+
Sbjct: 700 LCFPWLRPYTRNKLEERSKRCVFLGYSLTQTAYLCLDVDNNRLYTSRHVMFDESTYPFAA 759
Query: 970 --------PILFPTDHSSSSSA--EFYPLSTI----PIISHTAPPSSPVLAASGHSSPGP 1015
++ P + SSSSS +P S + P S PS P SP
Sbjct: 760 SIREQSQSSLVTPPESSSSSSPANSGFPCSVLRLQSPPASSPETPSPPQQQNDSPVSPRQ 819
Query: 1016 QDSPQQQQ---------PPSATLSADDPST-----STPAVVSPP---------------V 1046
SP PS ++S +P+ P S P
Sbjct: 820 TGSPTPSHHSQVRDSTLSPSPSVSNSEPTAPHENGPEPEAQSNPNSPFIGPLPNPNPETN 879
Query: 1047 PSSSAQSADTSASVVAAAPPVIV--------------NTHPMQTRAKNGIVKPRLQPTLL 1092
PSSS + S A PP N H M+TR+KN I KP+ + +L
Sbjct: 880 PSSSIEQRPVDKSTTTALPPNQTTIAATSNSRSQPPKNNHQMKTRSKNNITKPKTKTSLT 939
Query: 1093 LT----HL-EPTSVKQAMKDVKWYKAMQEEYTALMNNGTWTLVPLPANRTPVGCKWIYRI 1147
+ HL EP +V QA+KD KW AM +E+ A N TW LVP + VGC+W++++
Sbjct: 940 VALTQPHLSEPNTVTQALKDKKWRFAMSDEFDAQQRNHTWDLVPPNPTQHLVGCRWVFKL 999
Query: 1148 KENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILSLAISRGWPLQQLDV 1207
K P+G I+KYKARLVAKG++Q G DY+ETFSPV+K T+R++L +A+ + WPL+QLDV
Sbjct: 1000 KYLPNGLIDKYKARLVAKGFNQQYGVDYAETFSPVIKATTIRVVLDVAVKKNWPLKQLDV 1059
Query: 1208 NNAFLNGVLEEEVYMVQPPGFEHKDK-NLVCKLQKALYGLKQAPRAWFHRLKEVLIRFGF 1266
NNAFL G L EEVYM QPPGF KD+ + VC+L+KA+YGLKQAPRAW+ LK+ L+ GF
Sbjct: 1060 NNAFLQGTLTEEVYMAQPPGFVDKDRPSHVCRLRKAIYGLKQAPRAWYMELKQHLLNIGF 1119
Query: 1267 QASKCDPSLFTYYSSQGCIYMLVYVDDIIITGASMSLIQQLTAQLHSIFALKQLGQLDYF 1326
S D SLF Y +Y+LVYVDDII+TG+ + + + L F++K L YF
Sbjct: 1120 VNSLADTSLFIYSHGTTLLYLLVYVDDIIVTGSDHKSVSAVLSSLAERFSIKDPTDLHYF 1179
Query: 1327 LGVQVTHLSNGNLLLNQTKYISDLLTKVNMLDAAAISTPMQCGVKLSKNEGSALKDPTEY 1386
LG++ T + G L L Q KY++DLL K NMLDA ++TP+ KL+ + G+ L D +EY
Sbjct: 1180 LGIEATRTNTG-LHLMQRKYMTDLLAKHNMLDAKPVATPLPTSPKLTLHGGTKLNDASEY 1238
Query: 1387 RSVVGALQYATITRPEISFAVNKVCQFMAQPHEEHWKAVKRILRYLKGTLTHGILLQPCS 1446
RSVVG+LQY TRP+I+FAVN++ QFM QP +HW+A KR+LRYL GT THGI L S
Sbjct: 1239 RSVVGSLQYLAFTRPDIAFAVNRLSQFMHQPTSDHWQAAKRVLRYLAGTTTHGIFLNSSS 1298
Query: 1447 MTRPLPLIAYCDADWGSDPDDRRSTSGSCVFLGYNLISWTAKKQSLVARSSTEAEYRSLA 1506
P+ L A+ DADW D D ST+ ++LG N ISW++KKQ V+RSSTE+EYR++A
Sbjct: 1299 ---PIHLHAFSDADWAGDSADYVSTNAYVIYLGRNPISWSSKKQRGVSRSSTESEYRAVA 1355
Query: 1507 NTTAELLWVQSLLTELKIPFTI-PTVYCDNMSTVILTHNPILHTRTKHMEMDIFFVREKV 1565
N +E+ W+ SLLTEL I PT++CDN+ + NP+ H+R KH+ +D FVR +
Sbjct: 1356 NAASEIRWLCSLLTELHIRLPHGPTIFCDNIGATYICANPVFHSRMKHIALDYHFVRGMI 1415
Query: 1566 QDKSLLVQHVPSEHQKADIFTKALSPTRFLLLKDKLNV 1603
Q ++L V HV + Q AD TK+LS FL + K+ V
Sbjct: 1416 QSRALRVSHVSTNDQLADALTKSLSRPHFLSARSKIGV 1453
Score = 112 bits (281), Expect = 1e-24
Identities = 83/319 (26%), Positives = 148/319 (46%), Gaps = 37/319 (11%)
Query: 30 KLDDSNFYSWKQQVEGVIRSHKLQKFVQDPPMIPEKFLTEEDKITENVNPLFADWEQQDS 89
KL +N+ W Q+ ++ + L ++ + +IP + T ++ NP F W++QD
Sbjct: 32 KLTSTNYLMWSIQIHALLDGYDLAGYLDNSVVIPPETTTINSVVS--ANPSFTLWKRQDK 89
Query: 90 LLFTWLLSTLSPAILPNVIQCVHSYQVWEEIHSFFNAKSRAQSTQLRNELKNITKGTKTA 149
L+F+ L+ +SPA+ V + +S Q+W +++ + S QLR +++ +TKGTKT
Sbjct: 90 LIFSALIGAISPAVQSLVSRATNSSQIWSTLNNTYAKPSYGHIKQLRQQIQRLTKGTKTI 149
Query: 150 SEFLQRIKTIVNTLASIGEPVTFRDHLEAIFDGLSEEYSALMTVIYNQTTYFTISEVEAM 209
E++Q T ++ LA +G+P+ + +E I GL EEY ++ I + TI+E+
Sbjct: 150 DEYVQSHTTRLDQLAILGKPMEHEEQVEHILKGLPEEYKTVVDQIEGKDNTPTITEIHER 209
Query: 210 VISHEARFDRMRKKQLADTTPAVFLAQAQNSPSSSTAPAPPPPQAMWTQPAPAPSSTMAQ 269
+I+HE+ K L+D P PSSS P ++ + Q
Sbjct: 210 LINHES-------KLLSDEVP----------PSSS---------------FPMSANAVQQ 237
Query: 270 PP-ATSVAPSQPVDTAVDSGYGNRDEQNRNSGGYGR-GRGRGRGRNVQCTYCSKWGHDAA 327
+ +Q + + + N N Y + G+ + +C CS GH A
Sbjct: 238 RNFNNNCNQNQHKNRYQGNTHNNNTNTNSQPSTYNKSGQRTFKPYLGKCQICSVQGHSAR 297
Query: 328 SCWSRPSGVSSNANSSANS 346
C + + A+SSA+S
Sbjct: 298 RC-PQLQAMQLPASSSAHS 315
>At4g16870 retrotransposon like protein
Length = 1474
Score = 762 bits (1968), Expect = 0.0
Identities = 438/1009 (43%), Positives = 591/1009 (58%), Gaps = 98/1009 (9%)
Query: 685 WHSRLGHPHHDVLKQTLSLCNISVPSNKSVFSFCKACCLSKLHRLPSSSSNTVYNTPFEL 744
WHSRLGHP +L +S ++ V + S C C ++K H+LP S S+ +P E
Sbjct: 468 WHSRLGHPSSSILNTLISKFSLPVSVSASNKLACSDCFINKSHKLPFSISSIKSTSPLEY 527
Query: 745 VFADLWGPASLESSCGFLYFLTCVDACTKYTWVFPLKKKSDTCATFINFQAMVELQFGHK 804
+F+D+W L S + Y+L VD T+YTW++PL++KS +TFI F+A+VE +F K
Sbjct: 528 IFSDVWMSPIL-SPDNYKYYLVLVDHHTRYTWLYPLQQKSQVKSTFIAFKALVENRFQAK 586
Query: 805 LKSVQTDGGGEFKPLTSHFQKLGIIHRLTCPHTHHQNGSVERKHRHIVETGLALLSHASM 864
++++ +D GGEF L GI H + PHT NG ERKHRHIVETGL LL+ AS+
Sbjct: 587 IRTLYSDNGGEFIALREFLVSNGISHLTSPPHTPEHNGLSERKHRHIVETGLTLLTQASV 646
Query: 865 PLQFWDHAFLAATYLINRMSTTTLQGASPYFKLYGQYPDFKSLKVFGSACFPFLRPYNSN 924
P ++W +AF AA YLINRM T L SP+ KL+G P+++ L+VFG CFP+LRPY N
Sbjct: 647 PREYWPYAFAAAVYLINRMPTPVLSMESPFQKLFGSKPNYERLRVFGCLCFPWLRPYTHN 706
Query: 925 KLSLHSKECVFIGYSINHKGYKCLD-QSGRIYISKDVLFHEHRFPY-------------- 969
KL S+ CVF+GYS+ Y C D + R+Y S+ V+F E FP+
Sbjct: 707 KLEERSRRCVFLGYSLTQTAYLCFDVEHKRLYTSRHVVFDEASFPFSNLTSQNSLPTVTF 766
Query: 970 -----PILFPTDHSSS------SSA-----EFYPLSTIPIISHTAPP------------- 1000
P++ P SSS SS + P T P H++ P
Sbjct: 767 EQSSSPLVTPILSSSSVLPSCLSSPCTVLHQQQPPVTTPNSPHSSQPTTSPAPLSPHRST 826
Query: 1001 -----------SSPVLAASG--HSSP------GPQDSPQQQQPPSATLS----------A 1031
SSP+L++S +S P GP+ P+ Q PP LS
Sbjct: 827 TMDFQVPQVRSSSPLLSSSSSLNSEPTAPNENGPE--PEAQSPPIGPLSNPTHEAFIGPL 884
Query: 1032 DDPSTS-------TPAVVSPPVPSSSAQSADTSASVVAAAPPVIV---NTHPMQTRAKNG 1081
+P+ + TPA PV ++ + +V A+ N H M+TRAKN
Sbjct: 885 PNPNRNPTNEIEPTPAPHPKPVKPTTTTTTPNRTTVSDASHQPTAPQQNQHNMKTRAKNN 944
Query: 1082 IVKPRLQPTLLLT-----HLEPTSVKQAMKDVKWYKAMQEEYTALMNNGTWTLVPLPANR 1136
I KP + +L T EPT+V QA+KD KW AM +E+ A N TW LVP ++
Sbjct: 945 IKKPNTKFSLTATLPNRSPSEPTNVTQALKDKKWRFAMSDEFDAQQRNHTWDLVP-HESQ 1003
Query: 1137 TPVGCKWIYRIKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILSLAI 1196
VGCKW++++K P+G+I+KYKARLVAKG++Q G DY+ETFSPV+K T+RL+L +A+
Sbjct: 1004 LLVGCKWVFKLKYLPNGAIDKYKARLVAKGFNQQYGVDYAETFSPVIKSTTIRLVLDVAV 1063
Query: 1197 SRGWPLQQLDVNNAFLNGVLEEEVYMVQPPGFEHKDK-NLVCKLQKALYGLKQAPRAWFH 1255
+ W ++QLDVNNAFL G L EEVYM QPPGF KD+ VC+L+KA+YGLKQAPRAW+
Sbjct: 1064 KKDWEIKQLDVNNAFLQGTLTEEVYMAQPPGFIDKDRPTHVCRLRKAIYGLKQAPRAWYM 1123
Query: 1256 RLKEVLIRFGFQASKCDPSLFTYYSSQGCIYMLVYVDDIIITGASMSLIQQLTAQLHSIF 1315
LK+ L GF S D SLF Y +Y+LVYVDDII+TG+ S I + L F
Sbjct: 1124 ELKQHLFNIGFVNSLSDASLFIYCHGTTFVYVLVYVDDIIVTGSDKSSIDAVLTSLAERF 1183
Query: 1316 ALKQLGQLDYFLGVQVTHLSNGNLLLNQTKYISDLLTKVNMLDAAAISTPMQCGVKLSKN 1375
++K L YFLG++ T G L L Q KYI DLL K NM DA + TP+ KL+ +
Sbjct: 1184 SIKDPTDLHYFLGIEATRTKQG-LHLMQRKYIKDLLAKHNMADAKPVLTPLPTSPKLTLH 1242
Query: 1376 EGSALKDPTEYRSVVGALQYATITRPEISFAVNKVCQFMAQPHEEHWKAVKRILRYLKGT 1435
G+ L D +EYRSVVG+LQY TRP+I++AVN++ Q M QP E+HW+A KR+LRYL GT
Sbjct: 1243 GGTKLNDASEYRSVVGSLQYLAFTRPDIAYAVNRLSQLMPQPTEDHWQAAKRVLRYLAGT 1302
Query: 1436 LTHGILLQPCSMTRPLPLIAYCDADWGSDPDDRRSTSGSCVFLGYNLISWTAKKQSLVAR 1495
THGI L T PL L A+ DADW D DD ST+ ++LG N ISW++KKQ VAR
Sbjct: 1303 STHGIFL---DTTSPLNLHAFSDADWAGDSDDYVSTNAYVIYLGKNPISWSSKKQRGVAR 1359
Query: 1496 SSTEAEYRSLANTTAELLWVQSLLTELKIPFTI-PTVYCDNMSTVILTHNPILHTRTKHM 1554
SSTE+EYR++AN +E+ W+ SLL++L I I P+++CDN+ L NP+ H+R KH+
Sbjct: 1360 SSTESEYRAVANAASEVKWLCSLLSKLHIRLPIRPSIFCDNIGATYLCANPVFHSRMKHI 1419
Query: 1555 EMDIFFVREKVQDKSLLVQHVPSEHQKADIFTKALSPTRFLLLKDKLNV 1603
+D FVR +Q +L V HV + Q AD TK LS F + K+ V
Sbjct: 1420 AIDYHFVRNMIQSGALRVSHVSTRDQLADALTKPLSRAHFQSARFKIGV 1468
Score = 303 bits (776), Expect = 5e-82
Identities = 216/660 (32%), Positives = 319/660 (47%), Gaps = 95/660 (14%)
Query: 446 PTAQTPQFPRPPQQQVSTGMHPQAMLATVPAHHSSPVTWCPDSGATHHVTNNPGIFVDHV 505
P Q Q T P+A LA + ++ W DSGATHH+T++ H
Sbjct: 297 PQLQAMQPSSSSSASTFTPWQPRANLAMGAPYTAN--NWLLDSGATHHITSDLNALALHQ 354
Query: 506 PSSSQEQLLVGNGQGLPIQSIGSALLPSSFNPTASLILKKLLLVPSITKNLVSVSRFARD 565
P + + +++ +G L I GS LPS+ L L K+L VP I KNLVSV R
Sbjct: 355 PYNGDD-VMIADGTSLKITKTGSTFLPSN---ARDLTLNKVLYVPDIQKNLVSVYRLCNT 410
Query: 566 NNVFFSFHANHCSVHHQDTYELLLTGTVGDDGLYYFDGPLTKASSQPQSSSPLSSIQACL 625
N V F V +T LLL G D+ LY + KA++ + SP +++
Sbjct: 411 NQVSVEFFPASFQVKDLNTGTLLLQGRTKDE-LYEWPVTNPKATALFTTPSPKTTL---- 465
Query: 626 ASKSAANKASPAIDLQSATVSPIVPSLVPCNNADSVNVSSTVSDLVSCNSGSGISVYDLW 685
S + P+ + + +S S+ VS + S+ ++C+
Sbjct: 466 -SSWHSRLGHPSSSILNTLISKF-----------SLPVSVSASNKLACSD---------- 503
Query: 686 HSRLGHPHHDVLKQTLSLCNISVPSNKSVFSFCKACCLSKLHRLPSSSSNTVYNTPFELV 745
C ++K H+LP S S+ +P E +
Sbjct: 504 -----------------------------------CFINKSHKLPFSISSIKSTSPLEYI 528
Query: 746 FADLWGPASLESSCGFLYFLTCVDACTKYTWVFPLKKKSDTCATFINFQAMVELQFGHKL 805
F+D+W + + S + Y+L VD T+YTW++PL++KS +TFI F+A+VE +F K+
Sbjct: 529 FSDVW-MSPILSPDNYKYYLVLVDHHTRYTWLYPLQQKSQVKSTFIAFKALVENRFQAKI 587
Query: 806 KSVQTDGGGEFKPLTSHFQKLGIIHRLTCPHTHHQNGSVERKHRHIVETGLALLSHASMP 865
+++ +D GGEF L GI H + PHT NG ERKHRHIVETGL LL+ AS+P
Sbjct: 588 RTLYSDNGGEFIALREFLVSNGISHLTSPPHTPEHNGLSERKHRHIVETGLTLLTQASVP 647
Query: 866 LQFWDHAFLAATYLINRMSTTTLQGASPYFKLYGQYPDFKSLKVFGSACFPFLRPYNSNK 925
++W +AF AA YLINRM T L SP+ KL+G P+++ L+VFG CFP+LRPY NK
Sbjct: 648 REYWPYAFAAAVYLINRMPTPVLSMESPFQKLFGSKPNYERLRVFGCLCFPWLRPYTHNK 707
Query: 926 LSLHSKECVFIGYSINHKGYKCLD-QSGRIYISKDVLFHEHRFPYPILFPTDHSSSSSAE 984
L S+ CVF+GYS+ Y C D + R+Y S+ V+F E FP+ L T +S
Sbjct: 708 LEERSRRCVFLGYSLTQTAYLCFDVEHKRLYTSRHVVFDEASFPFSNL--TSQNS----- 760
Query: 985 FYPLSTIPIISHTAPPSSPVLAASGHSSPGPQDSP----QQQQPPSATLSADDPS--TST 1038
L T+ ++P +P+L++S P SP QQQPP T ++ S T++
Sbjct: 761 ---LPTVTFEQSSSPLVTPILSSSS-VLPSCLSSPCTVLHQQQPPVTTPNSPHSSQPTTS 816
Query: 1039 PAVVSP--------PVPSSSAQSADTSASVVAAAPPVIVNTHPMQTRAKNGIVKPRLQPT 1090
PA +SP VP + S S+S + P N + + A++ + P PT
Sbjct: 817 PAPLSPHRSTTMDFQVPQVRSSSPLLSSSSSLNSEPTAPNENGPEPEAQSPPIGPLSNPT 876
Score = 105 bits (261), Expect = 3e-22
Identities = 52/187 (27%), Positives = 102/187 (53%), Gaps = 2/187 (1%)
Query: 30 KLDDSNFYSWKQQVEGVIRSHKLQKFVQDPPMIPEKFLTEEDKITENVNPLFADWEQQDS 89
KL +N+ W Q+ ++ ++L + P LT + ++ NP + W++QD
Sbjct: 33 KLTSNNYLMWSLQIHALLDGYELAGHLDGSIETPAPTLTTNNVVS--ANPQYTLWKRQDR 90
Query: 90 LLFTWLLSTLSPAILPNVIQCVHSYQVWEEIHSFFNAKSRAQSTQLRNELKNITKGTKTA 149
L+F+ L+ +SP + P V + + Q+W+ + + + S QLR ++K + KGTKT
Sbjct: 91 LIFSALIGAISPPVQPLVSRATKASQIWKTLTNTYAKSSYDHIKQLRTQIKQLKKGTKTI 150
Query: 150 SEFLQRIKTIVNTLASIGEPVTFRDHLEAIFDGLSEEYSALMTVIYNQTTYFTISEVEAM 209
E++ T+++ LA +G+P+ + +E I +GL E+Y ++ I + +I+E+
Sbjct: 151 DEYVLSHTTLLDQLAILGKPMEHEEQVERILEGLPEDYKTVVDQIEGKDNTPSITEIHER 210
Query: 210 VISHEAR 216
+I+HEA+
Sbjct: 211 LINHEAK 217
>At1g53810
Length = 1522
Score = 746 bits (1925), Expect = 0.0
Identities = 446/1093 (40%), Positives = 612/1093 (55%), Gaps = 98/1093 (8%)
Query: 594 GDDGLYYFDG---PLTKASSQPQSSS----PLSSIQACLASKSAANKASPAIDLQSATVS 646
G D + DG P+T S +SS PL + C P I +VS
Sbjct: 351 GSDSIMVADGNFLPITHTGSGSIASSSGKIPLKEVLVC-----------PDIVKSLLSVS 399
Query: 647 PIVPSLVPCN---NADSVNVS--STVSDLVSCNSGSGI------------------SVYD 683
+ S PC+ +ADSV ++ +T LV + G+ + +
Sbjct: 400 KLT-SDYPCSVEFDADSVRINDKATKKLLVMGRNRDGLYSLEEPKLQVLYSTRQNSASSE 458
Query: 684 LWHSRLGHPHHDVLKQTLSLCNISVPSNKSVFSFCKACCLSKLHRLPSSSSNTVYNTPFE 743
+WH RLGH + +VL Q S +I + NK V + C+AC L K RLP S + P E
Sbjct: 459 VWHRRLGHANAEVLHQLASSKSIII-INKVVKTVCEACHLGKSTRLPFMLSTFNASRPLE 517
Query: 744 LVFADLWGPASLESSCGFLYFLTCVDACTKYTWVFPLKKKSDTCATFINFQAMVELQFGH 803
+ DLWGP+ S GF Y++ +D +++TW +PLK KSD +TF+ FQ +VE Q GH
Sbjct: 518 RIHCDLWGPSPTSSVQGFRYYVVFIDHYSRFTWFYPLKLKSDFFSTFVMFQKLVENQLGH 577
Query: 804 KLKSVQTDGGGEF--KPLTSHFQKLGIIHRLTCPHTHHQNGSVERKHRHIVETGLALLSH 861
K+K Q DGGGEF H Q GI ++CP+T QNG ERKHRHIVE GL+++
Sbjct: 578 KIKIFQCDGGGEFISSQFLKHLQDHGIQQNMSCPYTPQQNGMAERKHRHIVELGLSMIFQ 637
Query: 862 ASMPLQFWDHAFLAATYLINRMSTTTLQG-ASPYFKLYGQYPDFKSLKVFGSACFPFLRP 920
+ +PL++W +F A ++IN + T++L SPY KLYG+ P++ +L+VFG AC+P LR
Sbjct: 638 SKLPLKYWLESFFTANFVINLLPTSSLDNNESPYQKLYGKAPEYSALRVFGCACYPTLRD 697
Query: 921 YNSNKLSLHSKECVFIGYSINHKGYKCL-DQSGRIYISKDVLFHEHRFP----YPILFPT 975
Y S K S +CVF+GY+ +KGY+CL +GRIYIS+ V+F E+ P Y L P
Sbjct: 698 YASTKFDPRSLKCVFLGYNEKYKGYRCLYPPTGRIYISRHVVFDENTHPFESIYSHLHPQ 757
Query: 976 DHSSSSSAEF---------------YPLSTIPIISHTAPPSSPVLAASGHSSPGPQDSPQ 1020
D + A F YP+S+IP T ++P A+ + P D
Sbjct: 758 DKTPLLEAWFKSFHHVTPTQPDQSRYPVSSIPQPETTDLSAAPASVAAETAGPNASDDTS 817
Query: 1021 QQQP--------PSATLSADDPST-----STPAVVSPPVPSSSAQSADTSASVVAAAPPV 1067
Q P T D S S A S P P+ S+ ++ S + AP
Sbjct: 818 QDNETISVVSGSPERTTGLDSASIGDSYHSPTADSSHPSPARSSPASSPQGSPIQMAPAQ 877
Query: 1068 -----IVNTHPMQTRAKNGIVKPRLQPTLLLTHL----EPTSVKQAMKDVKWYKAMQEEY 1118
+ N H M TR K GI KP + +LLTH EP +V +A+K W AMQEE
Sbjct: 878 QVQAPVTNEHAMVTRGKEGISKPNKR-YVLLTHKVSIPEPKTVTEALKHPGWNNAMQEEM 936
Query: 1119 TALMNNGTWTLVPLPANRTPVGCKWIYRIKENPDGSINKYKARLVAKGYSQVQGFDYSET 1178
TWTLVP N +G W++R K + DGS++K KARLVAKG+ Q +G DY ET
Sbjct: 937 GNCKETETWTLVPYSPNMNVLGSMWVFRTKLHADGSLDKLKARLVAKGFKQEEGIDYLET 996
Query: 1179 FSPVVKPITVRLILSLAISRGWPLQQLDVNNAFLNGVLEEEVYMVQPPGFEHKDK-NLVC 1237
+SPVV+ TVRLIL +A W L+Q+DV NAFL+G L E VYM QP GF K K + VC
Sbjct: 997 YSPVVRTPTVRLILHVATVLKWELKQMDVKNAFLHGDLTETVYMRQPAGFVDKSKPDHVC 1056
Query: 1238 KLQKALYGLKQAPRAWFHRLKEVLIRFGFQASKCDPSLFTYYSSQGCIYMLVYVDDIIIT 1297
L K+LYGLKQ+PRAWF R L+ FGF S DPSLF Y S+ I +L+YVDD++IT
Sbjct: 1057 LLHKSLYGLKQSPRAWFDRFSNFLLEFGFICSLFDPSLFVYSSNNDVILLLLYVDDMVIT 1116
Query: 1298 GASMSLIQQLTAQLHSIFALKQLGQLDYFLGVQVTHLSNGNLLLNQTKYISDLLTKVNML 1357
G + + L A L+ F +K +GQ+ YFLG+Q+ +G L ++Q KY DLL +M
Sbjct: 1117 GNNSQSLTHLLAALNKEFRMKDMGQVHYFLGIQI-QTYDGGLFMSQQKYAEDLLITASMA 1175
Query: 1358 DAAAISTPMQCGVKLSKNEGSALKDPTEYRSVVGALQYATITRPEISFAVNKVCQFMAQP 1417
+ + + TP+ + N+ DPT +RS+ G LQY T+TRP+I FAVN VCQ M QP
Sbjct: 1176 NCSPMPTPLPLQLDRVSNQDEVFSDPTYFRSLAGKLQYLTLTRPDIQFAVNFVCQKMHQP 1235
Query: 1418 HEEHWKAVKRILRYLKGTLTHGILLQPCSMT------RPLPLIAYCDADWGSDPDDRRST 1471
+ +KRILRY+KGT++ GI S + L AY D+D+ + + RRS
Sbjct: 1236 SVSDFNLLKRILRYIKGTVSMGIQYNSNSSSVVSAYESDYDLSAYSDSDYANCKETRRSV 1295
Query: 1472 SGSCVFLGYNLISWTAKKQSLVARSSTEAEYRSLANTTAELLWVQSLLTELKIPF-TIPT 1530
G C F+G N+ISW++KKQ V+RSSTEAEYRSL+ T +E+ W+ S+L E+ + P
Sbjct: 1296 GGYCTFMGQNIISWSSKKQPTVSRSSTEAEYRSLSETASEIKWMSSILREIGVSLPDTPE 1355
Query: 1531 VYCDNMSTVILTHNPILHTRTKHMEMDIFFVREKVQDKSLLVQHVPSEHQKADIFTKALS 1590
++CDN+S V LT NP H RTKH ++D ++RE+V K+L+V+H+P Q ADIFTK+L
Sbjct: 1356 LFCDNLSAVYLTANPAFHARTKHFDVDHHYIRERVALKTLVVKHIPGHLQLADIFTKSLP 1415
Query: 1591 PTRFLLLKDKLNV 1603
F L+ KL V
Sbjct: 1416 FEAFTRLRFKLGV 1428
Score = 105 bits (262), Expect = 2e-22
Identities = 85/330 (25%), Positives = 136/330 (40%), Gaps = 66/330 (20%)
Query: 24 SHKLSIKLDDSNFYSWKQQVEGVIRSHKLQKFVQDPPMIPE--KFLTEEDKITENVNPLF 81
S+ +++ L+ N+ WK Q E + L FV P + +T + +E NP F
Sbjct: 12 SNCVTVTLNQQNYILWKSQFESFLSGQGLLGFVTGSISAPAQTRSVTHNNVTSEEPNPEF 71
Query: 82 ADWEQQDSLLFTWLLSTLSPAILPNVIQCVHSYQVWEEIHSFFNAKSRAQSTQLRNELKN 141
W Q D ++ +WLL + + IL V+ C S+QVW + + FN S ++ +L+ L+
Sbjct: 72 YTWHQTDQVVKSWLLGSFAEDILSVVVNCFTSHQVWLTLANHFNRVSSSRLFELQRRLQT 131
Query: 142 ITKGTKTASEFLQRIKTIVNTLASIGEPVTFRDHLEAIFDGLSEEYSALMTVIYNQTTYF 201
+ K T FL+ +K I + LAS+G PV + + + +GL EY + T I N
Sbjct: 132 LEKKDNTMEVFLKDLKHICDQLASVGSPVPEKMKIFSALNGLGREYEPIKTTIENS---- 187
Query: 202 TISEVEAMVISHEARFDRMRKKQLADTTPAVFLAQAQNSPSSSTAPAPPPPQAMWTQPAP 261
D+ P++ L + +S Q+ T+P
Sbjct: 188 ------------------------VDSNPSLSLDEV----ASKLRGYDDRLQSYVTEP-- 217
Query: 262 APSSTMAQPPATSVAPSQPVDTAVDSGYGNRDEQNRNSGGYGRGRG----RGRGRNVQ-- 315
T++ A +V S DSGY + + + + G G+ RGRG + Q
Sbjct: 218 ----TISPHVAFNVTHS-------DSGYYHNNNRGKGRSNSGSGKSSFSTRGRGFHQQIS 266
Query: 316 -------------CTYCSKWGHDAASCWSR 332
C C K GH A CW R
Sbjct: 267 PTSGSQAGNSGLVCQICGKAGHHALKCWHR 296
Score = 73.9 bits (180), Expect = 7e-13
Identities = 47/151 (31%), Positives = 74/151 (48%), Gaps = 6/151 (3%)
Query: 471 LATVPAHHSSPVTWCPDSGATHHVTNNPGIFVDHVPSSSQEQLLVGNGQGLPIQSIGSAL 530
+ V HH W PDS A+ HVTNN + P + ++V +G LPI GS
Sbjct: 315 ITDVTDHHGHE--WIPDSAASAHVTNNRHVLQQSQPYHGSDSIMVADGNFLPITHTGSGS 372
Query: 531 LPSSFNPTASLILKKLLLVPSITKNLVSVSRFARDNNVFFSFHANHCSVHHQDTYELLLT 590
+ SS + + LK++L+ P I K+L+SVS+ D F A+ ++ + T +LL+
Sbjct: 373 IASS---SGKIPLKEVLVCPDIVKSLLSVSKLTSDYPCSVEFDADSVRINDKATKKLLVM 429
Query: 591 GTVGDDGLYYFDGPLTKASSQPQSSSPLSSI 621
G DGLY + P + + +S S +
Sbjct: 430 GR-NRDGLYSLEEPKLQVLYSTRQNSASSEV 459
>At4g10690 retrotransposon like protein
Length = 1515
Score = 736 bits (1901), Expect = 0.0
Identities = 438/1094 (40%), Positives = 614/1094 (56%), Gaps = 103/1094 (9%)
Query: 594 GDDGLYYFDG---PLTKASSQP----QSSSPLSSIQACLASKSAANKASPAIDLQSATVS 646
GDD + +G P+T + P Q + PL + C P I +VS
Sbjct: 349 GDDSVIVGNGDFLPITHIGTIPLNISQGTLPLEDVLVC-----------PGITKSLLSVS 397
Query: 647 PIVPSLVPCN---NADSVNVSS--TVSDLVSCNSGSGISVY------------------D 683
+ PC+ ++DSV + T L N G+ V +
Sbjct: 398 KLTDDY-PCSFTFDSDSVVIKDKRTQQLLTQGNKHKGLYVLKDVPFQTYYSTRQQSSDDE 456
Query: 684 LWHSRLGHPHHDVLKQTLSLCNISVPSNKSVFSFCKACCLSKLHRLPSSSSNTVYNTPFE 743
+WH RLGHP+ +VL+ + I V NK+ + C+AC + K+ RLP +S V + P E
Sbjct: 457 VWHQRLGHPNKEVLQHLIKTKAIVV--NKTSSNMCEACQMGKVCRLPFVASEFVSSRPLE 514
Query: 744 LVFADLWGPASLESSCGFLYFLTCVDACTKYTWVFPLKKKSDTCATFINFQAMVELQFGH 803
+ DLWGPA + S+ GF Y++ +D +++TW +PLK KSD + F+ FQ +VE Q+ H
Sbjct: 515 RIHCDLWGPAPVTSAQGFQYYVIFIDNYSRFTWFYPLKLKSDFFSVFVLFQQLVENQYQH 574
Query: 804 KLKSVQTDGGGEFKP--LTSHFQKLGIIHRLTCPHTHHQNGSVERKHRHIVETGLALLSH 861
K+ Q DGGGEF +H GI ++CPHT QNG ER+HR++ E GL+L+ H
Sbjct: 575 KIAMFQCDGGGEFVSYKFVAHLASCGIKQLISCPHTPQQNGIAERRHRYLTELGLSLMFH 634
Query: 862 ASMPLQFWDHAFLAATYLINRMSTTTLQ-GASPYFKLYGQYPDFKSLKVFGSACFPFLRP 920
+ +P + W AF + +L N + ++TL SPY L+G P + +L+VFGSAC+P+LRP
Sbjct: 635 SKVPHKLWVEAFFTSNFLSNLLPSSTLSDNKSPYEMLHGTPPVYTALRVFGSACYPYLRP 694
Query: 921 YNSNKLSLHSKECVFIGYSINHKGYKCLDQ-SGRIYISKDVLFHEHRFPYPILFPTDHSS 979
Y NK S CVF+GY+ +KGY+CL +G++YI + VLF E +FPY ++ +
Sbjct: 695 YAKNKFDPKSLLCVFLGYNNKYKGYRCLHPPTGKVYICRHVLFDERKFPYSDIYSQFQTI 754
Query: 980 SSSAEF-------------------------YPLSTI---------PIISHTAPPSSPVL 1005
S S F +P +T+ P I+ TA +
Sbjct: 755 SGSPLFTAWQKGFSSTALSRETPSTNVEDIIFPSATVSSSVPTGCAPNIAETATAPDVDV 814
Query: 1006 AASGHS--SPGPQDSPQ-QQQPPSATLSADDPST-STPAVVSPPVPSSSAQSADTSA--- 1058
AA+ P P S QP +T + ST S A+ S P S S +
Sbjct: 815 AAAHDMVVPPSPITSTSLPTQPEESTSDQNHYSTDSETAISSAMTPQSINVSLFEDSDFP 874
Query: 1059 ---SVVAAAPPVIVNTHPMQTRAKNGIVKPRLQPTLLLT---HLEPTSVKQAMKDVKWYK 1112
SV+++ +HPM TRAK+GI KP + L + EP SVK+A+KD W
Sbjct: 875 PLQSVISSTTAAPETSHPMITRAKSGITKPNPKYALFSVKSNYPEPKSVKEALKDEGWTN 934
Query: 1113 AMQEEYTALMNNGTWTLVPLPANRTPVGCKWIYRIKENPDGSINKYKARLVAKGYSQVQG 1172
AM EE + TW LVP +GCKW+++ K N DGS+++ KARLVA+GY Q +G
Sbjct: 935 AMGEEMGTMHETDTWDLVPPEMVDRLLGCKWVFKTKLNSDGSLDRLKARLVARGYEQEEG 994
Query: 1173 FDYSETFSPVVKPITVRLILSLAISRGWPLQQLDVNNAFLNGVLEEEVYMVQPPGFEHKD 1232
DY ET+SPVV+ TVR IL +A W L+QLDV NAFL+ L+E V+M QPPGFE
Sbjct: 995 VDYVETYSPVVRSATVRSILHVATINKWSLKQLDVKNAFLHDELKETVFMTQPPGFEDPS 1054
Query: 1233 K-NLVCKLQKALYGLKQAPRAWFHRLKEVLIRFGFQASKCDPSLFTYYSSQGCIYMLVYV 1291
+ + VCKL+KA+Y LKQAPRAWF + L+++GF S DPSLF Y + +++L+YV
Sbjct: 1055 RPDYVCKLKKAIYDLKQAPRAWFDKFSSYLLKYGFICSFSDPSLFVYLKGRDVMFLLLYV 1114
Query: 1292 DDIIITGASMSLIQQLTAQLHSIFALKQLGQLDYFLGVQVTHLSNGNLLLNQTKYISDLL 1351
DD+I+TG + L+QQL L + F +K +G L YFLG+Q H N L L+Q KY SDLL
Sbjct: 1115 DDMILTGNNDVLLQQLLNILSTEFRMKDMGALHYFLGIQ-AHYHNDGLFLSQEKYTSDLL 1173
Query: 1352 TKVNMLDAAAISTPMQCGVKLSKNEGSALKDPTEYRSVVGALQYATITRPEISFAVNKVC 1411
M D +++ TP+Q + L + +PT +R + G LQY T+TRP+I FAVN VC
Sbjct: 1174 VNAGMSDCSSMPTPLQ--LDLLQGNNKPFPEPTYFRRLAGKLQYLTLTRPDIQFAVNFVC 1231
Query: 1412 QFMAQPHEEHWKAVKRILRYLKGTLTHGILLQPCSMTRPLPLIAYCDADWGSDPDDRRST 1471
Q M P + +KRIL YLKGT+T GI L S L Y D+DW D RRST
Sbjct: 1232 QKMHAPTMSDFHLLKRILHYLKGTMTMGINL---SSNTDSVLRCYSDSDWAGCKDTRRST 1288
Query: 1472 SGSCVFLGYNLISWTAKKQSLVARSSTEAEYRSLANTTAELLWVQSLLTELKIP-FTIPT 1530
G C FLGYN+ISW+AK+ V++SSTEAEYR+L+ +E+ W+ LL E+ +P IP
Sbjct: 1289 GGFCTFLGYNIISWSAKRHPTVSKSSTEAEYRTLSFAASEVSWIGFLLQEIGLPQQQIPE 1348
Query: 1531 VYCDNMSTVILTHNPILHTRTKHMEMDIFFVREKVQDKSLLVQHVPSEHQKADIFTKALS 1590
+YCDN+S V L+ NP LH+R+KH ++D ++VRE+V +L V+H+P+ Q ADIFTK+L
Sbjct: 1349 MYCDNLSAVYLSANPALHSRSKHFQVDYYYVRERVALGALTVKHIPASQQLADIFTKSLP 1408
Query: 1591 PTRFLLLKDKLNVV 1604
F L+ KL VV
Sbjct: 1409 QAPFCDLRFKLGVV 1422
Score = 80.9 bits (198), Expect = 6e-15
Identities = 65/266 (24%), Positives = 106/266 (39%), Gaps = 26/266 (9%)
Query: 67 LTEEDKITENVNPLFADWEQQDSLLFTWLLSTLSPAILPNVIQCVHSYQVWEEIHSFFNA 126
+T++D +E N F W + D L+ W+ +LS L VI + +VW + FN
Sbjct: 55 VTKDDIQSEEANQEFLKWTRIDQLVKAWIFGSLSEEALKVVIGLNSAQEVWLGLARRFNR 114
Query: 127 KSRAQSTQLRNELKNITKGTKTASEFLQRIKTIVNTLASIGEPVTFRDHLEAIFDGLSEE 186
S + L+ L +K KT +L +K I + L SIG PVT ++ + + +GL +E
Sbjct: 115 FSTTRKYDLQKRLGTCSKAGKTMDAYLSEVKNICDQLDSIGFPVTEQEKIFGVLNGLGKE 174
Query: 187 YSALMTVIYNQTTYFTISEVEAMVISHEARFDRMRKKQLADTTPAVFLAQAQNSPSSSTA 246
Y ++ TVI E V D + K D + + A ++ +P
Sbjct: 175 YESIATVI----------EHSLDVYPGPCFDDVVYKLTTFDDKLSTYTANSEVTPH---- 220
Query: 247 PAPPPPQAMWTQPAPAPSSTMAQPPATSVAPSQPVDTAVDSGYGNRDEQNRNSGGYGRGR 306
A +T + + + + + Y +R G G
Sbjct: 221 ------LAFYTDKSYSSRGN------NNSRGGRYGNFRGRGSYSSRGRGFHQQFGSGSNN 268
Query: 307 GRGRGRNVQCTYCSKWGHDAASCWSR 332
G G G C C K+GH A C++R
Sbjct: 269 GSGNGSKPTCQICRKYGHSAFKCYTR 294
Score = 67.8 bits (164), Expect = 5e-11
Identities = 49/141 (34%), Positives = 70/141 (48%), Gaps = 5/141 (3%)
Query: 477 HHSSPVTWCPDSGATHHVTNNPGIFVDHVPSSSQEQLLVGNGQGLPIQSIGSALLPSSFN 536
+ +S W PDS AT H+TN + S + ++VGNG LPI IG+ L S
Sbjct: 317 NQASSHEWLPDSAATAHITNTTDGLQNSQTYSGDDSVIVGNGDFLPITHIGTIPLNIS-- 374
Query: 537 PTASLILKKLLLVPSITKNLVSVSRFARDNNVFFSFHANHCSVHHQDTYELLLTGTVGDD 596
+L L+ +L+ P ITK+L+SVS+ D F+F ++ + + T +LL G
Sbjct: 375 -QGTLPLEDVLVCPGITKSLLSVSKLTDDYPCSFTFDSDSVVIKDKRTQQLLTQGN-KHK 432
Query: 597 GLYYF-DGPLTKASSQPQSSS 616
GLY D P S Q SS
Sbjct: 433 GLYVLKDVPFQTYYSTRQQSS 453
>At1g31210 putative reverse transcriptase
Length = 1415
Score = 725 bits (1871), Expect = 0.0
Identities = 435/1154 (37%), Positives = 625/1154 (53%), Gaps = 148/1154 (12%)
Query: 484 WCPDSGATHHVTNNPGIFVDHVPSSSQEQLLVGNGQGLPIQSIGSALLPSSFNPTASLIL 543
W PDS AT HVT++ + +LVG+G LPI GS + SS + L
Sbjct: 322 WHPDSAATAHVTSSTNGLQSATEYEGDDAVLVGDGTYLPITHTGSTTIKSS---NGKIPL 378
Query: 544 KKLLLVPSITKNLVSVSRFARDNNVFFSFHANHCSVHHQDTYELLLTGTVGDDGLYYFDG 603
++L+VP+I K+L+SVS+ D F AN + T +++ TG +GLY +
Sbjct: 379 NEVLVVPNIQKSLLSVSKLCDDYPCGVYFDANKVCIIDLQTQKVVTTGP-RRNGLYVLEN 437
Query: 604 PLTKASSQPQSSSPLSSIQACLASKS--------AANKA------SPAIDLQSATVSPIV 649
Q L S + C A++ A +KA S AI + + SP+
Sbjct: 438 ---------QEFVALYSNRQCAATEEVWHHRLGHANSKALQHLQNSKAIQINKSRTSPVC 488
Query: 650 PSLVPCNNADSVNVSSTVSDLVSCNSGSGISVYDLWHSRLGHPHHDVLKQTLSLCNISVP 709
PC S + +SD SR+ HP
Sbjct: 489 E---PCQMGKSSRLPFLISD-----------------SRVLHP----------------- 511
Query: 710 SNKSVFSFCKACCLSKLHRLPSSSSNTVYNTPFELVFADLWGPASLESSCGFLYFLTCVD 769
L ++H DLWGP+ + S+ G Y+ VD
Sbjct: 512 -------------LDRIH-------------------CDLWGPSPVVSNQGLKYYAIFVD 539
Query: 770 ACTKYTWVFPLKKKSDTCATFINFQAMVELQFGHKLKSVQTDGGGEF--KPLTSHFQKLG 827
++Y+W +PL KS+ + FI+FQ +VE Q K+K Q+DGGGEF L +H + G
Sbjct: 540 DYSRYSWFYPLHNKSEFLSVFISFQKLVENQLNTKIKVFQSDGGGEFVSNKLKTHLSEHG 599
Query: 828 IIHRLTCPHTHHQNGSVERKHRHIVETGLALLSHASMPLQFWDHAFLAATYLINRMSTTT 887
I HR++CP+T QNG ERKHRH+VE GL++L H+ P +FW +F A Y+INR+ ++
Sbjct: 600 IHHRISCPYTPQQNGLAERKHRHLVELGLSMLFHSHTPQKFWVESFFTANYIINRLPSSV 659
Query: 888 LQGASPYFKLYGQYPDFKSLKVFGSACFPFLRPYNSNKLSLHSKECVFIGYSINHKGYKC 947
L+ SPY L+G+ PD+ SL+VFGSAC+P LRP NK S +CVF+GY+ +KGY+C
Sbjct: 660 LKNLSPYEALFGEKPDYSSLRVFGSACYPCLRPLAQNKFDPRSLQCVFLGYNSQYKGYRC 719
Query: 948 L-DQSGRIYISKDVLFHEHRFPYPILFPTDHSSSSSAEFYPLSTIPI--------ISHTA 998
+G++YIS++V+F+E P+ P + P+ IS +
Sbjct: 720 FYPPTGKVYISRNVIFNESELPF---------KEKYQSLVPQYSTPLLQAWQHNKISEIS 770
Query: 999 PPSSPVLAASGHSSPGPQDSPQQQQPPSATLSADDPSTSTPAVVSPPVPSSSAQSADTS- 1057
P++PV S +P A T ++ P P+S+ + +D
Sbjct: 771 VPAAPVQLFS--------------KPIDLNTYAGSQVTEQ---LTDPEPTSNNEGSDEEV 813
Query: 1058 ---ASVVAAAPPVIVNTHPMQTRAKNGIVKPRLQPTLLLTHL---EPTSVKQAMKDVKWY 1111
A +AA ++N+H M TR+K GI KP + L+ + + EP ++ AMK W
Sbjct: 814 NPVAEEIAANQEQVINSHAMTTRSKAGIQKPNTRYALITSRMNTAEPKTLASAMKHPGWN 873
Query: 1112 KAMQEEYTALMNNGTWTLVPLPANRTPVGCKWIYRIKENPDGSINKYKARLVAKGYSQVQ 1171
+A+ EE + TW+LVP + + KW+++ K +PDGSI+K KARLVAKG+ Q +
Sbjct: 874 EAVHEEINRVHMLHTWSLVPPTDDMNILSSKWVFKTKLHPDGSIDKLKARLVAKGFDQEE 933
Query: 1172 GFDYSETFSPVVKPITVRLILSLAISRGWPLQQLDVNNAFLNGVLEEEVYMVQPPGFEHK 1231
G DY ETFSPVV+ T+RL+L ++ S+GWP++QLDV+NAFL+G L+E V+M QP GF
Sbjct: 934 GVDYLETFSPVVRTATIRLVLDVSTSKGWPIKQLDVSNAFLHGELQEPVFMYQPSGFIDP 993
Query: 1232 DK-NLVCKLQKALYGLKQAPRAWFHRLKEVLIRFGFQASKCDPSLFTYYSSQGCIYMLVY 1290
K VC+L KA+YGLKQAPRAWF L+ +GF SK DPSLF + +Y+L+Y
Sbjct: 994 QKPTHVCRLTKAIYGLKQAPRAWFDTFSNFLLDYGFVCSKSDPSLFVCHQDGKILYLLLY 1053
Query: 1291 VDDIIITGASMSLIQQLTAQLHSIFALKQLGQLDYFLGVQVTHLSNGNLLLNQTKYISDL 1350
VDDI++TG+ SL++ L L + F++K LG YFLG+Q+ +NG L L+QT Y +D+
Sbjct: 1054 VDDILLTGSDQSLLEDLLQALKNRFSMKDLGPPRYFLGIQIEDYANG-LFLHQTAYATDI 1112
Query: 1351 LTKVNMLDAAAISTPMQCGVKLSKNEGSALKDPTEYRSVVGALQYATITRPEISFAVNKV 1410
L + M D + TP+ +L +PT +RS+ G LQY TITRP+I FAVN +
Sbjct: 1113 LQQAGMSDCNPMPTPLP--QQLDNLNSELFAEPTYFRSLAGKLQYLTITRPDIQFAVNFI 1170
Query: 1411 CQFMAQPHEEHWKAVKRILRYLKGTLTHGILLQPCSMTRPLPLIAYCDADWGSDPDDRRS 1470
CQ M P + +KRILRY+KGT+ G+ P L L AY D+D + RRS
Sbjct: 1171 CQRMHSPTTSDFGLLKRILRYIKGTIGMGL---PIKRNSTLTLSAYSDSDHAGCKNTRRS 1227
Query: 1471 TSGSCVFLGYNLISWTAKKQSLVARSSTEAEYRSLANTTAELLWVQSLLTELKIPFTIPT 1530
T+G C+ LG NLISW+AK+Q V+ SSTEAEYR+L E+ W+ LL +L IP +PT
Sbjct: 1228 TTGFCILLGSNLISWSAKRQPTVSNSSTEAEYRALTYAAREITWISFLLRDLGIPQYLPT 1287
Query: 1531 -VYCDNMSTVILTHNPILHTRTKHMEMDIFFVREKVQDKSLLVQHVPSEHQKADIFTKAL 1589
VYCDN+S V L+ NP LH R+KH + D ++RE+V + QH+ + Q AD+FTK+L
Sbjct: 1288 QVYCDNLSAVYLSANPALHNRSKHFDTDYHYIREQVALGLIETQHISATFQLADVFTKSL 1347
Query: 1590 SPTRFLLLKDKLNV 1603
F+ L+ KL V
Sbjct: 1348 PRRAFVDLRSKLGV 1361
Score = 107 bits (268), Expect = 4e-23
Identities = 88/320 (27%), Positives = 135/320 (41%), Gaps = 53/320 (16%)
Query: 27 LSIKLDDSNFYSWKQQVEGVIRSHKLQKFVQDPPMIPE--KFLTEEDKITENVNPLFADW 84
+++KL DSN+ WK Q E ++ S KL FV P + + + +E NPL+ W
Sbjct: 17 VTLKLTDSNYLLWKTQFESLLSSQKLIGFVNGAVNAPSQSRLVVNGEVTSEEPNPLYESW 76
Query: 85 EQQDSLLFTWLLSTLSPAILPNVIQCVHSYQVWEEIHSFFNAKSRAQSTQLRNELKNITK 144
D L+ +WL TLS +L +V S Q+W + FN S A+ LR L+ ++K
Sbjct: 77 FCTDQLVRSWLFGTLSEEVLGHVHNLSTSRQIWVSLAENFNKSSVAREFSLRQNLQLLSK 136
Query: 145 GTKTASEFLQRIKTIVNTLASIGEPVTFRDHLEAIFDGLSEEYSALMTVIYNQTTYF--- 201
K S + + KTI + L+SIG+PV + +GL +Y + TVI + +
Sbjct: 137 KEKPFSVYCREFKTICDALSSIGKPVDESMKIFGFLNGLGRDYDPITTVIQSSLSKLPTP 196
Query: 202 TISEVEAMVISHEARFDRMRKKQLADTTPAVFLAQAQNSPSSSTAPAPPPPQAMWTQPAP 261
T ++V + V +++ ++ A TP + S S +P P Q
Sbjct: 197 TFNDVVSEVQGFDSKLQSY--EEAASVTPHLAF---NIERSESGSPQYNPNQ-------- 243
Query: 262 APSSTMAQPPATSVAPSQPVDTAVDSGYGNRDEQNRNSGGYG-RGRGRGR--------GR 312
G G R QN+ GGY RGRG + G
Sbjct: 244 -------------------------KGRG-RSGQNKGRGGYSTRGRGFSQHQSSPQVSGP 277
Query: 313 NVQCTYCSKWGHDAASCWSR 332
C C + GH A C++R
Sbjct: 278 RPVCQICGRTGHTALKCYNR 297
>At4g27210 putative protein
Length = 1318
Score = 707 bits (1826), Expect = 0.0
Identities = 398/954 (41%), Positives = 539/954 (55%), Gaps = 79/954 (8%)
Query: 683 DLWHSRLGHPHHDVLKQTLSLCNISVPSNKSVFSFCKACCLSKLHRLPSSSSNTVYNTPF 742
++WH RLGHPH +L+ P
Sbjct: 310 EVWHRRLGHPHPQILQ------------------------------------------PL 327
Query: 743 ELVFADLWGPASLESSCGFLYFLTCVDACTKYTWVFPLKKKSDTCATFINFQAMVELQFG 802
E V DLWGP ++ S GF Y+ +D ++++W++PLK KSD F+ F +VE Q
Sbjct: 328 ERVHCDLWGPTTITSVQGFRYYAVFIDHYSRFSWIYPLKLKSDFYNIFLAFHKLVENQLS 387
Query: 803 HKLKSVQTDGGGEF--KPLTSHFQKLGIIHRLTCPHTHHQNGSVERKHRHIVETGLALLS 860
K+ Q DGGGEF H Q GI +L+CPHT QNG ERKHRH+VE GL++L
Sbjct: 388 QKISVFQCDGGGEFVSHKFLQHLQSHGIQQQLSCPHTPQQNGLAERKHRHLVELGLSMLF 447
Query: 861 HASMPLQFWDHAFLAATYLINRMSTTTL-QGASPYFKLYGQYPDFKSLKVFGSACFPFLR 919
+ +P +FW AF A +LIN + T+ L + SPY KLY + PD+ SL+ FGSACFP LR
Sbjct: 448 QSHVPHKFWVEAFFTANFLINLLPTSALKESISPYEKLYDKKPDYTSLRSFGSACFPTLR 507
Query: 920 PYNSNKLSLHSKECVFIGYSINHKGYKCL-DQSGRIYISKDVLFHEHRFPY--------- 969
Y NK + S +CVF+GY+ +KGY+CL +GR+YIS+ V+F E +P+
Sbjct: 508 DYAENKFNPCSLKCVFLGYNEKYKGYRCLYPPTGRLYISRHVIFDESVYPFSHTYKHLHP 567
Query: 970 ----PILF--------PTDHSSSSSAEFYPLST---IPIISHTAPPSSPVLAASGHSSPG 1014
P+L P +S+S + PL T P + P P L S
Sbjct: 568 QPRTPLLAAWLRSSDSPAPSTSTSPSSRSPLFTSADFPPLPQRKTPLLPTLVPISSVSHA 627
Query: 1015 PQDSPQQQQPPSATLSADDPSTSTPAVVSPPVPSSSAQSADTSASVVAAAPPVIVNTHPM 1074
+ QQ + + D S S S ++ ASV N HPM
Sbjct: 628 SNITTQQSPDFDSERTTDFDSASIGDSSHSSQAGSDSEETIQQASVNVHQTHASTNVHPM 687
Query: 1075 QTRAKNGIVKPRLQPTLL---LTHLEPTSVKQAMKDVKWYKAMQEEYTALMNNGTWTLVP 1131
TRAK GI KP + L +++ EP +V A+K W AM EE TW+LVP
Sbjct: 688 VTRAKVGISKPNPRYVFLSHKVSYPEPKTVTAALKHPGWTGAMTEEIGNCSETQTWSLVP 747
Query: 1132 LPANRTPVGCKWIYRIKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLI 1191
++ +G KW++R K + DG++NK KAR+VAKG+ Q +G DY ET+SPVV+ TVRL+
Sbjct: 748 YKSDMHVLGSKWVFRTKLHADGTLNKLKARIVAKGFLQEEGIDYLETYSPVVRTPTVRLV 807
Query: 1192 LSLAISRGWPLQQLDVNNAFLNGVLEEEVYMVQPPGFEHKDK-NLVCKLQKALYGLKQAP 1250
L LA + W ++Q+DV NAFL+G L+E VYM QP GF K + VC L K++YGLKQ+P
Sbjct: 808 LHLATALNWDIKQMDVKNAFLHGDLKETVYMTQPAGFVDPSKPDHVCLLHKSIYGLKQSP 867
Query: 1251 RAWFHRLKEVLIRFGFQASKCDPSLFTYYSSQGCIYMLVYVDDIIITGASMSLIQQLTAQ 1310
RAWF + L+ FGF SK DPSLF Y + I +L+YVDD++ITG S + L A
Sbjct: 868 RAWFDKFSTFLLEFGFFCSKSDPSLFIYAHNNNLILLLLYVDDMVITGNSSQTLTSLLAA 927
Query: 1311 LHSIFALKQLGQLDYFLGVQVTHLSNGNLLLNQTKYISDLLTKVNMLDAAAISTPMQCGV 1370
L+ F + +GQL YFLG+QV NG L ++Q KY DLL +M + TP+ +
Sbjct: 928 LNKEFRMTDMGQLHYFLGIQVQRQQNG-LFMSQQKYAEDLLIAASMEHCTPLPTPLPVQL 986
Query: 1371 KLSKNEGSALKDPTEYRSVVGALQYATITRPEISFAVNKVCQFMAQPHEEHWKAVKRILR 1430
++ DPT +RS+ G LQY T+TRP+I FAVN VCQ M QP + +KRILR
Sbjct: 987 DRVPHQEELFSDPTYFRSIAGKLQYLTLTRPDIQFAVNFVCQKMHQPTISDFHLLKRILR 1046
Query: 1431 YLKGTLTHGILLQPCSMTRPLPLIAYCDADWGSDPDDRRSTSGSCVFLGYNLISWTAKKQ 1490
Y+KGT+T GI S P L AY D+DWG+ RRS G C F+G NL+SW++KK
Sbjct: 1047 YIKGTITMGI---SYSRDSPTLLQAYSDSDWGNCKQTRRSVGGLCTFMGTNLVSWSSKKH 1103
Query: 1491 SLVARSSTEAEYRSLANTTAELLWVQSLLTELKIPF-TIPTVYCDNMSTVILTHNPILHT 1549
V+RSSTEAEY+SL++ +E+LW+ +LL EL+IP P ++CDN+S V LT NP H
Sbjct: 1104 PTVSRSSTEAEYKSLSDAASEILWLSTLLRELRIPLPDTPELFCDNLSAVYLTANPAFHA 1163
Query: 1550 RTKHMEMDIFFVREKVQDKSLLVQHVPSEHQKADIFTKALSPTRFLLLKDKLNV 1603
RTKH ++D FVRE+V K+L+V+H+P Q ADIFTK+L F+ L+ KL V
Sbjct: 1164 RTKHFDIDFHFVRERVALKALVVKHIPGSEQIADIFTKSLPYEAFIHLRGKLGV 1217
Score = 69.7 bits (169), Expect = 1e-11
Identities = 44/116 (37%), Positives = 64/116 (54%), Gaps = 4/116 (3%)
Query: 484 WCPDSGATHHVTNNPGIFVDHVPSSSQEQLLVGNGQGLPIQSIGSALLPSSFNPTASLIL 543
W PDS AT HVTN+P P + ++V +G LPI GS L SS + ++ L
Sbjct: 177 WLPDSAATAHVTNSPRSLQQSQPYHGTDAIMVDDGNYLPITHTGSTNLASS---SGTVPL 233
Query: 544 KKLLLVPSITKNLVSVSRFARDNNVFFSFHANHCSVHHQDTYELLLTGTVGDDGLY 599
+L+ PSITK+L+S+S+ +D F + V+ + T +LLL G+ DGLY
Sbjct: 234 TDVLVCPSITKSLLSMSKLTQDFPCTVEFEYDGVRVNDKATKKLLLMGS-NRDGLY 288
Score = 50.1 bits (118), Expect = 1e-05
Identities = 32/102 (31%), Positives = 52/102 (50%), Gaps = 1/102 (0%)
Query: 134 QLRNELKNITKGTKTASEFLQRIKTIVNTLASIGEPVTFRDHLEAIFDGLSEEYSALMTV 193
+L+ L+N++K KT +L +K I + LAS+G PVT + + A +GL EY + T
Sbjct: 49 ELQRRLQNVSKRDKTMDAYLNDLKNICDQLASVGSPVTEKMKIFAALNGLGREYEPIKTT 108
Query: 194 IYNQTTYFTISEVEAMVISHEARFDRMRKKQLADTTPAVFLA 235
I N +E VI +D + L +TT + ++A
Sbjct: 109 IENSMDTQPGPSLE-NVIPKLTGYDDRLQGYLEETTISPYVA 149
>At4g28900 putative protein
Length = 1415
Score = 691 bits (1782), Expect = 0.0
Identities = 436/1167 (37%), Positives = 608/1167 (51%), Gaps = 181/1167 (15%)
Query: 484 WCPDSGATHHVTNNPGIFVDHVPSSSQEQLLVGNGQGLPIQSIGSALLPSSFNPTASLIL 543
W DSGAT H+TN+ P S ++ ++VGN LPI IGSA+L S+ +L L
Sbjct: 293 WVTDSGATSHITNSTSQLQSAQPYSGEDSVIVGNSDFLPITHIGSAVLTSN---QGNLPL 349
Query: 544 KKLLLVPSITKNLVSVSRFARDNNVFFSFHANHCSVHHQDTYELLLTGTVGDDGLYYFDG 603
+ +L+ P+ITK+L+SVS+ D F ++ V + T +LL GT +D LY +
Sbjct: 350 RDVLVCPNITKSLLSVSKLTSDYPCVIEFDSDGVIVKDKLTKQLLTKGTRHND-LYLLEN 408
Query: 604 PLTKASSQPQSSSPLSSIQACLASKSAANKASPAIDLQSATVSPIVPSLVPCNNADSVNV 663
P AC +S+ A
Sbjct: 409 P---------------KFMACYSSRQQAT------------------------------- 422
Query: 664 SSTVSDLVSCNSGSGISVYDLWHSRLGHPHHDVLKQTLSLCNISVPSNKSVFSFCKACCL 723
SD ++WH RLGHP+ DVL+Q L N ++ +K+ S C AC +
Sbjct: 423 ----SD-------------EVWHMRLGHPNQDVLQQLLR--NKAIVISKTSHSLCDACQM 463
Query: 724 SKLHRLPSSSSNTVYNTPFELVFADLWGPASLESSCGFLYFLTCVDACTKYTWVFPLKKK 783
K+ +LP +SS+ V + E V DLWGPA + SS GF Y++ +D +++TW +PL+ K
Sbjct: 464 GKICKLPFASSDFVSSRLLERVHCDLWGPAPVVSSQGFRYYVIFIDNYSRFTWFYPLRLK 523
Query: 784 SDTCATFINFQAMVELQFGHKLKSVQTDGGGEF--KPLTSHFQKLGIIHRLTCPHTHHQN 841
SD + F+ FQ MVE Q K+ S Q DGGGEF SH + GI ++CP+T QN
Sbjct: 524 SDFFSVFLTFQKMVENQCQQKIASFQCDGGGEFISNQFVSHLAECGIRQLISCPYTPQQN 583
Query: 842 GSVERKHRHIVETGLALLSHASMPLQFWDHAFLAATYLINRMSTTTLQG-ASPYFKLYGQ 900
G ERKHRHI E G +++ +P W AF + +L N + ++ L+ SPY L G+
Sbjct: 584 GIAERKHRHITELGSSMMFQGKVPQFLWVEAFYTSNFLCNLLPSSVLKDQKSPYEVLMGK 643
Query: 901 YPDFKSLKVFGSACFPFLRPYNSNKLSLHSKECVFIGYSINHKGYKCL-DQSGRIYISKD 959
P + SL+VFG AC+P LRPY SNK S CVF GY+ +KGYKC +G+IYI++
Sbjct: 644 APVYTSLRVFGCACYPNLRPYASNKFDPKSLLCVFTGYNEKYKGYKCFHPPTGKIYINRH 703
Query: 960 VLFHEHRFPYPILFPTDHSSSSS-------AEFYPLSTIPI------ISHTAP------- 999
VLF E +F + ++ S ++S + F P S IP IS+TA
Sbjct: 704 VLFDESKFLFSDIYSDKVSGTNSTLVSAWQSNFLPKS-IPATPEVLDISNTAASFSDEQG 762
Query: 1000 -----------------PSSPVLAASGHSSPGPQDSPQQQQPPSATLSADDPSTSTPAVV 1042
S P+ + S Q+SPQ + P S+ S +D S +
Sbjct: 763 EFSGAVGGGGCGCTADLDSVPIGNSLPSSPVTQQNSPQPETPISSAGSGNDAEDSELSEN 822
Query: 1043 SPPVPSSSAQSADTSASVVAAAPPVIVNTHPMQTRAKNGIVKPR---LQPTLLLTHLEPT 1099
S SS A T AA +HPM TR+K+GI KP T+ + P
Sbjct: 823 SENSESSVFSEATTETE---AADNTNDQSHPMITRSKSGIFKPNPKYAMFTVKSNYPVPK 879
Query: 1100 SVKQAMKDVKWYKAMQEEYTALMNNGTWTLVPLPANRTPVGCKWIYRIKENPDGSINKYK 1159
+VK A+KD W AM EEY + TW LVP + TP+GC+W+++ K DG++++ K
Sbjct: 880 TVKTALKDPGWTDAMGEEYDSFEETHTWDLVPPDSFITPLGCRWVFKTKLKADGTLDRLK 939
Query: 1160 ARLVAKGYSQVQGFDYSETFSPVVKPITVRLILSLAISRGWPLQQLDVNNAFLNGVLEEE 1219
ARLVAKGY Q +G DY ET+SPVV+ TVR IL +A W ++QLDV NAFL+G L+E
Sbjct: 940 ARLVAKGYEQEEGVDYMETYSPVVRTATVRTILHVATINKWEIKQLDVKNAFLHGDLKET 999
Query: 1220 VYMVQPPGFEHKDK-NLVCKLQKALYGLKQAPRAWFHRLKEVLIRFGFQASKCDPSLFTY 1278
VYM QPPGFE++D+ + VCKL KA+YGLKQAPRAWF + L+ FGF + DPSLF +
Sbjct: 1000 VYMYQPPGFENQDRPDYVCKLNKAIYGLKQAPRAWFDKFSTFLLEFGFICTYSDPSLFVF 1059
Query: 1279 YSSQGCIYMLVYVDDIIITGASMSLIQQLTAQLHSIFALKQLGQLDYFLGVQVTHLSNGN 1338
+ +++L+Y+DD+++TG
Sbjct: 1060 LKGRDLMFLLLYMDDMLLTG---------------------------------------- 1079
Query: 1339 LLLNQTKYISDLLTKVNMLDAAAISTPMQCGVKLSKNEGSALKDPTEYRSVVGALQYATI 1398
N KY DLL M D A + TP+ + + + DPT +RS+
Sbjct: 1080 ---NNKKYAMDLLVAAGMADCAPMPTPLPLQLDKVPGQQESFADPTYFRSL--------- 1127
Query: 1399 TRPEISFAVNKVCQFMAQPHEEHWKAVKRILRYLKGTLTHGILLQPCSMTRPLPLIAYCD 1458
AVN VCQ M P + +KR+LRYLKG + G+ L + L AY D
Sbjct: 1128 -------AVNLVCQKMHSPTVADFNLLKRVLRYLKGKVQMGLNLH---NNTDITLRAYSD 1177
Query: 1459 ADWGSDPDDRRSTSGSCVFLGYNLISWTAKKQSLVARSSTEAEYRSLANTTAELLWVQSL 1518
+DW + + RRS G C FLG N+ISW+AK+ V+RSSTEAEYR+L+ E+ W+ SL
Sbjct: 1178 SDWANCKETRRSVGGFCTFLGTNIISWSAKRHPTVSRSSTEAEYRTLSIAATEVKWISSL 1237
Query: 1519 LTELKI-PFTIPTVYCDNMSTVILTHNPILHTRTKHMEMDIFFVREKVQDKSLLVQHVPS 1577
L E+ I P +YCDN+S V LT NP +H R+K ++D +VRE+V +L+V+HVP+
Sbjct: 1238 LREIGIYQPAPPELYCDNLSAVYLTANPAMHNRSKAFDVDFHYVRERVALGALVVKHVPA 1297
Query: 1578 EHQKADIFTKALSPTRFLLLKDKLNVV 1604
HQ ADIFTK+L F L+ KL VV
Sbjct: 1298 SHQLADIFTKSLPQRPFFDLRYKLGVV 1324
Score = 101 bits (252), Expect = 3e-21
Identities = 59/195 (30%), Positives = 99/195 (50%), Gaps = 2/195 (1%)
Query: 12 SSSSSSSLSQAFSHKLSIKLDDSNFYSWKQQVEGVIRSHKLQKFVQDPPMIPE--KFLTE 69
+ +S SS + FSH +++KL +N+ WK Q E + + +L FV P + +
Sbjct: 2 ADNSDSSSALCFSHYVTLKLSTANYLLWKIQFETWLNNQRLLGFVTGANPCPNATRSIRN 61
Query: 70 EDKITENVNPLFADWEQQDSLLFTWLLSTLSPAILPNVIQCVHSYQVWEEIHSFFNAKSR 129
D++TE NP F W Q D + WLL +LS L +V S +VW + +N S
Sbjct: 62 GDQVTEATNPDFLTWVQNDQKIMGWLLGSLSEDALRSVYGLHTSREVWFSLAKKYNRVSA 121
Query: 130 AQSTQLRNELKNITKGTKTASEFLQRIKTIVNTLASIGEPVTFRDHLEAIFDGLSEEYSA 189
++ + L+ L ++K K+ E+L +K I + L SIG PV + + + +GL +EY
Sbjct: 122 SRKSDLQRRLNPVSKNEKSMLEYLNCVKQICDQLDSIGCPVPENEKIFGVLNGLGQEYML 181
Query: 190 LMTVIYNQTTYFTIS 204
+ T+I + +S
Sbjct: 182 VSTMIKGSMDTYPMS 196
Score = 34.7 bits (78), Expect = 0.46
Identities = 17/49 (34%), Positives = 20/49 (40%), Gaps = 6/49 (12%)
Query: 290 GNRDEQNRNSGGYG------RGRGRGRGRNVQCTYCSKWGHDAASCWSR 332
GNR N + G G G G C C+K+GH A CW R
Sbjct: 218 GNRGRNNYTTKGRGFPQQISSGSPSDSGTRPTCQICNKYGHSAYKCWKR 266
>At2g20460 putative retroelement pol polyprotein
Length = 1461
Score = 654 bits (1687), Expect = 0.0
Identities = 471/1544 (30%), Positives = 723/1544 (46%), Gaps = 227/1544 (14%)
Query: 78 NPLFADWEQQDSLLFTWLLSTLSPAILPNVIQCVHSYQVWEEIHSFFNAKSRAQSTQLRN 137
+P F W + +S++ +WLL+++SP I ++++ + +W ++ FN + ++ L
Sbjct: 119 DPNFRLWSRCNSMVKSWLLNSVSPQIYRSILRLNDATDIWRDLFDRFNLTNLPRTYNLTQ 178
Query: 138 ELKNITKGTKTASEFLQRIKTIVNTLAS---IGEPVTFRDHLEAIFDGLSEEYSALMTVI 194
E++++ +GT + SE+ +KT+ + L S + +P T +A+ E + +M +
Sbjct: 179 EIQDLRQGTMSLSEYYTLLKTLWDQLDSTEALDDPCTCG---KAVRLYQKAEKAKIMKFL 235
Query: 195 YNQTTYFTISEVEAMVISHEARFDRMRKKQLADTTPAVFLAQAQNSPSSSTAPAPPPPQA 254
+ I V +I+ KK L + NS P
Sbjct: 236 AGLNESYAI--VRRQIIA---------KKALPSLAEVYHILDQDNSQKGFFNVVAP---- 280
Query: 255 MWTQPAPAPSSTMAQPPATSVAPSQPVDTAVDSGYGNRDEQNRNSGGYGRGRGRGRGRNV 314
PA S ++ P TS P V S G +GR
Sbjct: 281 ----PAAFQVSEVSHSPITS-----PEIMYVQS-------------------GPNKGRPT 312
Query: 315 QCTYCSKWGHDAASCWSR---PSGVSSNANSSANSAQSGNSGSNFSVPGVNLGFSPMQFG 371
C++C++ GH A C+ + P G + SS + + SP +
Sbjct: 313 -CSFCNRVGHIAERCYKKHGFPPGFTPKGKSSDKPPKP-------QAVAAQVTLSPDKMT 364
Query: 372 GFPSMANFGFPPYAPYNSMLPQYGSSVLSPSSMPPWSPWCIPPTPYSHATDMQGMQAAGM 431
G F P N L SS L P + P + + A+ Q + +G+
Sbjct: 365 GQLETLAGNFSPDQIQN--LIALFSSQLQPQIVSPQT-----ASSQHEASSSQSVAPSGI 417
Query: 432 SFVRPWLPQGQFPAPTAQTPQFPRPPQQQVSTGMHPQAMLATVPAHHSSPVTWCPDSGAT 491
F P G + V + S TW DSGAT
Sbjct: 418 LF----------------------SPSTYCFIG------ILAVSHNSLSSDTWVIDSGAT 449
Query: 492 HHVTNNPGIFVDHVPSSSQEQLLVGNGQGLPIQSIGSALLPSSFNPTASLILKKLLLVPS 551
HHV+++ +F + +S + + G + I +G+ L+ +IL+ +L +P
Sbjct: 450 HHVSHDRKLF-QTLDTSIVSFVNLPTGPNVRISGVGTVLI------NKDIILQNVLFIPE 502
Query: 552 ITKNLVSVSRFARDNNVFFSFHANHCSVHHQDTYELLLTGTVGDDGLYYFDGPLTKASSQ 611
NL+S+S D F + C + QD + L G G Y
Sbjct: 503 FRLNLISISSLTTDLGTRVIFDPSCCQI--QDLTKGLTLGEGKRIGNLY----------- 549
Query: 612 PQSSSPLSSIQACLASKSAANKASPAIDLQSATVSPIVPSLVPCNNADSVNVSSTVSDLV 671
+D QS + SVN VS
Sbjct: 550 -------------------------VLDTQSPAI--------------SVNAVVDVSVWH 570
Query: 672 SCNSGSGISVYDLWHSRLGHPHHDVLKQTLSLCNISVPSNKSVFSFCKACCLSKLHRLPS 731
S D LG H K + C++ + + SF PS
Sbjct: 571 KRLGHPSFSRLDSLSEVLGTTRHKNKKS--AYCHVCHLAKQKKLSF------------PS 616
Query: 732 SSSNTVYNTPFELVFADLWGPASLESSCGFLYFLTCVDACTKYTWVFPLKKKSDTCATFI 791
+++ + N+ FEL+ D+WGP S+E+ G+ YFLT VD ++ TW++ LK KSD F
Sbjct: 617 ANN--ICNSTFELLHIDVWGPFSVETVEGYKYFLTIVDDHSRATWIYLLKSKSDVLTVFP 674
Query: 792 NFQAMVELQFGHKLKSVQTDGGGEFKPLTSHFQKLGIIHRLTCPHTHHQNGSVERKHRHI 851
F +VE Q+ ++KSV++D E T ++ GI+ +CP T QN VERKH+HI
Sbjct: 675 AFIDLVENQYDTRVKSVRSDNAKEL-AFTEFYKAKGIVSFHSCPETPEQNSVVERKHQHI 733
Query: 852 VETGLALLSHASMPLQFWDHAFLAATYLINRMSTTTLQGASPYFKLYGQYPDFKSLKVFG 911
+ AL+ ++M L +W L A +LINR + L +P+ L G+ PD+ LK FG
Sbjct: 734 LNVARALMFQSNMSLPYWGDCVLTAVFLINRTPSALLSNKTPFEVLTGKLPDYSQLKTFG 793
Query: 912 SACFPFLRPYNSNKLSLHSKECVFIGYSINHKGYKCLD-QSGRIYISKDVLFHEHRFPYP 970
C+ +K S+ CVF+GY KGYK LD +S ++IS++V FHE FP
Sbjct: 794 CLCYSSTSSKQRHKFLPRSRACVFLGYPFGFKGYKLLDLESNVVHISRNVEFHEELFP-- 851
Query: 971 ILFPTDHSSSSSAEFYPLSTIPIISHTAPPSSPVLAASGHSS--PGPQDSPQQQQPPSAT 1028
L + S++++++ + P P+ + + +S P PQ SP Q
Sbjct: 852 -LASSQQSATTASDVF------------TPMDPLSSGNSITSHLPSPQISPSTQ------ 892
Query: 1029 LSADDPSTSTPAVVSPPVPSSSAQSADTSASVVAAAPPVIVNTHPMQTRAKNGIVKPRLQ 1088
+ + A D V ++HP+ + + P
Sbjct: 893 ------------ISKRRITKFPAHLQDYHCYFVNKD-----DSHPISSSLSYSQISP--S 933
Query: 1089 PTLLLTHLE----PTSVKQAMKDVKWYKAMQEEYTALMNNGTWTLVPLPANRTPVGCKWI 1144
L + ++ P S +A +W A+ +E A+ TW + LP + VGCKW+
Sbjct: 934 HMLYINNISKIPIPQSYHEAKDSKEWCGAIDQEIGAMERTDTWEITSLPPGKKAVGCKWV 993
Query: 1145 YRIKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILSLAISRGWPLQQ 1204
+ +K + DGS+ ++KAR+VAKGY+Q +G DY+ETFSPV K TV+L+L ++ S+ W L Q
Sbjct: 994 FTVKFHADGSLERFKARIVAKGYTQKEGLDYTETFSPVAKMATVKLLLKVSASKKWYLNQ 1053
Query: 1205 LDVNNAFLNGVLEEEVYMVQPPGF-----EHKDKNLVCKLQKALYGLKQAPRAWFHRLKE 1259
LD++NAFLNG LEE +YM P G+ N+VC+L+K++YGLKQA R WF +
Sbjct: 1054 LDISNAFLNGDLEETIYMKLPDGYADIKGTSLPPNVVCRLKKSIYGLKQASRQWFLKFSN 1113
Query: 1260 VLIRFGFQASKCDPSLFTYYSSQGCIYMLVYVDDIIITGASMSLIQQLTAQLHSIFALKQ 1319
L+ GF+ D +LF I +LVYVDDI+I + Q LT L + F L++
Sbjct: 1114 SLLALGFEKQHGDHTLFVRCIGSEFIVLLVYVDDIVIASTTEQAAQSLTEALKASFKLRE 1173
Query: 1320 LGQLDYFLGVQVTHLSNGNLLLNQTKYISDLLTKVNMLDAAAISTPMQCGVKLSKNEGSA 1379
LG L YFLG++V S G + L+Q KY +LLT +MLD S PM ++LSKN+G
Sbjct: 1174 LGPLKYFLGLEVARTSEG-ISLSQRKYALELLTSADMLDCKPSSIPMTPNIRLSKNDGLL 1232
Query: 1380 LKDPTEYRSVVGALQYATITRPEISFAVNKVCQFMAQPHEEHWKAVKRILRYLKGTLTHG 1439
L+D YR +VG L Y TITRP+I+FAVNK+CQF + P H AV ++L+Y+KGT+ G
Sbjct: 1233 LEDKEMYRRLVGKLMYLTITRPDITFAVNKLCQFSSAPRTAHLAAVYKVLQYIKGTVGQG 1292
Query: 1440 ILLQPCSMTRPLPLIAYCDADWGSDPDDRRSTSGSCVFLGYNLISWTAKKQSLVARSSTE 1499
+ S L L Y DADWG+ PD RRST+G +F+G +LISW +KKQ V+RSS E
Sbjct: 1293 LFY---SAEDDLTLKGYTDADWGTCPDSRRSTTGFTMFVGSSLISWRSKKQPTVSRSSAE 1349
Query: 1500 AEYRSLANTTAELLWVQSLLTELKIPFTIPTVYCDNMSTVILTHNPILHTRTKHMEMDIF 1559
AEYR+LA + E+ W+ +LL L++ +P +Y D+ + V + NP+ H RTKH+E+D
Sbjct: 1350 AEYRALALASCEMAWLSTLLLALRVHSGVPILYSDSTAAVYIATNPVFHERTKHIEIDCH 1409
Query: 1560 FVREKVQDKSLLVQHVPSEHQKADIFTKALSPTRFLLLKDKLNV 1603
VREK+ + L + HV ++ Q ADI TK L P +F L K+++
Sbjct: 1410 TVREKLDNGQLKLLHVKTKDQVADILTKPLFPYQFAHLLSKMSI 1453
>At2g16670 putative retroelement pol polyprotein
Length = 1333
Score = 626 bits (1615), Expect = e-179
Identities = 362/965 (37%), Positives = 518/965 (53%), Gaps = 70/965 (7%)
Query: 682 YDLWHSRLGHPHHDVLKQTLSLCNISVPSNKSVFSFCKACCLSKLHR--LPSSSSNTVYN 739
Y+LWHSR+GHP V+ + ++SV S+ + C C +K R P S + T+
Sbjct: 389 YELWHSRMGHPAARVVS-LIPESSVSV-SSTHLNKACDVCHRAKQTRNSFPLSINKTL-- 444
Query: 740 TPFELVFADLWGPASLESSCGFLYFLTCVDACTKYTWVFPLKKKSDTCATFINFQAMVEL 799
FEL++ DLWGP S G YFLT +D ++ W++ L KS+ NF AM +
Sbjct: 445 RIFELIYCDLWGPYRTPSHTGARYFLTIIDDYSRGVWLYLLNDKSEAPCHLKNFFAMTDR 504
Query: 800 QFGHKLKSVQTDGGGEFKPLTSHFQKLGIIHRLTCPHTHHQNGSVERKHRHIVETGLALL 859
QF K+K+V++D G EF LT FQ+ G+IH +C T +N VERKHRH++ AL
Sbjct: 505 QFNVKIKTVRSDNGTEFLCLTKFFQEQGVIHERSCVATPERNDRVERKHRHLLNVARALR 564
Query: 860 SHASMPLQFWDHAFLAATYLINRMSTTTLQGASPYFKLYGQYPDFKSLKVFGSACFPFLR 919
A++P+QFW L A YLINR ++ L ++PY +L+ + P F L+VFGS C+ R
Sbjct: 565 FQANLPIQFWGECVLTAAYLINRTPSSVLNDSTPYERLHKKQPRFDHLRVFGSLCYAHNR 624
Query: 920 PYNSNKLSLHSKECVFIGYSINHKGYKCLD-QSGRIYISKDVLFHEHRFPYPILFPTDHS 978
+K + S+ CVF+GY KG++ D + ++S+DV+F E FP+ I H
Sbjct: 625 NRGGDKFAERSRRCVFVGYPHGQKGWRLFDLEQNEFFVSRDVVFSELEFPFRI----SHE 680
Query: 979 SSSSAEFYPLSTIPIIS----------HTAPPSSPVLAASG-----------HSSPGPQD 1017
+ E PI+ A P+ P+ +S HS+ P D
Sbjct: 681 QNVIEEEEEALWAPIVDGLIEEEVHLGQNAGPTPPICVSSPISPSATSSRSEHSTSSPLD 740
Query: 1018 SPQQQQPPSATLSADDPS---------------TSTPAVVSPPVPSSSAQSADTSASVVA 1062
+ P ++T SA PS T+ AV P VP QS A V
Sbjct: 741 TEVVPTPATSTTSASSPSSPTNLQFLPLSRAKPTTAQAVAPPAVPPPRRQSTRNKAPPVT 800
Query: 1063 AAPPVIVNTHPMQTRAKNGIVKPRLQPTLLLTHLEPTSVKQAMKDVKWYKAMQEEYTAL- 1121
V+ T ++ +K + +LQ D + + A Y A+
Sbjct: 801 LKDFVVNTTVCQESPSKLNSILYQLQKR---------------DDTRRFSASHTTYVAID 845
Query: 1122 --MNNGTWTLVPLPANRTPVGCKWIYRIKENPDGSINKYKARLVAKGYSQVQGFDYSETF 1179
N TWT+ LP + +G +W+Y++K N DGS+ +YKARLVA G Q +G DY ETF
Sbjct: 846 AQEENHTWTIEDLPPGKRAIGSQWVYKVKHNSDGSVERYKARLVALGNKQKEGEDYGETF 905
Query: 1180 SPVVKPITVRLILSLAISRGWPLQQLDVNNAFLNGVLEEEVYMVQPPGFEHKDKNLVCKL 1239
+PV K TVRL L +A+ R W + Q+DV+NAFL+G L EEVYM PPGFE N VC+L
Sbjct: 906 APVAKMATVRLFLDVAVKRNWEIHQMDVHNAFLHGDLREEVYMKLPPGFEASHPNKVCRL 965
Query: 1240 QKALYGLKQAPRAWFHRLKEVLIRFGFQASKCDPSLFTYYSSQGCIYMLVYVDDIIITGA 1299
+KALYGLKQAPR WF +L L R+GFQ S D SLFT I +L+YVDD+IITG
Sbjct: 966 RKALYGLKQAPRCWFEKLTTALKRYGFQQSLADYSLFTLVKGSVRIKILIYVDDLIITGN 1025
Query: 1300 SMSLIQQLTAQLHSIFALKQLGQLDYFLGVQVTHLSNGNLLLNQTKYISDLLTKVNMLDA 1359
S QQ L S F +K LG L YFLG++V + G + + Q KY D++++ +L
Sbjct: 1026 SQRATQQFKEYLASCFHMKDLGPLKYFLGIEVARSTTG-IYICQRKYALDIISETGLLGV 1084
Query: 1360 AAISTPMQCGVKLSKNEGSALKDPTEYRSVVGALQYATITRPEISFAVNKVCQFMAQPHE 1419
+ P++ KL + L DP YR +VG L Y +TR +++F+V+ + +FM +P E
Sbjct: 1085 KPANFPLEQNHKLGLSTSPLLTDPQRYRRLVGRLIYLAVTRLDLAFSVHILARFMQEPRE 1144
Query: 1420 EHWKAVKRILRYLKGTLTHGILLQPCSMTRPLPLIAYCDADWGSDPDDRRSTSGSCVFLG 1479
+HW A R++RYLK G+ L+ + + +CD+DW DP RRS +G V G
Sbjct: 1145 DHWAAALRVVRYLKADPGQGVFLR---RSGDFQITGWCDSDWAGDPMSRRSVTGYFVQFG 1201
Query: 1480 YNLISWTAKKQSLVARSSTEAEYRSLANTTAELLWVQSLLTELKIPFTIPTVY-CDNMST 1538
+ ISW KKQ V++SS EAEYR+++ +ELLW++ LL L + P + CD+ S
Sbjct: 1202 DSPISWKTKKQDTVSKSSAEAEYRAMSFLASELLWLKQLLFSLGVSHVQPMIMCCDSKSA 1261
Query: 1539 VILTHNPILHTRTKHMEMDIFFVREKVQDKSLLVQHVPSEHQKADIFTKALSPTRFLLLK 1598
+ + NP+ H RTKH+E+D FVR++ + +HV + Q ADIFTK L F +
Sbjct: 1262 IYIATNPVFHERTKHIEIDYHFVRDEFVKGVITPRHVGTTSQLADIFTKPLGRDCFSAFR 1321
Query: 1599 DKLNV 1603
KL +
Sbjct: 1322 IKLGI 1326
>At1g60020 hypothetical protein
Length = 1194
Score = 612 bits (1579), Expect = e-175
Identities = 381/997 (38%), Positives = 524/997 (52%), Gaps = 137/997 (13%)
Query: 463 TGMHPQAMLATVPAHHSSPVTWCPDSGATHHVTNNPGIFVDHVPSSSQEQLLVGNGQGLP 522
T P+A A ++++ W DSGATHH+T++ H P + E + + +G GL
Sbjct: 269 TPWQPRANAAIASPYNAN--NWLLDSGATHHITSDLNNLSLHQPYTGGEDVTIADGSGLS 326
Query: 523 IQSIGSALLPSSFNPTASLILKKLLLVPSITKNLVSVSRFARDNNVFFSFHANHCSVHHQ 582
I GSAL+ + P+ SL L +L VP+I KNL+SV R N V F H V
Sbjct: 327 ISHTGSALIST---PSRSLALTDVLYVPNIHKNLISVYRMCNANKVSVEFFPAHFQVKDL 383
Query: 583 DTYELLLTGTVGDDGLYYFDGPLTKASSQPQSSSPLSSIQACLASKSAANKASPAIDLQS 642
T LL G D+ ++ P+ + P+ SS ++ +P DL S
Sbjct: 384 KTGVQLLQGRTKDE---LYEWPV----NPPKPSSHFTT-------------TTPKTDLTS 423
Query: 643 ATVSPIVPSLVPCNNADSVNVSSTVSDLVSCNSGSGISVYDLWHSRLGHPHHDVLKQTLS 702
WHSRLGHP LK +S
Sbjct: 424 ------------------------------------------WHSRLGHPSLSTLKVVVS 441
Query: 703 LCNISVPSNKSVFSFCKACCLSKLHRLPSSSSNTVYNTPFELVFADLWGPASLESSCGFL 762
++ V ++ C C L+K H+LP ++ P E ++ DLW + + S F
Sbjct: 442 QFSLPVSNSLQKQFNCSDCLLNKTHKLPFHTNTITSTQPLEYLYIDLW-TSPIVSIDNFK 500
Query: 763 YFLTCVDACTKYTWVFPLKKKSDTCATFINFQAMVELQFGHKLKSVQTDGGGEFKPLTSH 822
Y+L VD T+Y+W +P+K+KS F+ F+A+V +F K+ + +D GGEF L S
Sbjct: 501 YYLVIVDHYTRYSWFYPIKQKSHVKDVFMTFKALVANKFQRKIIHLYSDNGGEFIALRSF 560
Query: 823 FQKLGIIHRLTCPHTHHQNGSVERKHRHIVETGLALLSHASMPLQFWDHAFLAATYLINR 882
GI H T PHT NG ERKHRHIVETGL LL ASMP +W +AF A YLINR
Sbjct: 561 LSSNGITHLTTPPHTPEHNGISERKHRHIVETGLTLLGQASMPKSYWSYAFTIAIYLINR 620
Query: 883 MSTTTLQGASPYFKLYGQYPDFKSLKVFGSACFPFLRPYNSNKLSLHSKECVFIGYSINH 942
MS+ + G SPY +L+GQ P++ L+VFG CFP+LRPY ++KL CVF+GYS
Sbjct: 621 MSSDVIGGISPYKRLFGQAPNYLKLRVFGCLCFPWLRPYTTHKLDDRPAPCVFLGYSQTQ 680
Query: 943 KGYKCLDQ-SGRIYISKDVLFHEHRFPY--PILFP-------TDHSSSSSAEFYPLSTIP 992
Y CL++ +GR+Y S+ V F E+ +P+ P L P +HS +++ P +P
Sbjct: 681 SAYLCLNRTTGRVYTSRHVQFVENTYPFTKPTLDPFTNLEESNNHSITTTVPSPPFVQLP 740
Query: 993 II------SHTAPP---------------SSPVLAASGHSS------------------- 1012
+ H PP SSPV+ +S S
Sbjct: 741 SVPPPTRDPHQPPPSQPAPSPSPLSPPSMSSPVMTSSPQFSSNRDSTTLHGDYSHVDYGL 800
Query: 1013 ------PGPQDSPQQQQPPSATLSADDPSTSTPAVVSPPVPSSSAQSADT-----SASVV 1061
PGP SP + PS S+ PS S +PP SS+ S+ T S
Sbjct: 801 SSPSNPPGPITSPTTSKSPSEPTSS--PSHSNQPNKTPPNSPSSSSSSPTPIPSPSPQSS 858
Query: 1062 AAAPPVIVNTHPMQTRAKNGIVKPRLQPTLLLT----HLEPTSVKQAMKDVKWYKAMQEE 1117
+ PP N H M+TRAKN I KP + TL T PT+V +A++D W AM EE
Sbjct: 859 NSPPPPPQNQHSMRTRAKNNITKPIKKLTLAATPKGKSKIPTTVAEALRDPNWRNAMSEE 918
Query: 1118 YTALMNNGTWTLVPLPANRTPVGCKWIYRIKENPDGSINKYKARLVAKGYSQVQGFDYSE 1177
+ A + N T+ LVP ++ VG +WI+ IK NPDGSIN+YKAR +AKG+ Q G DYS
Sbjct: 919 FNAGLRNSTYDLVPPKPHQNFVGTRWIFTIKYNPDGSINRYKARFLAKGFHQQHGLDYSN 978
Query: 1178 TFSPVVKPITVRLILSLAISRGWPLQQLDVNNAFLNGVLEEEVYMVQPPGFEHKDK-NLV 1236
TFSPV+K TV+ +L +A+SR W ++QLD+NNAFL G L E+VY+ QPPGF + D+ N V
Sbjct: 979 TFSPVIKSTTVQTVLDIAVSRSWDIRQLDINNAFLQGRLTEDVYVAQPPGFINPDRPNYV 1038
Query: 1237 CKLQKALYGLKQAPRAWFHRLKEVLIRFGFQASKCDPSLFTYYSSQGCIYMLVYVDDIII 1296
C L+KAL+GLKQAPRAW+ L+ L+ GF S + SLF ++ IY+LVYVDD +I
Sbjct: 1039 CHLKKALHGLKQAPRAWYQELRGFLLTCGFTNSVANTSLFIRQHNKDYIYILVYVDDFLI 1098
Query: 1297 TGASMSLIQQLTAQLHSIFALKQLGQLDYFLGVQVTHLSNGNLLLNQTKYISDLLTKVNM 1356
TG++ +LI Q L + F+LK LGQL YFL ++ T G L L Q +Y+ DLLTK M
Sbjct: 1099 TGSNSNLIAQFITCLANRFSLKDLGQLSYFLEIEATRTKAG-LHLMQRRYVLDLLTKTKM 1157
Query: 1357 LDAAAISTPMQCGVKLSKNEGSALKDPTEYRSVVGAL 1393
LDA +STPM KL+ G+ + +P YR ++G+L
Sbjct: 1158 LDAKTVSTPMSPTPKLTLTSGTPIDNPGGYRQILGSL 1194
Score = 95.9 bits (237), Expect = 2e-19
Identities = 55/216 (25%), Positives = 111/216 (50%), Gaps = 11/216 (5%)
Query: 10 MPSSSSSSSLSQAFSHKLSI---------KLDDSNFYSWKQQVEGVIRSHKLQKFVQDPP 60
M S+ S S+ + AFS ++ KL +N+ W +QV ++ + L ++
Sbjct: 1 MTSTPSGSTETIAFSETPTLLNVNMANITKLTPTNYIMWNRQVHALLDGYDLAGYIDGSV 60
Query: 61 MIPEKFLTEEDKITENVNPLFADWEQQDSLLFTWLLSTLSPAILPNVIQCVHSYQVWEEI 120
P + +T NP + W++QD L+++ +L T++ I P + + + ++WE++
Sbjct: 61 TAPSEMITTAG--VSAANPAYKFWKRQDKLIYSAILGTITTTIQPLLSRSNTAAEIWEKL 118
Query: 121 HSFFNAKSRAQSTQLRNELKNITKGTKTASEFLQRIKTIVNTLASIGEPVTFRDHLEAIF 180
S + S Q+R +K +KGTKT +E+ Q T + LA +G+P+ + +E +
Sbjct: 119 KSIYATPSWGHIQQMRQHIKQWSKGTKTITEYFQGHTTRFDELALLGKPLEHAEQIEFLL 178
Query: 181 DGLSEEYSALMTVIYNQTTYFTISEVEAMVISHEAR 216
GLSE+Y +++ + T++E+ +++ EA+
Sbjct: 179 GGLSEDYKSVVDQTEIRDKPPTLTELLEKLLNREAK 214
Score = 32.3 bits (72), Expect = 2.3
Identities = 22/69 (31%), Positives = 31/69 (44%), Gaps = 10/69 (14%)
Query: 225 LADTTPAVFLAQAQNSPSSSTAPAPPPPQAMWTQPAPAPSSTMAQPPATSVAPSQPVDTA 284
+ T P+ Q + P + P PPP +QPAP+PS P + + S PV T+
Sbjct: 727 ITTTVPSPPFVQLPSVPPPTRDPHQPPP----SQPAPSPS------PLSPPSMSSPVMTS 776
Query: 285 VDSGYGNRD 293
NRD
Sbjct: 777 SPQFSSNRD 785
>At1g70010 hypothetical protein
Length = 1315
Score = 609 bits (1570), Expect = e-174
Identities = 354/941 (37%), Positives = 522/941 (54%), Gaps = 41/941 (4%)
Query: 677 SGISVYDLWHSRLGHPHHDVLKQTLSLCNISVPSNKSVFSF-CKACCLSKLHRLPSSSSN 735
+ ++ +DLWH RLGHP L+ SL +S P K+ F C+ C +SK LP S N
Sbjct: 399 ASVTSHDLWHKRLGHPSVQKLQPMSSL--LSFPKQKNNTDFHCRVCHISKQKHLPFVSHN 456
Query: 736 TVYNTPFELVFADLWGPASLESSCGFLYFLTCVDACTKYTWVFPLKKKSDTCATFINFQA 795
+ PF+L+ D WGP S+++ G+ YFLT VD ++ TWV+ L+ KSD F
Sbjct: 457 NKSSRPFDLIHIDTWGPFSVQTHDGYRYFLTIVDDYSRATWVYLLRNKSDVLTVIPTFVT 516
Query: 796 MVELQFGHKLKSVQTDGGGEFKPLTSHFQKLGIIHRLTCPHTHHQNGSVERKHRHIVETG 855
MVE QF +K V++D E T + GI+ +CP T QN VERKH+HI+
Sbjct: 517 MVENQFETTIKGVRSDNAPELN-FTQFYHSKGIVPYHSCPETPQQNSVVERKHQHILNVA 575
Query: 856 LALLSHASMPLQFWDHAFLAATYLINRMSTTTLQGASPYFKLYGQYPDFKSLKVFGSACF 915
+L + +P+ +W L A YLINR+ L+ P+ L P + +KVFG C+
Sbjct: 576 RSLFFQSHIPISYWGDCILTAVYLINRLPAPILEDKCPFEVLTKTVPTYDHIKVFGCLCY 635
Query: 916 PFLRPYNSNKLSLHSKECVFIGYSINHKGYKCLD-QSGRIYISKDVLFHEHRFPYPILFP 974
P + +K S +K C FIGY KGYK LD ++ I +S+ V+FHE FP+
Sbjct: 636 ASTSPKDRHKFSPRAKACAFIGYPSGFKGYKLLDLETHSIIVSRHVVFHEELFPF---LG 692
Query: 975 TDHSSSSSAEFYPLSTIPIISHTAPPSSPVLAASGHSSPGPQDSPQQQQPPSATLSADDP 1034
+D S F L+ P + + S H +P S + P + +P
Sbjct: 693 SDLSQEEQNFFPDLNPTPPMQRQS---------SDHVNPSDSSSSVEILPSA------NP 737
Query: 1035 STSTPAVVSPPVPSSSAQSADTSASVVAAAPPVIVNTHPMQTR---AKNGIVKPRLQPTL 1091
+ + P P +S + A A + +V++ P + R + + I P L
Sbjct: 738 TNNVPE----PSVQTSHRKAKKPAYLQDYYCHSVVSSTPHEIRKFLSYDRINDPYLTFLA 793
Query: 1092 LLTHL-EPTSVKQAMKDVKWYKAMQEEYTALMNNGTWTLVPLPANRTPVGCKWIYRIKEN 1150
L EP++ +A K W AM E+ L TW + LPA++ +GC+WI++IK N
Sbjct: 794 CLDKTKEPSNYTEAEKLQVWRDAMGAEFDFLEGTHTWEVCSLPADKRCIGCRWIFKIKYN 853
Query: 1151 PDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILSLAISRGWPLQQLDVNNA 1210
DGS+ +YKARLVA+GY+Q +G DY+ETFSPV K +V+L+L +A L QLD++NA
Sbjct: 854 SDGSVERYKARLVAQGYTQKEGIDYNETFSPVAKLNSVKLLLGVAARFKLSLTQLDISNA 913
Query: 1211 FLNGVLEEEVYMVQPPGFEHKD-----KNLVCKLQKALYGLKQAPRAWFHRLKEVLIRFG 1265
FLNG L+EE+YM P G+ + N VC+L+K+LYGLKQA R W+ + L+ G
Sbjct: 914 FLNGDLDEEIYMRLPQGYASRQGDSLPPNAVCRLKKSLYGLKQASRQWYLKFSSTLLGLG 973
Query: 1266 FQASKCDPSLFTYYSSQGCIYMLVYVDDIIITGASMSLIQQLTAQLHSIFALKQLGQLDY 1325
F S CD + F S + +LVY+DDIII + + + L +Q+ S F L+ LG+L Y
Sbjct: 974 FIQSYCDHTCFLKISDGIFLCVLVYIDDIIIASNNDAAVDILKSQMKSFFKLRDLGELKY 1033
Query: 1326 FLGVQVTHLSNGNLLLNQTKYISDLLTKVNMLDAAAISTPMQCGVKLSKNEGSALKDPTE 1385
FLG+++ G + ++Q KY DLL + L S PM + + + G +
Sbjct: 1034 FLGLEIVRSDKG-IHISQRKYALDLLDETGQLGCKPSSIPMDPSMVFAHDSGGDFVEVGP 1092
Query: 1386 YRSVVGALQYATITRPEISFAVNKVCQFMAQPHEEHWKAVKRILRYLKGTLTHGILLQPC 1445
YR ++G L Y ITRP+I+FAVNK+ QF P + H +AV +IL+Y+KGT+ G+
Sbjct: 1093 YRRLIGRLMYLNITRPDITFAVNKLAQFSMAPRKAHLQAVYKILQYIKGTIGQGLFY--- 1149
Query: 1446 SMTRPLPLIAYCDADWGSDPDDRRSTSGSCVFLGYNLISWTAKKQSLVARSSTEAEYRSL 1505
S T L L Y +AD+ S D RRSTSG C+FLG +LI W ++KQ +V++SS EAEYRSL
Sbjct: 1150 SATSELQLKVYANADYNSCRDSRRSTSGYCMFLGDSLICWKSRKQDVVSKSSAEAEYRSL 1209
Query: 1506 ANTTAELLWVQSLLTELKIPFTIPT-VYCDNMSTVILTHNPILHTRTKHMEMDIFFVREK 1564
+ T EL+W+ + L EL++P + PT ++CDN + + + +N + H RTKH+E D VRE+
Sbjct: 1210 SVATDELVWLTNFLKELQVPLSKPTLLFCDNEAAIHIANNHVFHERTKHIESDCHSVRER 1269
Query: 1565 VQDKSLLVQHVPSEHQKADIFTKALSPTRFLLLKDKLNVVD 1605
+ + H+ +E Q AD FTK L P+ F L K+ +++
Sbjct: 1270 LLKGLFELYHINTELQIADPFTKPLYPSHFHRLISKMGLLN 1310
Score = 52.4 bits (124), Expect = 2e-06
Identities = 42/178 (23%), Positives = 89/178 (49%), Gaps = 16/178 (8%)
Query: 78 NPLFADWEQQDSLLFTWLLSTLSPAILPNVIQCVHSYQVWEEIHSFFNAKSRAQSTQLRN 137
+P W + +S++ +WLL+++S I +++ + +W+++++ F+ S + +LR
Sbjct: 25 DPYCKIWRRCNSMVKSWLLNSVSKEIYTSILYFPTAAAIWKDLYTRFHKSSLPRLYKLRQ 84
Query: 138 ELKNITKGTKTASEFLQRIKTIVNTLASI-GEPVTFRDHL-----EAIFD---GLSEEYS 188
++ ++ +G S + R +T+ L S+ P T D L + D GL++ Y
Sbjct: 85 QIHSLRQGNLDLSSYHTRTQTLWEELTSLQAVPRTVEDLLIERETNRVIDFLMGLNDCYD 144
Query: 189 ALMTVIYNQTTYFTISEVEAMVISHEARFDRMRKKQLADT---TPAVFLAQAQNSPSS 243
+ + I + T ++SEV M+ E + R +++ T T +VF Q+S S+
Sbjct: 145 TVRSQILMKKTLPSLSEVFNMIDQDETQ----RSARISTTPGMTSSVFPVSNQSSQSA 198
>At4g14460 retrovirus-related like polyprotein
Length = 1489
Score = 593 bits (1529), Expect = e-169
Identities = 465/1632 (28%), Positives = 717/1632 (43%), Gaps = 289/1632 (17%)
Query: 34 SNFYSWKQQVEGVIRSHKLQKFVQDPPMIPEKFLTEEDKITENVNPLFADWEQQDSLLFT 93
S+F+SW++ + + F+ P + + F W + + ++ T
Sbjct: 55 SDFHSWRRSILMALNVRNKLGFINGTITKPPEDHRD-----------FGAWSRCNDIVST 103
Query: 94 WLLSTLSPAILPNVIQCVHSYQVWEEIHSFFNAKSRAQSTQLRNELKNITKGTKTASEFL 153
WL++++ I +++ +W + S F + + +L I +G+
Sbjct: 104 WLMNSVDKKIGQSLLYIATVQGIWNNLLSRFKQDDAPRIFDIEQKLSKIEQGSMD----- 158
Query: 154 QRIKTIVNTLASIGEPVTFRDHLEAIFDGLSEEYSALMTVIYNQTTYFTISEVEAMVISH 213
I T L ++ E L G E +A+ Q S V +
Sbjct: 159 --ISTYYTALLTLWEEHRNYVELPVCTCGRCECDAAVKWEHLQQR-----SRVTKFLKEL 211
Query: 214 EARFDRMRKKQLADTTPAVFLAQAQNSPSSSTAPAPPPPQAMWTQPAPAPSSTMAQPPAT 273
FD+ R+ L P + +A N M TQ
Sbjct: 212 NEGFDQTRRHILM-LKPIPTIKEAFN---------------MVTQ----------DERQR 245
Query: 274 SVAPSQPVDTAVDSGYGNRDEQNRNSGGYGRGRGRGR-GRNVQCTYCSKWGHDAASCWSR 332
+V P VD+ + N N + Y R + CT+C K GH C+
Sbjct: 246 NVKPLTRVDSVA---FQNTSMINEDENAYVAAYNTVRPNQKPICTHCGKVGHTIQKCYK- 301
Query: 333 PSGVSSNANSSANSAQSGNSGSNFSVPGVNLGFSP-MQFGGFPSMANFGFPPYA----PY 387
+ ++GN+G + P L P M P M F PY
Sbjct: 302 -------VHGYPPGMKTGNTGYTYK-PNPQLHVQPRMPMMPQPRM-QFPAQPYTNSMQKA 352
Query: 388 NSMLPQYGSSVLSPSS----MPPWSPWCIPPTPY-SHATDMQGMQAAGMSFVRPWLPQGQ 442
N + Y + PS P +P+ P P+ +H + +Q + + Q Q
Sbjct: 353 NVVAQVYAETGAYPSEGYSQAPMMNPYGSYPMPHITHGGNNLSLQDFTPQQIEQMISQFQ 412
Query: 443 FPAPTAQTPQFPRPPQQQVSTGMHPQAMLATVPAHH-----SSPVTWCPDSGATHHVTNN 497
AQ Q P P ++ +P LATV H S+ T P + NN
Sbjct: 413 -----AQV-QVPEP----AASSSNPSP-LATVSEHGFMALTSTSGTIIPFPSTSLKYENN 461
Query: 498 PGIFVDHVPSSSQEQL-----LVGNGQGLPIQSIGSAL-----LPSSFNPTASLILKKLL 547
F +H S+ Q+ L ++ +G + S + + + + T LIL +L
Sbjct: 462 DLKFQNHTLSALQKFLPSDAWIIDSGASSHVCSDLAMFRELKSVSGTVHITQKLILHNVL 521
Query: 548 LVPSITKNLVSVSRFARDNNVFFSFHANHCSVHHQDTYELLLTGTVGDDGLYYFDGPLTK 607
VP NL+SVS + + F+ + C + EL +G LY+ L
Sbjct: 522 HVPDFKFNLMSVSSLVKTISCSAHFYVDCCLIQ-----ELSQGLMIGRGRLYHNLYILET 576
Query: 608 ASSQPQSSSPLSSIQACLASKSAANKASPAIDLQSATVSPIVPSLVPCNNADSVNVSSTV 667
++ P +S+P ACL + S N
Sbjct: 577 ENTSPSTSTP----AACLFTGSVLNDG--------------------------------- 599
Query: 668 SDLVSCNSGSGISVYDLWHSRLGHPHHDVLKQTLSLCNISVPSNKSVFSFCKACCLSKLH 727
LWH RLGHP VL+ KL
Sbjct: 600 ---------------HLWHQRLGHPSSVVLQ--------------------------KLK 618
Query: 728 RLPSSSSNTVYNTPFELVFADLWGPASLESSCGFLYFLTCVDACTKYTWVFPLKKKSDTC 787
RL S N + + PF+LV D+WGP S+ES GF YFLT VD CT+ TWV+ L+ K D
Sbjct: 619 RLAYISHNNLASNPFDLVHLDIWGPFSIESIEGFRYFLTVVDDCTRTTWVYMLRNKKDVS 678
Query: 788 ATFINFQAMVELQFGHKLKSVQTDGGGEFKPLTSHFQKLGIIHRLTCPHTHHQNGSVERK 847
+ F F +V QF K+K++++D E T ++ G++H +C +T QN VERK
Sbjct: 679 SVFPEFIKLVSTQFNAKIKAIRSDNAPEL-GFTEIVKEHGMLHHFSCAYTPQQNSVVERK 737
Query: 848 HRHIVETGLALLSHASMPLQFWDHAFLAATYLINRMSTTTLQGASPYFKLYGQYPDFKSL 907
H+HI+ ALL +++P+Q+W A +LINR+ + L SPY + + PD+ L
Sbjct: 738 HQHILNVARALLFQSNIPMQYWSDCVTTAVFLINRLPSPLLNNKSPYELILNKQPDYSLL 797
Query: 908 KVFGSACFPFLRPYNSNKLSLHSKECVFIGYSINHKGYKCLD-QSGRIYISKDVLFHEHR 966
K FG CF + K + ++ CVF+GY +KGYK LD +S + +S++V+F EH
Sbjct: 798 KNFGCLCFVSTNAHERTKFTPRARACVFLGYPSGYKGYKVLDLESHSVTVSRNVVFKEHV 857
Query: 967 FPYPILFPTDHSSSSSAEFYP------------LSTIPIISHTA--PPSSPVLAASGHSS 1012
FP F T + + + +P + T+P+I + P ++ A H+S
Sbjct: 858 FP----FKTSELLNKAVDMFPNSILPLPAPLHFVETMPLIDEDSLIPTTTDSRTADNHAS 913
Query: 1013 PGPQDSPQQQQPPSATLSADDPSTSTPAV----------------------VSPPVPSSS 1050
P P S T + D S + P +S P+ S
Sbjct: 914 SSSSALPSIIPPSSNTETQDIDSNAVPITRSKRTTRAPSYLSEYHCSLVPSISTLPPTDS 973
Query: 1051 AQSADTSASVVAAAPPVIVNTHPMQTRAKNGIVKPRLQPTLLL--THLEPTSVKQAMKDV 1108
+ + A+ P +P+ T P Q + T EP + QAMK
Sbjct: 974 SIPIHPLPEIFTASSPKKTTPYPISTVVSYDKYTPLCQSYIFAYNTETEPKTFSQAMKSE 1033
Query: 1109 KWYKAMQEEYTALMNNGTWTLVPLPANRTPVGCKWIYRIKENPDGSINKYKARLVAKGYS 1168
KW + EE A+ N TW++ LP ++ VGCKW++ IK NPDG++ +YKARLVA+G++
Sbjct: 1034 KWIRVAVEELQAMELNKTWSVESLPPDKNVVGCKWVFTIKYNPDGTVERYKARLVAQGFT 1093
Query: 1169 QVQGFDYSETFSPVVKPITVRLILSLAISRGWPLQQLDVNNAFLNGVLEEEVYMVQPPGF 1228
Q +G D+ +TFSPV K + +++L LA GW L Q+DV++AFL+G L+EE++M P G+
Sbjct: 1094 QQEGIDFLDTFSPVAKLTSAKMMLGLAAITGWTLTQMDVSDAFLHGDLDEEIFMSLPQGY 1153
Query: 1229 EHK-----DKNLVCKLQKALYGLKQAPRAWFHRLKEVLIRFGFQASKCDPSLFTYYSSQG 1283
N VC+L K++YGLKQA R W+ R
Sbjct: 1154 TPPAGTILPPNPVCRLLKSIYGLKQASRQWYKRF-------------------------- 1187
Query: 1284 CIYMLVYVDDIIITGASMSLIQQLTAQLHSIFALKQLGQLDYFLGVQVTHLSNGNLLLNQ 1343
+ LVY+DDI+I + + ++ L A L S F +K LG +FLG
Sbjct: 1188 -VAALVYIDDIMIASNNDAEVENLKALLRSEFKIKDLGPARFFLG--------------- 1231
Query: 1344 TKYISDLLTKVNMLDAAAISTPMQCGVKLSKNEGSALKDPTEYRSVVGALQYATITRPEI 1403
+L S PM + L ++ G+ L +PT YR ++G L Y TITRP+I
Sbjct: 1232 ------------LLGCKPSSIPMDPTLHLVRDMGTPLPNPTAYRKLIGRLLYLTITRPDI 1279
Query: 1404 SFAVNKVCQFMAQPHEEHWKAVKRILRYLKGTLTHGILLQPCSMTRPLPLIAYCDADWGS 1463
++AV+++ QF++ P + H +A ++LRY+K G++ S + L + DADW +
Sbjct: 1280 TYAVHQLSQFISAPSDIHLQAAHKVLRYIKANPGQGLMY---SADYEICLNGFSDADWAA 1336
Query: 1464 DPDDRRSTSGSCVFLGYNLISWTAKKQSLVARSSTEAEYRSLANTTAELLWVQSLLTELK 1523
D RRS SG C++LG +LISW +KKQ++ +RSSTE+EYRS+A T E++W+Q LL +L
Sbjct: 1337 CKDTRRSISGFCIYLGTSLISWKSKKQAVASRSSTESEYRSMAQATCEIIWLQQLLKDLH 1396
Query: 1524 IPFTIPT-VYCDNMSTVILTHNPILHTRTKHMEMDIFFVREKVQDKSLLVQHVPSEHQKA 1582
IP T P ++CDN S + + NP+ H RTKH+E+D VR++++ +L HVP+E+Q A
Sbjct: 1397 IPLTCPAKLFCDNKSALHSSLNPVFHERTKHIEIDCHTVRDQIKAGNLKALHVPTENQHA 1456
Query: 1583 DIFTKALSPTRF 1594
DI TKAL P F
Sbjct: 1457 DILTKALHPGPF 1468
>At4g27200 putative protein
Length = 819
Score = 510 bits (1313), Expect = e-144
Identities = 298/723 (41%), Positives = 407/723 (56%), Gaps = 49/723 (6%)
Query: 911 GSACFPFLRPYNSNKLSLHSKECVFIGYSINHKGYKCL-DQSGRIYISKDVLFHEHRFPY 969
GSACFP LR Y NK + S +CVF+GY+ +KGY+CL +GR+YIS+ V+F E +P+
Sbjct: 15 GSACFPTLRDYAENKFNPCSLKCVFLGYNEKYKGYRCLYPPTGRLYISRHVIFDESVYPF 74
Query: 970 -------------PILF--------PTDHSSSSSAEFYPLST---IPIISHTAPPSSPVL 1005
P+L P +S+S + PL T P + P P L
Sbjct: 75 SHTYKHLHPQPRTPLLAAWLRSSDSPAPSTSTSPSSRSPLFTSADFPPLPQRKTPLLPTL 134
Query: 1006 AASGHSSPGPQDSPQQQQPPSATLSADDPSTSTPAVVSPPVPSSSAQSADTSASVVAAAP 1065
S + QQ + + D S S S ++ ASV
Sbjct: 135 VPISSVSHASNITTQQSPDFDSERTTDFDSASIGDSSHSSQAGSDSEETIQQASVNVHQT 194
Query: 1066 PVIVNTHPMQTRAKNGIVKPRLQPTLL---LTHLEPTSVKQAMKDVKWYKAMQEEYTALM 1122
P N HPM TRAK GI KP + L +++ EP +V A+K W AM EE
Sbjct: 195 PASTNVHPMVTRAKVGISKPNPRYVFLSHKVSYPEPKTVTAALKHPGWTGAMTEEIGNCS 254
Query: 1123 NNGTWTLVPLPANRTPVGCKWIYRIKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPV 1182
TW+LVP ++ +G KW++R K + DG++NK KAR+VAKG+ Q +G DY ET+SPV
Sbjct: 255 ETQTWSLVPYKSDMHVLGSKWVFRTKLHADGTLNKLKARIVAKGFLQEEGIDYLETYSPV 314
Query: 1183 VKPITVRLILSLAISRGWPLQQLDVNNAFLNGVLEEEVYMVQPPGFEHKDKNLVCKLQKA 1242
V+ TVRL+L LA + W ++Q+DV NAFL+G L+E VYM QP +++ VC L K+
Sbjct: 315 VRTPTVRLVLHLATALNWDIKQMDVKNAFLHGDLKETVYMTQPAA----NRDHVCLLHKS 370
Query: 1243 LYGLKQAPRAWFHRLKEVLIRFGFQASKCDPSLFTYYSSQGCIYMLVYVDDIIITGASMS 1302
+YGLKQ+PRAWF + L+ FGF K DPSLF Y + I +L+
Sbjct: 371 IYGLKQSPRAWFDKFSTFLLEFGFFCRKSDPSLFIYAHNNNLILLLL-----------SQ 419
Query: 1303 LIQQLTAQLHSIFALKQLGQLDY-FLGVQVTHLSNGNLLLNQTKYISDLLTKVNMLDAAA 1361
+ L A L+ F + +GQ FLG+QV NG L ++Q KY DLL +M
Sbjct: 420 TLTSLLAALNKEFRMTDMGQHSLTFLGIQVQRQQNG-LFMSQQKYAEDLLIAASMEHCTP 478
Query: 1362 ISTPMQCGVKLSKNEGSALKDPTEYRSVVGALQYATITRPEISFAVNKVCQFMAQPHEEH 1421
+ TP+ + ++ DPT +RS+ G LQY T+TRP+I FAVN VCQ M QP
Sbjct: 479 LPTPLPVQLDRVPHQEELFSDPTYFRSIAGKLQYLTLTRPDIQFAVNFVCQKMHQPTISD 538
Query: 1422 WKAVKRILRYLKGTLTHGILLQPCSMTRPLPLIAYCDADWGSDPDDRRSTSGSCVFLGYN 1481
+ +KRILRY+KGT+T GI S P L AY D+DWG+ RRS G C F+G N
Sbjct: 539 FHLLKRILRYIKGTITMGI---SYSRDSPTLLQAYSDSDWGNCKQTRRSVGGLCTFMGTN 595
Query: 1482 LISWTAKKQSLVARSSTEAEYRSLANTTAELLWVQSLLTELKIPF-TIPTVYCDNMSTVI 1540
L+SW++KK V+RSSTEAEY+SL++ +E+LW+ +LL EL+IP P ++CDN+S V
Sbjct: 596 LVSWSSKKHPTVSRSSTEAEYKSLSDAASEILWLSTLLRELRIPLPDTPELFCDNLSAVY 655
Query: 1541 LTHNPILHTRTKHMEMDIFFVREKVQDKSLLVQHVPSEHQKADIFTKALSPTRFLLLKDK 1600
LT NP H RTKH ++D FVRE+V K+L+V+H+P Q ADIFTK+L F+ L+ K
Sbjct: 656 LTANPAFHARTKHFDIDFHFVRERVALKALVVKHIPGSEQIADIFTKSLPYEAFIHLRGK 715
Query: 1601 LNV 1603
L V
Sbjct: 716 LGV 718
>At4g10990 putative retrotransposon polyprotein
Length = 1203
Score = 462 bits (1189), Expect = e-130
Identities = 271/696 (38%), Positives = 387/696 (54%), Gaps = 48/696 (6%)
Query: 938 YSINHKGYKCLD-QSGRIYISKDVLFHEHRFPYPILFPTDHSSSSSAEFYPLSTIPIIS- 995
Y +KGYK LD +S I I+++V+FHE +FP F T S + +P S +P+ +
Sbjct: 335 YPSGYKGYKVLDLESHSISITRNVVFHETKFP----FKTSKFLKESVDMFPNSILPLPAP 390
Query: 996 ----HTAPPSSPVLAASGHSSPGPQDSPQQQQPP------------------SATLSADD 1033
+ P + A ++S S PP S ++
Sbjct: 391 LHFVESMPLDDDLRADDNNASTSNSASSASSIPPLPSTVNTQNTDALDIDTNSVPIARPK 450
Query: 1034 PSTSTPAVVSP----PVPSSSAQSADTSASVVAAA----PPVIVNTHPMQTRAKNGIVKP 1085
+ PA +S VP S+ S TS S+ + P I +PM T + P
Sbjct: 451 RNAKAPAYLSEYHCNSVPFLSSLSPTTSTSIETPSSSIPPKKITTPYPMSTAISYDKLTP 510
Query: 1086 RLQPTLLLTHLE--PTSVKQAMKDVKWYKAMQEEYTALMNNGTWTLVPLPANRTPVGCKW 1143
+ ++E P + QAMK KW +A EE AL N TW + L + VGCKW
Sbjct: 511 LFHSYICAYNVETEPKAFTQAMKSEKWTRAANEELHALEQNKTWIVESLTEGKNVVGCKW 570
Query: 1144 IYRIKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILSLAISRGWPLQ 1203
++ IK NPDGSI +YKARLVA+G++Q +G DY ETFSPV K +V+L+L LA + GW L
Sbjct: 571 VFTIKYNPDGSIERYKARLVAQGFTQQEGIDYMETFSPVAKFGSVKLLLGLAAATGWSLT 630
Query: 1204 QLDVNNAFLNGVLEEEVYMVQPPGFE-----HKDKNLVCKLQKALYGLKQAPRAWFHRLK 1258
Q+DV+NAFL+G L+EE+YM P G+ VC+L K+LYGLKQA R W+ RL
Sbjct: 631 QMDVSNAFLHGELDEEIYMSLPQGYTPPTGISLPSKPVCRLLKSLYGLKQASRQWYKRLS 690
Query: 1259 EVLIRFGFQASKCDPSLFTYYSSQGCIYMLVYVDDIIITGASMSLIQQLTAQLHSIFALK 1318
V + F S D ++F S I +LVYVDD++I S ++ L L S F +K
Sbjct: 691 SVFLGANFIQSPADNTMFVKVSCTSIIVVLVYVDDLMIASNDSSAVENLKELLRSEFKIK 750
Query: 1319 QLGQLDYFLGVQVTHLSNGNLLLNQTKYISDLLTKVNMLDAAAISTPMQCGVKLSKNEGS 1378
LG +FLG+++ S G + + Q KY +LL V + S PM + L+K G+
Sbjct: 751 DLGPARFFLGLEIARSSEG-ISVCQRKYAQNLLEDVGLSGCKPSSIPMDPNLHLTKEMGT 809
Query: 1379 ALKDPTEYRSVVGALQYATITRPEISFAVNKVCQFMAQPHEEHWKAVKRILRYLKGTLTH 1438
L + T YR +VG L Y ITRP+I+FAV+ + QF++ P + H +A ++LRYLKG
Sbjct: 810 LLPNATSYRELVGRLLYLCITRPDITFAVHTLSQFLSAPTDIHMQAAHKVLRYLKGNPGQ 869
Query: 1439 GILLQPCSMTRPLPLIAYCDADWGSDPDDRRSTSGSCVFLGYNLISWTAKKQSLVARSST 1498
G++ S + L L + DADWG+ D RRS +G C++LG +LI+W +KKQS+V+RSST
Sbjct: 870 GLMY---SASSELCLNGFSDADWGTCKDSRRSVTGFCIYLGTSLITWKSKKQSVVSRSST 926
Query: 1499 EAEYRSLANTTAELLWVQSLLTELKIPFTIPT-VYCDNMSTVILTHNPILHTRTKHMEMD 1557
E+EYRSLA T E++W+Q LL +L + T P ++CDN S + L NP+ H RTKH+E+D
Sbjct: 927 ESEYRSLAQATCEIIWLQQLLKDLHVTMTCPAKLFCDNKSALHLATNPVFHERTKHIEID 986
Query: 1558 IFFVREKVQDKSLLVQHVPSEHQKADIFTKALSPTR 1593
VR++++ L HVP+ +Q ADI TK L P +
Sbjct: 987 CHTVRDQIKAGKLKTLHVPTGNQLADILTKPLHPVQ 1022
Score = 67.4 bits (163), Expect = 6e-11
Identities = 33/85 (38%), Positives = 51/85 (59%), Gaps = 1/85 (1%)
Query: 762 LYFLTCVDACTKYTWVFPLKKKSDTCATFINFQAMVELQFGHKLKSVQTDGGGEFKPLTS 821
LYFLT VD CT+ TWV+ +K KS+ F F ++ Q+ K+K++++D E T
Sbjct: 251 LYFLTLVDDCTRTTWVYMMKNKSEVSNIFPVFVKLIFTQYNAKIKAIRSDNVKEL-AFTK 309
Query: 822 HFQKLGIIHRLTCPHTHHQNGSVER 846
++ G+IH+ +C +T QN VER
Sbjct: 310 FVKEQGMIHQFSCAYTPQQNSVVER 334
Score = 40.0 bits (92), Expect = 0.011
Identities = 40/140 (28%), Positives = 65/140 (45%), Gaps = 9/140 (6%)
Query: 484 WCPDSGATHHVTNNPGIFVDHVPSSSQEQLLVGNGQGLPIQSIGSALLPSSFNPTASLIL 543
W DSGA+ HV ++ +F + + S L NG + I G+ + T++LIL
Sbjct: 99 WIIDSGASSHVCSDLTMFRELIHVSGVTVTLP-NGTRVAITHTGTICI------TSTLIL 151
Query: 544 KKLLLVPSITKNLVSVSRFARDNNVFFSFHANHCSVHHQDTYELLLTGTVGDDGLYYFDG 603
+LLVP NL+SV + + F A+ C + + T L++ + LY +
Sbjct: 152 HNVLLVPDFKFNLISVCCLVKTLSYSAHFFADCCYI-QELTRGLMIGRGKTYNNLYILET 210
Query: 604 PLTKAS-SQPQSSSPLSSIQ 622
T S S P +SS ++Q
Sbjct: 211 QRTSFSPSLPAASSFTGTVQ 230
>At3g45520 copia-like polyprotein
Length = 1363
Score = 461 bits (1187), Expect = e-129
Identities = 316/957 (33%), Positives = 480/957 (50%), Gaps = 80/957 (8%)
Query: 684 LWHSRLGHPHHDVLKQTLSLCNISVPSNK--------SVFSFCKACCLSKLHRLPSSSSN 735
LWH RL H +S N+S+ K S+ C+ C + ++ + +
Sbjct: 447 LWHRRLCH---------MSQKNMSLLIKKGFLDKKKVSMLDTCEDCIYGRAKKIGFNLAQ 497
Query: 736 TVYNTPFELVFADLWGPASLESSCGFL-YFLTCVDACTKYTWVFPLKKKSDTCATFINFQ 794
E V +DLWG ++ S G YF++ +D T+ WV+ LK K + F+++
Sbjct: 498 HDTKKKLEYVHSDLWGAPTVPMSLGNCQYFISFIDDYTRKVWVYFLKTKDEAFEKFVSWI 557
Query: 795 AMVELQFGHKLKSVQTDGGGEF--KPLTSHFQKLGIIHRLTCPHTHHQNGSVERKHRHIV 852
++VE Q G ++K+++TD G EF + ++ G TC +T QNG VER +R I+
Sbjct: 558 SLVENQSGERVKTLRTDNGLEFCNRMFDGFCEEKGFQRHRTCAYTPQQNGVVERMNRTIM 617
Query: 853 ETGLALLSHASMPLQFWDHAFLAATYLINRMSTTTLQGASPYFKLYGQYPDFKSLKVFGS 912
E ++L + +P +FW A A LIN+ + + P + G+ P + L+ +G
Sbjct: 618 EKVRSMLCDSGLPKRFWAEATHTAVLLINKTPCSAINFEFPDKRWSGKAPIYSYLRRYG- 676
Query: 913 ACFPFLRPYNSNKLSLHSKECVFIGYSINHKGYKC-LDQSGRIYISKDVLFHEHRFPYPI 971
C F+ + KL+L +K+ V IGY KGYK L + + +S++V F E+ Y
Sbjct: 677 -CVTFVHT-DGGKLNLRAKKGVLIGYPSGVKGYKVWLIEEKKCVVSRNVSFQENAV-YKD 733
Query: 972 LFPTDHSSSSSAEFYPLSTIPIISHTAPPSSPVLAASGHSSPGPQDSPQQQQPPSATLSA 1031
L S + + S I + L A +S G + S Q P AT A
Sbjct: 734 LMQRKEQVSCEEDDHAGSYIDLD----------LEADKDNSSGGEQSQAQVTP--ATRGA 781
Query: 1032 DDPSTSTPAVVSPPVPSSSAQSADTSASVVAAAPPVIVNTHPMQTRAKNGIVKPR----- 1086
TSTP P + + D S ++ H ++ R + I PR
Sbjct: 782 ---VTSTP----PRYETDDIEETDVHQSPLSY--------HLVRDRERREIRAPRRFDDE 826
Query: 1087 ------LQPTLLLTHLEPTSVKQAMKDVKWYK---AMQEEYTALMNNGTWTLVPLPANRT 1137
L T +EP K+A++D W K AM EE + + N TWT V P +
Sbjct: 827 DYYAEALYTTEDGDAVEPADYKEAVRDENWDKWRLAMNEEIESQLKNDTWTTVTRPEKQR 886
Query: 1138 PVGCKWIYRIKEN-PDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILSLAI 1196
+G +WIY+ K+ P ++KARLVAKGY+Q +G DY E F+PVVK +++R++LS+
Sbjct: 887 IIGSRWIYKYKQGIPGVEEPRFKARLVAKGYAQREGVDYHEIFAPVVKHVSIRILLSIVA 946
Query: 1197 SRGWPLQQLDVNNAFLNGVLEEEVYMVQPPGFEHKDK-NLVCKLQKALYGLKQAPRAWFH 1255
L+QLDV AFL+G L+E++YM+ P G E K N VC L K+LYGLKQAPR W
Sbjct: 947 QENLELEQLDVKTAFLHGELKEKIYMMPPEGCESLFKENEVCLLNKSLYGLKQAPRQWNE 1006
Query: 1256 RLKEVLIRFGFQASKCDPSLFTY-YSSQGCIYMLVYVDDIIITGASMSLIQQLTAQLHSI 1314
+ + GF+ S D +T S +Y+L YVDD+++ +M I L +L
Sbjct: 1007 KFNHYMTEIGFKRSDYDSCAYTKKLSDDSTMYLLFYVDDMLVAANNMQAIDALKKELSIK 1066
Query: 1315 FALKQLGQLDYFLGVQVT-HLSNGNLLLNQTKYISDLLTKVNMLDAAAISTPMQCGVKLS 1373
F +K LG LG+++ G L L+Q Y++ +L NML++ TP+ +K+
Sbjct: 1067 FEMKDLGAAKKILGIEIIIDREAGVLWLSQESYLNKVLKTFNMLESKPALTPLGAHLKMK 1126
Query: 1374 KNEGSALKDPTEYR------SVVGALQYATI-TRPEISFAVNKVCQFMAQPHEEHWKAVK 1426
L EY S VG++ YA I TRP++++ V V +FM+QP +EHW VK
Sbjct: 1127 SATEEKLSTEEEYMNSVPYSSAVGSIMYAMIGTRPDLAYPVGVVSRFMSQPAKEHWLGVK 1186
Query: 1427 RILRYLKGTLTHGILLQPCSMTRPLPLIAYCDADWGSDPDDRRSTSGSCVFLGYNLISWT 1486
+LRY+KGT+ + + S + YCDAD+ +D D RRS +G LG N ISW
Sbjct: 1187 WVLRYIKGTVDTRLCYKRNS---DFSICGYCDADYAADLDKRRSITGLVFTLGGNTISWK 1243
Query: 1487 AKKQSLVARSSTEAEYRSLANTTAELLWVQSLLTELKIPFTIPTVYCDNMSTVILTHNPI 1546
+ Q +VA+SSTE EY SL E +W++ LL + ++CD+ S + L+ N +
Sbjct: 1244 SGLQRVVAQSSTECEYMSLTEAVKEAIWLKGLLKDFGYEQKNVEIFCDSQSAIALSKNNV 1303
Query: 1547 LHTRTKHMEMDIFFVREKVQDKSLLVQHVPSEHQKADIFTKALSPTRFLLLKDKLNV 1603
H RTKH+++ F+RE + D + V + +E ADIFTK L +F D L V
Sbjct: 1304 HHERTKHIDVKFHFIREIIADGKVEVSKISTEKNPADIFTKVLPVNKFQTALDFLRV 1360
>At2g06840 putative retroelement pol polyprotein
Length = 1102
Score = 451 bits (1161), Expect = e-126
Identities = 281/818 (34%), Positives = 406/818 (49%), Gaps = 103/818 (12%)
Query: 792 NFQAMVELQFGHKLKSVQTDGGGEFKPLTSHFQKLGIIHRLTCPHTHHQNGSVERKHRHI 851
NF AM + QF ++S+++D G EF TS+FQ+ GI+H +C T QN VERKHRHI
Sbjct: 357 NFCAMADRQFRKPVRSIRSDNGTEFMCHTSYFQEHGILHETSCVDTPQQNARVERKHRHI 416
Query: 852 VETGLALLSHASMPLQFWDHAFLAATYLINRMSTTTLQGASPYFKLYGQYPDFKSLKVFG 911
+ L + P + L+G +PY L+G+ P + L+ FG
Sbjct: 417 LNVARTCLFQGNFPTP-----------------SPVLKGKTPYEVLFGKQPSYDMLRTFG 459
Query: 912 SACFPFLRPYNSNKLSLHSKECVFIGYSINHKGYKCLDQSGRIYISKDVLFHEHRFPYPI 971
C+ +RP + +K + S++C+FIGY P+
Sbjct: 460 CLCYAHIRPRDKDKFASRSRKCIFIGY-----------------------------PHET 490
Query: 972 LFPTDHSSSSSAEFYPLSTIPIISHTAPPSSPVLAASGHSSPGPQDSPQ----QQQPPSA 1027
P H S P ST ++T P A HS P SP + S
Sbjct: 491 ATPNTHDSID-----PTSTSSDENNTPPEPVTPQAEQPHS-PSSISSPHIVHNKGSVHSR 544
Query: 1028 TLSADDPSTSTPAVVSPPVPSSSAQSADTSASVVAAAPPVIVNTHPMQTRAKNGIVKPRL 1087
L+ D S+S P +P + V V H + G+ +
Sbjct: 545 HLNEDHDSSS------PGLPELLGKGHRPKHPPVYLKDYVAHKVHSSPHTSSPGLSDSNV 598
Query: 1088 QPTLLLTHL----------EPTSVKQAMKDVKWYKAMQEEYTALMNNGTWTLVPLPANRT 1137
PT+ H+ E K + +W AMQ+E AL N TW + LP +
Sbjct: 599 SPTVSANHIAFMAAILDSNEQNHFKDDVLIKEWCDAMQKEIEALEANHTWDVTDLPHGKK 658
Query: 1138 PVGCKWIYRIKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILSLAIS 1197
+ KW+Y++K N DG++ ++KARLV G Q +G D+ ETF+PV K TVRL+L++A +
Sbjct: 659 AISSKWVYKLKFNSDGTLERHKARLVVMGNHQKEGIDFKETFAPVAKMTTVRLLLAVAAA 718
Query: 1198 RGWPLQQLDVNNAFLNGVLEEEVYMVQPPGFEHKDKNLVCKLQKALYGLKQAPRAWFHRL 1257
+ W + Q+DV+NAFL+G LE+E +LYGLKQAPR WF +L
Sbjct: 719 KDWDVFQMDVHNAFLHGDLEQE----------------------SLYGLKQAPRCWFAKL 756
Query: 1258 KEVLIRFGFQASKCDPSLFTYYSSQGCIYMLVYVDDIIITGASMSLIQQLTAQLHSIFAL 1317
L + GF S D SLF+ I+ LVYVDD II G ++ I LH F +
Sbjct: 757 STALRKLGFTQSYEDYSLFSLNRDGTVIHFLVYVDDFIIVGNNLKAIDHFKEHLHKCFHM 816
Query: 1318 KQLGQLDYFLGVQVTHLSNGNLLLNQTKYISDLLTKVNMLDAAAISTPMQCGVKLSKNEG 1377
K LG+L YFLG++V+ ++G L+Q KY D++ + +L + PM+ KL
Sbjct: 817 KDLGKLKYFLGLEVSRGADG-FCLSQQKYALDIINEAGLLGYKPSAVPMELHHKLGSISS 875
Query: 1378 SALKDPTEYRSVVGALQYATITRPEISFAVNKVCQFMAQPHEEHWKAVKRILRYLKGTLT 1437
+P +YR +V Y TITRP++S+AV+ + QFM P E HW A R++RYLKG+
Sbjct: 876 PVFDNPAQYRRLVDRFIYLTITRPDLSYAVHILSQFMQTPLEAHWHATLRLVRYLKGSPD 935
Query: 1438 HGILLQPCSMTRPLPLIAYCDADWGSDPDDRRSTSGSCVFLGYNLISWTAKKQSLVARSS 1497
GILL+ R L L AYCD+D+ P RRS S ++LG ISW KKQ V+ SS
Sbjct: 936 QGILLR---SDRALSLTAYCDSDYNPCPRTRRSLSAYVLYLGDTPISWKTKKQDTVSSSS 992
Query: 1498 TEAEYRSLANTTAELLWVQSLLTELKIPFTIP-TVYCDNMSTVILTHNPILHTRTKHMEM 1556
EAEYR++A T E+ W+++L+T L + T P ++CD+ + + + NP+ H RTKH+E
Sbjct: 993 AEAEYRAMAYTLKEIKWLKALMTTLGVDHTQPILLFCDSQAAIHIAANPVFHERTKHIEK 1052
Query: 1557 DIFFVREKVQDKSLLVQHVPSEHQKADIFTKALSPTRF 1594
D VR+ V DK + H+ + D+ TKAL F
Sbjct: 1053 DCHQVRDAVTDKVISTPHIST----TDLLTKALPRPTF 1086
Score = 36.2 bits (82), Expect = 0.16
Identities = 37/136 (27%), Positives = 59/136 (43%), Gaps = 28/136 (20%)
Query: 487 DSGATHHVTNNPGIFV---DHVPSSSQEQLLVGNGQGLPIQSIGSALLPSSFNPTASLIL 543
DSGA+HH+T + I V D +PS+ + +G+ + LL SS+ L
Sbjct: 196 DSGASHHMTGDCSILVDVFDIIPSAVTKP----DGKASCATKCVTLLLSSSYK------L 245
Query: 544 KKLLLVPSITKNLVSVSRFARDNNVFFSFHANHCSVHHQDTYELLLTGTVGDDGLYYFDG 603
+ +L VP L+SVS+ + F + L+ G V + +YYF G
Sbjct: 246 QDVLFVPDFDCTLISVSKLLKQTGPF--------------SRTLIGAGEV-RERVYYFTG 290
Query: 604 PLTKASSQPQSSSPLS 619
L ++ + S S S
Sbjct: 291 VLVASAHKTSSDSTSS 306
>At2g21460 putative retroelement pol polyprotein
Length = 1333
Score = 435 bits (1119), Expect = e-122
Identities = 290/930 (31%), Positives = 466/930 (49%), Gaps = 45/930 (4%)
Query: 684 LWHSRLGHPHHDVLKQTLSLCNISVPSNKSVFSFCKACCLSKLHRLPSSSSNTVYNTPFE 743
LWH RLGH + L + S C+ C K R+ + + E
Sbjct: 418 LWHRRLGHMSQKNMDLLLKK-GLLDKKKVSKLETCEDCIYGKAKRIGFNLAQHDTREKLE 476
Query: 744 LVFADLWGPASLESSCGFL-YFLTCVDACTKYTWVFPLKKKSDTCATFINFQAMVELQFG 802
V +DLWG S+ S G YF++ +D T+ ++ LK K + F+ + +VE Q
Sbjct: 477 YVHSDLWGAPSVPFSLGKCQYFISFIDDYTRKVRIYFLKTKDEAFDKFVEWANLVENQTD 536
Query: 803 HKLKSVQTDGGGEF--KPLTSHFQKLGIIHRLTCPHTHHQNGSVERKHRHIVETGLALLS 860
++K+++TD G EF + + GI+ TC +T QNG ER +R ++E ++LS
Sbjct: 537 KRIKTLRTDNGLEFCNRSFDEFCSQKGILWHRTCAYTPQQNGVAERMNRTLMEKVRSMLS 596
Query: 861 HASMPLQFWDHAFLAATYLINRMSTTTLQGASPYFKLYGQYPDFKSLKVFGSACFPFLRP 920
+ +P +FW A LIN+ ++ L P + G+ P + L+ FG C F+
Sbjct: 597 DSGLPKKFWAEATHTTAILINKTPSSALNYEVPDKRWSGKSPIYSYLRRFG--CIAFVHT 654
Query: 921 YNSNKLSLHSKECVFIGYSINHKGYKC-LDQSGRIYISKDVLFHEHRFPYPILFPTDHSS 979
+ KL+ +K+ + +GY I KGYK L + + +S++V+F E+ Y + + +
Sbjct: 655 -DDGKLNPRAKKGILVGYPIGVKGYKIWLLEEKKCVVSRNVIFQENA-SYKDMMQSKDAE 712
Query: 980 SSSAEFYPLSTIPI-ISHTAPPSSPVLAASGHSSPGPQDSPQQQQPPSATLSADDPSTST 1038
E P S + + + H V+ + G SP P + ++ ++ T
Sbjct: 713 KDENEAPPSSYLDLDLDH-----EEVITSGGDDPIVEAQSPFNPSPATTQTYSEGVNSET 767
Query: 1039 PAVVSPPVPSSSAQSADTSASVVAAAPPVIVNTHPMQTRAKNGIVKPRLQPTLLLTHLEP 1098
+ SP S D + A PV + A L T +EP
Sbjct: 768 DIIQSP---LSYQLVRDRDRRTIRA--PVRFDDEDYLAEA--------LYTTEDSGEIEP 814
Query: 1099 TSVKQAMKDVKWYK---AMQEEYTALMNNGTWTLVPLPANRTPVGCKWIYRIKEN-PDGS 1154
+A + + W K AM EE + + N TWT+V P ++ +G +WIY+ K P
Sbjct: 815 ADYSEAKRSMNWNKWKLAMNEEMESQIKNHTWTVVKRPQHQKVIGSRWIYKFKLGIPGVE 874
Query: 1155 INKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILSLAISRGWPLQQLDVNNAFLNG 1214
++KARLVAKGY+Q +G DY E F+PVVK +++R+++S+ L+QLDV AFL+G
Sbjct: 875 EGRFKARLVAKGYAQRKGIDYHEIFAPVVKHVSIRILMSIVAQEDLELEQLDVKTAFLHG 934
Query: 1215 VLEEEVYMVQPPGFEHKDK-NLVCKLQKALYGLKQAPRAWFHRLKEVLIRFGFQASKCDP 1273
L+E++YMV P G+E K + VC L K+LYGLKQAP+ W + + GF S D
Sbjct: 935 ELKEKIYMVPPEGYEEMFKEDEVCLLNKSLYGLKQAPKQWNEKFNAYMSEIGFIRSLYDS 994
Query: 1274 SLFTYYSSQGC-IYMLVYVDDIIITGASMSLIQQLTAQLHSIFALKQLGQLDYFLGVQVT 1332
+ S G +Y+L+YVDD+++ + I QL +L F +K LG LG+++
Sbjct: 995 CAYIKELSDGSRVYLLLYVDDMLVAAKNKEDISQLKEELSQRFDMKDLGAAKRILGMEII 1054
Query: 1333 HLSNGNLL-LNQTKYISDLLTKVNMLDAAAISTPMQCGVKL------SKNEGSALKDPTE 1385
N L L+Q Y++ +L NM ++ + TP+ +K+ + +
Sbjct: 1055 RNREENTLWLSQNGYLNKILETYNMAESKHVVTPLGAHLKMRAATVEKQEQDEDYMKSIP 1114
Query: 1386 YRSVVGALQYATI-TRPEISFAVNKVCQFMAQPHEEHWKAVKRILRYLKGTLTHGILLQP 1444
Y S VG++ YA I TRP++++ V + ++M+QP EHW VK +LRY+KG+L G LQ
Sbjct: 1115 YSSAVGSIMYAMIGTRPDLAYPVGIISRYMSQPAREHWLGVKWVLRYIKGSL--GTKLQ- 1171
Query: 1445 CSMTRPLPLIAYCDADWGSDPDDRRSTSGSCVFLGYNLISWTAKKQSLVARSSTEAEYRS 1504
+ ++ YCDAD + D RRS +G LG + ISW + +Q +VA S+TEAEY S
Sbjct: 1172 YKRSSDFKVVGYCDADHAACKDRRRSITGLVFTLGGSTISWKSGQQRVVALSTTEAEYMS 1231
Query: 1505 LANTTAELLWVQSLLTELKIPFTIPTVYCDNMSTVILTHNPILHTRTKHMEMDIFFVREK 1564
L E +W++ LL E ++CD+ S + L+ N + H RTKH+++ ++R+
Sbjct: 1232 LTEAVKEAVWMKGLLKEFGYEQKSVEIFCDSQSAIALSKNNVHHERTKHIDVRYQYIRDI 1291
Query: 1565 VQDKSLLVQHVPSEHQKADIFTKALSPTRF 1594
+ + V + +E ADIFTK + +F
Sbjct: 1292 IANGDGDVVKIDTEKNPADIFTKIVPVNKF 1321
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.131 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,008,501
Number of Sequences: 26719
Number of extensions: 1775596
Number of successful extensions: 14142
Number of sequences better than 10.0: 368
Number of HSP's better than 10.0 without gapping: 208
Number of HSP's successfully gapped in prelim test: 170
Number of HSP's that attempted gapping in prelim test: 8612
Number of HSP's gapped (non-prelim): 3279
length of query: 1615
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1502
effective length of database: 8,299,349
effective search space: 12465622198
effective search space used: 12465622198
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0365.13


