

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0348.15
(242 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
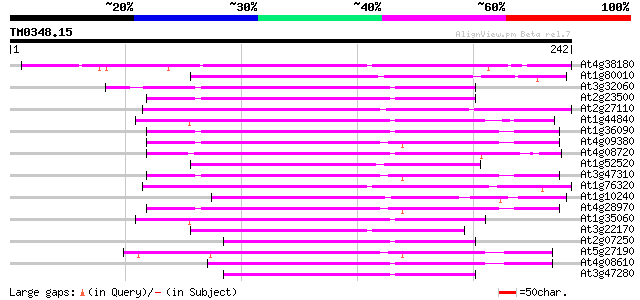
Score E
Sequences producing significant alignments: (bits) Value
At4g38180 unknown protein (At4g38180) 79 3e-15
At1g80010 hypothetical protein 68 4e-12
At3g32060 hypothetical protein 67 7e-12
At2g23500 Mutator-like transposase 66 1e-11
At2g27110 Mutator-like transposase 66 2e-11
At1g44840 hypothetical protein 64 6e-11
At1g36090 hypothetical protein 63 1e-10
At4g09380 putative protein 62 4e-10
At4g08720 putative protein 62 4e-10
At1g52520 F6D8.26 62 4e-10
At3g47310 putative protein 61 5e-10
At1g76320 putative phytochrome A signaling protein 61 5e-10
At1g10240 unknown protein 61 5e-10
At4g28970 putative protein 59 2e-09
At1g35060 hypothetical protein 58 4e-09
At3g22170 far-red impaired response protein, putative 57 7e-09
At2g07250 putative transposase of FARE2.1 (CDS1) 56 2e-08
At5g27190 putative protein 56 2e-08
At4g08610 predicted transposon protein 56 2e-08
At3g47280 putative protein 56 2e-08
>At4g38180 unknown protein (At4g38180)
Length = 788
Score = 78.6 bits (192), Expect = 3e-15
Identities = 63/245 (25%), Positives = 111/245 (44%), Gaps = 15/245 (6%)
Query: 6 KVQVDSMNNNWVPPRHMLATLKENNPGNLSTI--TQV----YNRIKKVKELDCGPLTEMQ 59
K +D++ + PR +++ L + G +S + T+V Y R + K ++ +
Sbjct: 197 KTLIDTLQAAGMGPRRIMSALIKEY-GGISKVGFTEVDCRNYMRNNRQKSIEGEIQLLLD 255
Query: 60 YLLKKLAE-ANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIP 118
YL + A+ N+ + + ED V +FW+ P AI F F V D TY++N++++P
Sbjct: 256 YLRQMNADNPNFFYSVQGSEDQSV-GNVFWADPKAIMDFTHFGDTVTFDTTYRSNRYRLP 314
Query: 119 LLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTDRELALLS 178
G+ G ++ E +VW L A A P I TD + + +
Sbjct: 315 FAPFTGVNHHGQPILFGCAFIINETEASFVWLFNTW--LAAMSAHPPVSITTDHDAVIRA 372
Query: 179 AVRNIFPGATHLLCLFHINKNVEAKCK-LWVDTTDFKALEMQKWNEVVYAETTTQFEEEW 237
A+ ++FPGA H C +HI K + K +++ F++ + K V E+ FE W
Sbjct: 373 AIMHVFPGARHRFCKWHILKKCQEKLSHVFLKHPSFES-DFHKC--VNLTESVEDFERCW 429
Query: 238 RDMCD 242
+ D
Sbjct: 430 FSLLD 434
>At1g80010 hypothetical protein
Length = 696
Score = 68.2 bits (165), Expect = 4e-12
Identities = 45/163 (27%), Positives = 74/163 (44%), Gaps = 7/163 (4%)
Query: 79 DSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPLLEMVGLTSTGLTYSIAFCY 138
D G + +FW A ++ F V++ D T +N +++PL+ VG+ G T +
Sbjct: 268 DDGSLRNVFWIDARARAAYSHFGDVLLFDTTCLSNAYELPLVAFVGINHHGDTILLGCGL 327
Query: 139 MTRERTPDYVWALECMKSLLADPARLPGVIVTDRELALLSAVRNIFPGATHLLCLFHINK 198
+ + YVW + + R P + +T++ A+ +AV +FP A H L L H+
Sbjct: 328 LADQSFETYVWLFRAWLTCML--GRPPQIFITEQCKAMRTAVSEVFPRAHHRLSLTHVLH 385
Query: 199 NVEAKCKLWVDTTDFKALEMQKWNEVVY-AETTTQFEEEWRDM 240
N+ C+ V D M N VVY +FE W +M
Sbjct: 386 NI---CQSVVQLQDSDLFPM-ALNRVVYGCLKVEEFETAWEEM 424
>At3g32060 hypothetical protein
Length = 487
Score = 67.4 bits (163), Expect = 7e-12
Identities = 43/160 (26%), Positives = 80/160 (49%), Gaps = 9/160 (5%)
Query: 42 NRIKKVKELDCGPLTEMQYLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFP 101
+R + K+++C Y+LKK+ + + K E D +F + +I+ F
Sbjct: 212 SREESYKDINC-----YLYMLKKVNDGTVTYLKLDENDK--FQYVFVALGASIEGFRVMR 264
Query: 102 HVVIMDCTYKTNKFQIPLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADP 161
V+I+D T+ N + L+ Y IAF + E + W E +K+++ D
Sbjct: 265 KVLIVDATHLKNGYGGVLVFASAQDPNRHHYIIAFAVLDGENDASWEWFFEKLKTVVPDT 324
Query: 162 ARLPGVIVTDRELALLSAVRNIFPGATHLLCLFHINKNVE 201
+ L V +TDR +L+ A+RN++ A H C++H+++NV+
Sbjct: 325 SEL--VFMTDRNASLIKAIRNVYTAAHHGYCIWHLSQNVK 362
>At2g23500 Mutator-like transposase
Length = 784
Score = 66.2 bits (160), Expect = 1e-11
Identities = 40/142 (28%), Positives = 72/142 (50%), Gaps = 4/142 (2%)
Query: 60 YLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPL 119
Y+LKK+ + + K E D +F + +I+ F V+I+D T+ N + L
Sbjct: 355 YMLKKVNDGTVTYLKLDENDK--FQYVFVALGASIEGFRVMRKVLIVDATHLKNGYGGVL 412
Query: 120 LEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTDRELALLSA 179
+ Y IAF + E + W E +K+++ D + L V +TDR +L+ A
Sbjct: 413 VFASAQDPNRHHYIIAFAVLDGENDASWEWFFEKLKTVVPDTSEL--VFMTDRNASLIKA 470
Query: 180 VRNIFPGATHLLCLFHINKNVE 201
+RN++ A H C++H+++NV+
Sbjct: 471 IRNVYTAAHHGYCIWHLSQNVK 492
>At2g27110 Mutator-like transposase
Length = 851
Score = 65.9 bits (159), Expect = 2e-11
Identities = 38/185 (20%), Positives = 82/185 (43%), Gaps = 4/185 (2%)
Query: 58 MQYLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQI 117
++Y + AE + ++ + +FW+ + + F V +D Y+ N+F++
Sbjct: 201 LEYFKRMQAENPGFFYAVQLDEDNQMSNVFWADSRSRVAYTHFGDTVTLDTRYRCNQFRV 260
Query: 118 PLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTDRELALL 177
P G+ G + E ++W + + + D + P +VTD++ A+
Sbjct: 261 PFAPFTGVNHHGQAILFGCALILDESDTSFIWLFKTFLTAMRD--QPPVSLVTDQDRAIQ 318
Query: 178 SAVRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNEVVYAETTTQFEEEW 237
A +FPGA H + + + + E + KL + + +++ +N + + ET +FE W
Sbjct: 319 IAAGQVFPGARHCINKWDVLR--EGQEKLAHVCLAYPSFQVELYNCINFTETIEEFESSW 376
Query: 238 RDMCD 242
+ D
Sbjct: 377 SSVID 381
>At1g44840 hypothetical protein
Length = 926
Score = 64.3 bits (155), Expect = 6e-11
Identities = 48/185 (25%), Positives = 83/185 (43%), Gaps = 14/185 (7%)
Query: 55 LTEMQYLLKKLAEANYVHFKRH----EEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTY 110
L E +LLK H + +E + +F + +I F ++++D T+
Sbjct: 522 LAEYLHLLKLTNPGTITHIETERDVEDESKERFLYMFLAFGASIAGFRHLRRILVVDGTH 581
Query: 111 KTNKFQIPLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVT 170
K++ LL G + Y + F + E + W ++ ++AD L I++
Sbjct: 582 LKGKYKGVLLTSSGQDANFQVYPLGFAVVDSENDESWTWFFTKLERIIADSKTL--TILS 639
Query: 171 DRELALLSAVRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNEVVYAETT 230
DR ++L AV+ +FP A H C+ H+ +N++ K K KAL Q YA T
Sbjct: 640 DRHSSILVAVKRVFPQANHGACIIHLCRNIQTKYK-------NKAL-TQLVKNAGYAFTG 691
Query: 231 TQFEE 235
T+F+E
Sbjct: 692 TKFKE 696
>At1g36090 hypothetical protein
Length = 645
Score = 63.2 bits (152), Expect = 1e-10
Identities = 42/178 (23%), Positives = 85/178 (47%), Gaps = 10/178 (5%)
Query: 60 YLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPL 119
Y+L+K+ + + E LF + I+ F V+++D T+ + L
Sbjct: 329 YMLEKVNPGTVTYVELEGEKK--FKYLFIALGACIEGFRAMRKVIVVDATHLKTVYGGML 386
Query: 120 LEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTDRELALLSA 179
+ Y +AF + E+ ++W LE +K++ +D RL V ++DR ++ A
Sbjct: 387 VIATAHDPNHHHYPLAFGIIDSEKDVSWIWFLEKLKTVYSDVPRL--VFISDRHQSIKKA 444
Query: 180 VRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNEVVYAETTTQFEEEW 237
V+ ++P A H C++H+ +N+ + K+ D K+ + +A T ++FE+E+
Sbjct: 445 VKTVYPNALHAACIWHLCQNMRDRVKIDKDGA------AVKFRDCAHAYTESEFEKEF 496
>At4g09380 putative protein
Length = 960
Score = 61.6 bits (148), Expect = 4e-10
Identities = 41/179 (22%), Positives = 87/179 (47%), Gaps = 12/179 (6%)
Query: 60 YLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPL 119
Y+L+K+ + + E LF + I+ F V+++D T+ + L
Sbjct: 369 YMLEKVNPGTVTYVELEGEKK--FKYLFIALGACIEGFRAMRKVIVVDATHLKTVYGGML 426
Query: 120 LEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVI-VTDRELALLS 178
+ Y +AF + E+ ++W LE +K++ +D +PG++ ++DR ++
Sbjct: 427 VIATAQDPNHHHYPLAFGIIDSEKDVSWIWFLENLKTVYSD---VPGLVFISDRHQSIKK 483
Query: 179 AVRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNEVVYAETTTQFEEEW 237
AV+ ++P A H C++H+ +N+ + K+ D K+ + +A T ++FE+E+
Sbjct: 484 AVKTVYPNALHAACIWHLCQNMRDRVKIDKDGA------AVKFRDCAHAYTESEFEKEF 536
>At4g08720 putative protein
Length = 848
Score = 61.6 bits (148), Expect = 4e-10
Identities = 48/180 (26%), Positives = 87/180 (47%), Gaps = 11/180 (6%)
Query: 60 YLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPL 119
Y+LKK+ + + K E SG LF + +I+ F V+I+D T+ N + L
Sbjct: 349 YMLKKVNDGTVTYMKLDE--SGKFQYLFIALGASIEGFQAMRKVIIVDATHLKNGYGGVL 406
Query: 120 LEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTDRELALLSA 179
+ Y IA + E + W E + S++ D L V ++DR +L+
Sbjct: 407 VFASARDPNRHHYIIAVGVLDGENDASWGWFFEKLLSVVPDTPEL--VFMSDRNSSLIKG 464
Query: 180 VRNIFPGATHLLCLFHINKNVEA-KCKLWVDTTDFKALEMQKWNEVVYAETTTQFEEEWR 238
+RN + A H C++H+++NV+A + D ++ +E+ + +Y T ++FE E+R
Sbjct: 465 IRNAYTAAHHGYCVWHLSQNVKAHSTNINRDVLAWRFMELSR----IY--TWSEFEREYR 518
>At1g52520 F6D8.26
Length = 703
Score = 61.6 bits (148), Expect = 4e-10
Identities = 36/125 (28%), Positives = 61/125 (48%), Gaps = 3/125 (2%)
Query: 79 DSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPLLEMVGLTSTGLTYSIAFCY 138
D G + +FW+ + + F V+ +D +Y + KF+IPL+ G+ G T ++ +
Sbjct: 273 DEGQLRNVFWADAFSKVSCSYFGDVIFIDSSYISGKFEIPLVTFTGVNHHGKTTLLSCGF 332
Query: 139 MTRERTPDYVWALECMKSLLADPARLPGVIVTDRELALLSAVRNIFPGATHLLCLFHINK 198
+ E Y W L+ S++ R P IVTDR L +A+ +FP + L HI +
Sbjct: 333 LAGETMESYHWLLKVWLSVM---KRSPQTIVTDRCKPLEAAISQVFPRSHQRFSLTHIMR 389
Query: 199 NVEAK 203
+ K
Sbjct: 390 KIPEK 394
>At3g47310 putative protein
Length = 735
Score = 61.2 bits (147), Expect = 5e-10
Identities = 41/179 (22%), Positives = 86/179 (47%), Gaps = 12/179 (6%)
Query: 60 YLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPL 119
Y+L+K+ + + E LF + I+ F T V+++D T+ + L
Sbjct: 318 YMLEKVNPGTVTYVELEGEKK--FKYLFIALGACIEGFRTMRKVIVVDATHLKTVYGGML 375
Query: 120 LEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVI-VTDRELALLS 178
+ Y +AF + E ++W LE +K++ +D +PG++ ++DR ++
Sbjct: 376 VIATAQDPNHHHYPLAFGIIDSENDVSWIWFLEKLKTVYSD---VPGLVFISDRHQSIKK 432
Query: 179 AVRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNEVVYAETTTQFEEEW 237
V+ ++P A H C++H+ +N+ + K+ D K+ + +A T ++FE+E+
Sbjct: 433 VVKTVYPNALHAACIWHLCQNMRDRVKIDKDGA------AVKFRDCAHAYTESEFEKEF 485
>At1g76320 putative phytochrome A signaling protein
Length = 670
Score = 61.2 bits (147), Expect = 5e-10
Identities = 38/186 (20%), Positives = 85/186 (45%), Gaps = 6/186 (3%)
Query: 58 MQYLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQI 117
+++L++ E F + ++ +FW I+ + +F VV + +Y +K+++
Sbjct: 167 LEFLMRMQEENPKFFFAVDFSEDHLLRNVFWVDAKGIEDYKSFSDVVSFETSYFVSKYKV 226
Query: 118 PLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTDRELALL 177
PL+ VG+ + + + YVW ++ L+A + P V++TD+ A+
Sbjct: 227 PLVLFVGVNHHVQPVLLGCGLLADDTVYTYVWLMQSW--LVAMGGQKPKVMLTDQNNAIK 284
Query: 178 SAVRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNEVVYAE-TTTQFEEE 236
+A+ + P H CL+H+ + W + ++ M+K + +Y + +F+
Sbjct: 285 AAIAAVLPETRHCYCLWHVLDQLPRNLDYW---SMWQDTFMKKLFKCIYRSWSEEEFDRR 341
Query: 237 WRDMCD 242
W + D
Sbjct: 342 WLKLID 347
>At1g10240 unknown protein
Length = 680
Score = 61.2 bits (147), Expect = 5e-10
Identities = 40/159 (25%), Positives = 71/159 (44%), Gaps = 16/159 (10%)
Query: 88 WSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPLLEMVGLTSTGLTYSIAFCYMTRERTPDY 147
WS+ ++I+ + F V+ D T++ + ++PL VG+ + G+ + E +
Sbjct: 262 WSYASSIQSYELFGDAVVFDTTHRLSAVEMPLGIWVGVNNYGVPCFFGCVLLRDENLRSW 321
Query: 148 VWALECMKSLLADPARLPGVIVTDRELALLSAVRNIFPGATHLLCLFHINKNVEAKCKLW 207
WAL+ + A P I+TD + L A+ P H LC++ V K W
Sbjct: 322 SWALQAFTGFMNGKA--PQTILTDHNMCLKEAIAGEMPATKHALCIW----MVVGKFPSW 375
Query: 208 VDT------TDFKALEMQKWNEVVYAETTTQFEEEWRDM 240
+ D+KA ++ + + E+ +FE WRDM
Sbjct: 376 FNAGLGERYNDWKA----EFYRLYHLESVEEFELGWRDM 410
>At4g28970 putative protein
Length = 914
Score = 59.3 bits (142), Expect = 2e-09
Identities = 40/179 (22%), Positives = 86/179 (47%), Gaps = 12/179 (6%)
Query: 60 YLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPL 119
Y+L+K+ + + E LF + I+ F V+++D T+ + L
Sbjct: 369 YMLEKVNPGTVTYVELEGEKK--FKYLFIALGACIEGFRAMRKVIVVDATHLKTVYGGML 426
Query: 120 LEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVI-VTDRELALLS 178
+ Y +AF + E+ ++W LE +K++ +D +PG++ ++DR ++
Sbjct: 427 VIATAQDPNHHHYPLAFGIIDSEKDVSWIWFLEKLKTVYSD---VPGLVFISDRHQSIKK 483
Query: 179 AVRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNEVVYAETTTQFEEEW 237
AV+ ++P A H C++H+ +N+ + + D K+ + +A T ++FE+E+
Sbjct: 484 AVKTVYPNALHAACIWHLCQNMRDRVTIDKDGA------AVKFRDCAHAYTESEFEKEF 536
>At1g35060 hypothetical protein
Length = 873
Score = 58.2 bits (139), Expect = 4e-09
Identities = 38/155 (24%), Positives = 72/155 (45%), Gaps = 6/155 (3%)
Query: 55 LTEMQYLLKKLAEANYVHFKRH----EEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTY 110
L E +LLK H + +E + +F + +I+ F V+++D T+
Sbjct: 473 LAEYLHLLKLTNPGTITHIETEPDIEDERKERFLYMFLAFGASIQGFKHLRRVLVVDGTH 532
Query: 111 KTNKFQIPLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVT 170
K++ LL G + Y +AF + E + W ++ ++AD L I++
Sbjct: 533 LKGKYKGVLLTASGQDANFQVYPLAFAVVDSENDDAWTWFFTKLERIIADNNTL--TILS 590
Query: 171 DRELALLSAVRNIFPGATHLLCLFHINKNVEAKCK 205
DR ++ V+ +FP A H C+ H+ +N++A+ K
Sbjct: 591 DRHESIKVGVKKVFPQAHHGACIIHLCRNIQARFK 625
>At3g22170 far-red impaired response protein, putative
Length = 814
Score = 57.4 bits (137), Expect = 7e-09
Identities = 33/118 (27%), Positives = 56/118 (46%), Gaps = 2/118 (1%)
Query: 79 DSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPLLEMVGLTSTGLTYSIAFCY 138
D + +FW + + +F VV +D TY NK+++PL VG+ +
Sbjct: 260 DDQRVKNVFWVDAKSRHNYGSFCDVVSLDTTYVRNKYKMPLAIFVGVNQHYQYMVLGCAL 319
Query: 139 MTRERTPDYVWALECMKSLLADPARLPGVIVTDRELALLSAVRNIFPGATHLLCLFHI 196
++ E Y W +E L A + P V++T+ ++ + S V IFP H L L+H+
Sbjct: 320 ISDESAATYSWLMETW--LRAIGGQAPKVLITELDVVMNSIVPEIFPNTRHCLFLWHV 375
>At2g07250 putative transposase of FARE2.1 (CDS1)
Length = 633
Score = 56.2 bits (134), Expect = 2e-08
Identities = 33/109 (30%), Positives = 55/109 (50%), Gaps = 2/109 (1%)
Query: 93 AIKLFNTFPHVVIMDCTYKTNKFQIPLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALE 152
+I+ + V+ +D + T+KF+ LL Y IAF + E + W L+
Sbjct: 318 SIRGYKLMRKVISIDGAHLTSKFKGTLLGASAQDGNFNLYPIAFAIVDSENDASWDWFLK 377
Query: 153 CMKSLLADPARLPGVIVTDRELALLSAVRNIFPGATHLLCLFHINKNVE 201
C+ +++ D L V V+DR ++ S + +P A H LC FH+ KN+E
Sbjct: 378 CLLNIIPDENDL--VFVSDRAASIASGLSGNYPLAHHGLCTFHLQKNLE 424
>At5g27190 putative protein
Length = 779
Score = 55.8 bits (133), Expect = 2e-08
Identities = 49/190 (25%), Positives = 83/190 (42%), Gaps = 16/190 (8%)
Query: 50 LDCGP---LTEMQYLLKKLAEANYVHFKRHEEDSGVIME-LFWSHPNAIKLFNTFPHVVI 105
L C P T++ L L E N E D+ + LF + I+ F VVI
Sbjct: 336 LHCSPEKSYTDLYSYLHVLKEVNPGTISYVEVDAQQKFKYLFVALGACIEGFKVMRKVVI 395
Query: 106 MDCTYKTNKFQIPLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLP 165
+D T+ + L+ Y IA + RE + W +K+++ D +P
Sbjct: 396 VDATFLKTVYGGMLVFATAQDPNHHNYIIASAVIDRENDASWSWFFNKLKTVIPD---VP 452
Query: 166 GVI-VTDRELALLSAVRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNEV 224
G++ V+DR +++ ++ +FP A H C++H+++NV+ + K E N +
Sbjct: 453 GLVFVSDRHQSIIKSIMQVFPNARHGHCVWHLSQNVKVRVK--------TEKEEAAANFI 504
Query: 225 VYAETTTQFE 234
A TQFE
Sbjct: 505 ACAHVYTQFE 514
>At4g08610 predicted transposon protein
Length = 907
Score = 55.8 bits (133), Expect = 2e-08
Identities = 40/149 (26%), Positives = 67/149 (44%), Gaps = 10/149 (6%)
Query: 86 LFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPLLEMVGLTSTGLTYSIAFCYMTRERTP 145
LF S I+ F VVI+D T+ + L+ Y IA + RE
Sbjct: 395 LFVSLGVCIEGFKVMRKVVIVDATFLKTVYGDMLVFATAQDPNHHNYIIASAVIDRENDA 454
Query: 146 DYVWALECMKSLLADPARLPGVIVTDRELALLSAVRNIFPGATHLLCLFHINKNVEAKCK 205
+ W +K+++ D L V V+DR +++ ++ ++FP A H C++H+++NV+ + K
Sbjct: 455 SWSWFFNKLKTVIPDELGL--VFVSDRHQSIIKSIMHVFPNARHGHCVWHLSQNVKVRVK 512
Query: 206 LWVDTTDFKALEMQKWNEVVYAETTTQFE 234
E N + A TQFE
Sbjct: 513 --------TEKEEAAANFIACAHVYTQFE 533
>At3g47280 putative protein
Length = 739
Score = 55.8 bits (133), Expect = 2e-08
Identities = 33/109 (30%), Positives = 55/109 (50%), Gaps = 2/109 (1%)
Query: 93 AIKLFNTFPHVVIMDCTYKTNKFQIPLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALE 152
+I+ + V+ +D + T+KF+ LL Y IAF + E + W L+
Sbjct: 355 SIRGYKLMRKVISIDGAHLTSKFKGTLLGASAQDGNFNLYPIAFAIVDSENDASWDWFLK 414
Query: 153 CMKSLLADPARLPGVIVTDRELALLSAVRNIFPGATHLLCLFHINKNVE 201
C+ +++ D L V V+DR ++ S + +P A H LC FH+ KN+E
Sbjct: 415 CLLNIIPDENDL--VFVSDRAASIASRLSGNYPLAHHGLCTFHLQKNLE 461
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.135 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,674,830
Number of Sequences: 26719
Number of extensions: 221351
Number of successful extensions: 689
Number of sequences better than 10.0: 85
Number of HSP's better than 10.0 without gapping: 73
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 565
Number of HSP's gapped (non-prelim): 91
length of query: 242
length of database: 11,318,596
effective HSP length: 96
effective length of query: 146
effective length of database: 8,753,572
effective search space: 1278021512
effective search space used: 1278021512
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0348.15


