

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0346.9
(569 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
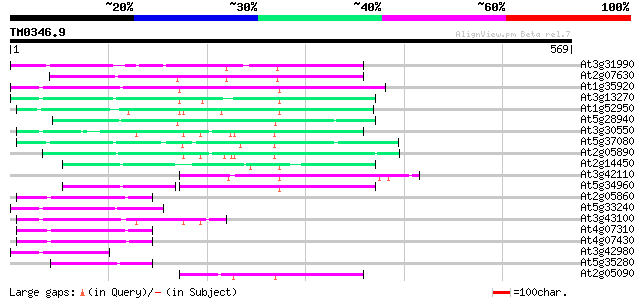
Score E
Sequences producing significant alignments: (bits) Value
At3g31990 hypothetical protein 119 6e-27
At2g07630 putative replication protein A1 115 5e-26
At1g35920 hypothetical protein 114 2e-25
At3g13270 hypothetical protein 106 4e-23
At1g52950 replication protein A1, putative 96 4e-20
At5g28940 93 5e-19
At3g30550 putative replication protein 83 5e-16
At5g37080 putative protein 81 1e-15
At2g05890 putative replication protein A1 81 2e-15
At2g14450 putative replication protein A1 75 1e-13
At3g42110 putative protein 69 5e-12
At5g34960 putative protein 67 2e-11
At2g05860 putative replication protein A1 64 3e-10
At5g33240 putative protein 63 4e-10
At3g43100 putative protein 62 7e-10
At4g07310 61 1e-09
At4g07430 61 2e-09
At3g42980 putative protein 60 4e-09
At5g35280 putative protein 55 1e-07
At2g05090 putative replication protein A1 53 4e-07
>At3g31990 hypothetical protein
Length = 450
Score = 119 bits (297), Expect = 6e-27
Identities = 97/369 (26%), Positives = 162/369 (43%), Gaps = 33/369 (8%)
Query: 1 MSGKIYVISTINPSKETWNIVAKVQRLWLSPSLYGSKLPFSLDMVLVDSKGDKIHATVRK 60
M+ I + I P K W + +V W + ++ + + DMVL D +G KIHA+V++
Sbjct: 1 MAQPIAYLDDIKPYKTYWRVQVRVLHTWKTFTMQFGE---TFDMVLSDVRGKKIHASVKR 57
Query: 61 TLIYRFQSLLTEGRVYQISFFGVGESGRDFRTTGHAFKINFQIHTFVCVVTNTPINHSPY 120
+ RF+ + G I FG+ + ++ T H +K+ F T H
Sbjct: 58 EHLNRFERPIVRGEWRAIENFGLTYATGQYKATDHRYKMGFMAQT-----------HVVG 106
Query: 121 SFMAISEIMFNDPDASFLIDVIGILTGASGEHEFEKDGVSQKKITLELDQDGIHNVYGAT 180
+ + E+ D SF V A+GE V K+ D I N + +
Sbjct: 107 QIVNVGEM--ETIDGSFAEQVFTACETANGEMIICL--VRFAKLKTYKDVRSISNAFNTS 162
Query: 181 KILFNPAIDEATKIRERNDPASQVLSQLQDSGMIYVEE----DLLRQTDGKTIEQIKDLT 236
+IL N I E + ++ L+ ++ V E D Q KTI+++ ++
Sbjct: 163 QILINTDIPEILEFKDALPKDGLALTLIESKPKSEVAELPTGDFYLQFAKKTIKEVSEMF 222
Query: 237 EKSYCVVLGTMKYIPDGMDWYYPACK-CSKKVYPA-----DGMYFCEACNRHVASPLYKF 290
+ VL T+ I WYY ACK C+KKV ++CE C V + L ++
Sbjct: 223 D-----VLCTIYDIDRDWSWYYIACKKCNKKVTKVVTTSLKAQFWCETCRAPVNNVLARY 277
Query: 291 RIQLRVMNSKNTATFVLFDQEANSVVKKPCAILVDASNKAGPDSPNVPSDILDLIDRTFL 350
++ ++VM+S +LFD A+ +V P L+D S D N+P I +LI +T+
Sbjct: 278 KLHVKVMDSSGEMKLMLFDAMASEIVGCPANNLLDGSFDEIEDPDNIPGSIKNLIGKTYQ 337
Query: 351 FKIEVSNSS 359
+ + V N +
Sbjct: 338 YLVCVENEN 346
>At2g07630 putative replication protein A1
Length = 458
Score = 115 bits (289), Expect = 5e-26
Identities = 91/375 (24%), Positives = 159/375 (42%), Gaps = 58/375 (15%)
Query: 41 SLDMVLVDSKGDKIHATVRKTLIYRFQSLLTEGRVYQISFFGVGESGRDFRTTGHAFKIN 100
+ DMVL D +G KIHA+V++ + RF+ L+ G I FG+ + F+ T H +K+
Sbjct: 6 TFDMVLSDVRGKKIHASVKREHLNRFERLIVPGEWRAIENFGLTYATGQFKATDHRYKMG 65
Query: 101 FQIHTFVCVVTNTPINHSPY-SFMAISEIMFNDPDASFLIDVIGILTGASGEHEFEKDGV 159
F T V V P++ S + S A ++++ + ++LIDV+G + +
Sbjct: 66 FMAQTRV--VRIDPLSDSYFLSLTAFNDVLNGGLNQNYLIDVVGQIVNVGEMETIDVHNQ 123
Query: 160 SQKKITLEL---------------------------------------------DQDGIH 174
KKI EL D I
Sbjct: 124 PTKKIDFELRDQKDERLPCTLWGSFAEQVFTACEAANGEMIICLVRFAKLKTYKDVRSIS 183
Query: 175 NVYGATKILFNPAIDEATKIRERNDPASQVLSQLQDSGMIYVEE----DLLRQTDGKTIE 230
N + ++IL NP + E + ++ L+ ++ ++E D Q KTI+
Sbjct: 184 NAFNTSQILINPDLPEILEFKDALPKDCLALTLIESKPKSKIDEFPTGDFYLQFAKKTIK 243
Query: 231 QIKDLTEKSYCVVLGTMKYIPDGMDWYYPACK-CSKKVYPA-----DGMYFCEACNRHVA 284
++ ++ + VL T+ I WYY CK C+KKV +CE C +
Sbjct: 244 EVSEMFDVGRVKVLCTIYDIDRDWSWYYIVCKKCNKKVTKVVMTSLKAQLWCETCRAPIT 303
Query: 285 SPLYKFRIQLRVMNSKNTATFVLFDQEANSVVKKPCAILVDASNKAGPDSPNVPSDILDL 344
+ L ++++ ++VM+S +LFD ++ +V P L+D S D +P I +L
Sbjct: 304 NVLARYKLHVKVMDSSGEMKLMLFDAMSSEIVGCPANNLLDGSFDEIEDPDILPDSIKNL 363
Query: 345 IDRTFLFKIEVSNSS 359
I +T+ + + V N +
Sbjct: 364 IGKTYQYLVCVENEN 378
>At1g35920 hypothetical protein
Length = 567
Score = 114 bits (284), Expect = 2e-25
Identities = 101/431 (23%), Positives = 177/431 (40%), Gaps = 55/431 (12%)
Query: 1 MSGKIYVISTINPSKETWNIVAKVQRLWLSPSLYGSKLPFSLDMVLVDSKGDKIHATVRK 60
M+ + + P K W + K+ W Y S ++++V+ D G K+HATV+K
Sbjct: 1 MAATFAYLKDVRPYKNAWRVQVKILHSWKQ---YTSNTRETIELVISDEHGKKMHATVKK 57
Query: 61 TLIYRFQSLLTEGRVYQISFFGVGESGRDFRTTGHAFKINFQIHTFVCVVTNTPINHSPY 120
L+ +F L G I FG+ + FR T H +K+ FQ+ T V + ++ S +
Sbjct: 58 ELVSKFVHKLIVGEWVFIEIFGLTYASGQFRPTNHLYKMAFQVRTEVMGCAS--VSDSNF 115
Query: 121 SFMA-ISEIMFNDPDASFLIDVIGILTGASGEHEFEKDGVSQKKITLEL-DQ-------- 170
+A S+I + + L+D IG + E + + KI E+ DQ
Sbjct: 116 LTLASFSKIQSGELNPHMLVDAIGQIITVGELEELKANNKRTTKIDFEIRDQMDERMQVT 175
Query: 171 ------------------------DGIHNVYGATKILFNPAIDEATKIRER--NDPASQV 204
+ N + A+++ NP E + ND A V
Sbjct: 176 LWGTYAQEVYRACQESEGKNTASVKSLSNSFDASQVHVNPDFPEVHDFSQTLLNDGAICV 235
Query: 205 L-SQLQDSGMIYVEEDLLRQTDGKTIEQIKDLTEKSYCVVLGTMKYIPDGMDWYYPACK- 262
+++ M+ V+ + TIE + TE VL T+ + WYY +CK
Sbjct: 236 FRARVPRFEMVAVKRTDYSEYTRNTIENLLSSTEVGKVRVLCTIYAVDTDWAWYYISCKT 295
Query: 263 CSKKVYPADG------------MYFCEACNRHVASPLYKFRIQLRVMNSKNTATFVLFDQ 310
C+KKV ++C+ C V + + ++ I +VM+S A +LFD
Sbjct: 296 CNKKVNHIHAGVNGVNNKGKKPRFWCDTCTSVVTNVVSRYMIYAKVMDSTGEAKLLLFDS 355
Query: 311 EANSVVKKPCAILVDASNKAGPDSPNVPSDILDLIDRTFLFKIEVSNSSNSIYEPSYRVK 370
+ ++ + A + + S D ++P + +LI +TFLF + V + + Y+V
Sbjct: 356 ICSEIIGESAASVFNGSVDEIDDPEDLPDSVKNLIGKTFLFLVCVEKDNILDGKEIYKVS 415
Query: 371 KFCYDPEIINQ 381
K +I +
Sbjct: 416 KVLLKDGLIEE 426
>At3g13270 hypothetical protein
Length = 570
Score = 106 bits (264), Expect = 4e-23
Identities = 93/432 (21%), Positives = 174/432 (39%), Gaps = 75/432 (17%)
Query: 1 MSGKIYVISTINPSKETWNIVAKVQRLWLSPSLYGSKLPFSLDMVLVDSKGDKIHATVRK 60
M+ ++ + P K +W + K W Y + +L++V D G KIH +VRK
Sbjct: 1 MAATFGYLTDVRPYKTSWRVQVKTLHAWRQ---YTANTGETLEVVFSDETGKKIHCSVRK 57
Query: 61 TLIYRFQSLLTEGRVYQISFFGVGESGRDFRTTGHAFKINFQIHTFVCVVTNTPINHSPY 120
L+ ++ ++LT G I F + +G FR T H +K++F T V+ + ++ S Y
Sbjct: 58 DLVSKYANMLTVGEWVFIETFSLNYAGGSFRPTSHLYKMSFVNGT--SVIPSPSVSDSNY 115
Query: 121 SFMAISE-IMFNDPDASFLIDVIGILTGASGEHEFEKDGVSQKKITLELDQ--------- 170
+A E I + + + L+DVIG + E E + KI ++
Sbjct: 116 LTLATFEKIQSGELNPTMLVDVIGQIVMIGELEELEANNKPTTKIDFDIRDASDERIGVT 175
Query: 171 -------------------------DGIHNVYGATKILFNPAIDEATKI----------- 194
+ N + A+++ NP E
Sbjct: 176 LWGTFAQQVYRGCNEAGARPVVCVLRNLSNSFDASQVFVNPQFPEVDAFLRTLPQDGILL 235
Query: 195 --RERNDPASQVLSQLQDSGMIYVEEDLLRQTDGKTIEQIKDLTEKSYCVVLGTMKYIPD 252
RER V+S+ DS + Y TI+Q+ + + ++ T+ I
Sbjct: 236 TYRERIPKLEIVVSKKSDSCLEYPR---------NTIQQLLNAVDVGKARLMCTIYAIDT 286
Query: 253 GMDWYYPACK-CSKKVYPADG------------MYFCEACNRHVASPLYKFRIQLRVMNS 299
WYY +C+ C+KKV ++C++C V + + ++ I +VM++
Sbjct: 287 DWAWYYISCRACNKKVTHIHSGVHGVNNKGKKPRFWCDSCKTVVTNVIARYMIYAKVMDN 346
Query: 300 KNTATFVLFDQEANSVVKKPCAILVDASNKAGPDSPNVPSDILDLIDRTFLFKIEVSNSS 359
+ +LFDQ ++ + +++ S D ++P + +L +TFLF + + +
Sbjct: 347 TGESKLLLFDQICTEIIGETAPSVLNGSVDEIDDPDDLPDALKNLYGKTFLFLVSIEKEN 406
Query: 360 NSIYEPSYRVKK 371
+ Y+V K
Sbjct: 407 IWDGKEIYKVTK 418
>At1g52950 replication protein A1, putative
Length = 566
Score = 96.3 bits (238), Expect = 4e-20
Identities = 91/427 (21%), Positives = 171/427 (39%), Gaps = 76/427 (17%)
Query: 8 ISTINPSKETWNIVAKVQRLWLSPSLYGSKLPFSLDMVLVDSKGDKIHATVRKTLIYRFQ 67
I + P K +W I K+ W + Y S +M+L D G+KI A ++K + + Q
Sbjct: 10 IKNLKPFKTSWCIQVKILHAW---NHYTKGSGMSYEMMLADEDGNKIQAGIKKEHLLKLQ 66
Query: 68 SLLTEGRVYQISFFGVGESGRDFRTTGHAFKINFQIHTFVCVVTNTPINHSP-------Y 120
+ G I F V ++ +R+T H ++IN Q ++T ++SP
Sbjct: 67 RYVKIGHWTIIEEFSVTKASGLYRSTTHPYRINIQ--------SSTRFSNSPTISDEIWL 118
Query: 121 SFMAISEIMFNDPDASFLIDVIGILTGASGEHEFEKDGVSQKKITLELDQ---------- 170
+ ++++ D + L++VIG L + G KKI +L
Sbjct: 119 DLVNFNDVLSGTLDQNKLVNVIGQLVNVGEIQLLDVQGKPTKKIDFQLRDTDDNRLPCSL 178
Query: 171 -----DGIH------------------------------NVYGATKILFNPAIDEATKIR 195
D IH N + A+++ NP + E + +
Sbjct: 179 WGKFADQIHKVSKESVGGIVICLIRWAKLGHYKEIRSISNAFDASEVFINPILTEVEEFK 238
Query: 196 ERNDPASQVLSQLQDSGM---IYVEEDLLRQTDGKTIEQIKDLTEKSYCVVLGTMKYIPD 252
L+ ++ + V E+ +Q + KTI ++K E ++ ++ I
Sbjct: 239 NLLPSDGLALTIMEPKPRFQPLRVREERSKQFERKTIAELKASFEVEKVKIICSILSIDL 298
Query: 253 GMDWYYPA-CKCSKKVYPADG---------MYFCEACNRHVASPLYKFRIQLRVMNSKNT 302
WYY A KC+KKV+ + +Y C+ C V S ++ + L +M++
Sbjct: 299 DYSWYYYAHIKCNKKVFKSKKTLSSGAKKIIYRCDKCATDVTSIEARYWLHLDIMDNTGE 358
Query: 303 ATFVLFDQEANSVVKKPCAILVDASNKAGPDSPNVPSDILDLIDRTFLFKIEVSNSSNSI 362
+ +LFD ++ P L+D +N+ + +P + ++I +T+ F + V + S
Sbjct: 359 SKLMLFDSFVEPIIGCPATELLDVTNEQIDEPQPLPDVVKNIIGKTYQFLVCVEQDNISR 418
Query: 363 YEPSYRV 369
Y+V
Sbjct: 419 GNDEYKV 425
>At5g28940
Length = 485
Score = 92.8 bits (229), Expect = 5e-19
Identities = 87/389 (22%), Positives = 149/389 (37%), Gaps = 63/389 (16%)
Query: 44 MVLVDSKGDKIHATVRKTLIYRFQSLLTEGRVYQISFFGVGESGRDFRTTGHAFKINFQI 103
MVL D +G KIHA+V+K L+ +++ + + I F +G+ FR T H FK+ F
Sbjct: 1 MVLADQRGCKIHASVKKALVKKYERHIALDQWRIIENFFLGQVRGQFRVTNHQFKMAFVN 60
Query: 104 HTFVCVVTNTPINHSPYSFMAISEIMFNDPDASFLIDVIGILTGASGEHEFEKDGVSQKK 163
TFV T N + S I+ + + +L+DV+G + + + S KK
Sbjct: 61 STFVSPCP-TVTNDAYLSLPTFESIIVGELNLFYLVDVLGQVVNVGKVEDIVVNNESTKK 119
Query: 164 ITLEL---------------------------------------------DQDGIHNVYG 178
+ EL ++ I N +
Sbjct: 120 LEFELRNEMNDRLPCTLWGKFADQVIQECENSTGEIVVCLIRFAKINTFRGENQITNAFD 179
Query: 179 ATKILFNPAIDEATKIRER--NDPASQVLSQLQDSGMIYVEED-LLRQTDGKTIEQIKDL 235
A+ + NP E + D + +S+ + D Q K+I +
Sbjct: 180 ASVVTINPEFPEVEHFKNSLPKDGLALTISETNPRRELSFGSDGYFDQYPIKSISCLLHF 239
Query: 236 TEKSYCVVLGTMKYIPDGMDWYYPACK-CSKKVY------------PADGMYFCEACNRH 282
E C V+ + I W+Y ACK CSKKV P Y+C CN +
Sbjct: 240 LEVQKCKVMCKIHTIEGDFGWFYVACKKCSKKVEMNNKDKTAIADKPTKTTYWCPKCNLN 299
Query: 283 VASPLYKFRIQLRVMNSKNTATFVLFDQEANSVVKKPCAILVDASNKAGPDSPNVPSDIL 342
+ + +F++ + V++ ++FD A+ ++ L+D D +P I
Sbjct: 300 ITQVVPRFKLHVGVIDQTGDIKCIMFDSAASKMIGHSAFDLLDGVYD-DDDPERMPPSIK 358
Query: 343 DLIDRTFLFKIEVSNSSNSIYEPSYRVKK 371
+L+ +TF + + N + S +Y V K
Sbjct: 359 NLVGKTFQLLVCIENDNLSGRNDTYMVGK 387
>At3g30550 putative replication protein
Length = 509
Score = 82.8 bits (203), Expect = 5e-16
Identities = 100/419 (23%), Positives = 164/419 (38%), Gaps = 85/419 (20%)
Query: 8 ISTINPSKETWNIVAKVQRLWLSPSLYGSKLPFSLDMVLVDSKGDKIHATVRKTLIYRFQ 67
+ ++ P K W I K+ +W Y K S++++LVD GDK++A+VR+ I +F+
Sbjct: 12 LKSLKPYKNAWRIQVKLLHVWRQ---YSVKAGESIEIILVDEAGDKMYASVRREQIKKFE 68
Query: 68 SLLTEGRVYQISFFGVGESGRDFRTTGHAFKINFQIHTFVCVVTNTPI-NHSPYSFMAIS 126
LTEG V++I TT + Q H F T +P N S F++++
Sbjct: 69 RCLTEG-VWKII------------TTITLNPTSGQYHWFCFQTTVSPCDNVSDALFLSLA 115
Query: 127 E---IMFNDPDASFLIDVIGILTGASGEHEFEKDGVSQKKITLEL-DQDGIH---NVYGA 179
+ I+ +++ L ++IG + S KKI L DQ ++G
Sbjct: 116 KFDVILSRSANSNILHNIIGQVVNRSEIQNLNASNKPTKKIDFHLRDQHDTRLACTLWGK 175
Query: 180 TKILFNPAIDEAT-----------KIRERNDPASQVLSQLQDSGMIYVEEDLLR------ 222
+ + A E+T KI ND S +S D ++V+ L
Sbjct: 176 YAEIVDQACQESTDGIVVCLIRFAKINLYNDTRS--VSNSFDVSQVFVDHTLAELGLFKQ 233
Query: 223 --QTDG---------------------------KTIEQIKDLTEKSYCVVLGTMKYIPDG 253
TDG +TI+++ ++ C + T+ I
Sbjct: 234 SIPTDGLTLGSSGSFHKRLYAPRTGDDDGDYPRQTIKEVLTFSDVGKCKTVCTVSAIDTD 293
Query: 254 MDWYYPACKC-SKKV------------YPADGMYFCEACNRHVASPLYKFRIQLRVMNSK 300
WYY C+ +KKV P ++CE CN S + KF + L +M+
Sbjct: 294 WPWYYFCCRAHNKKVVKEEAIKFEDVKLPQKPRFWCEICNGFAKSVVPKFWLHLHIMDQT 353
Query: 301 NTATFVLFDQEANSVVKKPCAILVDASNKAGPDSPNVPSDILDLIDRTFLFKIEVSNSS 359
A +LFD A ++ L+ S D +P I L +TF F + V +
Sbjct: 354 GEAKCMLFDSHAKEILGITAHDLLAGSFDEIEDPTVLPDVINCLKGKTFQFLLCVQREN 412
>At5g37080 putative protein
Length = 566
Score = 81.3 bits (199), Expect = 1e-15
Identities = 86/432 (19%), Positives = 172/432 (38%), Gaps = 60/432 (13%)
Query: 8 ISTINPSKETWNIVAKVQRLWLSPSLYGSKLPFSLDMVLVDSKGDKIHATVRKTLIYRFQ 67
+S + P K W + K+ LW + +LD+++ D G K+H T+RK I +F
Sbjct: 38 LSDVKPFKTNWCVRVKILHLWRQTKDASRE---TLDVIICDENGSKMHITIRKHYIGKFH 94
Query: 68 SLLTEGRVYQISFFGVGESGRDFRTTGHAFKINFQIHTFVCVVTNTPINHSPYSFMAISE 127
+ G I F + ++ + +++ F +T V+ P++ S + +A E
Sbjct: 95 RSIKVGDWKIIDNFTLSPLTGKYKISSLTYRMAFNHNT--DVIKCDPVSDSVFLDLADFE 152
Query: 128 -IMFNDPDASFLIDVIGILTGASGEHEFEKDGVSQKKITLELDQDGI---------HNVY 177
+ D + LID++G + E ++Q K +LD + + V+
Sbjct: 153 GVKTESYDENVLIDILGQVVRVGKVDEI----MAQNKPNKKLDNERLPCTLWGVFAEKVF 208
Query: 178 GATKILFNPAIDEATKIRERNDPASQVLSQLQDSGMIYVEEDLLRQTDGKTIEQI----- 232
A K + + DE T + R + ++ + V E ++ +E
Sbjct: 209 SALKSIKH---DEKTVVLIRYAKINNFKGEISITSAFDVSEVIINPVHVPEVELFVKSLP 265
Query: 233 --------------KDLTEKSYCVVLGTMKYIPDGMDWYYPAC-KCSKKV---------- 267
+DL + +LG++ I W+Y C KC++K
Sbjct: 266 SDGLALTIQDNYVNRDLIKVGQWRILGSIFAIDTDWGWFYFGCPKCNRKTELVKESTSTG 325
Query: 268 ----YPADGMYFCEACNRHVASPLYKFRIQLRVMNSKNTATFVLFDQEANSVVKKPCAIL 323
P ++C+ C + + ++++ +RVM+ ++F+ A ++ K L
Sbjct: 326 KMVKTPMKPKFWCDKCQESITNVEARYKLHVRVMDQTAEIKLMVFENNATKLIGKSSEEL 385
Query: 324 VDASNKAGPDSPNVPSDIL-DLIDRTFLFKIEVSNSSNSIYEPSYRVKKFCYDPEIINQF 382
VD + + P + SD++ +L +TF F + V ++ + Y+V K + +I +
Sbjct: 386 VDGQYE---EDPTIISDVITNLCGKTFHFLVSVEKANIYGGKDIYKVTKVHFGTDIAKEE 442
Query: 383 ISNSPHLMYDES 394
YD S
Sbjct: 443 SGLLEDTQYDPS 454
>At2g05890 putative replication protein A1
Length = 456
Score = 80.9 bits (198), Expect = 2e-15
Identities = 100/425 (23%), Positives = 166/425 (38%), Gaps = 67/425 (15%)
Query: 34 YGSKLPFSLDMVLVDSKGDKIHATVRKTLIYRFQSLLTEGRVYQISFFGVGESGRDFRTT 93
Y K S+ M+LVD GDK++A VR+ I +F+ +TEG I+ + + +R +
Sbjct: 7 YSVKAGESIKMILVDKAGDKMYAAVRREQIKKFERCVTEGVWKIITTITLNPTSDQYRIS 66
Query: 94 GHAFKINFQIHTFVCVVTNTPINHSPYSFMAISEIMFNDPDASFLIDVIGILTGASGEHE 153
+KI F T V +T + S I+ +++ L DV+G + S +
Sbjct: 67 DLKYKIGFVFKTTVS-PCDTVSDSLFLSLAKFDVILSGSTNSNILHDVMGQVIDRSEIQD 125
Query: 154 FEKDGVSQKKITLEL-DQDGIH---NVYGATKILFNPAIDEAT-----------KIRERN 198
+ KKI L DQ ++G + + A E+T KI N
Sbjct: 126 LNANNKPTKKIDFHLRDQHDTRLACILWGKYAEIVDKACQESTDGIVVCLIRFAKINLYN 185
Query: 199 DPASQVLSQLQDSGMIYV-----EEDLLRQ---TDG------------------------ 226
D S +S D ++V E DL +Q TDG
Sbjct: 186 DTRS--VSNFFDVSQVFVDPTLAELDLFKQSIPTDGLTLGSSGSFHKRLYAPRTGDDDGD 243
Query: 227 ---KTIEQIKDLTEKSYCVVLGTMKYIPDGMDWYYPACKC-SKKV------------YPA 270
+TI+++ ++ C + T+ I YY C+ +KKV P
Sbjct: 244 YPRQTIKEVLTSSDVGKCKTVCTVSAIDTDWPCYYFCCRAHNKKVVKEEAIKLEDVKQPQ 303
Query: 271 DGMYFCEACNRHVASPLYKFRIQLRVMNSKNTATFVLFDQEANSVVKKPCAILVDASNKA 330
++CE CN S + KF + L +M+ + A +LFD A ++ L+D S
Sbjct: 304 KPRFWCEICNGFAKSVVAKFCLHLHIMDQTDEARCMLFDSHAKEILGTTAPQLLDGSFDE 363
Query: 331 GPDSPNVPSDILDLIDRTFLFKIEVSNSSNSIYEPSYRVKKFCYDPEIINQFISNSPHLM 390
D +P I L +TF F + + + S+ V + Y I+++ +
Sbjct: 364 IEDPTVLPDVINGLKGKTFQFLLCIQRENIFGGYDSFTVAR-VYTGNIVDEIVQEDSDAY 422
Query: 391 YDESN 395
D S+
Sbjct: 423 VDPSS 427
>At2g14450 putative replication protein A1
Length = 527
Score = 74.7 bits (182), Expect = 1e-13
Identities = 81/336 (24%), Positives = 133/336 (39%), Gaps = 47/336 (13%)
Query: 54 IHATVRKTLIYRFQSLLTEGRVYQISFFGVGESGRDFRTTGHAFKINFQIHTFVCVVTNT 113
+HA V+K L+ +F L G I FG+ + FR T H +K+ F + T V +
Sbjct: 1 MHAIVKKELVSKFVHKLLVGEWVFIEIFGLTYASGQFRPTNHLYKMAFHVRTKVMGCAS- 59
Query: 114 PINHSPYSFMA-ISEIMFNDPDASFLIDVIGILTGASGEHEFEKDGVSQKKITLELDQDG 172
++ S + +A S+I + + L+D IG + E E + KI E+
Sbjct: 60 -VSDSNFLTLAPFSKIQSGELNPHMLVDAIGQIITVGELEELEANNKPTTKIDFEI---- 114
Query: 173 IHNVYGATKILFNPAIDEATKIRERNDPASQVLSQLQDSGMIYVEEDLLRQTDGKTIEQI 232
+DE ++ A QV Q+S Q G T
Sbjct: 115 ------------RDQMDERMQVTLWGTYAQQVYRACQESEGKNSHVLRWLQLKGLTTVST 162
Query: 233 KDLTEKSYCV----VLGTMKYIPDGMDWYYPACK-CSKKVYPADG------------MYF 275
+ + K C+ VL T+ I WYY CK C+KKV ++
Sbjct: 163 QGIPLK-ICLARLRVLCTIYAIDTDWAWYYINCKTCNKKVNHIHAGVNGVNNKGKKPRFW 221
Query: 276 CEACNRHVASPLYKFRIQLRVMNSKNTATFVLFDQEANSVVKKPCAILVDASNKAGPDSP 335
CE C V + V++S A +LFD + ++ + +++ S D
Sbjct: 222 CETCKSFVTN----------VVSSTGEAKLLLFDSICSEIIGEYATSVLNGSVDEIEDPE 271
Query: 336 NVPSDILDLIDRTFLFKIEVSNSSNSIYEPSYRVKK 371
++P + +LI +TFLF + V + S + Y+V K
Sbjct: 272 DLPDSVKNLISKTFLFLVWVEKDNISDGKEIYKVSK 307
>At3g42110 putative protein
Length = 448
Score = 69.3 bits (168), Expect = 5e-12
Identities = 69/278 (24%), Positives = 118/278 (41%), Gaps = 41/278 (14%)
Query: 173 IHNVYGATKILFNPAIDEATKIRER--NDPASQVLSQLQDSGMIYVEEDL-----LRQTD 225
+ N + T++ NP EA ++ +D S + + Q + V +D+ LR +
Sbjct: 139 LSNSFDVTQVHVNPPSSEAVFFQQALPSDGLSLISRERQPRWEMVVNKDVDWLDYLRNS- 197
Query: 226 GKTIEQIKDLTEKSYCVVLGTMKYIPDGMDWYYPACK-CSKKVYPADG------------ 272
I +K E VL T+ I WYY +CK C+KKV
Sbjct: 198 ---ISDLKCSIEIGKARVLCTIYAIDTDWAWYYISCKTCNKKVNHIHAGVHGVNNKGKKP 254
Query: 273 MYFCEACNRHVASPLYKFRIQLRVMNSKNTATFVLFDQEANSVVKKPCAILVDASNKAGP 332
++C+ C V + + ++ + VM++ A F+LFD + + + ++D S
Sbjct: 255 RFWCDTCKTVVTNVIPRYMLYANVMDNTGEAKFLLFDSICSELTGESATSVLDGSLDEVV 314
Query: 333 DSPNVPSDILDLIDRTFLFKIEVSNSSNSIYEPSYRV-KKFC-----------YDPEIIN 380
D N+P + +LI +TFLF + V S + Y+V K C Y +I+N
Sbjct: 315 DPENLPEPVKNLIGKTFLFLVWVEKEHISDGKEVYKVWKVLCKGGLLEEQLLEYSADIVN 374
Query: 381 QF---ISNSPHLMYDESNPFYFLATPTSVLKQESINPE 415
++ P LM + S+ TP+S K+ NP+
Sbjct: 375 PASIESADQPQLMLENSHDTSDFGTPSS--KRIYANPD 410
>At5g34960 putative protein
Length = 1033
Score = 67.4 bits (163), Expect = 2e-11
Identities = 56/215 (26%), Positives = 96/215 (44%), Gaps = 16/215 (7%)
Query: 173 IHNVYGATKILFNPAIDEATKIRER--NDPASQVL-SQLQDSGMIYVEEDLLRQTDGKTI 229
+ N + A+++ NP EA + ND A V +++ M+ V+ TI
Sbjct: 191 LSNSFDASQVHVNPDFPEAHHFSQTLPNDGAICVYRTRVPRFEMVAVKRIDYSVYTRNTI 250
Query: 230 EQIKDLTEKSYCVVLGTMKYIPDGMDWYYPACK-CSKKVYPADG------------MYFC 276
E + TE VL T+ I WYY +CK C+KKV ++C
Sbjct: 251 EDLLSSTEVGKVRVLCTIYAIDTDWAWYYISCKTCNKKVNHIHAGVNGVNNKGKKPKFWC 310
Query: 277 EACNRHVASPLYKFRIQLRVMNSKNTATFVLFDQEANSVVKKPCAILVDASNKAGPDSPN 336
+ C V + + ++ I +VM+S + A VLFD ++ + +++ S D +
Sbjct: 311 DTCKSVVTNVVSRYMIYAKVMDSTSEAKLVLFDSICFEIIGESATSVLNGSVNEIEDPED 370
Query: 337 VPSDILDLIDRTFLFKIEVSNSSNSIYEPSYRVKK 371
+P + +LI +TFLF + V + S + Y+V K
Sbjct: 371 LPDSVKNLIGKTFLFLVWVEKDNISDGKEIYKVSK 405
Score = 47.0 bits (110), Expect = 3e-05
Identities = 33/116 (28%), Positives = 54/116 (46%), Gaps = 3/116 (2%)
Query: 54 IHATVRKTLIYRFQSLLTEGRVYQISFFGVGESGRDFRTTGHAFKINFQIHTFVCVVTNT 113
+HATV+K L+ +F L G I F + + FR T H +K+ FQ+ T V +
Sbjct: 1 MHATVKKELVSKFVHKLIVGEWVFIEIFRLTYASSQFRPTNHLYKMAFQVRTEVMGCAS- 59
Query: 114 PINHSPYSFMA-ISEIMFNDPDASFLIDVIGILTGASGEHEFEKDGVSQKKITLEL 168
++ S + +A S+I + + L+D IG + E E + KI E+
Sbjct: 60 -VSDSNFLTLAPFSKIQSGELNPHMLVDAIGQIITVGELEELEANNKPTTKIDFEI 114
>At2g05860 putative replication protein A1
Length = 238
Score = 63.5 bits (153), Expect = 3e-10
Identities = 43/139 (30%), Positives = 70/139 (49%), Gaps = 6/139 (4%)
Query: 8 ISTINPSKETWNIVAKVQRLWLSPSLYGSKLPFSLDMVLVDSKGDKIHATVRKTLIYRFQ 67
+S + S W I +V R +L + SK+ + ++L D G I ATV + ++
Sbjct: 7 LSLLKSSIRGWCIRGRVVRTFLVSLVPSSKV---MGLILADEHGMTIEATVGYKMSDHYK 63
Query: 68 SLLTEGRVYQISFFGVGESGRDFRTTGHAFKINFQIHTFVCVVTNTP-INHSPYSFMAIS 126
+ EG I+ FGV E+ R T H+FKI F + T V + + P I H Y + S
Sbjct: 64 DFINEGEWVTITNFGVVENSGSVRATTHSFKIGFSVDTVVRLTSPVPAIPH--YRLASFS 121
Query: 127 EIMFNDPDASFLIDVIGIL 145
I+ ++ D S L+D++G +
Sbjct: 122 SIIDDEIDKSVLVDLVGAI 140
>At5g33240 putative protein
Length = 192
Score = 63.2 bits (152), Expect = 4e-10
Identities = 43/156 (27%), Positives = 72/156 (45%), Gaps = 4/156 (2%)
Query: 1 MSGKIYVISTINPSKETWNIVAKVQRLWLSPSLYGSKLPFSLDMVLVDSKGDKIHATVRK 60
M+ + + P K +W + KV W Y + +L +VLV+S+G KIHA+V+K
Sbjct: 1 MAASFAFLRDVRPYKTSWRVQVKVLHSWCQ---YTNMTGETLKLVLVNSQGVKIHASVKK 57
Query: 61 TLIYRFQSLLTEGRVYQISFFGVGESGRDFRTTGHAFKINFQIHTFVCVVTNTPINHSPY 120
L+ ++ + L I F + + FR TGH +K++F T V + ++ N S
Sbjct: 58 DLVSKYVNNLPLNEWTFIETFALNHASGQFRPTGHLYKMSFVTGTQV-IRCDSVSNSSYL 116
Query: 121 SFMAISEIMFNDPDASFLIDVIGILTGASGEHEFEK 156
S + I D + + L+ + L EF K
Sbjct: 117 SLATFANIQTGDSNPAILVGTLTHLGTMEHNKEFWK 152
>At3g43100 putative protein
Length = 269
Score = 62.4 bits (150), Expect = 7e-10
Identities = 59/231 (25%), Positives = 102/231 (43%), Gaps = 27/231 (11%)
Query: 8 ISTINPSKETWNIVAKVQRLWLSPSLYGSKLPFSLDMVLVDSKGDKIHATVRKTLIYRFQ 67
+ ++ P K W I K+ +W Y K S++M+LVD GDK++A VR+ I +F+
Sbjct: 12 LKSLKPYKNAWRIQVKLLHVWRQ---YSVKAGESIEMILVDEAGDKMYAAVRREQIKKFE 68
Query: 68 SLLTEGRVYQISFFGVGESGRDFRTTGHAFKINFQIHTFVCVVTNTPINHSPYSFMAISE 127
LTEG I+ + + +R + +KI F T V + S F+++++
Sbjct: 69 RCLTEGVWKIITTITLNPTSGKYRISDLKYKIGFVFKTTVSPCDSV----SDALFLSLAK 124
Query: 128 ---IMFNDPDASFLIDVIGILTGASGEHEFEKDGVSQKKITLEL-DQDGIH---NVYGAT 180
I+ +++ L DV+G + S + + KKI L DQ ++G
Sbjct: 125 FDVILSGSANSNILHDVMGQVVDRSEIQDLNANNKPTKKIDFHLKDQHDTRLACTLWGKY 184
Query: 181 KILFNPAIDEAT-----------KIRERNDPASQVLSQLQDSGMIYVEEDL 220
+ + A E+T KI ND S+ +S D ++V+ L
Sbjct: 185 AEIVDQACQESTDGIVVCLIRFAKINLYND--SRSVSNSFDVSQVFVDPTL 233
>At4g07310
Length = 242
Score = 61.2 bits (147), Expect = 1e-09
Identities = 42/139 (30%), Positives = 70/139 (50%), Gaps = 6/139 (4%)
Query: 8 ISTINPSKETWNIVAKVQRLWLSPSLYGSKLPFSLDMVLVDSKGDKIHATVRKTLIYRFQ 67
+S + PS W I +V R +L + SK+ + ++L + G I ATV + ++
Sbjct: 7 LSLLKPSIRGWCIRGRVVRTFLVSLVPSSKV---MGLILAEEHGMTIEATVGYKMSDHYK 63
Query: 68 SLLTEGRVYQISFFGVGESGRDFRTTGHAFKINFQIHTFVCVVTNTP-INHSPYSFMAIS 126
+ EG I+ FGV E+ R T H+FKI F + V + + P I H Y + S
Sbjct: 64 DFINEGEWVTITNFGVVENSGSVRATTHSFKIGFFVDIVVRLTSPLPAIPH--YRLASFS 121
Query: 127 EIMFNDPDASFLIDVIGIL 145
I+ ++ D S L+D++G +
Sbjct: 122 SIIDDEIDKSVLVDLVGAI 140
>At4g07430
Length = 242
Score = 60.8 bits (146), Expect = 2e-09
Identities = 42/139 (30%), Positives = 69/139 (49%), Gaps = 6/139 (4%)
Query: 8 ISTINPSKETWNIVAKVQRLWLSPSLYGSKLPFSLDMVLVDSKGDKIHATVRKTLIYRFQ 67
+S + PS W I +V R +L + SK+ + ++L D G I TV + ++
Sbjct: 7 LSLLKPSITGWCIRGRVVRTFLVSLVPSSKV---MGLILADEHGMTIETTVGYKMSDHYK 63
Query: 68 SLLTEGRVYQISFFGVGESGRDFRTTGHAFKINFQIHTFVCVVTNTP-INHSPYSFMAIS 126
+ EG I+ FGV E+ R T H FKI F + T V + + P I H+ + S
Sbjct: 64 HFINEGEWVTITSFGVVENSCSVRATTHTFKIGFSVDTVVRLTSLVPAILHN--RLASFS 121
Query: 127 EIMFNDPDASFLIDVIGIL 145
I+ ++ D S L+D++G +
Sbjct: 122 SIIDDEIDKSVLVDLVGAI 140
>At3g42980 putative protein
Length = 303
Score = 59.7 bits (143), Expect = 4e-09
Identities = 35/101 (34%), Positives = 50/101 (48%), Gaps = 3/101 (2%)
Query: 1 MSGKIYVISTINPSKETWNIVAKVQRLWLSPSLYGSKLPFSLDMVLVDSKGDKIHATVRK 60
M+ + I P K +W I KV W Y S +L++VLVD+ G KIHA+V+K
Sbjct: 1 MAASFAFLRDIRPYKTSWRIQVKVFHSWRQ---YTSMTGETLELVLVDAHGVKIHASVKK 57
Query: 61 TLIYRFQSLLTEGRVYQISFFGVGESGRDFRTTGHAFKINF 101
L+ F + L I F + + FR TGH +K+ F
Sbjct: 58 DLVSNFVNNLLLNEWRFIETFALNHASGQFRPTGHLYKMTF 98
>At5g35280 putative protein
Length = 205
Score = 55.1 bits (131), Expect = 1e-07
Identities = 32/105 (30%), Positives = 56/105 (52%), Gaps = 3/105 (2%)
Query: 42 LDMVLVDSKGDKIHATVRKTLIYRFQSLLTEGRVYQISFFGVGESGRDFRTTGHAFKINF 101
+ ++L D G I ATV + ++ + EG I+ FGV E+ R T H+FKI F
Sbjct: 1 MGLILADEHGMTIEATVGYKMSDHYKDFINEGEWVTITNFGVVENSGSVRATTHSFKIGF 60
Query: 102 QIHTFVCVVTNTPINHSP-YSFMAISEIMFNDPDASFLIDVIGIL 145
+ T V + +P++ P Y + S I+ ++ D S L++++G +
Sbjct: 61 SVDTVVKL--TSPVHAIPHYRLASFSSIIDDEIDKSVLVNLVGAI 103
>At2g05090 putative replication protein A1
Length = 521
Score = 53.1 bits (126), Expect = 4e-07
Identities = 39/209 (18%), Positives = 89/209 (41%), Gaps = 25/209 (11%)
Query: 173 IHNVYGATKILFNPAIDEATKIRERNDPASQVLSQLQDSGMIYVEEDLLRQTD------- 225
I + + + ++ +P + ++ P+ + +QDS YV D+++
Sbjct: 165 ITSAFDVSDVIIHPVHVPEVDLFVKSLPSDGLALTIQDS---YVNRDIIKTKSVDFFDYP 221
Query: 226 GKTIEQIKDLTEKSYCVVLGTMKYIPDGMDWYYPAC-KCSKKVY--------------PA 270
KTI + + + +LG++ I W+Y C KC++K+ P
Sbjct: 222 AKTIVDLLNGRDVGQWRILGSIFAIDTDWGWFYFGCPKCNRKIELVKESTSTVKRIQAPT 281
Query: 271 DGMYFCEACNRHVASPLYKFRIQLRVMNSKNTATFVLFDQEANSVVKKPCAILVDASNKA 330
++C+ + + ++++ +RVM+ ++F+ A ++++K L+D +
Sbjct: 282 KPKFWCDKYQESITNVEARYKLHIRVMDQTGEIKLMVFENNATNLIRKSSEELLDGQYEE 341
Query: 331 GPDSPNVPSDILDLIDRTFLFKIEVSNSS 359
D P I +L +TF F + V ++
Sbjct: 342 IEDPTIKPDVITNLCGKTFHFLVSVEKAN 370
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.134 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,510,238
Number of Sequences: 26719
Number of extensions: 614230
Number of successful extensions: 1776
Number of sequences better than 10.0: 90
Number of HSP's better than 10.0 without gapping: 56
Number of HSP's successfully gapped in prelim test: 34
Number of HSP's that attempted gapping in prelim test: 1659
Number of HSP's gapped (non-prelim): 114
length of query: 569
length of database: 11,318,596
effective HSP length: 105
effective length of query: 464
effective length of database: 8,513,101
effective search space: 3950078864
effective search space used: 3950078864
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0346.9


