

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0346.7
(449 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
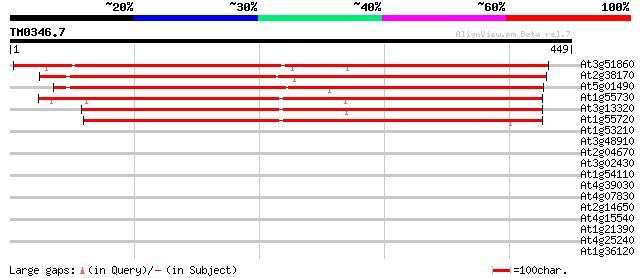
Score E
Sequences producing significant alignments: (bits) Value
At3g51860 Ca2+/H+-exchanging like protein 551 e-157
At2g38170 Ca2+ antiporter like protein 535 e-152
At5g01490 cation/proton antiporter (CAX4) 470 e-133
At1g55730 H+/Ca2+ antiporter, putative 360 e-100
At3g13320 low affinity calcium antiporter CAX2 353 1e-97
At1g55720 H+/Ca2+ antiporter, putative 335 3e-92
At1g53210 unknown protein 40 0.003
At3g48910 hypothetical protein 33 0.42
At2g04670 putative retroelement pol polyprotein 32 0.93
At3g02430 hypothetical protein 31 1.6
At1g54110 hypothetical protein 31 1.6
At4g39030 enhanced disease susceptibility 5 gene (EDS5) 30 2.1
At4g07830 putative reverse transcriptase 30 2.1
At2g14650 putative retroelement pol polyprotein 30 2.7
At4g15540 unknown protein 30 3.5
At1g21390 hypothetical protein 30 3.5
At4g25240 Pollen-specific protein SKU5 homologue (SKS1) 28 7.9
At1g36120 putative reverse transcriptase gb|AAD22339.1 28 7.9
>At3g51860 Ca2+/H+-exchanging like protein
Length = 459
Score = 551 bits (1420), Expect = e-157
Identities = 291/444 (65%), Positives = 356/444 (79%), Gaps = 21/444 (4%)
Query: 4 SVVSPSPSMNEDLESNTTNEETSHE--------NLSRSMVKRSDSVLVTNNINARFQMLR 55
S+V P ++ E+ +N T + +S E N+S S +++ + + + + L+
Sbjct: 3 SIVEPWAAIAENGNANVTAKGSSRELRHGRTAHNMSSSSLRKKSDLRLVQKVPCK--TLK 60
Query: 56 IFMTNLRVVLLGTKLVVLFPAVPLAVAADFYKFGRPWIFAFSLLGLAPLAERVSFLTEQI 115
++NL+ V+LGTKL +LF A+PLA+ A+ Y +GRP IF SL+GL PLAERVSFLTEQ+
Sbjct: 61 NILSNLQEVILGTKLTLLFLAIPLAILANSYNYGRPLIFGLSLIGLTPLAERVSFLTEQL 120
Query: 116 AYFTGPTVGGLLNATCGNATEMIIAILALRKNKVNVVKFSLLGSVLSNLLLVLGSSLLCG 175
A++TGPTVGGLLNATCGNATE+IIAILAL NKV VVK+SLLGS+LSNLLLVLG+SL G
Sbjct: 121 AFYTGPTVGGLLNATCGNATELIIAILALANNKVAVVKYSLLGSILSNLLLVLGTSLFFG 180
Query: 176 GLANLKREQRYDRKQADVNSLLLLLGLLCHLLPLLFKYSLAGGDYSIATS-----TLQLS 230
G+AN++REQR+DRKQADVN LLL+GLLCHLLPLL KY+ G ++TS +L LS
Sbjct: 181 GIANIRREQRFDRKQADVNFFLLLMGLLCHLLPLLLKYAATG---EVSTSMINKMSLTLS 237
Query: 231 RASSIVMLLAYVAYIFFQLKTHRRLFDAQEVIIDIEEDD---EEKAVIGFWSAFTWLVGM 287
R SSIVML+AY+AY+ FQL THR+LF+AQ+ D +D+ EE VIGFWS F WLVGM
Sbjct: 238 RTSSIVMLIAYIAYLIFQLWTHRQLFEAQQDDDDAYDDEVSVEETPVIGFWSGFAWLVGM 297
Query: 288 TLVISLLSEYVVGTIEAASDSWGISVSFISIILLPIVGNAAEHAGSIIFAFKNKLDISLG 347
T+VI+LLSEYVV TIE ASDSWG+SVSFISIILLPIVGNAAEHAG+IIFAFKNKLDISLG
Sbjct: 298 TIVIALLSEYVVDTIEDASDSWGLSVSFISIILLPIVGNAAEHAGAIIFAFKNKLDISLG 357
Query: 348 VAMGSATQISMFVVPLSVIVAWIMGIKMDLDFNLLETGCLAFAIIVTAFTLQDGTSHYMK 407
VA+GSATQIS+FVVPLSVIVAWI+GIKMDL+FN+LET LA AII+TAFTLQDGTSHYMK
Sbjct: 358 VALGSATQISLFVVPLSVIVAWILGIKMDLNFNILETSSLALAIIITAFTLQDGTSHYMK 417
Query: 408 GVVLTLCYLVIAACFFVNKTAQIN 431
G+VL LCY++IAACFFV++ Q N
Sbjct: 418 GLVLLLCYVIIAACFFVDQIPQPN 441
>At2g38170 Ca2+ antiporter like protein
Length = 463
Score = 535 bits (1378), Expect = e-152
Identities = 275/409 (67%), Positives = 333/409 (81%), Gaps = 8/409 (1%)
Query: 25 TSHENLSRSMVKRSDSVLVTNNINARFQMLRIFMTNLRVVLLGTKLVVLFPAVPLAVAAD 84
T+H S S+ K+SD ++ ++ L+ F++NL+ V+LGTKL +LFPA+P A+
Sbjct: 33 TAHNMSSSSLRKKSDLRVIQK---VPYKGLKDFLSNLQEVILGTKLAILFPAIPAAIICT 89
Query: 85 FYKFGRPWIFAFSLLGLAPLAERVSFLTEQIAYFTGPTVGGLLNATCGNATEMIIAILAL 144
+ +PWIF SLLGL PLAERVSFLTEQ+A++TGPT+GGLLNATCGNATE+IIAILAL
Sbjct: 90 YCGVSQPWIFGLSLLGLTPLAERVSFLTEQLAFYTGPTLGGLLNATCGNATELIIAILAL 149
Query: 145 RKNKVNVVKFSLLGSVLSNLLLVLGSSLLCGGLANLKREQRYDRKQADVNSLLLLLGLLC 204
NKV VVK+SLLGS+LSNLLLVLG+SL CGG+AN++REQR+DRKQADVN LLLLG LC
Sbjct: 150 TNNKVAVVKYSLLGSILSNLLLVLGTSLFCGGIANIRREQRFDRKQADVNFFLLLLGFLC 209
Query: 205 HLLPLLFKYSLAGGDYSIATST---LQLSRASSIVMLLAYVAYIFFQLKTHRRLFDAQEV 261
HLLPLL Y L G+ S A + L +SR SIVML++Y+AY+ FQL THR+LFDAQE
Sbjct: 210 HLLPLLVGY-LKNGEASAAVLSDMQLSISRGFSIVMLISYIAYLVFQLWTHRQLFDAQEQ 268
Query: 262 IIDIEED-DEEKAVIGFWSAFTWLVGMTLVISLLSEYVVGTIEAASDSWGISVSFISIIL 320
+ ++D ++E AVI FWS F WLVGMTLVI+LLSEYVV TIE ASD W +SVSFISIIL
Sbjct: 269 EDEYDDDVEQETAVISFWSGFAWLVGMTLVIALLSEYVVATIEEASDKWNLSVSFISIIL 328
Query: 321 LPIVGNAAEHAGSIIFAFKNKLDISLGVAMGSATQISMFVVPLSVIVAWIMGIKMDLDFN 380
LPIVGNAAEHAG++IFAFKNKLDISLGVA+GSATQI +FVVPL++IVAWI+GI MDL+F
Sbjct: 329 LPIVGNAAEHAGAVIFAFKNKLDISLGVALGSATQIGLFVVPLTIIVAWILGINMDLNFG 388
Query: 381 LLETGCLAFAIIVTAFTLQDGTSHYMKGVVLTLCYLVIAACFFVNKTAQ 429
LETGCLA +II+TAFTLQDG+SHYMKG+VL LCY +IA CFFV+K Q
Sbjct: 389 PLETGCLAVSIIITAFTLQDGSSHYMKGLVLLLCYFIIAICFFVDKLPQ 437
>At5g01490 cation/proton antiporter (CAX4)
Length = 446
Score = 470 bits (1210), Expect = e-133
Identities = 239/399 (59%), Positives = 314/399 (77%), Gaps = 11/399 (2%)
Query: 36 KRSDSVLVTNNINARFQMLRIFMTNLRVVLLGTKLVVLFPAVPLAVAADFYKFGRPWIFA 95
KRSD L++ R++ +R +TNL+ VLLGTKL +LFPAVPLAV A Y R W+FA
Sbjct: 45 KRSDLKLISR---VRWEFMRRILTNLQEVLLGTKLFILFPAVPLAVVAHRYDCPRAWVFA 101
Query: 96 FSLLGLAPLAERVSFLTEQIAYFTGPTVGGLLNATCGNATEMIIAILALRKNKVNVVKFS 155
SLLGL PLAER+SFLTEQIA+ TGPTVGGL+NATCGNATEMIIAILA+ + K+ +VK S
Sbjct: 102 LSLLGLTPLAERISFLTEQIAFHTGPTVGGLMNATCGNATEMIIAILAVGQRKMRIVKLS 161
Query: 156 LLGSVLSNLLLVLGSSLLCGGLANLKREQRYDRKQADVNSLLLLLGLLCHLLPLLFKYSL 215
LLGS+LSNLL VLG+SL GG++NL++ Q +D +Q D+NS+LL L LLC LP++ ++++
Sbjct: 162 LLGSILSNLLFVLGTSLFLGGISNLRKHQSFDPRQGDMNSMLLYLALLCQTLPMIMRFTM 221
Query: 216 AGGDYSIATSTLQLSRASSIVMLLAYVAYIFFQLKTHRR------LFDAQEVIID-IEED 268
+Y + + LSRASS VML+AY+A++ F L + L ++V D + +
Sbjct: 222 EAEEYD-GSDVVVLSRASSFVMLIAYLAFLIFHLFSSHLSPPPPPLPQREDVHDDDVSDK 280
Query: 269 DEEKAVIGFWSAFTWLVGMTLVISLLSEYVVGTIEAASDSWGISVSFISIILLPIVGNAA 328
+EE AVIG WSA WL+ MTL+++LLS+Y+V TI+ A+DSWG+SV FI IILLPIVGNAA
Sbjct: 281 EEEGAVIGMWSAIFWLIIMTLLVALLSDYLVSTIQDAADSWGLSVGFIGIILLPIVGNAA 340
Query: 329 EHAGSIIFAFKNKLDISLGVAMGSATQISMFVVPLSVIVAWIMGIKMDLDFNLLETGCLA 388
EHAG++IFAF+NKLDI+LG+A+GSATQI++FVVP++V+VAW MGI+MDL+FNLLET C A
Sbjct: 341 EHAGAVIFAFRNKLDITLGIALGSATQIALFVVPVTVLVAWTMGIEMDLNFNLLETACFA 400
Query: 389 FAIIVTAFTLQDGTSHYMKGVVLTLCYLVIAACFFVNKT 427
+I+VT+ LQDGTS+YMKG+VL LCY+VIAACFFV+ +
Sbjct: 401 LSILVTSLVLQDGTSNYMKGLVLLLCYVVIAACFFVSNS 439
>At1g55730 H+/Ca2+ antiporter, putative
Length = 441
Score = 360 bits (924), Expect = e-100
Identities = 197/416 (47%), Positives = 275/416 (65%), Gaps = 15/416 (3%)
Query: 24 ETSHENLSR----SMVKRSDSVLVTNNINARFQMLRIFMTN------LRVVLLGTKLVVL 73
+ H++L R S +++ S++ +++A F+ N ++V+L KL +L
Sbjct: 21 DVEHKSLFRRDTDSPERKAASLMEQGSLSASFRECSTKTPNNSVLQSFKIVILSNKLNLL 80
Query: 74 FPAVPLAVAADFYKFGRPWIFAFSLLGLAPLAERVSFLTEQIAYFTGPTVGGLLNATCGN 133
P PLA+ + + WIF SL+G+ PLAER+ + TEQ+A +TG TVGGLLNAT GN
Sbjct: 81 LPFGPLAILLHYLTDNKGWIFLLSLVGITPLAERLGYATEQLACYTGSTVGGLLNATFGN 140
Query: 134 ATEMIIAILALRKNKVNVVKFSLLGSVLSNLLLVLGSSLLCGGLANLKREQRYDRKQADV 193
TE+II+I AL+ + VV+ +LLGS+LSN+LLVLG + CGGL ++EQ +D+ A V
Sbjct: 141 VTELIISIFALKSGMIRVVQLTLLGSILSNMLLVLGCAFFCGGLVFSQKEQVFDKGNAVV 200
Query: 194 NSLLLLLGLLCHLLPLLFKYSLAGGDYSIATSTLQLSRASSIVMLLAYVAYIFFQLKTHR 253
NS LLL+ ++ L P + Y+ + + +S L LSR SS +ML+AY AY+FFQLK+
Sbjct: 201 NSGLLLMAVMGLLFPAVLHYTHS--EVHAGSSELALSRFSSCIMLVAYAAYLFFQLKSQP 258
Query: 254 RLFDAQEVIIDIEE---DDEEKAVIGFWSAFTWLVGMTLVISLLSEYVVGTIEAASDSWG 310
+ + E DD+E I W A WL +T +SLLS Y+V IE AS SW
Sbjct: 259 SSYTPLTEETNQNEETSDDDEDPEISKWEAIIWLSILTAWVSLLSGYLVDAIEGASVSWK 318
Query: 311 ISVSFISIILLPIVGNAAEHAGSIIFAFKNKLDISLGVAMGSATQISMFVVPLSVIVAWI 370
I +SFIS+ILLPIVGNAAEHAG+I+FA K+KLD+SLGVA+GS+ QISMF VP V++ W+
Sbjct: 319 IPISFISVILLPIVGNAAEHAGAIMFAMKDKLDLSLGVAIGSSIQISMFAVPFCVVIGWM 378
Query: 371 MGIKMDLDFNLLETGCLAFAIIVTAFTLQDGTSHYMKGVVLTLCYLVIAACFFVNK 426
MG +MDL+F L ET L +IV AF LQ+GTS+Y KG++L LCYL++AA FFV++
Sbjct: 379 MGAQMDLNFQLFETATLFITVIVVAFFLQEGTSNYFKGLMLILCYLIVAASFFVHE 434
>At3g13320 low affinity calcium antiporter CAX2
Length = 441
Score = 353 bits (905), Expect = 1e-97
Identities = 182/372 (48%), Positives = 258/372 (68%), Gaps = 5/372 (1%)
Query: 58 MTNLRVVLLGTKLVVLFPAVPLAVAADFYKFGRPWIFAFSLLGLAPLAERVSFLTEQIAY 117
+ ++++V+ KL +L P PLA+ + + W+F +L+G+ PLAER+ + TEQ+A
Sbjct: 65 LNSIKIVIFCNKLNLLLPFGPLAILVHYMIDSKGWVFLLTLVGITPLAERLGYATEQLAC 124
Query: 118 FTGPTVGGLLNATCGNATEMIIAILALRKNKVNVVKFSLLGSVLSNLLLVLGSSLLCGGL 177
+TGPTVGGLLNAT GN TE+II+I AL+ + VV+ +LLGS+LSN+LLVLG + CGGL
Sbjct: 125 YTGPTVGGLLNATFGNVTELIISIFALKNGMIRVVQLTLLGSILSNMLLVLGCAFFCGGL 184
Query: 178 ANLKREQRYDRKQADVNSLLLLLGLLCHLLPLLFKYSLAGGDYSIATSTLQLSRASSIVM 237
+++Q +D+ A VNS LLL+ ++ L P + Y+ + + +S L LSR SS +M
Sbjct: 185 VFYQKDQVFDKGIATVNSGLLLMAVMGILFPAVLHYTHS--EVHAGSSELALSRFSSCIM 242
Query: 238 LLAYVAYIFFQLKTHRRLFDAQEVIIDIEED---DEEKAVIGFWSAFTWLVGMTLVISLL 294
L+AY AY+FFQLK+ + + + E+ ++E I W A WL +T +SLL
Sbjct: 243 LIAYAAYLFFQLKSQSNSYSPLDEESNQNEETSAEDEDPEISKWEAIIWLSILTAWVSLL 302
Query: 295 SEYVVGTIEAASDSWGISVSFISIILLPIVGNAAEHAGSIIFAFKNKLDISLGVAMGSAT 354
S Y+V IE AS SW I ++FIS ILLPIVGNAAEHAG+I+FA K+KLD+SLGVA+GS+
Sbjct: 303 SGYLVDAIEGASVSWNIPIAFISTILLPIVGNAAEHAGAIMFAMKDKLDLSLGVAIGSSI 362
Query: 355 QISMFVVPLSVIVAWIMGIKMDLDFNLLETGCLAFAIIVTAFTLQDGTSHYMKGVVLTLC 414
QISMF VP V++ W+MG +MDL+F L ET L +IV AF LQ+G+S+Y KG++L LC
Sbjct: 363 QISMFAVPFCVVIGWMMGQQMDLNFQLFETAMLFITVIVVAFFLQEGSSNYFKGLMLILC 422
Query: 415 YLVIAACFFVNK 426
YL++AA FFV++
Sbjct: 423 YLIVAASFFVHE 434
>At1g55720 H+/Ca2+ antiporter, putative
Length = 401
Score = 335 bits (859), Expect = 3e-92
Identities = 177/378 (46%), Positives = 249/378 (65%), Gaps = 13/378 (3%)
Query: 60 NLRVVLLGTKLVVLFPAVPLAVAADFYKFGRPWIFAFSLLGLAPLAERVSFLTEQIAYFT 119
+ ++V+L KL +L P PLA+ + + W F SL+G+ PLAER+ + TEQ++ +T
Sbjct: 21 SFKIVILSNKLNLLLPFGPLAILVHYLTDNKGWFFLLSLVGITPLAERLGYATEQLSCYT 80
Query: 120 GPTVGGLLNATCGNATEMIIAILALRKNKVNVVKFSLLGSVLSNLLLVLGSSLLCGGLAN 179
G TVGGLLNAT GN E+II+I+AL+ + VV+ +LLGS+LSN+LLVLG + CGGL
Sbjct: 81 GATVGGLLNATFGNVIELIISIIALKNGMIRVVQLTLLGSILSNILLVLGCAFFCGGLVF 140
Query: 180 LKREQRYDRKQADVNSLLLLLGLLCHLLPLLFKYSLAGGDYSIATSTLQLSRASSIVMLL 239
++Q +D++ A V+S +LL+ ++ L P Y+ + + +S L LSR S +ML+
Sbjct: 141 PGKDQVFDKRNAVVSSGMLLMAVMGLLFPTFLHYTHS--EVHAGSSELALSRFISCIMLV 198
Query: 240 AYVAYIFFQLKTHRRLFDAQEVIIDIEEDDEEKAVIGFWSAFTWLVGMTLVISLLSEYVV 299
AY AY+FFQLK+ + + + +D+E I W A WL T +SLLS Y+V
Sbjct: 199 AYAAYLFFQLKSQPSFYTEKTNQNEETSNDDEDPEISKWEAIIWLSIFTAWVSLLSGYLV 258
Query: 300 GTIEAASDSWGISVSFISIILLPIVGNAAEHAGSIIFAFKNKLDISLGVAMGSATQISMF 359
IE S SW I +SFIS+ILLPIVGNAAEHAG+I+FA K+KLD+SLGVA+GS+ QISMF
Sbjct: 259 DAIEGTSVSWKIPISFISVILLPIVGNAAEHAGAIMFAMKDKLDLSLGVAIGSSIQISMF 318
Query: 360 VVPLSVIVAWIMGIKMDLDFNLLETGCLAFAIIVTAFTLQ-----------DGTSHYMKG 408
VP V++ W+MG +MDL+ L ET L +IV AF LQ +GTS+Y K
Sbjct: 319 AVPFCVVIGWMMGAQMDLNLQLFETATLLITVIVVAFFLQLVNLEANYETNEGTSNYFKR 378
Query: 409 VVLTLCYLVIAACFFVNK 426
++L LCYL++AA FFV++
Sbjct: 379 LMLILCYLIVAASFFVHE 396
>At1g53210 unknown protein
Length = 585
Score = 40.0 bits (92), Expect = 0.003
Identities = 28/124 (22%), Positives = 58/124 (46%), Gaps = 8/124 (6%)
Query: 267 EDDEEKAVIGFWSAFTWLVGMTLVI-----SLLSEYVVGTIEAASDSWGISVSFISIILL 321
E+DEE + T + L++ + ++ +V T+ S + GI FIS I L
Sbjct: 414 ENDEEGGEVADPKWITIKAALLLLLGAAIAAAFADPLVDTVNNFSAATGIPSFFISFIAL 473
Query: 322 PIVGNAAEHAGSIIFAFKNKL---DISLGVAMGSATQISMFVVPLSVIVAWIMGIKMDLD 378
P+ N++E +IIFA + K+ ++ G T ++ + + + + ++ G+ +
Sbjct: 474 PLATNSSEAVSAIIFASRKKIRTASLTFSELCGGVTMNNILCLSVFLAIVYVRGLTWNFS 533
Query: 379 FNLL 382
+L
Sbjct: 534 SEVL 537
>At3g48910 hypothetical protein
Length = 224
Score = 32.7 bits (73), Expect = 0.42
Identities = 16/60 (26%), Positives = 31/60 (51%)
Query: 3 VSVVSPSPSMNEDLESNTTNEETSHENLSRSMVKRSDSVLVTNNINARFQMLRIFMTNLR 62
+ ++SP P S + E+ SH++ ++++ SDS + + LRIF+ N+R
Sbjct: 160 IDLISPCPEARSRSVSRSYQEQKSHDHQLETVIELSDSETDDEEHCKKARELRIFLQNIR 219
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 31.6 bits (70), Expect = 0.93
Identities = 24/97 (24%), Positives = 40/97 (40%), Gaps = 18/97 (18%)
Query: 261 VIIDIEEDDEEKAVIGFWSAFTWLVGMTLVISLL--------SEYVVGTIEAASDSWGIS 312
V + I D + K FW AF +G + +S SE + T+E
Sbjct: 1116 VPVSIVSDRDSKFTFAFWRAFQAKMGTKVQMSTAYHPQTDGQSERTIQTLE--------- 1166
Query: 313 VSFISIILLPIVGNAAEHAGSIIFAFKNKLDISLGVA 349
+ + +L G+ A+H + FA+ N S+G+A
Sbjct: 1167 -DMLRMCVLDWGGHWADHLSLVEFAYNNSYQASIGMA 1202
>At3g02430 hypothetical protein
Length = 219
Score = 30.8 bits (68), Expect = 1.6
Identities = 19/69 (27%), Positives = 32/69 (45%), Gaps = 1/69 (1%)
Query: 380 NLLETGCL-AFAIIVTAFTLQDGTSHYMKGVVLTLCYLVIAACFFVNKTAQINQHPRTSF 438
NLL TG L AF ++ FT H + + L +L+ A+CF + T + T +
Sbjct: 53 NLLPTGTLLAFQLLTPVFTSNGVCDHATRFLTAVLLFLLAASCFVSSFTDSVKADDGTIY 112
Query: 439 GAKPSFEAI 447
+F+ +
Sbjct: 113 FGFVTFKGM 121
>At1g54110 hypothetical protein
Length = 808
Score = 30.8 bits (68), Expect = 1.6
Identities = 37/169 (21%), Positives = 72/169 (41%), Gaps = 20/169 (11%)
Query: 91 PWI---FAFSLLGLAPLAERVSFLTEQIA--YFTGPTVGGLLNATCGNATEMIIAILALR 145
PW+ F S++ +A + L Y P++ GL GN+ +++ +AL
Sbjct: 643 PWVLGGFIMSIVWFYMIANELVALLVTFGGIYGINPSILGLTVLAWGNSMGDLVSNIALS 702
Query: 146 KNKVNVVKFSLLG---SVLSNLLLVLGSSLLCGGLANLKREQRYDRKQADVNSLLLLLGL 202
N + V+ +L G + N L+ LG S+ G + K + Y + NSL LG
Sbjct: 703 MNGGDGVQIALSGCYAGPMFNTLVGLGMSMFLGAWS--KSPETY--MIPEDNSLFYTLGF 758
Query: 203 LCHLLPLLFKYSLAGGDYSIATSTLQLSRASSIVMLLAYVAYIFFQLKT 251
L + L + + ++ ++ I ++ Y+ ++ F+L +
Sbjct: 759 LI--------FGLIWSLVMLPRNEMRPNKVMGIGLITLYLIFVTFRLSS 799
>At4g39030 enhanced disease susceptibility 5 gene (EDS5)
Length = 543
Score = 30.4 bits (67), Expect = 2.1
Identities = 29/106 (27%), Positives = 52/106 (48%), Gaps = 18/106 (16%)
Query: 113 EQIAYFTGPTVGGLLNATCGNATEMIIAILALRKNKVNVVKFSLLGSVLSN-------LL 165
++I FTGP +G + CG +I ++ + + + + G+VL + L
Sbjct: 100 KEIVKFTGPAMGMWI---CGPLMSLIDTVVIGQGSSIELAALGP-GTVLCDHMSYVFMFL 155
Query: 166 LVLGSSLLCGGLANLKREQRYDRKQADVN-SLLLLLGLLCHLLPLL 210
V S+++ LA + D+K+A S+LL +GL+C L+ LL
Sbjct: 156 SVATSNMVATSLA------KQDKKEAQHQISVLLFIGLVCGLMMLL 195
>At4g07830 putative reverse transcriptase
Length = 611
Score = 30.4 bits (67), Expect = 2.1
Identities = 24/97 (24%), Positives = 40/97 (40%), Gaps = 18/97 (18%)
Query: 261 VIIDIEEDDEEKAVIGFWSAFTWLVGMTLVISLL--------SEYVVGTIEAASDSWGIS 312
V + I D + K FW AF +G + +S SE + T+E
Sbjct: 301 VPVSIVSDRDSKFTSAFWRAFQAEMGTKVQMSTAYHPQTDGQSERTIQTLE--------- 351
Query: 313 VSFISIILLPIVGNAAEHAGSIIFAFKNKLDISLGVA 349
+ + +L G+ A+H + FA+ N S+G+A
Sbjct: 352 -DMLRMCVLDWRGHWADHLSLVEFAYNNSYQASIGMA 387
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 30.0 bits (66), Expect = 2.7
Identities = 24/97 (24%), Positives = 40/97 (40%), Gaps = 18/97 (18%)
Query: 261 VIIDIEEDDEEKAVIGFWSAFTWLVGMTLVISLL--------SEYVVGTIEAASDSWGIS 312
V + I D + K FW AF +G + +S SE + T+E
Sbjct: 1036 VPVSIVSDRDSKFTSAFWRAFQAEMGTKVQMSTAYHPQTYGQSERTIQTLE--------- 1086
Query: 313 VSFISIILLPIVGNAAEHAGSIIFAFKNKLDISLGVA 349
+ + +L G+ A+H + FA+ N S+G+A
Sbjct: 1087 -DMLRMCVLDWGGHWADHLSLVEFAYNNSYPASIGMA 1122
>At4g15540 unknown protein
Length = 270
Score = 29.6 bits (65), Expect = 3.5
Identities = 18/53 (33%), Positives = 25/53 (46%), Gaps = 5/53 (9%)
Query: 53 MLRIFMTNLRVV-----LLGTKLVVLFPAVPLAVAADFYKFGRPWIFAFSLLG 100
MLR T +++ + G ++VL+ L VAA F F WI LLG
Sbjct: 96 MLRSSATQAKIIGTIVSISGALVIVLYKGPKLLVAASFTSFESSWIIGGLLLG 148
>At1g21390 hypothetical protein
Length = 260
Score = 29.6 bits (65), Expect = 3.5
Identities = 26/92 (28%), Positives = 41/92 (44%), Gaps = 14/92 (15%)
Query: 6 VSPSPSMNEDLESNTTNEETSHENLSRSMVKRSDSVLVTNNINARFQMLRIFMTNLR--- 62
VSP PS++E E+ ++ +S S KRS S N+I +R + +F +
Sbjct: 171 VSPRPSISE--ETVKVEDKEWWNRMSESSTKRSGSSSSNNSIRSRSSLRYVFKLKNQSFH 228
Query: 63 ------VVLLGTKLVVLFP---AVPLAVAADF 85
+LLG L++ F +P ADF
Sbjct: 229 LSYLTYAILLGFALLIRFDESFCLPRKEKADF 260
>At4g25240 Pollen-specific protein SKU5 homologue (SKS1)
Length = 589
Score = 28.5 bits (62), Expect = 7.9
Identities = 16/42 (38%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query: 172 LLCGGLANLKREQRYDRKQADVNSLLLLLGLLCHLLPLLFKY 213
L CG L NL++EQ + + +N L L+ LL LL +F++
Sbjct: 548 LYCGALKNLQKEQHHSAATSILNGHLKLM-LLMVLLASVFRF 588
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 28.5 bits (62), Expect = 7.9
Identities = 26/117 (22%), Positives = 46/117 (39%), Gaps = 22/117 (18%)
Query: 261 VIIDIEEDDEEKAVIGFWSAFTWLVGMTLVISLL--------SEYVVGTIEAASDSWGIS 312
V + I + K FW AF +G + +S SE + T+E
Sbjct: 969 VPVSILSHRDSKFTSAFWRAFQVEMGTKVQMSTAYHPQTDGQSERTIQTLE--------- 1019
Query: 313 VSFISIILLPIVGNAAEHAGSIIFAFKNKLDISLGVAMGSATQISMFVVPLSVIVAW 369
+ + +L G+ A+H + FA+ N S+G+A A ++ P ++ W
Sbjct: 1020 -DMLQMCVLDWGGHWADHLSLVKFAYNNSYQASIGMAPFEA----LYGRPCRTLLCW 1071
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.139 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,770,868
Number of Sequences: 26719
Number of extensions: 333117
Number of successful extensions: 1240
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 1214
Number of HSP's gapped (non-prelim): 18
length of query: 449
length of database: 11,318,596
effective HSP length: 103
effective length of query: 346
effective length of database: 8,566,539
effective search space: 2964022494
effective search space used: 2964022494
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0346.7


