

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0346.11
(564 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
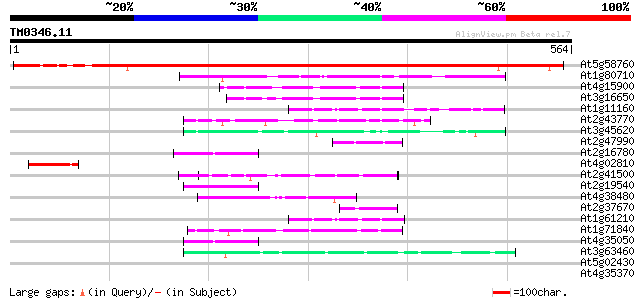
Score E
Sequences producing significant alignments: (bits) Value
At5g58760 putative protein 719 0.0
At1g80710 unknown protein 87 2e-17
At4g15900 PRL1 protein 55 8e-08
At3g16650 PP1/PP2A phosphatases pleiotropic regulator PRL2 55 8e-08
At1g11160 hypothetical protein 54 2e-07
At2g43770 putative splicing factor 53 5e-07
At3g45620 unknown protein 47 2e-05
At2g47990 unknown protein 46 6e-05
At2g16780 putative WD-40 repeat protein, MSI2 46 6e-05
At4g02810 unknown protein 45 8e-05
At2g41500 putative U4/U6 small nuclear ribonucleoprotein 45 8e-05
At2g19540 putative WD-40 repeat protein 44 2e-04
At4g38480 unknown protein 44 3e-04
At2g37670 putative WD-40 repeat protein 44 3e-04
At1g61210 44 3e-04
At1g71840 unknown protein (At1g71840) 43 4e-04
At4g35050 WD-40 repeat protein (MSI3) 43 5e-04
At3g63460 putative protein 42 7e-04
At5g02430 putative protein 42 0.001
At4g35370 putative protein 42 0.001
>At5g58760 putative protein
Length = 576
Score = 719 bits (1856), Expect = 0.0
Identities = 364/585 (62%), Positives = 446/585 (76%), Gaps = 47/585 (8%)
Query: 5 TRRVPFPKVVIERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEKLEVGL 64
+RR P++VI RDTDSE SSSEEEEE E+D E E+E+E V GK E L
Sbjct: 6 SRRKRDPEIVIARDTDSELSSSEEEEE--EEDNYPFSESEEEDE-AVKNGGKIE-----L 57
Query: 65 DANR-KGKTPITLPLRKVCKVCKKPGHESGFKGATYIDCPMKPCFLCKTPGHTT------ 117
+ N+ KGK PIT+ K+ KKPGHE+GFKGATYIDCPMKPCFLCK PG
Sbjct: 58 EKNKAKGKAPITV------KLIKKPGHEAGFKGATYIDCPMKPCFLCKMPGKAPAISYRF 111
Query: 118 -LTCP--HRFSTEHGVVPAPRKKTCKPFEYVFERQLRPGIPSIKPKYVIPDQVNCAVIRY 174
LTC T+HG++P + T P ++VF+RQL+P IP IKPKYVIPDQV+CAVIRY
Sbjct: 112 LLTCELFDDVVTDHGILPTSHRNTKNPIDFVFKRQLQPRIPPIKPKYVIPDQVHCAVIRY 171
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNPTNDC 234
HSRR+TCLEFHPTKNNI+LSGDKKGQ+GVWDF +V+EK VYGNIHS +NNM+F+PTND
Sbjct: 172 HSRRVTCLEFHPTKNNILLSGDKKGQIGVWDFGKVYEKNVYGNIHSVQVNNMRFSPTNDD 231
Query: 235 MVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVLVADNFG 294
MVYSASSDGTI +TDLETG SS+L+NLNP+GWQG ++WKMLYGMDINSEKG+VL ADNFG
Sbjct: 232 MVYSASSDGTIGYTDLETGTSSTLLNLNPDGWQGANSWKMLYGMDINSEKGVVLAADNFG 291
Query: 295 FLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDIRRMETGS 354
FLH++D R+NN TG ILIHK+ +KV G+ CNP++P++LL+CGNDH+AR+WD+R+++ +
Sbjct: 292 FLHMIDHRTNNSTGEPILIHKQGSKVCGLDCNPVQPELLLSCGNDHFARIWDMRKLQPKA 351
Query: 355 SIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKMDSPSREIVHSHDFNRYL 414
S++DL H RVV+SAYFSP SG KILTT DNR+R+WDSIFG +D PSREIVHS+DFNR+L
Sbjct: 352 SLHDLAHKRVVNSAYFSPSSGTKILTTCQDNRIRIWDSIFGNLDLPSREIVHSNDFNRHL 411
Query: 415 TPFKAEWDPKDPSESLAVVGRYISENYNGAALHPIDFIDVSTGKLVAEVMDPNITTISPV 474
TPFKAEWDPKD SESL V+GRYISENYNG ALHPIDFID S G+LVAEVMDPNITTI+PV
Sbjct: 412 TPFKAEWDPKDTSESLIVIGRYISENYNGTALHPIDFIDASNGQLVAEVMDPNITTITPV 471
Query: 475 NKLHPNDDILASGSSS-----------------SLFIWKPKEKSEPVHEKDESKIVVCGR 517
NKLHP DD+LASGSS L + +E V EK + KI++C
Sbjct: 472 NKLHPRDDVLASGSSRFGIHYPTATIVTVGGHFFLLTDLSLDNTEMVEEKKDKKIIICYG 531
Query: 518 AEKKRGK--KRGADTDESDNEGFNSK---LKKSKFQ-KTSKSQKT 556
KK+GK KRG+D ++ +++ F+SK +K +K+Q KT+K KT
Sbjct: 532 DSKKKGKKQKRGSDDEDDEDDIFSSKGKNIKVNKYQAKTTKKTKT 576
>At1g80710 unknown protein
Length = 516
Score = 87.4 bits (215), Expect = 2e-17
Identities = 86/339 (25%), Positives = 155/339 (45%), Gaps = 42/339 (12%)
Query: 171 VIRYHSRRITCLEFHPTKN-NIILSGDKKGQLGVWDFVRVHEK----VVYGNIHSCLLNN 225
V R RI ++F P +N ++ +GDK G +G W+ +E+ + HS +++
Sbjct: 209 VARVVPGRIFVVQFLPCENVKMVAAGDKLGNVGFWNLDCGNEEDNDGIYLFTPHSAPVSS 268
Query: 226 MKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDI--NSE 283
+ F + V S+S DG I D+E + + ST + ++ + N E
Sbjct: 269 IVFQQNSLSRVISSSYDGLIRLMDVEKSVFDLVY----------STDEAIFSLSQRPNDE 318
Query: 284 KGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYAR 343
+ L D +G ++ D+R+ + L H++ ++ I NP P ++ T D A
Sbjct: 319 QSLYFGQD-YGVFNVWDLRAGKSVFHWEL-HER--RINSIDFNPQNPHVMATSSTDGTAC 374
Query: 344 VWDIRRM--ETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKMDSPS 401
+WD+R M + ++ + H+R V SAYFSP SG + TTSLDN + + + +
Sbjct: 375 LWDLRSMGAKKPKTLSTVNHSRAVHSAYFSP-SGLSLATTSLDNYI----GVLSGANFEN 429
Query: 402 REIVHSHDFNRYLTPFKAEWDPKDPSESLAVVGRYISENYNGAALHPIDFIDVSTGKLVA 461
+++ ++ +R+++ FKA W D S Y G ID I+ + V
Sbjct: 430 TCMIYHNNTSRWISKFKAVWGWDD------------SYIYVGNLSKKIDVINPKLKRTVM 477
Query: 462 EVMDPNITTISPVNKLHPNDDILASGSSS--SLFIWKPK 498
E+ +P I HP + +GS++ +++W K
Sbjct: 478 ELHNPLQRAIPCRIHCHPYNVGTLAGSTAGGQVYVWTTK 516
>At4g15900 PRL1 protein
Length = 486
Score = 55.5 bits (132), Expect = 8e-08
Identities = 48/188 (25%), Positives = 88/188 (46%), Gaps = 25/188 (13%)
Query: 212 KVVYGNIHSCLLNNMKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPST 271
+V+ G H + ++ F+P+N+ + S+D TI D+ TG+ + + E
Sbjct: 170 RVIQG--HLGWVRSVAFDPSNEWFC-TGSADRTIKIWDVATGVLKLTLTGHIE------- 219
Query: 272 WKMLYGMDINSEKGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEP- 330
+ G+ +++ + A + + D+ N +I + + G++C + P
Sbjct: 220 --QVRGLAVSNRHTYMFSAGDDKQVKCWDLEQNK------VIRSYHGHLSGVYCLALHPT 271
Query: 331 -DILLTCGNDHYARVWDIRRMETGSSIYDLK-HNRVVSSAYFSPVSGNKILTTSLDNRLR 388
D+LLT G D RVWDIR T I+ L H+ V S + P + +++T S D ++
Sbjct: 272 LDVLLTGGRDSVCRVWDIR---TKMQIFALSGHDNTVCSVFTRP-TDPQVVTGSHDTTIK 327
Query: 389 VWDSIFGK 396
WD +GK
Sbjct: 328 FWDLRYGK 335
>At3g16650 PP1/PP2A phosphatases pleiotropic regulator PRL2
Length = 479
Score = 55.5 bits (132), Expect = 8e-08
Identities = 47/180 (26%), Positives = 85/180 (47%), Gaps = 22/180 (12%)
Query: 219 HSCLLNNMKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGM 278
H + ++ F+P+N+ + S+D TI D+ TG+ + L G G + G+
Sbjct: 169 HLGWVRSVAFDPSNEWFC-TGSADRTIKIWDVATGV----LKLTLTGHIG-----QVRGL 218
Query: 279 DINSEKGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEP--DILLTC 336
+++ + A + + D+ N +I + + G++C + P D++LT
Sbjct: 219 AVSNRHTYMFSAGDDKQVKCWDLEQNK------VIRSYHGHLHGVYCLALHPTLDVVLTG 272
Query: 337 GNDHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGK 396
G D RVWDIR T I+ L H+ V S P + +++T S D+ ++ WD +GK
Sbjct: 273 GRDSVCRVWDIR---TKMQIFVLPHDSDVFSVLARP-TDPQVITGSHDSTIKFWDLRYGK 328
Score = 35.8 bits (81), Expect = 0.065
Identities = 28/88 (31%), Positives = 39/88 (43%), Gaps = 18/88 (20%)
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWD-------FVRVHEKVVYGNIHSCLLNNMK 227
H + CL HPT +++L+G + VWD FV H+ V+ ++
Sbjct: 253 HLHGVYCLALHPTL-DVVLTGGRDSVCRVWDIRTKMQIFVLPHDSDVF---------SVL 302
Query: 228 FNPTNDCMVYSASSDGTISFTDLETGIS 255
PT D V + S D TI F DL G S
Sbjct: 303 ARPT-DPQVITGSHDSTIKFWDLRYGKS 329
>At1g11160 hypothetical protein
Length = 974
Score = 54.3 bits (129), Expect = 2e-07
Identities = 61/220 (27%), Positives = 93/220 (41%), Gaps = 33/220 (15%)
Query: 281 NSEKGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDH 340
NSE+ LVL + G + L D+ + K A H+ N V H P + L + +D
Sbjct: 16 NSEEVLVLAGASSGVIKLWDLEES-KMVRAFTGHRSNCSAVEFH--PFG-EFLASGSSDT 71
Query: 341 YARVWDIRRMETGSSIYDLK-HNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKMDS 399
RVWD R+ I K H R +S+ FSP G +++ LDN ++VWD GK+
Sbjct: 72 NLRVWDTRKK---GCIQTYKGHTRGISTIEFSP-DGRWVVSGGLDNVVKVWDLTAGKL-- 125
Query: 400 PSREIVHSHDFNRYLTPFKAEWDPKDPSESLAVVGRYISENYNGAALHPIDFIDVSTGKL 459
H+F + P ++ P E L G+A + F D+ T +L
Sbjct: 126 -------LHEFKCHEGPIRSL--DFHPLEFLLA---------TGSADRTVKFWDLETFEL 167
Query: 460 VAEVMDPNITTISPVNKLHPNDDILASGSSSSL--FIWKP 497
+ P T + + HP+ L G L + W+P
Sbjct: 168 IGTTR-PEATGVRAI-AFHPDGQTLFCGLDDGLKVYSWEP 205
Score = 34.3 bits (77), Expect = 0.19
Identities = 24/81 (29%), Positives = 41/81 (49%), Gaps = 9/81 (11%)
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVR---VHEKVVYGNIHSCLLNNMKFNPT 231
H+R I+ +EF P ++SG + VWD +HE H + ++ F+P
Sbjct: 90 HTRGISTIEFSPD-GRWVVSGGLDNVVKVWDLTAGKLLHEF----KCHEGPIRSLDFHPL 144
Query: 232 NDCMVYSASSDGTISFTDLET 252
+ ++ + S+D T+ F DLET
Sbjct: 145 -EFLLATGSADRTVKFWDLET 164
>At2g43770 putative splicing factor
Length = 343
Score = 52.8 bits (125), Expect = 5e-07
Identities = 67/267 (25%), Positives = 110/267 (41%), Gaps = 54/267 (20%)
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEK-----VVYGNIHSCLLNNMKFN 229
H + ++F+P +I SG ++ +W RVH V+ G+ ++ L +
Sbjct: 52 HPSAVYTMKFNPA-GTLIASGSHDREIFLW---RVHGDCKNFMVLKGHKNAIL----DLH 103
Query: 230 PTND-CMVYSASSDGTISFTDLETGIS-------SSLMNLNPEGWQGPSTWKMLYGMDIN 281
T+D + SAS D T+ D+ETG SS +N +GP
Sbjct: 104 WTSDGSQIVSASPDKTVRAWDVETGKQIKKMAEHSSFVNSCCPTRRGPP----------- 152
Query: 282 SEKGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHY 341
L++ + G L DMR AI ++ + + D + T G D+
Sbjct: 153 ----LIISGSDDGTAKLWDMRQRG----AIQTFPDKYQITAVSFSDAA-DKIFTGGVDND 203
Query: 342 ARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKMDSPS 401
+VWD+R+ E ++ H ++ SP G+ +LT +DN+L VWD + +P
Sbjct: 204 VKVWDLRKGEATMTLEG--HQDTITGMSLSP-DGSYLLTNGMDNKLCVWDM---RPYAPQ 257
Query: 402 REIV-----HSHDFNRYLTPFKAEWDP 423
V H H+F + L K W P
Sbjct: 258 NRCVKIFEGHQHNFEKNL--LKCSWSP 282
>At3g45620 unknown protein
Length = 481
Score = 47.4 bits (111), Expect = 2e-05
Identities = 78/345 (22%), Positives = 136/345 (38%), Gaps = 68/345 (19%)
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNP-TND 233
H + +EF+ T ++++SG Q+ +W+++ K+ Y + H + KF P T+D
Sbjct: 54 HEGCVNAVEFNST-GDVLVSGSDDRQIMLWNWLSGSRKLSYPSGHCENVFQTKFIPFTDD 112
Query: 234 CMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVLVADNF 293
+ ++ +DG + + + + + G +K+ + + +
Sbjct: 113 RTIITSGADGQVRLGQI---LENGKVETKRLGRHHGRVYKLAV---LPGDPNVFYSCGED 166
Query: 294 GFLHLVDMRSNNKT--------GNAILIHKKNTKVV--GIHCNPIEPDILLTCGNDHYAR 343
GF+ D+RSN+ T H ++++ I +P L G+D YAR
Sbjct: 167 GFVQHFDIRSNSATMVLYSSPFTQGCRRHHSSSRIRLNSIAIDPRNSYYLAVGGSDEYAR 226
Query: 344 VWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDS--IFGKMDSPS 401
V+D RR++ V+ A PV+ T LR +S I G S +
Sbjct: 227 VYDTRRVQLAPVC-----RHVLPDA---PVN------TFCPRHLRETNSVHITGLAYSKA 272
Query: 402 REIVHSHDFNRYLTPFKAEWDPKDPSESLAVVGRYISENYNGAALHPIDFIDVSTGKLVA 461
E++ S++ Y+ E G P + VS KL
Sbjct: 273 GELLVSYNDELI----------------------YLFEKNMGYGSSP---VSVSPEKL-Q 306
Query: 462 EVMDP-------NITTISPVNKLHPNDDILASGSS-SSLFIWKPK 498
E+ +P N T+ VN PND+ + SGS +FIWK K
Sbjct: 307 EMEEPQVYIGHRNAQTVKGVNFFGPNDEYVTSGSDCGHIFIWKKK 351
>At2g47990 unknown protein
Length = 530
Score = 45.8 bits (107), Expect = 6e-05
Identities = 23/71 (32%), Positives = 38/71 (53%), Gaps = 3/71 (4%)
Query: 325 CNPIEPDILLTCGNDHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLD 384
C+P+ +L+T DH +VWD R+ T + I ++ H V + P G ++ T+
Sbjct: 186 CSPVNDSMLVTGSYDHTVKVWD-ARVHTSNWIAEINHGLPVEDVVYLPSGG--LIATAGG 242
Query: 385 NRLRVWDSIFG 395
N ++VWD I G
Sbjct: 243 NSVKVWDLIGG 253
Score = 34.3 bits (77), Expect = 0.19
Identities = 18/60 (30%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query: 333 LLTCGNDHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDS 392
L++ G+D + WD+ S + L H V SPV+ + ++T S D+ ++VWD+
Sbjct: 151 LVSGGDDGVVKYWDVAGATVISDL--LGHKDYVRCGDCSPVNDSMLVTGSYDHTVKVWDA 208
>At2g16780 putative WD-40 repeat protein, MSI2
Length = 415
Score = 45.8 bits (107), Expect = 6e-05
Identities = 27/88 (30%), Positives = 46/88 (51%), Gaps = 3/88 (3%)
Query: 165 DQVNCAVIRY--HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCL 222
D+V A+ Y H I + +H N+ S + G+L +WD R ++ +H
Sbjct: 204 DKVLNAMFVYEGHESAIADVSWHMKNENLFGSAGEDGRLVIWD-TRTNQMQHQVKVHERE 262
Query: 223 LNNMKFNPTNDCMVYSASSDGTISFTDL 250
+N + FNP N+ ++ +ASSD T++ DL
Sbjct: 263 VNYLSFNPFNEWVLATASSDSTVALFDL 290
Score = 29.3 bits (64), Expect = 6.1
Identities = 14/98 (14%), Positives = 44/98 (44%), Gaps = 1/98 (1%)
Query: 172 IRYHSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNPT 231
++ H R + L F+P ++ + + ++D +++ + + H + ++++P
Sbjct: 256 VKVHEREVNYLSFNPFNEWVLATASSDSTVALFDLRKLNAPLHVMSSHEGEVFQVEWDPN 315
Query: 232 NDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGP 269
++ ++ S+ D + DL + + + + GP
Sbjct: 316 HETVLASSGEDRRLMVWDLNR-VGEEQLEIELDAEDGP 352
>At4g02810 unknown protein
Length = 271
Score = 45.4 bits (106), Expect = 8e-05
Identities = 23/50 (46%), Positives = 31/50 (62%), Gaps = 1/50 (2%)
Query: 20 DSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEKLEVGLDANRK 69
D+E+ EEEEE E++EEE E E+EEE+ +G NE E G N+K
Sbjct: 192 DAEEEEEEEEEEEEEEEEEEEEEEEEEEEDEEGIVGNNENFE-GKSGNKK 240
Score = 38.9 bits (89), Expect = 0.008
Identities = 29/86 (33%), Positives = 46/86 (52%), Gaps = 8/86 (9%)
Query: 16 ERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEKLEVGLDANRKGKTPIT 75
E D D + + EEEEE +EEE E E+EEE E ++E+ VG + N +GK+
Sbjct: 184 EEDEDDQYDAEEEEEE----EEEEEEEEEEEEEEEEEEEEEDEEGIVGNNENFEGKS--- 236
Query: 76 LPLRKVCKVCKKPGHESGFKGATYID 101
+KV K+ +E+G + T ++
Sbjct: 237 -GNKKVSNRPKRRCNENGCEPKTMLN 261
>At2g41500 putative U4/U6 small nuclear ribonucleoprotein
Length = 554
Score = 45.4 bits (106), Expect = 8e-05
Identities = 54/224 (24%), Positives = 97/224 (43%), Gaps = 25/224 (11%)
Query: 170 AVIRYHSRRITCLEFHPTKNNIIL-SGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKF 228
AV++ H R T + F P + + S D+ +L W + G++ L + F
Sbjct: 292 AVLKDHKERATDVVFSPVDDCLATASADRTAKL--WKTDGTLLQTFEGHLDR--LARVAF 347
Query: 229 NPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVL 288
+P+ + + S D T D+ TG L +G S + +YG+ + L
Sbjct: 348 HPSGKYLG-TTSYDKTWRLWDINTGAELLLQ-------EGHS--RSVYGIAFQQDGALAA 397
Query: 289 VADNFGFLHLVDMRSNNKTGNAILIHKKNTK-VVGIHCNPIEPDILLTCGNDHYARVWDI 347
+ D+R TG +IL+ + + K V ++ +P L + G D+ R+WD+
Sbjct: 398 SCGLDSLARVWDLR----TGRSILVFQGHIKPVFSVNFSP-NGYHLASGGEDNQCRIWDL 452
Query: 348 RRMETGSSIYDL-KHNRVVSSAYFSPVSGNKILTTSLDNRLRVW 390
R + S+Y + H +VS + P G + T S D ++ +W
Sbjct: 453 RMRK---SLYIIPAHANLVSQVKYEPQEGYFLATASYDMKVNIW 493
Score = 42.7 bits (99), Expect = 5e-04
Identities = 48/212 (22%), Positives = 90/212 (41%), Gaps = 40/212 (18%)
Query: 191 IILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNPTNDCMVYSAS--------SD 242
I+ + G +W+ +V + H ++ F+P +DC+ +++ +D
Sbjct: 269 ILATCSLSGVTKLWEMPQVTNTIAVLKDHKERATDVVFSPVDDCLATASADRTAKLWKTD 328
Query: 243 GTISFTDLETGISSSLMNLNPEG-WQGPSTWKMLYGM-DINSEKGLVLVADNFGFLHLVD 300
GT+ T + + +P G + G +++ + + DIN
Sbjct: 329 GTLLQTFEGHLDRLARVAFHPSGKYLGTTSYDKTWRLWDIN------------------- 369
Query: 301 MRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDIRRMETGSSIYDLK 360
TG +L+ + +++ V + + +CG D ARVWD+R TG SI +
Sbjct: 370 ------TGAELLLQEGHSRSVYGIAFQQDGALAASCGLDSLARVWDLR---TGRSILVFQ 420
Query: 361 -HNRVVSSAYFSPVSGNKILTTSLDNRLRVWD 391
H + V S FSP +G + + DN+ R+WD
Sbjct: 421 GHIKPVFSVNFSP-NGYHLASGGEDNQCRIWD 451
>At2g19540 putative WD-40 repeat protein
Length = 469
Score = 43.9 bits (102), Expect = 2e-04
Identities = 22/76 (28%), Positives = 35/76 (45%)
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNPTNDC 234
H+ + L++ P + N+ S G + VWD + H+ +N + +N C
Sbjct: 268 HTASVEDLQWSPAEENVFASCSVDGSVAVWDIRLGKSPALSFKAHNADVNVISWNRLASC 327
Query: 235 MVYSASSDGTISFTDL 250
M+ S S DGT S DL
Sbjct: 328 MLASGSDDGTFSIRDL 343
Score = 38.5 bits (88), Expect = 0.010
Identities = 36/177 (20%), Positives = 77/177 (43%), Gaps = 26/177 (14%)
Query: 182 LEFHPTKNNIILSGDKKGQLGVWDFVR---VHEKVVYGNIHSCLLNNMKFNPTNDCMVYS 238
+++ P +LSGD K + +W+ + + + H+ + +++++P + + S
Sbjct: 229 IDWSPATAGRLLSGDCKSMIHLWEPASGSWAVDPIPFAG-HTASVEDLQWSPAEENVFAS 287
Query: 239 ASSDGTISFTDLETGISSSL------MNLNPEGWQGPSTWKMLYGMDINSEKGLVLVADN 292
S DG+++ D+ G S +L ++N W ++ + G S+ G + D
Sbjct: 288 CSVDGSVAVWDIRLGKSPALSFKAHNADVNVISWNRLASCMLASG----SDDGTFSIRD- 342
Query: 293 FGFLHLVDMRSNNKTGNAILIHKKNTK--VVGIHCNPIEPDILLTCGNDHYARVWDI 347
L L+ K G+A++ H + K + I + E L D+ +WD+
Sbjct: 343 ---LRLI------KGGDAVVAHFEYHKHPITSIEWSAHEASTLAVTSGDNQLTIWDL 390
Score = 33.9 bits (76), Expect = 0.25
Identities = 50/245 (20%), Positives = 85/245 (34%), Gaps = 62/245 (25%)
Query: 219 HSCLLNNMKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKML--- 275
H C+ N ++ P N + S + G + D+ + +++ L EG G S
Sbjct: 159 HGCV-NRIRAMPQNSHICVSWADSGHVQVWDMSSHLNA-LAESETEGKDGTSPVLNQAPL 216
Query: 276 ----------YGMDIN-SEKGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIH 324
Y +D + + G +L D +HL + S + + I V +
Sbjct: 217 VNFSGHKDEGYAIDWSPATAGRLLSGDCKSMIHLWEPASGSWAVDPIPFAGHTASVEDLQ 276
Query: 325 CNPIEPDILLTCGNDHYARVWDIR---------------------------RMETGS--- 354
+P E ++ +C D VWDIR + +GS
Sbjct: 277 WSPAEENVFASCSVDGSVAVWDIRLGKSPALSFKAHNADVNVISWNRLASCMLASGSDDG 336
Query: 355 --SIYDLK--------------HNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKMD 398
SI DL+ H ++S +S + + TS DN+L +WD K +
Sbjct: 337 TFSIRDLRLIKGGDAVVAHFEYHKHPITSIEWSAHEASTLAVTSGDNQLTIWDLSLEKDE 396
Query: 399 SPSRE 403
E
Sbjct: 397 EEEAE 401
>At4g38480 unknown protein
Length = 471
Score = 43.5 bits (101), Expect = 3e-04
Identities = 44/165 (26%), Positives = 79/165 (47%), Gaps = 14/165 (8%)
Query: 190 NIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNP-TNDCMVYSASSDGTISFT 248
+I+LSG Q+ +WD+ K+ + + H + KF P ++D + ++++D + ++
Sbjct: 68 DILLSGSDDRQVILWDWQTASVKLSFDSGHFNNIFQAKFMPFSDDRTIVTSAADKQVRYS 127
Query: 249 D-LETG-ISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVLVADNFGFLHLVDMRSNNK 306
LE+G + +SL+ + QGP K+ ++ S ++ H D+R+
Sbjct: 128 KILESGQVETSLLGKH----QGP-VHKL--AVEPGSPFSFYTCGEDGAVKHF-DLRTRVA 179
Query: 307 TGNAILIHKKNTKVVGIHC---NPIEPDILLTCGNDHYARVWDIR 348
T K VV +H +P P +L G D YARV+DIR
Sbjct: 180 TNLFTCKEAKFNLVVYLHAIAVDPRNPGLLAVAGMDEYARVYDIR 224
Score = 28.9 bits (63), Expect = 8.0
Identities = 23/84 (27%), Positives = 39/84 (46%), Gaps = 11/84 (13%)
Query: 331 DILLTCGNDHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNK-ILTTSLDNRLR- 388
DILL+ +D +WD + S +D H + A F P S ++ I+T++ D ++R
Sbjct: 68 DILLSGSDDRQVILWDWQTASVKLS-FDSGHFNNIFQAKFMPFSDDRTIVTSAADKQVRY 126
Query: 389 --------VWDSIFGKMDSPSREI 404
V S+ GK P ++
Sbjct: 127 SKILESGQVETSLLGKHQGPVHKL 150
>At2g37670 putative WD-40 repeat protein
Length = 903
Score = 43.5 bits (101), Expect = 3e-04
Identities = 23/59 (38%), Positives = 33/59 (54%), Gaps = 3/59 (5%)
Query: 332 ILLTCGNDHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVW 390
+LL+ D R+WDI ET + + HN V+ FSPV N L+ SLD ++R+W
Sbjct: 519 LLLSSSMDKTVRLWDI---ETKTCLKLFAHNDYVTCIQFSPVDENYFLSGSLDAKIRIW 574
Score = 38.5 bits (88), Expect = 0.010
Identities = 80/400 (20%), Positives = 140/400 (35%), Gaps = 94/400 (23%)
Query: 169 CAVIRYHSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKF 228
C + H+ +TC++F P N LSG ++ +W H
Sbjct: 539 CLKLFAHNDYVTCIQFSPVDENYFLSGSLDAKIRIWSIQDRH------------------ 580
Query: 229 NPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVL 288
+ ++DL ++++ P+G G I S KG+
Sbjct: 581 ---------------VVEWSDLHEMVTAAC--YTPDG----------QGALIGSHKGICR 613
Query: 289 VAD----NFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARV 344
D + +D++SN K+ + K+ +P+ P +L D R+
Sbjct: 614 AYDTEDCKLSQTNQIDVQSNKKS-------QAKRKITSFQFSPVNPSEVLVTSADSRIRI 666
Query: 345 WDIRRMETGSS-IYDLKHNRVVSSAYFSPVS--GNKILTTSLDNRLRVWDSIFGKMDSPS 401
D GS I+ K R S + S G I+ S D+++ +W + F + S
Sbjct: 667 LD------GSEVIHKFKGFRNTCSQLSASYSQDGKYIICASEDSQVYLWKNDFHRTRSTL 720
Query: 402 REIVHSHDFNRYLTPFKAEWDPKDPSESLAVVGRYISENYNGAALHPIDFIDVSTGKLVA 461
H H + KD S ++ G E P+ K ++
Sbjct: 721 TTQSHEH------------FHCKDVSAAVPWHGHVRGEP------PPVQIHSKRHSKRIS 762
Query: 462 EVMDPNITTISPVNKLHPNDDILASGSSSSLFIWKPKEKSEPVHEKDESKIVVCGRAEKK 521
P+ T SP ++ A+G ++S K P+ +K +K + + E++
Sbjct: 763 TSSQPSSTISSPT-----KEETSATGPTTS--NRNKKSGLPPMPKKAATKSQI--QPEEE 813
Query: 522 RGKKRGADTDESDNEGFNSKLKKSKFQKTSKSQKTKWKLS 561
G + G + ES NS + S S S T +LS
Sbjct: 814 AGPELG--SSESFRSSMNSSEQHSSRFGESPSINTSSRLS 851
Score = 29.3 bits (64), Expect = 6.1
Identities = 31/130 (23%), Positives = 59/130 (44%), Gaps = 13/130 (10%)
Query: 306 KTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDI--RRMETGSSIYDLKHNR 363
+T + + N V I +P++ + L+ D R+W I R + S ++++
Sbjct: 535 ETKTCLKLFAHNDYVTCIQFSPVDENYFLSGSLDAKIRIWSIQDRHVVEWSDLHEM---- 590
Query: 364 VVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKMDSPSREIVHSH---DFNRYLTPFKAE 420
V++A ++P G L S R +D+ K+ ++ V S+ R +T F +
Sbjct: 591 -VTAACYTP-DGQGALIGSHKGICRAYDTEDCKLSQTNQIDVQSNKKSQAKRKITSF--Q 646
Query: 421 WDPKDPSESL 430
+ P +PSE L
Sbjct: 647 FSPVNPSEVL 656
>At1g61210
Length = 282
Score = 43.5 bits (101), Expect = 3e-04
Identities = 36/118 (30%), Positives = 58/118 (48%), Gaps = 9/118 (7%)
Query: 281 NSEKGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDH 340
+S + LVL + G + L D+ K A H+ N V H P + L + +D
Sbjct: 97 DSAEVLVLAGASSGVIKLWDVEEA-KMVRAFTGHRSNCSAVEFH--PFG-EFLASGSSDA 152
Query: 341 YARVWDIRRMETGSSIYDLK-HNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKM 397
++WDIR+ I K H+R +S+ F+P G +++ LDN ++VWD GK+
Sbjct: 153 NLKIWDIRKK---GCIQTYKGHSRGISTIRFTP-DGRWVVSGGLDNVVKVWDLTAGKL 206
Score = 33.5 bits (75), Expect = 0.32
Identities = 24/81 (29%), Positives = 40/81 (48%), Gaps = 9/81 (11%)
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVR---VHEKVVYGNIHSCLLNNMKFNPT 231
HSR I+ + F P ++SG + VWD +HE H + ++ F+P
Sbjct: 171 HSRGISTIRFTPD-GRWVVSGGLDNVVKVWDLTAGKLLHEF----KFHEGPIRSLDFHPL 225
Query: 232 NDCMVYSASSDGTISFTDLET 252
+ ++ + S+D T+ F DLET
Sbjct: 226 -EFLLATGSADRTVKFWDLET 245
>At1g71840 unknown protein (At1g71840)
Length = 407
Score = 43.1 bits (100), Expect = 4e-04
Identities = 48/221 (21%), Positives = 98/221 (43%), Gaps = 22/221 (9%)
Query: 179 ITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNI---HSCLLNNMKFNPTNDCM 235
I + +HP + +I+L+G + L +W+ +K Y N+ H+ + F P +
Sbjct: 158 IEWVRWHP-RGHIVLAGSEDCSLWMWNA----DKEAYLNMFSGHNLNVTCGDFTPDGK-L 211
Query: 236 VYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVLVADNFGF 295
+ + S D ++ + +T S ++ +P +G L +DINS L + G
Sbjct: 212 ICTGSDDASLIVWNPKTCESIHIVKGHPYHTEG------LTCLDINSNSSLAISGSKDGS 265
Query: 296 LHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDIRRMETGSS 355
+H+V++ K +++ H + + V + + T G D +WD++ +
Sbjct: 266 VHIVNI-VTGKVVSSLNSHTDSVECVKFSPSSATIPLAATGGMDKKLIIWDLQH---STP 321
Query: 356 IYDLKHNRVVSSAYFSPVSGNKILTTSLDN-RLRVWDSIFG 395
+ +H V+S + + +K L T N + +WDS+ G
Sbjct: 322 RFICEHEEGVTSLTW--IGTSKYLATGCANGTVSIWDSLLG 360
>At4g35050 WD-40 repeat protein (MSI3)
Length = 424
Score = 42.7 bits (99), Expect = 5e-04
Identities = 24/76 (31%), Positives = 39/76 (50%), Gaps = 1/76 (1%)
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNPTNDC 234
H I + +H NI S QL +WD +R ++ +H +N + FNP N+
Sbjct: 217 HQSIIEDVAWHMKNENIFGSAGDDCQLVIWD-LRTNQMQHQVKVHEREINYLSFNPFNEW 275
Query: 235 MVYSASSDGTISFTDL 250
++ +ASSD T++ DL
Sbjct: 276 VLATASSDSTVALFDL 291
>At3g63460 putative protein
Length = 1097
Score = 42.4 bits (98), Expect = 7e-04
Identities = 67/341 (19%), Positives = 133/341 (38%), Gaps = 34/341 (9%)
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVY------GNIHSCLLNNMKF 228
H + LEF+ +N++ SG G++ +WD ++ E + G+ ++ + +
Sbjct: 121 HKGPVRGLEFNAISSNLLASGADDGEICIWDLLKPSEPSHFPLLKGSGSATQGEISFISW 180
Query: 229 NPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVL 288
N ++ S S +GT DL ++N + S + + ++ ++ +
Sbjct: 181 NRKVQQILASTSYNGTTVIWDLRK--QKPIINFADSVRRRCSV--LQWNPNVTTQIMVAS 236
Query: 289 VADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDIR 348
D+ L L DMR+ H++ V+ + P + LLTC D+ WD
Sbjct: 237 DDDSSPTLKLWDMRNIMSPVREFTGHQRG--VIAMEWCPSDSSYLLTCAKDNRTICWDTN 294
Query: 349 RMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKMDSPSREIVHSH 408
E + + N ++ P I +S D ++ +++ ++ SR V +
Sbjct: 295 TAEIVAEL--PAGNNWNFDVHWYPKIPGVISASSFDGKIGIYN-----IEGCSRYGVEEN 347
Query: 409 DFNRYLTPFKA-EWDPKDPSESLAVVGRYISENYNGAALHPIDFIDVSTGKLVAEVMDPN 467
+F P KA +W + S G+ +S + A T +++EV
Sbjct: 348 NFG--TAPLKAPKWYKRPVGASFGFGGKLVSCHARAPA--------KGTSSILSEVEQSL 397
Query: 468 ITTISPVNKLHPNDDILASGSSSSLFIWKPKEKSEPVHEKD 508
++ S N D+ +S K E++E EK+
Sbjct: 398 VSRTSEFEAAIENGDM----TSLRGLCEKKSEETESEEEKE 434
Score = 35.4 bits (80), Expect = 0.085
Identities = 56/284 (19%), Positives = 116/284 (40%), Gaps = 26/284 (9%)
Query: 191 IILSGDKKGQLGVWDFVRV-----HEKVVYGN--IHSCLLNNMKFNPTNDCMVYSASSDG 243
+I G G + +W+ + + E + G+ +H + ++FN + ++ S + DG
Sbjct: 86 LIAGGLVDGNIDLWNPLSLIGSQPSENALVGHLSVHKGPVRGLEFNAISSNLLASGADDG 145
Query: 244 TISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVLVADNF-GFLHLVDMR 302
I DL S L G +T + + N + +L + ++ G + D+R
Sbjct: 146 EICIWDLLKPSEPSHFPLLK--GSGSATQGEISFISWNRKVQQILASTSYNGTTVIWDLR 203
Query: 303 SNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYA---RVWDIRRMETGSSIYDL 359
N ++ V + NP ++ +D + ++WD+R + + +
Sbjct: 204 KQKPIINFADSVRRRCSV--LQWNPNVTTQIMVASDDDSSPTLKLWDMRNIMSPVREF-T 260
Query: 360 KHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKMDSPSREIVHSHDFNRYLTPFKA 419
H R V + + P + +LT + DNR WD+ ++ + E+ +++N F
Sbjct: 261 GHQRGVIAMEWCPSDSSYLLTCAKDNRTICWDTNTAEIVA---ELPAGNNWN-----FDV 312
Query: 420 EWDPKDPS--ESLAVVGRYISENYNGAALHPIDFIDVSTGKLVA 461
W PK P + + G+ N G + + ++ + T L A
Sbjct: 313 HWYPKIPGVISASSFDGKIGIYNIEGCSRYGVEENNFGTAPLKA 356
>At5g02430 putative protein
Length = 905
Score = 42.0 bits (97), Expect = 0.001
Identities = 52/208 (25%), Positives = 86/208 (41%), Gaps = 37/208 (17%)
Query: 226 MKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINSEKG 285
MKF+P + + SA D I +++ +M++N EG P M D +SE
Sbjct: 417 MKFSPDSHLLA-SAGEDCAIHVWEVQ---ECEIMSMN-EGSLTPIHPSMSGSTDKSSEGD 471
Query: 286 LVLVADNFGFLHLVDMRSNNKTGNAI--LIHKKNTKVVGIHCNPI--------------- 328
V+ + S +K GN I +H T V + PI
Sbjct: 472 AAEVSQD---KKKKGKTSMSKKGNQIPDYVHAPET-VFSLSDKPICSFTGHLDDVLDLSW 527
Query: 329 -EPDILLTCGNDHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRL 387
+LL+ D R+WDI ET S + HN V+ F+P+ + ++ SLD ++
Sbjct: 528 SRSQLLLSSSMDKTVRLWDI---ETQSCLKLFAHNDYVTCVQFNPLDEDYFISGSLDAKI 584
Query: 388 RVWDSIFGKMDSPSREIVHSHDFNRYLT 415
R+W + +R++V +D +T
Sbjct: 585 RIW-------NISNRQVVEWNDLKEMVT 605
Score = 30.0 bits (66), Expect = 3.6
Identities = 9/38 (23%), Positives = 22/38 (57%)
Query: 168 NCAVIRYHSRRITCLEFHPTKNNIILSGDKKGQLGVWD 205
+C + H+ +TC++F+P + +SG ++ +W+
Sbjct: 551 SCLKLFAHNDYVTCVQFNPLDEDYFISGSLDAKIRIWN 588
Score = 29.6 bits (65), Expect = 4.7
Identities = 11/27 (40%), Positives = 21/27 (77%)
Query: 365 VSSAYFSPVSGNKILTTSLDNRLRVWD 391
+++ FSP++ +++L TS D+R+RV D
Sbjct: 654 ITAFQFSPINPSEVLVTSADSRIRVLD 680
>At4g35370 putative protein
Length = 414
Score = 41.6 bits (96), Expect = 0.001
Identities = 46/207 (22%), Positives = 80/207 (38%), Gaps = 14/207 (6%)
Query: 188 KNNIILSGDKKGQLGVWDFVRVHEK---VVYGNIHSCLLNNMKFNPTNDCMVYSASSDGT 244
K N + G + + +WD V+E V G I L + FN N +V S S D
Sbjct: 179 KGNFVAIGTMESSIEIWDLDLVNEVLPCVQLGRIAGQTLFFLTFNWENRNIVASGSEDKK 238
Query: 245 ISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVLVADNFGFLHLVDMRSN 304
+ D+ TG M + K ++ + N+ VL++ + V ++
Sbjct: 239 VKVWDVATGKCKVTMEHHE---------KKVHAVAWNNYTPEVLLSGSRD--RTVVLKDG 287
Query: 305 NKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDIRRMETGSSIYDLKHNRV 364
N+ L KV + +P + D + +D R + S H+
Sbjct: 288 RDPSNSGLKWSTEAKVEKLAWDPHSEHSFVVSLKDGTVKGFDTRASDLSPSFIIHAHDSE 347
Query: 365 VSSAYFSPVSGNKILTTSLDNRLRVWD 391
VSS ++ + N + T S D +++WD
Sbjct: 348 VSSISYNIHAPNLLATGSADESVKLWD 374
Score = 38.9 bits (89), Expect = 0.008
Identities = 41/166 (24%), Positives = 74/166 (43%), Gaps = 10/166 (6%)
Query: 182 LEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNPTNDCMVYSASS 241
L F+ NI+ SG + ++ VWD KV + H ++ + +N ++ S S
Sbjct: 220 LTFNWENRNIVASGSEDKKVKVWDVATGKCKVTMEH-HEKKVHAVAWNNYTPEVLLSGSR 278
Query: 242 DGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVLVADNFGFLHLVDM 301
D T+ D +S L W + + L D +SE V+ + G + D
Sbjct: 279 DRTVVLKDGRDPSNSGLK------WSTEAKVEKL-AWDPHSEHSFVVSLKD-GTVKGFDT 330
Query: 302 RSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDI 347
R+++ + + I IH +++V I N P++L T D ++WD+
Sbjct: 331 RASDLSPSFI-IHAHDSEVSSISYNIHAPNLLATGSADESVKLWDL 375
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.134 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,526,060
Number of Sequences: 26719
Number of extensions: 716311
Number of successful extensions: 7180
Number of sequences better than 10.0: 299
Number of HSP's better than 10.0 without gapping: 159
Number of HSP's successfully gapped in prelim test: 146
Number of HSP's that attempted gapping in prelim test: 4646
Number of HSP's gapped (non-prelim): 1396
length of query: 564
length of database: 11,318,596
effective HSP length: 104
effective length of query: 460
effective length of database: 8,539,820
effective search space: 3928317200
effective search space used: 3928317200
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0346.11


