

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0340.18
(967 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
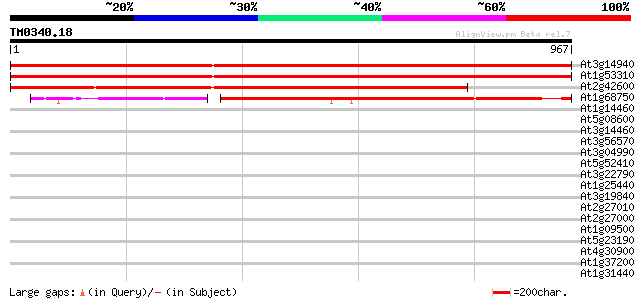
Score E
Sequences producing significant alignments: (bits) Value
At3g14940 phosphoenolpyruvate carboxylase (PPC) 1713 0.0
At1g53310 putative phosphoenolpyrovate carboxylase gb|AAC24594.1... 1711 0.0
At2g42600 phosphoenolpyruvate carboxylase like protein 1354 0.0
At1g68750 putative phosphoenolpyruvate carboxylase 546 e-155
At1g14460 hypothetical protein 34 0.46
At5g08600 putative protein 33 0.59
At3g14460 disease resistance protein, putative 33 0.78
At3g56570 putative protein 33 1.0
At3g04990 hypothetical protein 32 1.3
At5g52410 unknown protein 32 1.7
At3g22790 unknown protein 31 3.0
At1g25440 unknown protein 31 3.9
At3g19840 unknown protein 30 5.0
At2g27010 putative cytochrome P450 30 5.0
At2g27000 putative cytochrome P450 30 5.0
At1g09500 putative cinnamyl alcohol dehydrogenase 30 5.0
At5g23190 cytochrome P450-like protein 30 6.6
At4g30900 hypothetical protein 30 6.6
At1g37200 hypothetical protein 30 6.6
At1g31440 unknown protein 30 6.6
>At3g14940 phosphoenolpyruvate carboxylase (PPC)
Length = 968
Score = 1713 bits (4437), Expect = 0.0
Identities = 845/969 (87%), Positives = 914/969 (94%), Gaps = 3/969 (0%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MA RN+EKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDL+ETVQE+
Sbjct: 1 MAGRNIEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLRETVQEL 60
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYE K EP +LE LG+++TSLD GDSIV++K+F+HMLNLANLAEEVQIAHRRRIK
Sbjct: 61 YELSAEYEGKREPSKLEELGSVLTSLDPGDSIVISKAFSHMLNLANLAEEVQIAHRRRIK 120
Query: 121 -LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRR 179
LKKGDF DE +ATTESDIEETFK+LV ++ KSP+E+FD LKNQTVDLVLTAHPTQSVRR
Sbjct: 121 KLKKGDFVDESSATTESDIEETFKRLVSDLGKSPEEIFDALKNQTVDLVLTAHPTQSVRR 180
Query: 180 SLLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAG 239
SLLQKHGRIR+CL QLYAKDITPDDKQELDE+LQREIQAAFRTDEIRRT PTPQDEMRAG
Sbjct: 181 SLLQKHGRIRDCLAQLYAKDITPDDKQELDESLQREIQAAFRTDEIRRTPPTPQDEMRAG 240
Query: 240 MSYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVT 299
MSYFHETIWKGVPKFLRRVDTALKNIGI+ERVPYNAPLIQFSSWMGGDRDGNPRVTPEVT
Sbjct: 241 MSYFHETIWKGVPKFLRRVDTALKNIGIDERVPYNAPLIQFSSWMGGDRDGNPRVTPEVT 300
Query: 300 RDVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWK 359
RDVCLLARMMAANLYY+QIE+LMFELSMWRC DE RVRA+EL +S KDA AKHYIEFWK
Sbjct: 301 RDVCLLARMMAANLYYNQIENLMFELSMWRCTDEFRVRADELHRNSRKDA-AKHYIEFWK 359
Query: 360 NIPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSL 419
IPP+EPYRV+LG+VR++LY TRERSR LL+N SDI EE TFTNVE+FLEPLELCYRSL
Sbjct: 360 TIPPTEPYRVILGDVRDKLYHTRERSRQLLSNGISDIPEEATFTNVEQFLEPLELCYRSL 419
Query: 420 CACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGS-YLEWS 478
C+CGD IADGSLLDFLRQVSTFGLSLVRLDIRQES+RHTDVLDAITKHL IGS Y +WS
Sbjct: 420 CSCGDSPIADGSLLDFLRQVSTFGLSLVRLDIRQESERHTDVLDAITKHLDIGSSYRDWS 479
Query: 479 EEKRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSD 538
EE RQ+WLL+ELSGKRPLFGPDLP+TEEI DVLDTF VI+ELPSD FGAYIISMAT+PSD
Sbjct: 480 EEGRQEWLLAELSGKRPLFGPDLPKTEEISDVLDTFKVISELPSDCFGAYIISMATSPSD 539
Query: 539 VLAVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYS 598
VLAVELLQRECHVKNPLRVVPLFEKLADLE APAA+ARLFS++WY+NRINGKQEVMIGYS
Sbjct: 540 VLAVELLQRECHVKNPLRVVPLFEKLADLEAAPAAVARLFSIDWYKNRINGKQEVMIGYS 599
Query: 599 DSGKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPD 658
DSGKDAGR SAAW+LYKAQEEL+KVAK+YGVKLTMFHGRGGTVGRGGGPTHLAILSQPPD
Sbjct: 600 DSGKDAGRLSAAWELYKAQEELVKVAKKYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPD 659
Query: 659 TIHGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAV 718
T++GSLRVTVQGEVIEQSFGE HLCFRTLQR+TAATLEHGM+PPISPKPEWRAL+DEMAV
Sbjct: 660 TVNGSLRVTVQGEVIEQSFGEAHLCFRTLQRFTAATLEHGMNPPISPKPEWRALLDEMAV 719
Query: 719 IATEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWT 778
+ATEEYRS+VF+EPRFVEYFRLATPELEYGRMNIGSRP+KRKPSGGIE+LRAIPWIFAWT
Sbjct: 720 VATEEYRSVVFQEPRFVEYFRLATPELEYGRMNIGSRPSKRKPSGGIESLRAIPWIFAWT 779
Query: 779 QTRFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAA 838
QTRFHLPVWLGFGAAF++ I KD+RNL+MLQ+MY QWPFFRVTIDL+EMVFAKGDPGIAA
Sbjct: 780 QTRFHLPVWLGFGAAFRYAIKKDVRNLHMLQDMYKQWPFFRVTIDLIEMVFAKGDPGIAA 839
Query: 839 LYDRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLN 898
LYD+LLVSEDLW+FGE+LR F+ETK L+LQ A HKDLLEGDPYLKQRLRLRDSYITTLN
Sbjct: 840 LYDKLLVSEDLWAFGEKLRANFDETKNLVLQTAGHKDLLEGDPYLKQRLRLRDSYITTLN 899
Query: 899 VCQAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKG 958
VCQAYTLKRIRD NYNV LRPHISKE + SK A ELV LNPTSEYAPGLEDTLILTMKG
Sbjct: 900 VCQAYTLKRIRDANYNVTLRPHISKEIMQSSKSAQELVKLNPTSEYAPGLEDTLILTMKG 959
Query: 959 IAAGMQNTG 967
IAAG+QNTG
Sbjct: 960 IAAGLQNTG 968
>At1g53310 putative phosphoenolpyrovate carboxylase gb|AAC24594.1;
similar to ESTs gb|R30301.1, gb|AI992716.1,
emb|Z29154.1, gb|W43080.1, emb|Z30862.1
Length = 967
Score = 1711 bits (4432), Expect = 0.0
Identities = 839/968 (86%), Positives = 912/968 (93%), Gaps = 2/968 (0%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MANR LEKMASID LRQLVP KVSEDDKLVEYDALLLDRFLDILQDLHGEDL+ETVQE+
Sbjct: 1 MANRKLEKMASIDVHLRQLVPGKVSEDDKLVEYDALLLDRFLDILQDLHGEDLRETVQEL 60
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YE SAEYE KHEP++LE LG+++TSLD GDSIV+AK+F+HMLNLANLAEEVQIA+RRRIK
Sbjct: 61 YEHSAEYEGKHEPKKLEELGSVLTSLDPGDSIVIAKAFSHMLNLANLAEEVQIAYRRRIK 120
Query: 121 -LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRR 179
LKKGDF DE +ATTESD+EETFKKLVG++ KSP+E+FD LKNQTVDLVLTAHPTQSVRR
Sbjct: 121 KLKKGDFVDESSATTESDLEETFKKLVGDLNKSPEEIFDALKNQTVDLVLTAHPTQSVRR 180
Query: 180 SLLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAG 239
SLLQKHGRIR+CL QLYAKDITPDDKQELDEALQREIQAAFRTDEI+RT PTPQDEMRAG
Sbjct: 181 SLLQKHGRIRDCLAQLYAKDITPDDKQELDEALQREIQAAFRTDEIKRTPPTPQDEMRAG 240
Query: 240 MSYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVT 299
MSYFHETIWKGVPKFLRRVDTALKNIGI ERVPYNAPLIQFSSWMGGDRDGNPRVTPEVT
Sbjct: 241 MSYFHETIWKGVPKFLRRVDTALKNIGIEERVPYNAPLIQFSSWMGGDRDGNPRVTPEVT 300
Query: 300 RDVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWK 359
RDVCLLARMMAA +Y++QIEDLMFE+SMWRCNDELR RA+E+ ++S KDA AKHYIEFWK
Sbjct: 301 RDVCLLARMMAATMYFNQIEDLMFEMSMWRCNDELRARADEVHANSRKDA-AKHYIEFWK 359
Query: 360 NIPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSL 419
+IP +EPYRV+LG+VR++LY TRER+ LL+N +SD+ E TF N+E+FLEPLELCYRSL
Sbjct: 360 SIPTTEPYRVILGDVRDKLYHTRERAHQLLSNGHSDVPVEATFINLEQFLEPLELCYRSL 419
Query: 420 CACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSE 479
C+CGDR IADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAIT HL IGSY EWSE
Sbjct: 420 CSCGDRPIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITTHLDIGSYREWSE 479
Query: 480 EKRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDV 539
E+RQ+WLLSELSGKRPLFG DLP+TEEI DVLDTFHVIAELP+D+FGAYIISMATAPSDV
Sbjct: 480 ERRQEWLLSELSGKRPLFGSDLPKTEEIADVLDTFHVIAELPADSFGAYIISMATAPSDV 539
Query: 540 LAVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSD 599
LAVELLQREC VK PLRVVPLFEKLADLE APAA+ARLFSV+WY+NRINGKQEVMIGYSD
Sbjct: 540 LAVELLQRECRVKQPLRVVPLFEKLADLEAAPAAVARLFSVDWYKNRINGKQEVMIGYSD 599
Query: 600 SGKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDT 659
SGKDAGR SAAWQLYKAQEEL+KVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDT
Sbjct: 600 SGKDAGRLSAAWQLYKAQEELVKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDT 659
Query: 660 IHGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVI 719
I+GSLRVTVQGEVIEQSFGEEHLCFRTLQR+TAATLEHGM PPISPKPEWRAL+DEMAV+
Sbjct: 660 INGSLRVTVQGEVIEQSFGEEHLCFRTLQRFTAATLEHGMRPPISPKPEWRALLDEMAVV 719
Query: 720 ATEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQ 779
ATEEYRS+VF+EPRFVEYFRLATPELEYGRMNIGSRP+KRKPSGGIE+LRAIPWIFAWTQ
Sbjct: 720 ATEEYRSVVFQEPRFVEYFRLATPELEYGRMNIGSRPSKRKPSGGIESLRAIPWIFAWTQ 779
Query: 780 TRFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAAL 839
TRFHLPVWLGFG+A +HVI KD+RNL+MLQ+MY WPFFRVTIDL+EMVFAKGDPGIAAL
Sbjct: 780 TRFHLPVWLGFGSAIRHVIEKDVRNLHMLQDMYQHWPFFRVTIDLIEMVFAKGDPGIAAL 839
Query: 840 YDRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNV 899
YD+LLVSE+LW FGE+LR FEETK+L+LQ A HKDLLEGDPYLKQRLRLRDSYITTLNV
Sbjct: 840 YDKLLVSEELWPFGEKLRANFEETKKLILQTAGHKDLLEGDPYLKQRLRLRDSYITTLNV 899
Query: 900 CQAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGI 959
CQAYTLKRIRDP+Y+V LRPHISKE + SKPA EL+ LNPTSEYAPGLEDTLILTMKGI
Sbjct: 900 CQAYTLKRIRDPSYHVTLRPHISKEIAESSKPAKELIELNPTSEYAPGLEDTLILTMKGI 959
Query: 960 AAGMQNTG 967
AAG+QNTG
Sbjct: 960 AAGLQNTG 967
>At2g42600 phosphoenolpyruvate carboxylase like protein
Length = 793
Score = 1354 bits (3505), Expect = 0.0
Identities = 669/789 (84%), Positives = 731/789 (91%), Gaps = 5/789 (0%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MA RNLEKMASIDAQLR L P KVSEDDKL+EYDALLLDRFLDILQDLHGED++E VQE
Sbjct: 1 MAARNLEKMASIDAQLRLLAPGKVSEDDKLIEYDALLLDRFLDILQDLHGEDVREFVQEC 60
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YE++A+Y+ ++LE LGN++TSLD GDSIVV KSF++ML+LANLAEEVQIA+RRRIK
Sbjct: 61 YEVAADYDGNRNTEKLEELGNMLTSLDPGDSIVVTKSFSNMLSLANLAEEVQIAYRRRIK 120
Query: 121 -LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRR 179
LKKGDFADE +ATTESDIEET K+L+ ++ K+P+EVFD LKNQTVDLVLTAHPTQSVRR
Sbjct: 121 KLKKGDFADEASATTESDIEETLKRLL-QLNKTPEEVFDALKNQTVDLVLTAHPTQSVRR 179
Query: 180 SLLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAG 239
SLLQK GRIR+CLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRT PTPQDEMRAG
Sbjct: 180 SLLQKFGRIRDCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTPPTPQDEMRAG 239
Query: 240 MSYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVT 299
MSYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVT
Sbjct: 240 MSYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVT 299
Query: 300 RDVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWK 359
RDVCLLARMMAANLY+SQIEDLMFE+SMWRCN+ELRVRAE + AKHYIEFWK
Sbjct: 300 RDVCLLARMMAANLYFSQIEDLMFEMSMWRCNEELRVRAERQRCAKRD---AKHYIEFWK 356
Query: 360 NIPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSL 419
IP +EPYR +LG+VR++LY TRER+R LL++ SD+ E+ FT+V++FLEPLELCYRSL
Sbjct: 357 QIPANEPYRAILGDVRDKLYNTRERARQLLSSGVSDVPEDAVFTSVDQFLEPLELCYRSL 416
Query: 420 CACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSE 479
C CGDR IADGSLLDFLRQVSTFGL+LV+LDIRQES+RH+DVLDAIT HLGIGSY EWSE
Sbjct: 417 CDCGDRPIADGSLLDFLRQVSTFGLALVKLDIRQESERHSDVLDAITTHLGIGSYKEWSE 476
Query: 480 EKRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDV 539
+KRQ+WLLSELSGKRPLFGPDLP+TEE+ DVLDTF VI+ELPSD+FGAYIISMATAPSDV
Sbjct: 477 DKRQEWLLSELSGKRPLFGPDLPKTEEVADVLDTFKVISELPSDSFGAYIISMATAPSDV 536
Query: 540 LAVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSD 599
LAVELLQREC + +PLRVVPLFEKLADLE+APAA+ARLFS+EWYRNRINGKQEVMIGYSD
Sbjct: 537 LAVELLQRECGITDPLRVVPLFEKLADLESAPAAVARLFSIEWYRNRINGKQEVMIGYSD 596
Query: 600 SGKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDT 659
SGKDAGR SAAWQLYK QEEL+KVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDT
Sbjct: 597 SGKDAGRLSAAWQLYKTQEELVKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDT 656
Query: 660 IHGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVI 719
IHG LRVTVQGEVIEQSFGEEHLCFRTLQR+TAATLEHGMHPP+SPKPEWR LMDEMA+I
Sbjct: 657 IHGQLRVTVQGEVIEQSFGEEHLCFRTLQRFTAATLEHGMHPPVSPKPEWRVLMDEMAII 716
Query: 720 ATEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQ 779
ATEEYRS+VFKEPRFVEYFRLATPELEYGRMNIGSRP+KRKPSGGIE+LRAIPWIFAWTQ
Sbjct: 717 ATEEYRSVVFKEPRFVEYFRLATPELEYGRMNIGSRPSKRKPSGGIESLRAIPWIFAWTQ 776
Query: 780 TRFHLPVWL 788
TRF P L
Sbjct: 777 TRFTYPCGL 785
>At1g68750 putative phosphoenolpyruvate carboxylase
Length = 981
Score = 546 bits (1408), Expect = e-155
Identities = 291/620 (46%), Positives = 396/620 (62%), Gaps = 49/620 (7%)
Query: 364 SEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLCACG 423
S PYR+VLGEV+ +L +TR L+ + ++++ ++ LEPL LCY SL + G
Sbjct: 395 SAPYRIVLGEVKEKLVKTRRLLELLIEGLPCEYDPKNSYETSDQLLEPLLLCYESLQSSG 454
Query: 424 DRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEEKRQ 483
R +ADG L D +R+VSTFG+ LV+LD+RQE+ RH++ LDAIT +L +G+Y EW EEK+
Sbjct: 455 ARVLADGRLADLIRRVSTFGMVLVKLDLRQEAARHSEALDAITTYLDMGTYSEWDEEKKL 514
Query: 484 QWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVLAVE 543
++L EL GKRPL + ++K+VLDTF V AEL S++ GAY+ISMA+ SDVLAVE
Sbjct: 515 EFLTRELKGKRPLVPQCIKVGPDVKEVLDTFRVAAELGSESLGAYVISMASNASDVLAVE 574
Query: 544 LLQRECHVK-----------NPLRVVPLFEKLADLETAPAALARLFSVEWYRNRI----N 588
LLQ++ + LRVVPLFE + DL A ++ +L S++WYR I N
Sbjct: 575 LLQKDARLALTSEHGKPCPGGTLRVVPLFETVNDLRAAGPSIRKLLSIDWYREHIQKNHN 634
Query: 589 GKQEVMIGYSDSGKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPT 648
G QEVM+GYSDSGKDAGRF+AAW+LYKAQE ++ E+G+K+T+FHGRGG++GRGGGPT
Sbjct: 635 GHQEVMVGYSDSGKDAGRFTAAWELYKAQENVVAACNEFGIKITLFHGRGGSIGRGGGPT 694
Query: 649 HLAILSQPPDTIHGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPE 708
+LAI SQPP ++ GSLR T QGE+++ FG R L+ YT A L + PP P+ E
Sbjct: 695 YLAIQSQPPGSVMGSLRSTEQGEMVQAKFGIPQTAVRQLEVYTTAVLLATLKPPQPPREE 754
Query: 709 -WRALMDEMAVIATEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIET 767
WR LM+E++ I+ + YRS V++ P F+ YF ATP+ E G +NIGSRP +RK S GI
Sbjct: 755 KWRNLMEEISGISCQHYRSTVYENPEFLSYFHEATPQAELGFLNIGSRPTRRKSSSGIGH 814
Query: 768 LRAIPWIFAWTQTRFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEM 827
LRAIPW+FAWTQTRF LP WLG GA K V K + + L+EMY +WPFF+ T++L+EM
Sbjct: 815 LRAIPWVFAWTQTRFVLPAWLGVGAGLKGVSEKG--HADDLKEMYKEWPFFQSTLELIEM 872
Query: 828 VFAKGDPGIAALYDRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRL 887
V AK D + YD LVSE G +LR T++ +L ++ H+ LL+ + LK+ +
Sbjct: 873 VLAKADIPMTKHYDEQLVSEKRRGLGTELRKELMTTEKYVLVISGHEKLLQDNKSLKKLI 932
Query: 888 RLRDSYITTLNVCQAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPG 947
R Y+ +N+ Q LKR+R N KLR
Sbjct: 933 DSRLPYLNAMNMLQVEILKRLRRDEDNNKLR----------------------------- 963
Query: 948 LEDTLILTMKGIAAGMQNTG 967
D L++T+ GIAAGM+NTG
Sbjct: 964 --DALLITINGIAAGMRNTG 981
Score = 174 bits (442), Expect = 2e-43
Identities = 110/314 (35%), Positives = 169/314 (53%), Gaps = 41/314 (13%)
Query: 36 LLLDRFLDILQDLHGEDLKETVQEVYELSAEYERKHEPQRLEVLGNL--------ITSLD 87
LL F D+LQ G E V+ + + A+ +E NL I+ +
Sbjct: 23 LLGSLFHDVLQREVGNPFMEKVERI-RILAQSALNLRMAGIEDTANLLEKQLTSEISKMP 81
Query: 88 AGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIKLKKGDFADEGNATTESDIEETFKKLVG 147
+++ +A++F H LNL +A+ H ++ L+ G
Sbjct: 82 LEEALTLARTFTHSLNLMGIAD----THHSQL-LQSG----------------------- 113
Query: 148 EMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRSLLQKHGRIRNCLTQLYAKDITPDDKQE 207
SP E++ + Q V++VLTAHPTQ RR+L KH RI + L D++ +D++
Sbjct: 114 ---ISPDELYKTVCKQEVEIVLTAHPTQINRRTLQYKHIRIAHLLEYNTRSDLSVEDRET 170
Query: 208 LDEALQREIQAAFRTDEIRRTAPTPQDEMRAGMSYFHETIWKGVPKFLRRVDTALKNIGI 267
L E L REI + ++TDE+RR PTP DE RAG++ +++WK VP++LRRV +LK
Sbjct: 171 LIEDLVREITSLWQTDELRRQKPTPVDEARAGLNIVEQSLWKAVPQYLRRVSNSLKKF-T 229
Query: 268 NERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTRDVCLLARMMAANLYYSQIEDLMFELSM 327
+ +P ++F SWMGGDRDGNP VT +VT++V LL+R MA +LY +++ L FELS
Sbjct: 230 GKPLPLTCTPMKFGSWMGGDRDGNPNVTAKVTKEVSLLSRWMAIDLYIREVDSLRFELST 289
Query: 328 WRCNDELRVRAEEL 341
RC+D A+++
Sbjct: 290 DRCSDRFSRLADKI 303
>At1g14460 hypothetical protein
Length = 1102
Score = 33.9 bits (76), Expect = 0.46
Identities = 16/40 (40%), Positives = 23/40 (57%)
Query: 900 CQAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLN 939
CQ Y ++RD + V+LR S E +DV A +L+ LN
Sbjct: 591 CQKYIFNKVRDGDIVVRLRKIASDENLDVESQALDLIALN 630
>At5g08600 putative protein
Length = 822
Score = 33.5 bits (75), Expect = 0.59
Identities = 28/102 (27%), Positives = 46/102 (44%), Gaps = 4/102 (3%)
Query: 312 NLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKNIPPSEPYRVVL 371
N Y ++ + FEL+ C DE V +E+ + D +H KN+ P
Sbjct: 72 NNRYERVVNYEFELAE-DCEDE-NVESEDDDDDDDDDDDDRHS-RMLKNVTEL-PISAFQ 127
Query: 372 GEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLE 413
GE +N+ E N D+LE + T +++F+EPL+
Sbjct: 128 GESKNKRVVFTEPYPESEFNPTRDVLEGKSLTTIDDFMEPLQ 169
>At3g14460 disease resistance protein, putative
Length = 1424
Score = 33.1 bits (74), Expect = 0.78
Identities = 27/100 (27%), Positives = 45/100 (45%), Gaps = 5/100 (5%)
Query: 425 RAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEEKRQQ 484
R +A+ L L Q G ++ I + ++ +L+ KH+ + E+SE + Q
Sbjct: 92 RVVAEAGGLGGLFQNLMAGREAIQKKIEPKMEKVVRLLEHHVKHIEVIGLKEYSETREPQ 151
Query: 485 WLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDN 524
W + S RP DLPQ + V D ++ L SD+
Sbjct: 152 W--RQASRSRP---DDLPQGRLVGRVEDKLALVNLLLSDD 186
>At3g56570 putative protein
Length = 531
Score = 32.7 bits (73), Expect = 1.0
Identities = 36/156 (23%), Positives = 68/156 (43%), Gaps = 26/156 (16%)
Query: 31 VEYDALLLDRFLDILQDLHGEDLKETVQEVYELSAEYERKHE-------PQRLEVLGNLI 83
++ D L+ D+ L G +L + V+E + L E +++ PQ ++ I
Sbjct: 104 IQEDLPLVWSLEDLDSLLSGTELHKLVKEDHVLIYEDWKENILPLTSSLPQNVDSDSFGI 163
Query: 84 TSLDAGDSIVVAKSF-------AHMLNLANL------AEEVQIAHRRRIKLKKGDFADEG 130
A S++ ++SF + M+ LA+L AE+V H + + D D
Sbjct: 164 KEYLAAKSLIASRSFEIDDYHGSGMVPLADLFNHKTGAEDVHFTHESDSEADESDNDDAA 223
Query: 131 NATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVD 166
N TT+ D + ++ SP++ F+E+ + D
Sbjct: 224 NETTDEDEPSS------KISSSPEQSFEEVPGENTD 253
>At3g04990 hypothetical protein
Length = 227
Score = 32.3 bits (72), Expect = 1.3
Identities = 31/157 (19%), Positives = 67/157 (41%), Gaps = 6/157 (3%)
Query: 87 DAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIKLKKGDFAD-EGNATTESDIEETFKKL 145
DA + ++ A +LN + E R + LK+ + + +S E K
Sbjct: 7 DAAEIMICAGQLKGLLNHLRMGEANIEKSSRELDLKEKELQILSSDLEQKSHAFEAEKSE 66
Query: 146 VGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRSLLQKHGRIRNCLTQL-----YAKDI 200
VG++KK +E +EL+++ L + V+R L K ++ + +L A+ +
Sbjct: 67 VGDLKKLVEECTEELRSKRNLLTVKLDSLIRVQRELELKDNQLVQVMAELKRRYSEARHV 126
Query: 201 TPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMR 237
++ DE ++ + + D+I+ + + + R
Sbjct: 127 QKRKREMEDETATKKKELSMTVDQIQESGKQLEKKSR 163
>At5g52410 unknown protein
Length = 510
Score = 32.0 bits (71), Expect = 1.7
Identities = 33/168 (19%), Positives = 62/168 (36%), Gaps = 2/168 (1%)
Query: 52 DLKETVQEVYELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEV 111
DL + LS R +P + SL GD+ V + +AE V
Sbjct: 154 DLSAGEHGITALSFGRTRLFQPSKAVTKAQTAVSLAIGDAFEVVGEELARIEAEAMAENV 213
Query: 112 QIAHRRRIKLKKGDF--ADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVL 169
AH + + D + E E +I + +KL E K + E + +T+ L
Sbjct: 214 VCAHNELVAQVEKDINASFEKELLREKEIVDAVEKLAEEAKSELARLRVEKEEETLALER 273
Query: 170 TAHPTQSVRRSLLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQ 217
++ +L + + L L + ++E + LQ++++
Sbjct: 274 ERTSIETEMEALARIRNELEEQLQSLASNKAEMSYEKERFDRLQKQVE 321
>At3g22790 unknown protein
Length = 1694
Score = 31.2 bits (69), Expect = 3.0
Identities = 35/165 (21%), Positives = 66/165 (39%), Gaps = 13/165 (7%)
Query: 105 ANLAEEVQIAHRRRIKLKKGDFADEGNATTESDIEETFKKL--VGEMKKSPQEVFDELKN 162
ANL ++QI KL E N+ E+ + +L V E K +E F LKN
Sbjct: 663 ANLLSQLQIMTENMQKLL------EKNSLLETSLSGANIELQCVKEKSKCFEEFFQLLKN 716
Query: 163 QTVDLVLTAHPTQSVRRSLLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRT 222
+L+ S ++ +K G + T+L K +++ E++ + T
Sbjct: 717 DKAELIKERESLISQLNAVKEKLGVLEKKFTELEGKYADLQREKQFKNLQVEELRVSLAT 776
Query: 223 DE-----IRRTAPTPQDEMRAGMSYFHETIWKGVPKFLRRVDTAL 262
++ R+ T +++ +S+ E +F +D A+
Sbjct: 777 EKQERASYERSTDTRLADLQNNVSFLREECRSRKKEFEEELDRAV 821
>At1g25440 unknown protein
Length = 417
Score = 30.8 bits (68), Expect = 3.9
Identities = 23/74 (31%), Positives = 39/74 (52%), Gaps = 10/74 (13%)
Query: 27 DDKLVEYDALLLDRFLDILQDLHGEDLKETVQEVYELSAEYERKHEPQRLEVLGNLITSL 86
++++ E A+ +D F D +D+ G TV +ELS +YE H+ EV+ N+ +S
Sbjct: 226 EEEVEEIKAMSMDIFDDDRKDVDG-----TVP--FELSFDYESSHKTSEEEVMKNVESS- 277
Query: 87 DAGDSIVVAKSFAH 100
G+ +V K H
Sbjct: 278 --GECVVKVKEEEH 289
>At3g19840 unknown protein
Length = 830
Score = 30.4 bits (67), Expect = 5.0
Identities = 19/66 (28%), Positives = 35/66 (52%), Gaps = 1/66 (1%)
Query: 125 DFADEGNATTESDIEETFKKLVGEMKKSPQEVFD-ELKNQTVDLVLTAHPTQSVRRSLLQ 183
D DE + ++ + + FK+++ E +P ++ EL D A P+ SVRRSL +
Sbjct: 440 DSEDEDSGPSKEECSKQFKEMLKERGIAPFSKWEKELPKIIFDPRFKAIPSHSVRRSLFE 499
Query: 184 KHGRIR 189
++ + R
Sbjct: 500 QYVKTR 505
>At2g27010 putative cytochrome P450
Length = 498
Score = 30.4 bits (67), Expect = 5.0
Identities = 14/43 (32%), Positives = 23/43 (52%)
Query: 360 NIPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTF 402
N+PPS P ++G + + L RS L+++Y +L H F
Sbjct: 34 NLPPSPPSLPIIGHLHHLLSLFMHRSLQKLSSKYGPLLYLHVF 76
>At2g27000 putative cytochrome P450
Length = 514
Score = 30.4 bits (67), Expect = 5.0
Identities = 14/43 (32%), Positives = 23/43 (52%)
Query: 360 NIPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTF 402
N+PPS P ++G + + L RS L+++Y +L H F
Sbjct: 38 NLPPSPPSLPIIGHLHHLLSLFMHRSLQKLSSKYGPLLYLHVF 80
>At1g09500 putative cinnamyl alcohol dehydrogenase
Length = 325
Score = 30.4 bits (67), Expect = 5.0
Identities = 21/72 (29%), Positives = 35/72 (48%), Gaps = 5/72 (6%)
Query: 116 RRRIKLKKGDFADEGNATTESD----IEETFKKLVGEMKKSPQ-EVFDELKNQTVDLVLT 170
+ R+KL K D DEG+ D + T + +K PQ E+ + N T++++ T
Sbjct: 55 KERLKLFKADLLDEGSFELAIDGCETVFHTASPVAITVKTDPQVELINPAVNGTINVLRT 114
Query: 171 AHPTQSVRRSLL 182
SV+R +L
Sbjct: 115 CTKVSSVKRVIL 126
>At5g23190 cytochrome P450-like protein
Length = 559
Score = 30.0 bits (66), Expect = 6.6
Identities = 31/127 (24%), Positives = 57/127 (44%), Gaps = 13/127 (10%)
Query: 119 IKLKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVR 178
I+ +K + + EG T SD+ F L E +S + F L++ V+ +L T SV
Sbjct: 287 IRTRKKELSLEGETTKRSDLLTVFMGLRDEKGESFSDKF--LRDICVNFILAGRDTSSVA 344
Query: 179 RS----LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQD 234
S LL+K+ + + K + D + A + + + F +EI++ D
Sbjct: 345 LSWFFWLLEKNPEVEEKIMVEMCKILRQRD--DHGNAEKSDYEPVFGPEEIKK-----MD 397
Query: 235 EMRAGMS 241
++A +S
Sbjct: 398 YLQAALS 404
>At4g30900 hypothetical protein
Length = 131
Score = 30.0 bits (66), Expect = 6.6
Identities = 19/60 (31%), Positives = 28/60 (46%), Gaps = 5/60 (8%)
Query: 370 VLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLCACGDRAIAD 429
V+G++R+ R R L Y D + T +E L+L +R+LC C DR D
Sbjct: 14 VVGDMRDAWPSARVRKNVALIRTYHDFKGDKQGT-----VEFLKLIFRALCLCWDRQTQD 68
>At1g37200 hypothetical protein
Length = 1564
Score = 30.0 bits (66), Expect = 6.6
Identities = 26/99 (26%), Positives = 48/99 (48%), Gaps = 5/99 (5%)
Query: 529 IISMATAPSDVLAVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRIN 588
+I T+ +D+ ++ LLQ E H + + K+A+L A+A L + W+ +R
Sbjct: 183 LIEQRTSEADLWSLTLLQNESHRSYVEKFKAIKSKIANL-NEEVAIAALRNGLWFSSRF- 240
Query: 589 GKQEVMIGYSDSGKDAGRFSAAWQLYKAQEELIKVAKEY 627
++E+ + S DA A K +EEL +A ++
Sbjct: 241 -REELTVRQPISLDDA--LHKALHFAKGEEELAVLALKF 276
>At1g31440 unknown protein
Length = 439
Score = 30.0 bits (66), Expect = 6.6
Identities = 37/155 (23%), Positives = 68/155 (43%), Gaps = 32/155 (20%)
Query: 116 RRRIKLKKGDFADE------GNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVD-LV 168
RRR KLK+ D ++E + + ++++ + K L E K+ EV D+ +N T L
Sbjct: 171 RRRSKLKESDISEEAYIKLKNSESRLAELKSSMKTLGKEATKAMLEVDDQQQNVTSQRLR 230
Query: 169 LTAHPTQSVRRSLLQKHGRIRNCL-------------TQLYAKDITPDDKQELDEALQRE 215
+S R+ L ++ + + L+ +D +QE + E
Sbjct: 231 ALVEAERSYHRNALDILDKLHSEMIAEEEAIGSSPKSLPLHIEDSASLPQQEPNSNSSGE 290
Query: 216 IQA-------AFRTDEIRR-----TAPTPQDEMRA 238
I++ A R +EI+ T P+P+DEM++
Sbjct: 291 IKSNPLGKIKASRREEIKSNPQEVTKPSPKDEMKS 325
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,773,696
Number of Sequences: 26719
Number of extensions: 956252
Number of successful extensions: 2538
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 2515
Number of HSP's gapped (non-prelim): 25
length of query: 967
length of database: 11,318,596
effective HSP length: 109
effective length of query: 858
effective length of database: 8,406,225
effective search space: 7212541050
effective search space used: 7212541050
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0340.18


