

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0340.11
(420 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
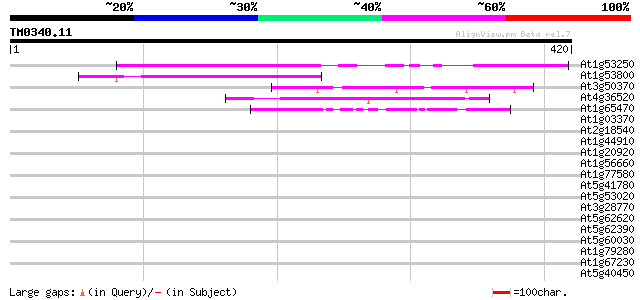
Score E
Sequences producing significant alignments: (bits) Value
At1g53250 hypothetical protein; similar to ESTs gb|T22311.1, gb|... 201 5e-52
At1g53800 unknown protein 114 1e-25
At3g50370 putative protein 47 2e-05
At4g36520 trichohyalin like protein 45 1e-04
At1g65470 hypothetical protein, 3' partial 43 4e-04
At1g03370 unknown protein 41 0.001
At2g18540 putative vicilin storage protein (globulin-like) 41 0.001
At1g44910 splicing factor like protein 40 0.003
At1g20920 putative RNA helicase 40 0.003
At1g56660 hypothetical protein 39 0.005
At1g77580 unknown protein 39 0.007
At5g41780 putative protein 38 0.009
At5g53020 putative protein 38 0.012
At3g28770 hypothetical protein 37 0.016
At5g62620 unknown protein 37 0.020
At5g62390 KED - like protein 36 0.035
At5g60030 KED - like protein 36 0.035
At1g79280 hypothetical protein 36 0.035
At1g67230 unknown protein 36 0.045
At5g40450 unknown protein 35 0.059
>At1g53250 hypothetical protein; similar to ESTs gb|T22311.1,
gb|AA585734.1, emb|F19896.1
Length = 363
Score = 201 bits (512), Expect = 5e-52
Identities = 129/338 (38%), Positives = 181/338 (53%), Gaps = 66/338 (19%)
Query: 81 VVGLSDSLEIESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALR 140
V ++ E +S S + +KE RR KIGLANKG+VPWNKGRKHS +TR RIK+RT EAL
Sbjct: 75 VKAMNKDTEADSDSDRKIKEEERRRKIGLANKGKVPWNKGRKHSEDTRRRIKQRTIEALT 134
Query: 141 EPKVRKKMAEHPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGG 200
PKVRKKM++H + HS++ KEKI S+++VW ER +SKR +E+ SW + IA AA+KGG
Sbjct: 135 NPKVRKKMSDHQQPHSNETKEKIRASVKQVWAERSRSKRLKEKFMSSWSENIAEAARKGG 194
Query: 201 CGQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGS 260
G+ ELDWDSY KIK+ +QL AEEK + K ++ +AKEA K +
Sbjct: 195 SGEAELDWDSYEKIKQDFSSEQLQLAEEKARAK------------EQTKMIAKEAAKART 242
Query: 261 GEKELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKD 320
++RR EK K +E K +E +I+ K K
Sbjct: 243 --------------------EKMRRAAEKKKEREE---KDRREGKIR---------KPKQ 270
Query: 321 LRERIKGRGNIKSQLCKNANEDGEAALEITQQFKLDTKLTKINISKNINSEVAREGGFSN 380
RE I + KL +LTKI+ K ++A
Sbjct: 271 EREN----------------------PTIASRSKLKKRLTKIHKKKTSLGKIAIGTDRVV 308
Query: 381 SIYPTYNKLDLEVIKREKLRKEASFADRIQAARLKKGN 418
S+ KLDL++I++E+ R + S AD+IQAA+ ++G+
Sbjct: 309 SVAAKLEKLDLDLIRKERTRGDISLADQIQAAKNQRGS 346
>At1g53800 unknown protein
Length = 568
Score = 114 bits (285), Expect = 1e-25
Identities = 61/184 (33%), Positives = 99/184 (53%), Gaps = 14/184 (7%)
Query: 52 ELQINVDDDSDNQSETSVDSSDYLNGG--DGVVGLSDSLEIESLSRKAVKERMRRFKIGL 109
E + +++ S S SS NG DG + D +E++RR +I
Sbjct: 67 ETKYPAQKENERSSSLSSASSKSSNGSADDGEEQVDD------------REKLRRMRISK 114
Query: 110 ANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRR 169
AN+G PWNKGRKHS ET ++I+ RT A+++PK++ K+A + + + KI +R
Sbjct: 115 ANRGNTPWNKGRKHSPETLQKIRERTKIAMQDPKIKMKLANLGHAQNKETRMKIGEGVRM 174
Query: 170 VWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEK 229
W R + ++ +E W+ +A AAK+G +EEL WDSYN + +Q +L+ L E++
Sbjct: 175 RWARRKERRKVQETCHFEWQNLLAEAAKQGYTDEEELQWDSYNILDQQNQLEWLESVEQR 234
Query: 230 EKEK 233
+ K
Sbjct: 235 KAIK 238
>At3g50370 putative protein
Length = 2152
Score = 47.4 bits (111), Expect = 2e-05
Identities = 58/229 (25%), Positives = 99/229 (42%), Gaps = 44/229 (19%)
Query: 197 KKGGCGQEELDWDSYNKIKEQLELQQLLQAEEK-----EKEKLMAIARAEKFIQSWSECL 251
KK Q E + +LE Q +Q EE+ E+E+++ +AR E+ E
Sbjct: 452 KKEALKQTEFHDPVRESFEAELERVQKMQEEERRRIIEEQERVIELARTEE------EER 505
Query: 252 AKEAKKGGSGEKELDWDSYEK-IQEEMFLLNQIRRTTE--KAKAKERARTKAEKEARIKA 308
+ A++ ++ L+ ++ E + E L RR E K+K +E+ R E+E R +A
Sbjct: 506 LRLAREQDERQRRLEEEAREAAFRNEQERLEATRRAEELRKSKEEEKHRLFMEEERRKQA 565
Query: 309 IKKVMLNQKKKDLRERIKGRGNIKSQLCKNAN----------------------EDGEAA 346
K QK +L E+I R ++ C +++ ED E
Sbjct: 566 AK-----QKLLELEEKISRRQAEAAKGCSSSSTISEDKFLDIVKEKDSADVVDWEDSERM 620
Query: 347 LE-ITQQFKLDTKLTKINISKNINSEVAREG--GFSNSIYPTYNKLDLE 392
++ IT LD + + N S+ +R+G GF + PT+ K D+E
Sbjct: 621 VDRITTSSTLDLSVPMRSFESNATSQFSRDGSFGFPDRQKPTWRKEDIE 669
>At4g36520 trichohyalin like protein
Length = 1432
Score = 44.7 bits (104), Expect = 1e-04
Identities = 50/203 (24%), Positives = 91/203 (44%), Gaps = 26/203 (12%)
Query: 162 KISYSLRRVWQE-RLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLEL 220
K++ L+R+ +E R+K R RE+ + + + K + + L
Sbjct: 624 KLNEPLKRMEEETRIKEARLREE-------------------NDRRERVAVEKAENEKRL 664
Query: 221 QQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDW---DSYEKIQEEM 277
+ L+ EEKE++ A +AE ++ E ++ ++EL+ +++EK +E
Sbjct: 665 KAALEQEEKERKIKEAREKAENERRAVEAREKAEQERKMKEQQELELQLKEAFEKEEENR 724
Query: 278 FLLNQIRRTTEKAKAKERARTKAEKEARIK-AIKKVMLNQKKKDLRERIKGRGNIKSQLC 336
+ EK + + AR K E E RIK A +K L Q+ K E+ + IK +
Sbjct: 725 RMREAFALEQEKERRIKEAREKEENERRIKEAREKAELEQRLKATLEQEEKERQIKERQE 784
Query: 337 KNANEDGEAALEITQQFKLDTKL 359
+ NE A E+ +Q + + KL
Sbjct: 785 REENE--RRAKEVLEQAENERKL 805
Score = 33.9 bits (76), Expect = 0.17
Identities = 56/259 (21%), Positives = 107/259 (40%), Gaps = 23/259 (8%)
Query: 91 ESLSRKAVKERMRR-FKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMA 149
E+ ++ RMR F + + R+ K + E RIK +A E +++ +
Sbjct: 715 EAFEKEEENRRMREAFALEQEKERRI---KEAREKEENERRIKEAREKAELEQRLKATLE 771
Query: 150 EHPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDW- 208
+ + +IKE+ QER +++R +++ EQ K Q+E +
Sbjct: 772 QEEK--ERQIKER---------QEREENERRAKEVL---EQAENERKLKEALEQKENERR 817
Query: 209 --DSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELD 266
++ K + + +L++ ++ EEKEK + A RAE + + +E + KE +
Sbjct: 818 LKETREKEENKKKLREAIELEEKEKRLIEAFERAEIERRLKEDLEQEEMRMRLQEAKERE 877
Query: 267 WDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEK--EARIKAIKKVMLNQKKKDLRER 324
E + + Q + E++ KER + EK E +A + N+ D E
Sbjct: 878 RLHRENQEHQENERKQHEYSGEESDEKERDACEMEKTCETTKEAHGEQSSNESLSDTLEE 937
Query: 325 IKGRGNIKSQLCKNANEDG 343
+ N S + E+G
Sbjct: 938 NESIDNDVSVNKQKKEEEG 956
Score = 32.0 bits (71), Expect = 0.66
Identities = 52/248 (20%), Positives = 101/248 (39%), Gaps = 13/248 (5%)
Query: 114 RVPWNKG-RKHSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRRVWQ 172
++P + G RK S+ + + +A EPK+ K + H +D + + R
Sbjct: 477 KLPKHSGTRKLSSRHKRHENKLAEKAPEEPKIEK--SRHVEMGND-LPDHGGIVKHRNLL 533
Query: 173 ERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKE 232
+ ++K F E+ ++ + K ++LD ++ K E Q+ + E+
Sbjct: 534 KPEENKLFTEKPAKQKKELLCEEKTKR-IQNQQLDKKTHQKAAETN--QECVYDWEQNAR 590
Query: 233 KLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRRTTEKAKA 292
KL E ++ E K + E + K EE + + R E +
Sbjct: 591 KLREALGNESTLEVSVELNGNGKKMEMRSQSETKLNEPLKRMEEETRIKEARLREENDRR 650
Query: 293 KERARTKAEKEARIK-AIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANEDGEAALEITQ 351
+ A KAE E R+K A+++ +K K+ RE+ + + A E E ++ +
Sbjct: 651 ERVAVEKAENEKRLKAALEQEEKERKIKEAREKAE-----NERRAVEAREKAEQERKMKE 705
Query: 352 QFKLDTKL 359
Q +L+ +L
Sbjct: 706 QQELELQL 713
>At1g65470 hypothetical protein, 3' partial
Length = 618
Score = 42.7 bits (99), Expect = 4e-04
Identities = 50/195 (25%), Positives = 93/195 (47%), Gaps = 24/195 (12%)
Query: 181 REQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIARA 240
RE+ W ++ AA+K G E+D S+ Q ++ + + K +EKL+ + +
Sbjct: 194 REETEKLWRSDLSKAAEKLGKILSEVDIRSFMDNMMQKNSSEMAEKDSKREEKLL-LKQL 252
Query: 241 EKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKA 300
EK + C A++ KK E+++ EK+Q+E + ++ +KA E + K
Sbjct: 253 EK-----NRCEAEKEKK--RMERQV---LKEKLQQE-----KEQKLLQKAIVDENNKEKE 297
Query: 301 EKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANEDGEAALEITQQFKLDTKLT 360
E E+R K IKK ++ +K+ + R K + +K QL + I ++F +K +
Sbjct: 298 ETESR-KRIKK-QQDESEKEQKRREKEQAELKKQL------QVQKQASIMERFLKKSKDS 349
Query: 361 KINISKNINSEVARE 375
+ K +SEV +
Sbjct: 350 SLTQPKLPSSEVTAQ 364
Score = 33.9 bits (76), Expect = 0.17
Identities = 20/60 (33%), Positives = 35/60 (58%), Gaps = 1/60 (1%)
Query: 89 EIESLSRKAVKERMRRFKIG-LANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKK 147
E + + R+ +KE++++ K L K V N K E+R+RIK++ E+ +E K R+K
Sbjct: 262 EKKRMERQVLKEKLQQEKEQKLLQKAIVDENNKEKEETESRKRIKKQQDESEKEQKRREK 321
>At1g03370 unknown protein
Length = 1859
Score = 41.2 bits (95), Expect = 0.001
Identities = 48/195 (24%), Positives = 88/195 (44%), Gaps = 29/195 (14%)
Query: 217 QLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKEL----DWDSYEK 272
+LEL+Q + +E E K+ A+ AE F +EAK+ GE+ L W+ +K
Sbjct: 588 RLELEQSILKKEMEAAKIKAVESAESF---------REAKE--RGERSLKDIHSWEG-QK 635
Query: 273 IQEEMFLLNQIRRTT----EKAKAKER-----ARTKAEKEARIKAIKKVMLNQKKKDLRE 323
I + L Q + T E KAK R A K E+ A+ K + L +K+ +
Sbjct: 636 IMLQEELKGQREKVTVLQKEVTKAKNRQNQIEAALKQERTAKGKLSAQASLIRKE---TK 692
Query: 324 RIKGRGNIKSQLCKNANEDGEAALEITQQFKLDTKLTKINISKNINSEVAREGGFSNSIY 383
++ G ++ + K E + I +L+ +++++ + + + +A + G S S
Sbjct: 693 ELEALGKVEEERIKGKAET-DVKYYIDNIKRLEREISELKLKSDYSRIIALKKGSSESKA 751
Query: 384 PTYNKLDLEVIKREK 398
L + +KRE+
Sbjct: 752 TKRESLGMPKVKRER 766
>At2g18540 putative vicilin storage protein (globulin-like)
Length = 699
Score = 40.8 bits (94), Expect = 0.001
Identities = 48/219 (21%), Positives = 91/219 (40%), Gaps = 25/219 (11%)
Query: 89 EIESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALR----EPKV 144
E+ L R+ ++ER RR + + + + A RE KRR E + E
Sbjct: 416 ELSKLMRE-IEERKRR------EEEEIERRRKEEEEARKREEAKRREEEEAKRREEEETE 468
Query: 145 RKKMAEHPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQE 204
RKK E ++ +++ +R +ER K + EQ E+
Sbjct: 469 RKKREEEEARKREEERKREEEEAKRREEERKKREEEAEQARKREEE-------------R 515
Query: 205 ELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKE 264
E + + K +E+ + ++ + E K +E+ R E+ + E +E + E+E
Sbjct: 516 EKEEEMAKKREEERQRKEREEVERKRREE-QERKRREEEARKREEERKREEEMAKRREQE 574
Query: 265 LDWDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKE 303
E+++ ++ + +R E AK +E+ R K E+E
Sbjct: 575 RQRKEREEVERKIREEQERKREEEMAKRREQERQKKERE 613
Score = 37.4 bits (85), Expect = 0.016
Identities = 67/293 (22%), Positives = 117/293 (39%), Gaps = 42/293 (14%)
Query: 129 ERIKRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSW 188
E KRR E + R+K E R + + + + RR +E + KR E+
Sbjct: 425 EERKRREEEEIER---RRKEEEEARKREEAKRREEEEAKRREEEETERKKREEEEARKRE 481
Query: 189 EQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWS 248
E+ +E + K +E+ Q + EE+EKE+ MA R E+
Sbjct: 482 EERKREE-------EEAKRREEERKKREEEAEQARKREEEREKEEEMAKKREEE------ 528
Query: 249 ECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKE----A 304
++ K+ E++ + K +EE + R+ E+ K +E + E+E
Sbjct: 529 ----RQRKEREEVERKRREEQERKRREE-----EARKREEERKREEEMAKRREQERQRKE 579
Query: 305 RIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANEDGEAALEITQQFKLDTKLTKINI 364
R + +K+ Q++K E K R + + K E+ E + K + ++ KI
Sbjct: 580 REEVERKIREEQERKREEEMAKRR---EQERQKKEREEMERKKREEEARKREEEMAKIRE 636
Query: 365 SKNINSEVAREGGFSNSIYPTYNKLDLEVIKREKLRK-EASFADRIQAARLKK 416
+ E RE + + E ++RE+ RK E A R + R KK
Sbjct: 637 EERQRKE--RED-------VERKRREEEAMRREEERKREEEAAKRAEEERRKK 680
Score = 30.0 bits (66), Expect = 2.5
Identities = 41/194 (21%), Positives = 76/194 (39%), Gaps = 32/194 (16%)
Query: 89 EIESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRR-----TSEALREPK 143
E E + RK +E+ R+ + A K R K + A+ RE+ ++R +RE +
Sbjct: 533 EREEVERKRREEQERKRREEEARK-REEERKREEEMAKRREQERQRKEREEVERKIREEQ 591
Query: 144 VRKKMAEHPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQ 203
RK+ E + + ++K + R +E KR E + +
Sbjct: 592 ERKREEEMAKRREQERQKKEREEMERKKREEEARKREEEMAKIR---------------E 636
Query: 204 EELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEK 263
EE ++ + ++ ++ EE+ K + A RAE +E +K E+
Sbjct: 637 EERQRKEREDVERKRREEEAMRREEERKREEEAAKRAE-----------EERRKKEEEEE 685
Query: 264 ELDWDSYEKIQEEM 277
+ W K EE+
Sbjct: 686 KRRWPPQPKPPEEI 699
>At1g44910 splicing factor like protein
Length = 958
Score = 39.7 bits (91), Expect = 0.003
Identities = 38/139 (27%), Positives = 64/139 (45%), Gaps = 13/139 (9%)
Query: 225 QAEEKEKEKLMAIARA-EKFIQSWSECLA----KEAKKGGSGEKELDWDSYEKIQEEMFL 279
+ EEKE KL +A + ++ E +++K+ +E E + + +F
Sbjct: 737 EKEEKEARKLQRLAEEFTNLLHTFKEITVASNWEDSKQLVEESQEYRSIGDESVSQGLFE 796
Query: 280 LNQIRRTTEKAKAKERAR----TKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQL 335
I EKAK KER R + EKE K +K ++++ RER K +G +S
Sbjct: 797 -EYITSLQEKAKEKERKRDEEKVRKEKERDEKEKRKDKDKERREKEREREKEKGKERS-- 853
Query: 336 CKNANEDGEAALEITQQFK 354
K DGE A+++++ K
Sbjct: 854 -KREESDGETAMDVSEGHK 871
>At1g20920 putative RNA helicase
Length = 1166
Score = 39.7 bits (91), Expect = 0.003
Identities = 49/232 (21%), Positives = 94/232 (40%), Gaps = 34/232 (14%)
Query: 119 KGRKHSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRRVWQERLKSK 178
K R+ + ER K + SE RE RKK + D ++ +R +ER + +
Sbjct: 24 KSRRDRDRSNERKKDKGSEKRREKDRRKKRVKSSDSEDDYDRDDDEEREKRKEKERERRR 83
Query: 179 RFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIA 238
R ++++ E+ +K +++++ + + E ++ + E+++ K
Sbjct: 84 RDKDRVKRRSER------RKSSDSEDDVEEEDERDKRRVNEKERGHREHERDRGKDRKRD 137
Query: 239 RAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERART 298
R +E +K E+E D + E+ +EE EK + KER R
Sbjct: 138 R------------EREERKDKEREREKDRERREREREE----------REKERVKERERR 175
Query: 299 KAEKEARIKAIKKVMLNQKKKDLRERIKGRGN------IKSQLCKNANEDGE 344
+ E R + ++ ++ RER + GN +K L + E GE
Sbjct: 176 EREDGERDRREREKERGSRRNRERERSREVGNEESDDDVKRDLKRRRKEGGE 227
Score = 28.1 bits (61), Expect = 9.5
Identities = 37/148 (25%), Positives = 57/148 (38%), Gaps = 11/148 (7%)
Query: 124 SAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRRV--WQERLKSKRFR 181
S+ + KR++ E E K +K E K+ E++ RRV WQE + K
Sbjct: 240 SSRHEDSPKRKSVEDNGEKKEKKTREEELEDEQKKLDEEVEKRRRRVQEWQELKRKKEEA 299
Query: 182 EQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAE 241
E S N K G E + D E+ ++ EE + E
Sbjct: 300 ES--ESKGDADGNEPKAGKAWTLEGESDDEEGHPEEKSETEMDVDEETKPEN----DGDA 353
Query: 242 KFIQSWSECLAKEAKKGGSG---EKELD 266
K + +E A ++ GG G E+E+D
Sbjct: 354 KMVDLENETAATVSESGGDGAVDEEEID 381
>At1g56660 hypothetical protein
Length = 522
Score = 38.9 bits (89), Expect = 0.005
Identities = 69/331 (20%), Positives = 135/331 (39%), Gaps = 59/331 (17%)
Query: 91 ESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMAE 150
E ++KA KE+ + + K + G+K+ + +E+ + T E ++PK KK E
Sbjct: 147 EEKNKKADKEK--KHEDVSQEKEELEEEDGKKN--KKKEKDESGTEEKKKKPKKEKKQKE 202
Query: 151 HPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDS 210
+ + DK ++K K+ +KG G +L+ +
Sbjct: 203 ESKSNEDK---------------KVKGKK-----------------EKGEKG--DLEKED 228
Query: 211 YNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSY 270
K KE E Q ++ ++ +K K + EK E A+E KK EK+ +S
Sbjct: 229 EEKKKEHDETDQEMKEKDSKKNK-----KKEK-----DESCAEEKKKKPDKEKKEKDEST 278
Query: 271 E----KIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIK 326
E K++ + + + E K KE T+ E + K+ KKK +++ K
Sbjct: 279 EKEDKKLKGKKGKGEKPEKEDEGKKTKEHDATEQEMDDEAADHKE----GKKKKNKDKAK 334
Query: 327 GRGNIKSQLCKNANEDGEAALEITQQFKLDTKLTKINISKNINSEVAREGGFSNSIYPTY 386
+ + ++C+ +D + T+Q K K K S+ +V + N +
Sbjct: 335 KKETVIDEVCEKETKDKDDDEGETKQKKNKKKEKK---SEKGEKDVKEDKKKENPLETEV 391
Query: 387 NKLDLEVIKREKLRKEASFADRIQAARLKKG 417
D+++ + E +KE + + ++++ G
Sbjct: 392 MSRDIKLEEPEAEKKEEDDTEEKKKSKVEGG 422
Score = 34.7 bits (78), Expect = 0.10
Identities = 54/274 (19%), Positives = 105/274 (37%), Gaps = 28/274 (10%)
Query: 91 ESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMAE 150
E +K KE+ + + ++ KG+ E + K+ E ++ + A+
Sbjct: 261 EEKKKKPDKEKKEKDESTEKEDKKLKGKKGKGEKPEKEDEGKKTKEHDATEQEMDDEAAD 320
Query: 151 HPRFHSDKIKEKISYS---LRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELD 207
H K K+K + V ++ K K E +K +E+
Sbjct: 321 HKEGKKKKNKDKAKKKETVIDEVCEKETKDKDDDE----------GETKQKKNKKKEKKS 370
Query: 208 WDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDW 267
+KE + + L+ E ++ + AEK + +E K +GG E+
Sbjct: 371 EKGEKDVKEDKKKENPLETEVMSRDIKLEEPEAEKKEEDDTEEKKKSKVEGGESEEGKKK 430
Query: 268 DSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAE--KEARIKAIKKVMLNQKKKDLRERI 325
+K + + ++ T++ K E K + K+ +I+ K +K KD++++
Sbjct: 431 KKKDKKKNK-------KKDTKEPKMTEDEEEKKDDSKDVKIEG-SKAKEEKKDKDVKKKK 482
Query: 326 KGR--GNIKSQLCKNANEDG---EAALEITQQFK 354
G G +K++L K + G E EI Q K
Sbjct: 483 GGNDIGKLKTKLAKIDEKIGALMEEKAEIENQIK 516
>At1g77580 unknown protein
Length = 779
Score = 38.5 bits (88), Expect = 0.007
Identities = 41/156 (26%), Positives = 77/156 (49%), Gaps = 15/156 (9%)
Query: 219 ELQQLLQAEEKEKEKLMAIARA--EKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQ-E 275
EL++ L+ E EKE+L + + EK + LA E + S KEL+ + EK++ E
Sbjct: 396 ELEEKLEKLEAEKEELKSEVKCNREKAVVHVENSLAAEIEVLTSRTKELE-EQLEKLEAE 454
Query: 276 EMFLLNQIRRTTEKAKAKERARTKAEKE---ARIKAIKKVM--LNQKKKDLRERIKGRGN 330
++ L ++++ E+A A+ E E RIK +++ + L +K +L+ +K
Sbjct: 455 KVELESEVKCNREEAVAQVENSLATEIEVLTCRIKQLEEKLEKLEVEKDELKSEVKCNRE 514
Query: 331 IKSQLCKNANEDGEAALEITQQFKLDTKLTKINISK 366
++S L E ++ +L+ KL K+ + K
Sbjct: 515 VESTL------RFELEAIACEKMELENKLEKLEVEK 544
>At5g41780 putative protein
Length = 537
Score = 38.1 bits (87), Expect = 0.009
Identities = 51/251 (20%), Positives = 104/251 (41%), Gaps = 34/251 (13%)
Query: 125 AETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSL------RRVWQERLKSK 178
+ET+ R+KR E + K K+ + +K+ +K+ + R+ + E +KSK
Sbjct: 182 SETQMRLKRLEEETEKRAKAEMKIVKEKEALWNKV-QKLEAGVDTFRKKRKEFNEEMKSK 240
Query: 179 RFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLM--- 235
Q + I K ++L++ ++KEQ ++ Q L E K+++KL+
Sbjct: 241 ITENQKLHTKIAVIDEIEDKS----KKLEY----QVKEQEDIIQRLSMEIKDQKKLLKEQ 292
Query: 236 -----AIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLN-------QI 283
+ +K ++ WS +EL D K+++ + +L+ QI
Sbjct: 293 KDAIDKFSEDQKLMKRWSFGSKLNTNLLEKKMEELAEDFRMKMEDHIRILHRRIHVAEQI 352
Query: 284 RRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANEDG 343
++ + K R T+ E+ +A+ + + K+ + + + G K E G
Sbjct: 353 HLESKSSYIKTRDNTQTEENRGNRAVSETQFKKIKEMVEQGLAG----PEMAIKKLEESG 408
Query: 344 EAALEITQQFK 354
E +T+ K
Sbjct: 409 ELGNRVTRLAK 419
Score = 28.9 bits (63), Expect = 5.5
Identities = 51/211 (24%), Positives = 84/211 (39%), Gaps = 35/211 (16%)
Query: 144 VRKKMAEHPRFHSDKIKEKISYSLRRVW---QERLKSKRFREQIFLSWEQCIANAAKKGG 200
+ KKM E K+++ I RR+ Q L+SK S+ + N +
Sbjct: 320 LEKKMEELAEDFRMKMEDHIRILHRRIHVAEQIHLESKS-------SYIKTRDNTQTEEN 372
Query: 201 CGQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSEC--LAKEAKKG 258
G + + KIKE +E Q L E MAI + E+ + + LAKE
Sbjct: 373 RGNRAVSETQFKKIKEMVE--QGLAGPE------MAIKKLEESGELGNRVTRLAKEIDSA 424
Query: 259 GSGEKELDWDSYEKIQ-----------EEMFLLNQIRRTTEKAKAKERARTKAEKEARIK 307
KE D + +++ +E L ++ + +AK E K ++
Sbjct: 425 RKWVKEKDNNMKHEVETLEAKLECREAQESLLKEKLSKL--EAKLAEEGTEKLSLSKAMR 482
Query: 308 AIKKVMLNQKKKDLRERIKGRGNIKS--QLC 336
IKK+ +N K+K+ G G ++ QLC
Sbjct: 483 KIKKLEINVKEKEFELLSLGEGKREAIRQLC 513
>At5g53020 putative protein
Length = 720
Score = 37.7 bits (86), Expect = 0.012
Identities = 51/214 (23%), Positives = 94/214 (43%), Gaps = 40/214 (18%)
Query: 216 EQLELQQLLQAEEKEKEKLMAIARAEKFIQSWS----------ECLAKEAKKGGSGEKEL 265
E EL+++ + K EK+++I ++K Q+W E L KE + ++
Sbjct: 43 EITELKKVRNDDAKANEKVVSIIASQK--QNWLRERYGLRLQIEALMKELRNIEKRKRHS 100
Query: 266 DWDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERI 325
+ E+++E+ LL + +KA +E+ + + +E +KA K+V +DLRE
Sbjct: 101 LLELQERLKEKEGLLE----SKDKAIEEEKRKCELLEERLVKAEKEV------QDLRET- 149
Query: 326 KGRGNIKSQLCKNANEDGEAALEITQQFKLDTKLTKINISKNINSEVAREGGFSNSIYPT 385
+ E + E+ +Q K T L + + + +E++R +
Sbjct: 150 ------------QERDVQEHSSELWRQKK--TFLELASSQRQLEAELSRANKQIEAKGHE 195
Query: 386 YNKLDLEVIKREKLRKEASFADRIQAARLKKGNL 419
L LE+ K+RK+ DRI A +KK L
Sbjct: 196 LEDLSLEI---NKMRKDLEQKDRILAVMMKKSKL 226
>At3g28770 hypothetical protein
Length = 2081
Score = 37.4 bits (85), Expect = 0.016
Identities = 52/265 (19%), Positives = 117/265 (43%), Gaps = 19/265 (7%)
Query: 118 NKGRKHSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRRVWQER--- 174
NK +K S ++ + + + ++ K +++ + + DK +E+ R+ +E+
Sbjct: 996 NKEKKESEDSASKNREKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEES 1055
Query: 175 --LKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKE 232
LK+K+ +E+ ++ + +KK +E D S K +++ E ++ +++ ++KE
Sbjct: 1056 RDLKAKK-KEEETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKE 1114
Query: 233 KLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRRTTEKAKA 292
+ +K E KK EK+ + K+ ++ + + EK++
Sbjct: 1115 E-------DKKDMEKLEDQNSNKKKEDKNEKKKS--QHVKLVKKESDKKEKKENEEKSET 1165
Query: 293 KERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANEDGEAALEITQQ 352
KE +K++K K KK +Q+KK +E + + + +L KN ED + + +
Sbjct: 1166 KEIESSKSQKNEVDKKEKKSSKDQQKKKEKEM---KESEEKKLKKN-EEDRKKQTSVEEN 1221
Query: 353 FKLDTKLTKINISKNINSEVAREGG 377
K + N K+ ++ G
Sbjct: 1222 KKQKETKKEKNKPKDDKKNTTKQSG 1246
Score = 34.3 bits (77), Expect = 0.13
Identities = 68/351 (19%), Positives = 134/351 (37%), Gaps = 44/351 (12%)
Query: 59 DDSDNQSETSVDSSDYLNGGDGVVGLSDSLEIESLSRKAVKERMRRFKIGLANKGRVPWN 118
+ DN+ +S ++ D G D E + KE+ G V N
Sbjct: 800 ETKDNKKLSSTENRDEAKERSGEDNKEDKEESKDYQSVEAKEKNEN--------GGVDTN 851
Query: 119 KGRKHSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKE------KISYSLRRVWQ 172
G K ++ + + +A +E ++KK E R KE + +++
Sbjct: 852 VGNKEDSKDLKDDRSVEVKANKEESMKKKREEVQRNDKSSTKEVRDFANNMDIDVQKGSG 911
Query: 173 ERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKE 232
E +K K+ ++ G +E + D+ N +Q ++K+K+
Sbjct: 912 ESVKYKKDEKKE-----------------GNKEENKDTINTSSKQKG------KDKKKKK 948
Query: 233 KLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRRTTEKAKA 292
K + +K + E + E KK +KE K++EE + EK ++
Sbjct: 949 KESKNSNMKKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEEN------KDNKEKKES 1002
Query: 293 KERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANEDGEAALEITQQ 352
++ A EK+ + K KK+ + + K R S+ K+ E E+ ++
Sbjct: 1003 EDSASKNREKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKK 1062
Query: 353 FKLDTKLTKINIS-KNINSEVAREGGFSNSIYPTYNKLDLEVIKREKLRKE 402
+ +TK K + + K+ E +E + S+ +K + + + K RK+
Sbjct: 1063 KEEETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKK 1113
Score = 33.5 bits (75), Expect = 0.23
Identities = 63/353 (17%), Positives = 139/353 (38%), Gaps = 41/353 (11%)
Query: 63 NQSETSVDSSDYLNGGDGVVGLSDSLEIESLSRKAVKERMRRFKIGLANKGRVPWNKGRK 122
N + + + + G +G +S++ E+L K K+ ++ + + G K
Sbjct: 577 NDGDHTKEKREETQGNNG-----ESVKNENLENKEDKKELKDDE-----------SVGAK 620
Query: 123 HSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRRVWQERLKSKR-FR 181
+ ET KR ++ + + K+ ++ ++D KEK + ++SK +
Sbjct: 621 TNNETSLEEKREQTQKGHDNSINSKIVDNKGGNADSNKEKEVHVGDSTNDNNMESKEDTK 680
Query: 182 EQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAE 241
++ + +++KG G+E + DS K + + Q ++K + A+
Sbjct: 681 SEVEVKKND---GSSEKGEEGKEN-NKDSMEDKKLENKESQTDSKDDKSVDDKQEEAQIY 736
Query: 242 KFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAE 301
+ + + KK S E + + +++ + + ++ +EK + E+ +K
Sbjct: 737 GGESKDDKSVEAKGKKKESKENKKTKTNENRVRNKEENVQGNKKESEKVEKGEKKESKDA 796
Query: 302 KEARIKAIKKVMLNQKKKDLRERIKGRGN---------IKSQLCKNANEDG--------- 343
K K KK+ + + + +ER G N +S K NE+G
Sbjct: 797 KSVETKDNKKLSSTENRDEAKER-SGEDNKEDKEESKDYQSVEAKEKNENGGVDTNVGNK 855
Query: 344 EAALEITQQFKLDTKLTKINISKNINSEVAR-EGGFSNSIYPTYNKLDLEVIK 395
E + ++ ++ K K K EV R + + + N +D++V K
Sbjct: 856 EDSKDLKDDRSVEVKANKEESMKKKREEVQRNDKSSTKEVRDFANNMDIDVQK 908
>At5g62620 unknown protein
Length = 681
Score = 37.0 bits (84), Expect = 0.020
Identities = 35/133 (26%), Positives = 61/133 (45%), Gaps = 24/133 (18%)
Query: 105 FKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKEKIS 164
FK GL++ + P + KH+++ + +R + L+ ++ +E P +
Sbjct: 49 FKTGLSSLSQDPLTRPEKHNSQRELQERRAPTRPLKSLLYQESQSESP-----------A 97
Query: 165 YSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQ----EELDWDSYNKIKEQLEL 220
LRR + L S RF + F N + K G + ++ W+ KI E+LE
Sbjct: 98 QGLRRRTRI-LSSLRFDPETF--------NPSSKDGSVELHKSAKVAWEVGRKIWEELES 148
Query: 221 QQLLQAEEKEKEK 233
+ L+A EKEK+K
Sbjct: 149 GKTLKALEKEKKK 161
>At5g62390 KED - like protein
Length = 446
Score = 36.2 bits (82), Expect = 0.035
Identities = 46/178 (25%), Positives = 81/178 (44%), Gaps = 36/178 (20%)
Query: 200 GCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGG 259
G G+ + W + K ++ E L A +KEK K AIA AE E KK
Sbjct: 151 GSGERKYRWTTEIKGGKKDEEGLKLAALKKEKAKAKAIAAAE-----------AEKKKNK 199
Query: 260 SGEKELDWDSYEKIQEEMFLLNQ---IRRTT--------EKAKAKERARTKAEKEARIKA 308
+ +K +W + K + E ++ I+ TT E+ + KE+ TK++K+ + +
Sbjct: 200 NKKKSYNWTTEVKSERENGEVSHTYIIKATTGGEKKKKHEEKEKKEKIETKSKKKEKTRV 259
Query: 309 IKKVMLNQKKKD----------LRERIKGR-GNIKSQLCKNANEDGE-AALEITQQFK 354
+ V+ ++++D LR+ R G ++++ KN E AA+ I + FK
Sbjct: 260 V--VIEEEEEEDDESSEHGAIVLRKAFSRRNGAVRTKKGKNKEMPPEYAAVMIQRAFK 315
>At5g60030 KED - like protein
Length = 292
Score = 36.2 bits (82), Expect = 0.035
Identities = 35/152 (23%), Positives = 68/152 (44%), Gaps = 8/152 (5%)
Query: 211 YNKIKEQLELQQLLQAE---EKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDW 267
Y + +++ ++++ A+ EK EKL A R+E+ + E K+ KK E +D
Sbjct: 111 YGRERDEKKMKKSKDADVVDEKVNEKLEAEQRSEERRERKKE---KKKKKNNKDEDVVDE 167
Query: 268 DSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKG 327
EK+++E ++ R +K+K EKE K + +KKK+ E +
Sbjct: 168 KVKEKLEDEQKSADRKERKKKKSKKNNDEDVVDEKEKLEDEQKSAEIKEKKKNKDEDVVD 227
Query: 328 RGNIKSQLCKNANEDGEAALEITQQFKLDTKL 359
+ + ++ GE E ++ K D ++
Sbjct: 228 EK--EKEKLEDEQRSGERKKEKKKKRKSDEEI 257
>At1g79280 hypothetical protein
Length = 2111
Score = 36.2 bits (82), Expect = 0.035
Identities = 61/295 (20%), Positives = 122/295 (40%), Gaps = 33/295 (11%)
Query: 119 KGRKHSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRRVWQ--ERLK 176
K R S +K + +E K +K+ H ++ E LR ++ +
Sbjct: 1322 KARMESENFENLLKTKQTELDLCMKEMEKLRMETDLHKKRVDE-----LRETYRNIDIAD 1376
Query: 177 SKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLMA 236
R ++++ E+ A A C + L+ + + E+ EL + + +++L
Sbjct: 1377 YNRLKDEVRQLEEKLKAKDAHAEDCKKVLLEKQNKISLLEK-ELTNCKKDLSEREKRLDD 1435
Query: 237 IARAEKFIQSWSECLAKEAKKGGSGEKELDWDS--YEKIQEEMFLLNQI----------- 283
+A+ +QS +E +K L+ YEK ++E+ NQ
Sbjct: 1436 AQQAQATMQSEFNKQKQELEKNKKIHYTLNMTKRKYEKEKDELSKQNQSLAKQLEEAKEE 1495
Query: 284 --RRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANE 341
+RTT A ++ + + EKE RI+ + K ++Q K ++R++ + +L K +E
Sbjct: 1496 AGKRTTTDAVVEQSVKEREEKEKRIQILDK-YVHQLKDEVRKKTEDLKKKDEELTKERSE 1554
Query: 342 DGEAALEITQQFKLDTKLTKINISK-NINSEVAREGGFSNSIYPTYNKLDLEVIK 395
E+ LTKI K ++ E+A+ + ++ T+ +LE +K
Sbjct: 1555 RKSVEKEV------GDSLTKIKKEKTKVDEELAKLERYQTAL--THLSEELEKLK 1601
Score = 30.4 bits (67), Expect = 1.9
Identities = 47/223 (21%), Positives = 89/223 (39%), Gaps = 30/223 (13%)
Query: 115 VPWNKGRKHSAETRERIKRRTSEALRE---------PKVRKKMAEHPRFHSDKIKEKISY 165
VP K H E E +R E E K R ++ R DK+ + ++
Sbjct: 665 VPGRKNFLHLLEDSEEATKRAQEKAFERIRILEEDFAKARSEVIAI-RSERDKLAMEANF 723
Query: 166 SLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQ 225
+ ++ +S+R RE++ N+ +L D K++E E L
Sbjct: 724 AREKLEGIMKESERKREEM---------NSVLARNIEFSQLIIDHQRKLRESSE--SLHA 772
Query: 226 AEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRR 285
AEE ++ M ++ ++ E L+ K+ L Y ++Q + + +
Sbjct: 773 AEEISRKLSMEVS----VLKQEKELLSNAEKRASDEVSALSQRVY-RLQATLDTV----Q 823
Query: 286 TTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGR 328
+TE+ + + RA + ++E IK +++ KK+ ER R
Sbjct: 824 STEEVREETRAAERRKQEEHIKQLQREWAEAKKELQEERSNAR 866
>At1g67230 unknown protein
Length = 1132
Score = 35.8 bits (81), Expect = 0.045
Identities = 47/207 (22%), Positives = 84/207 (39%), Gaps = 24/207 (11%)
Query: 213 KIKEQLEL----QQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWD 268
++KEQ+E Q+LLQ E ++ + A+ E F + W E ++AK G E + D
Sbjct: 492 ELKEQIEKCRSQQELLQKEAEDLK-----AQRESFEKEWEELDERKAKIG--NELKNITD 544
Query: 269 SYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGR 328
EK++ + L + K K+ A E+E + K + + R + +
Sbjct: 545 QKEKLERHIHLEEE-----RLKKEKQAANENMERELETLEVAKASFAETMEYERSMLSKK 599
Query: 329 GNIKSQLCKNANEDGEAALEITQQFKLDTKLTKINISKNINSEVAREGGFSNSIYPTYNK 388
+ + E + LE Q L+ K ++ K + E RE SN Y
Sbjct: 600 AESERSQLLHDIEMRKRKLESDMQTILEEKERELQAKKKLFEE-EREKELSNINY----- 653
Query: 389 LDLEVIKREKLRKEASFADRIQAARLK 415
L + R ++ + RI+ +L+
Sbjct: 654 --LRDVARREMMDMQNERQRIEKEKLE 678
>At5g40450 unknown protein
Length = 2910
Score = 35.4 bits (80), Expect = 0.059
Identities = 33/148 (22%), Positives = 63/148 (42%), Gaps = 3/148 (2%)
Query: 227 EEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRRT 286
E+K++E + A+ + E A + E+E + +QE++ +++
Sbjct: 2580 EDKKEETVDALITNVQVQDQPKEDFEAAAIEKEISEQEHKLNDLTDVQEDIGTYVKVQVP 2639
Query: 287 TEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANEDGEAA 346
++ K A+KE +K + K ++ + IK +++ + + N D EAA
Sbjct: 2640 DDEIKGDGHDSVAAQKEETSSIEEKREVEHVKAEMEDAIKHEVSVEEKNNTSENIDHEAA 2699
Query: 347 LEITQQFKLDTKLTKINI---SKNINSE 371
EI Q+ T + K I K IN E
Sbjct: 2700 KEIEQEEGKQTNIVKEEIREEEKEINQE 2727
Score = 35.0 bits (79), Expect = 0.077
Identities = 49/223 (21%), Positives = 87/223 (38%), Gaps = 46/223 (20%)
Query: 213 KIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLA----------KEAKKGGSGE 262
K KE++ + Q+ ++ K + +A S +E + KEA E
Sbjct: 1090 KAKEEVPMLQIKNEDDATKIHETRVEQARDIGPSLTEICSINQNQPEEQVKEACSKEEQE 1149
Query: 263 KELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLR 322
KE+ +S E I E + L+ + A+E T E ++ K V+L +K++
Sbjct: 1150 KEISTNS-ENIVNETYALHSVEA------AEEETATNGESLDDVETTKSVLLEVRKEEEE 1202
Query: 323 ERIKGRGNIKSQLCKNANEDGEAALEITQQFKLDTKLTKINISKNINSE--VAREGGF-- 378
+K D E L+ ++ +L+T T + +K +N+E A E
Sbjct: 1203 AEMK--------------TDAEPRLDAIEKEELETVKTVVQDAKIVNNEETTAHESESLK 1248
Query: 379 --------SNSIYPTYNKLDLEVIKRE---KLRKEASFADRIQ 410
+ + T N D E I RE +EA ++I+
Sbjct: 1249 GDNHQEKNAEPVEATQNLDDAEQISREVTVDTEREADITEKIE 1291
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.130 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,030,444
Number of Sequences: 26719
Number of extensions: 385675
Number of successful extensions: 1932
Number of sequences better than 10.0: 170
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 149
Number of HSP's that attempted gapping in prelim test: 1661
Number of HSP's gapped (non-prelim): 285
length of query: 420
length of database: 11,318,596
effective HSP length: 102
effective length of query: 318
effective length of database: 8,593,258
effective search space: 2732656044
effective search space used: 2732656044
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0340.11


