

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0320b.3
(224 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
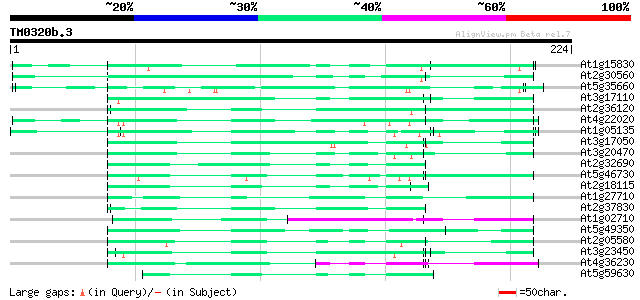
Score E
Sequences producing significant alignments: (bits) Value
At1g15830 hypothetical protein 60 1e-09
At2g30560 putative glycine-rich protein 58 4e-09
At5g35660 unknown protein 57 8e-09
At3g17110 hypothetical protein 55 3e-08
At2g36120 unknown protein 55 3e-08
At4g22020 glycine-rich protein 55 4e-08
At1g05135 unknown protein 55 4e-08
At3g17050 unknown protein 54 7e-08
At3g20470 glycine-rich protein, putative 54 9e-08
At2g32690 unknown protein 51 4e-07
At5g46730 unknown protein 49 2e-06
At2g18115 putative protein 49 3e-06
At1g27710 unknown protein 48 4e-06
At2g37830 hypothetical protein 48 5e-06
At1g02710 hypothetical protein 47 6e-06
At5g49350 unknown protein 47 8e-06
At2g05580 unknown protein 47 8e-06
At3g23450 hypothetical protein 47 1e-05
At4g36230 putative glycine-rich cell wall protein 46 2e-05
At5g59630 glycine-rich protein - like 45 4e-05
>At1g15830 hypothetical protein
Length = 483
Score = 59.7 bits (143), Expect = 1e-09
Identities = 55/176 (31%), Positives = 65/176 (36%), Gaps = 42/176 (23%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+GGG V G GGG G +V+ GCGGG A
Sbjct: 338 GTGGGEAGAGMQVMQGWGGGGSGAATQVMQ---------------------GCGGGDAGA 376
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
+ A V V+ GCGGG GGGDGGG G G G GGG
Sbjct: 377 ITQVMQGWGGGGAGAVTQVMQ--GCGGGG-------GGGDGGGGQ------GTGIGGGGG 421
Query: 160 GDYG---GGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGG---GGGANSG 209
G+ G GGG P+ +G + SDGGGG GGG +G
Sbjct: 422 GEQGTGVGGGGDTCTQVTHGGGGAPLTMIGGGGGEQGVTGSDGGGGRGRGGGKVAG 477
Score = 49.3 bits (116), Expect = 2e-06
Identities = 51/169 (30%), Positives = 60/169 (35%), Gaps = 57/169 (33%)
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEG--GGNGG 59
G GGG+ G + V GW GGG AV ++G GG GG
Sbjct: 368 GCGGGDAG------AITQVMQGW----------------GGGGAGAVTQVMQGCGGGGGG 405
Query: 60 GGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVL 119
G GGG + + GGG GG + VG + V L
Sbjct: 406 GDGGGGQGTGI------------------GGGGGGEQGTGVGGGGDTCTQVTHGGGGAPL 447
Query: 120 ATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDK 168
IG GGG + V G DGGG G G GGG GGG K
Sbjct: 448 TMIGGGGGEQGVT----GSDGGG--------GRG---RGGGKVAGGGKK 481
Score = 47.4 bits (111), Expect = 6e-06
Identities = 48/171 (28%), Positives = 53/171 (30%), Gaps = 41/171 (23%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G G T GG G GG G + + V I GG G G +
Sbjct: 299 GGGRGSTGEGVTDGGGRTGNKGGNGGSIKIGVGTNGITGGTG-------GGEAGAGMQVM 351
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G A M GCGGG G GGG Q GCG GGG
Sbjct: 352 QGWGGGGSGAATQVMQ-------GCGGGDAGAITQVMQGWGGGGAGAVTQVMQGCGGGGG 404
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGV 210
G GGGG + GGGGGG +GV
Sbjct: 405 GGDGGGGQGTGI---------------------------GGGGGGEQGTGV 428
Score = 31.2 bits (69), Expect = 0.46
Identities = 24/64 (37%), Positives = 30/64 (46%), Gaps = 7/64 (10%)
Query: 151 GGGCG--VGGGGDYGGGGDKVVVAAIMVVV-ATPVVTVGRWWRRRRRR*SDGGGG----G 203
GGGC + GG + GGGD V++ + A V V R RR+ DGGG
Sbjct: 20 GGGCPMTISGGRLFTGGGDPVIITGGRLSTGAAGGVNVDRSKGGGRRKTGDGGGDPVVIS 79
Query: 204 GGAN 207
GG N
Sbjct: 80 GGEN 83
>At2g30560 putative glycine-rich protein
Length = 171
Score = 58.2 bits (139), Expect = 4e-09
Identities = 56/167 (33%), Positives = 64/167 (37%), Gaps = 52/167 (31%)
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGG 61
GGGG GGR G GGGG A GG +GGGG
Sbjct: 17 GGGGSGGGR-----------------------------GGGGGGGAKGGCGGGGKSGGGG 47
Query: 62 GGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLAT 121
GGG +V S + R + GG GGG+ G + A +
Sbjct: 48 GGGGYMVAPGSNRSSYISRDNFESDPKGGSGGGGKGGGGGGGISGGGAGGKS-------- 99
Query: 122 IGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYG--GGG 166
GCGGG GGG GGG + GGGCG GGGG G GGG
Sbjct: 100 -GCGGGKS------GGGGGGG------KNGGGCGGGGGGKGGKSGGG 133
Score = 55.5 bits (132), Expect = 2e-08
Identities = 50/170 (29%), Positives = 57/170 (33%), Gaps = 60/170 (35%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
GSGGGGK GGG GGGGGGG + GGCGGG +
Sbjct: 8 GSGGGGKGGGGGGSGGGRGGGGGGGAK-----------------------GGCGGGGKSG 44
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G + + ++ K + GGG GGG GG G G G
Sbjct: 45 GGGGGGGYMVAPGSNRSSYISRDNFESDPKGGSG--GGGKGGGGG------GGISGGGAG 96
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G G GG K GGGGGGG N G
Sbjct: 97 GKSGCGGGK-----------------------------SGGGGGGGKNGG 117
Score = 39.3 bits (90), Expect = 0.002
Identities = 32/103 (31%), Positives = 35/103 (33%), Gaps = 41/103 (39%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
GSGGGGK GGG GGG GG GCGGG++
Sbjct: 76 GSGGGGKGGGG---GGGISGGGAGGK------------------------SGCGGGKSGG 108
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGG 142
G + GCGGG GGG GGG
Sbjct: 109 GGGGGKNGG--------------GCGGGGGGKGGKSGGGSGGG 137
Score = 39.3 bits (90), Expect = 0.002
Identities = 33/94 (35%), Positives = 37/94 (39%), Gaps = 20/94 (21%)
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPV 182
G GGG GGG GGG GGCG GG GGGG +VA P
Sbjct: 13 GKGGGGGGSGGGRGGGGGGGA-------KGGCGGGGKSGGGGGGGGYMVA--------PG 57
Query: 183 VTVGRWWRRRR-----RR*SDGGGGGGGANSGVS 211
+ R + S GGG GGG G+S
Sbjct: 58 SNRSSYISRDNFESDPKGGSGGGGKGGGGGGGIS 91
Score = 30.8 bits (68), Expect = 0.60
Identities = 22/58 (37%), Positives = 22/58 (37%), Gaps = 26/58 (44%)
Query: 152 GGCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GG G GGGG GGGG GR GGGGGGGA G
Sbjct: 5 GGSGSGGGGKGGGGGGS---------------GGGR-----------GGGGGGGAKGG 36
Score = 30.0 bits (66), Expect = 1.0
Identities = 15/29 (51%), Positives = 17/29 (57%), Gaps = 7/29 (24%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVV 68
G GGGGK GG +GGG GGG +V
Sbjct: 120 GGGGGGK-------GGKSGGGSGGGGYMV 141
>At5g35660 unknown protein
Length = 343
Score = 57.0 bits (136), Expect = 8e-09
Identities = 68/216 (31%), Positives = 76/216 (34%), Gaps = 84/216 (38%)
Query: 3 GGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGGG 62
GGGG+GG GW W GGGG GGG GGGGG
Sbjct: 95 GGGGHGG-------------GWKGGGGHGGGW------KGGGGHGGGWKGGGGGRGGGGG 135
Query: 63 GGDRVVVV----VSVVAIEWWRR-LWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVV 117
GG + V S ++ +R W+ GGG GGG G
Sbjct: 136 GGAELETVEIDSASDEDVQNYRGGHGGGWKEGGGQGGGWKGGGGQ--------------- 180
Query: 118 VLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVG-------GGGDYGGGGDKVV 170
GGG K GGG GGGW + GGG G G GGG GGGG
Sbjct: 181 -------GGGWKG-----GGGQGGGW-----KGGGGQGGGWKGGGGQGGGWKGGGGQ--- 220
Query: 171 VAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGA 206
G W+ R GGGGGGGA
Sbjct: 221 ---------------GGGWKGGGGR---GGGGGGGA 238
Score = 56.6 bits (135), Expect = 1e-08
Identities = 65/203 (32%), Positives = 71/203 (34%), Gaps = 55/203 (27%)
Query: 3 GGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGGG 62
GGGG GG GW + W GGGG+ GGG GGGGG
Sbjct: 196 GGGGQGG-------------GWKGGGGQGGGW------KGGGGQG-GGWKGGGGRGGGGG 235
Query: 63 GGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLATI 122
GG + A E + W GGG GGG G
Sbjct: 236 GGAELENEEINGASEEIQ----WQGGGGGHGGGWQGGGGGHGGGWQGGGGGHGGGWQGGG 291
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPV 182
G GGG K GGG GGGW + GG G GGGG + GGG
Sbjct: 292 GRGGGWKG-----GGGHGGGWQGGGGRGGGWKGGGGGGGWRGGGG--------------- 331
Query: 183 VTVGRWWRRRRRR*SDGGGGGGG 205
G WR GGGGGGG
Sbjct: 332 ---GGGWR--------GGGGGGG 343
Score = 52.0 bits (123), Expect = 3e-07
Identities = 66/220 (30%), Positives = 73/220 (33%), Gaps = 81/220 (36%)
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGG- 60
GGGGG G L V + + + G GGG K EGGG GGG
Sbjct: 131 GGGGGGGAELETVEIDSASDEDVQNYRG----------GHGGGWK------EGGGQGGGW 174
Query: 61 -GGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVL 119
GGGG W+ GGG GGG G
Sbjct: 175 KGGGGQ-----------------GGGWKGGGGQGGGWKGGGGQ----------------- 200
Query: 120 ATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVA 179
GGG K GGG GGGW Q GG G GG G GGGG ++ I
Sbjct: 201 -----GGGWKG-----GGGQGGGWKGGGGQGGGWKGGGGRGGGGGGGAELENEEI----- 245
Query: 180 TPVVTVGRWWRRRRRR*SDGGGG------GGGANSGVSWQ 213
+W GGGG GGG G WQ
Sbjct: 246 NGASEEIQW--------QGGGGGHGGGWQGGGGGHGGGWQ 277
Score = 50.4 bits (119), Expect = 7e-07
Identities = 63/211 (29%), Positives = 68/211 (31%), Gaps = 88/211 (41%)
Query: 3 GGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGGG 62
GGGG GG GW + W GGGG+ GG GGGG
Sbjct: 186 GGGGQGG-------------GWKGGGGQGGGW------KGGGGQG------GGWKGGGGQ 220
Query: 63 GGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLATI 122
GG W+ GGG GGG G A + A I
Sbjct: 221 GGG--------------------WKGGGGRGGGGG---GGAELENEEINGAS-----EEI 252
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPV 182
GG GGG GGGW GGG G GGG GGGG
Sbjct: 253 QWQGG--------GGGHGGGWQ------GGGGGHGGGWQGGGGGH--------------- 283
Query: 183 VTVGRWWRRRRRR*SDGGGGGGGANSGVSWQ 213
G W+ R GGG GG G WQ
Sbjct: 284 ---GGGWQGGGGR---GGGWKGGGGHGGGWQ 308
Score = 50.1 bits (118), Expect = 1e-06
Identities = 59/181 (32%), Positives = 67/181 (36%), Gaps = 50/181 (27%)
Query: 40 GSGGGGKAVAATVEGGGNGGG--GGGGDRVVVVVSVVAIEWWR----RLWWWWR*GGGCG 93
G GGG K GGG+GGG GGGG W+ W GGG G
Sbjct: 88 GHGGGWKG------GGGHGGGWKGGGGHG----------GGWKGGGGHGGGWKGGGGGRG 131
Query: 94 GGRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGG 153
GG G +V A+ V G GGG K GGG GGGW Q GG
Sbjct: 132 GGGGG--GAELETVEIDSASDEDVQNYRGGHGGGWKE-----GGGQGGGWKGGGGQGGGW 184
Query: 154 CGVGG-GGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGVSW 212
G GG GG + GGG + G W+ GGG GG G W
Sbjct: 185 KGGGGQGGGWKGGGGQ-----------------GGGWKGGG---GQGGGWKGGGGQGGGW 224
Query: 213 Q 213
+
Sbjct: 225 K 225
Score = 37.0 bits (84), Expect = 0.008
Identities = 30/84 (35%), Positives = 33/84 (38%), Gaps = 27/84 (32%)
Query: 125 GGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVG--GGGDYGGGGDKVVVAAIMVVVATPV 182
GGG K GG GGG W+ GGG G G GGG +GGG
Sbjct: 80 GGGWKGGGGHGGGWKGGGGHGGGWKGGGGHGGGWKGGGGHGGG----------------- 122
Query: 183 VTVGRWWRRRRRR*SDGGGGGGGA 206
W+ GGGGGGGA
Sbjct: 123 ------WKGGGG--GRGGGGGGGA 138
>At3g17110 hypothetical protein
Length = 175
Score = 55.1 bits (131), Expect = 3e-08
Identities = 51/170 (30%), Positives = 60/170 (35%), Gaps = 57/170 (33%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G G + + GGGNGGGGGGG GGG GGG
Sbjct: 30 GRGSGSGSGYGSGSGGGNGGGGGGGG-----------------------GGGGGGGGGGG 66
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G + S G G G + + GG +GGG GGG G GGG
Sbjct: 67 DGSGSGS----------------GYGSGYGSGSGYGGGNNGGGGG------GGGRGGGGG 104
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G GG G+ + G + R GGGGGGG G
Sbjct: 105 GGNGGSGNGSGYGS------------GSGYGSGAGRGGGGGGGGGGGGGG 142
Score = 48.5 bits (114), Expect = 3e-06
Identities = 43/129 (33%), Positives = 48/129 (36%), Gaps = 23/129 (17%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
GSGGG GGG GGGGGGGD + + GGG GGG
Sbjct: 42 GSGGGNGGGGGGGGGGGGGGGGGGGDGSGSGSGYGSGYGSGSGYGGGNNGGGGGGGGRGG 101
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G G GS G G G G+ + GGG G GGG
Sbjct: 102 GGGGGNG----------------GSGNGS-------GYGSGSGYGSGAGRGGGGGGGGGG 138
Query: 160 GDYGGGGDK 168
G GGGG +
Sbjct: 139 GGGGGGGSR 147
Score = 45.1 bits (105), Expect = 3e-05
Identities = 43/127 (33%), Positives = 45/127 (34%), Gaps = 49/127 (38%)
Query: 40 GSG-GGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAA 98
GSG G G + GG NGGGGGGG R GGG GGG
Sbjct: 72 GSGYGSGYGSGSGYGGGNNGGGGGGGGR----------------------GGGGGGGNGG 109
Query: 99 VVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGG 158
+ G G GS A GGG GGG GGG G GG
Sbjct: 110 SGNGSGYGS---------------GSGYGSGAGR---GGGGGGG--------GGGGGGGG 143
Query: 159 GGDYGGG 165
GG G G
Sbjct: 144 GGSRGNG 150
>At2g36120 unknown protein
Length = 255
Score = 55.1 bits (131), Expect = 3e-08
Identities = 57/170 (33%), Positives = 60/170 (34%), Gaps = 53/170 (31%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
GSG GG A E GG+GGGGGGG GGG GGG A
Sbjct: 107 GSGEGGGAGYGGGEAGGHGGGGGGGA-----------------------GGGGGGGGGAH 143
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGG A A GGG GGG GG G GGG
Sbjct: 144 GG---------------------GYGGGQGAGA---GGGYGGGGA------GGHGGGGGG 173
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G+ GGGG G + GGGGGGG SG
Sbjct: 174 GNGGGGGGGSGEGGAHGGGYGAGGGAGEGYGGGAGAGGHGGGGGGGGGSG 223
Score = 52.0 bits (123), Expect = 3e-07
Identities = 54/170 (31%), Positives = 56/170 (32%), Gaps = 76/170 (44%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGGNGGGGGGG GG GGG A
Sbjct: 158 GYGGGGAGGHGGGGGGGNGGGGGGGSGE---------------------GGAHGGGYGAG 196
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G A G GGG+ A GG GGG GGG G GGG
Sbjct: 197 GG------------------AGEGYGGGAGA-----GGHGGGG--------GGGGGSGGG 225
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G GGGG + GGG GGG SG
Sbjct: 226 G--GGGGGYAAASGY----------------------GHGGGAGGGEGSG 251
Score = 48.1 bits (113), Expect = 4e-06
Identities = 51/172 (29%), Positives = 55/172 (31%), Gaps = 69/172 (40%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG GGGGGGG GGG GGG
Sbjct: 82 GGGGGG-------HGGGAGGGGGGGP-----------------------GGGYGGGSGEG 111
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G A G GGG GGG GG + G G G GGG
Sbjct: 112 GGAGYGGGEAGGHG---------GGGGGGAGGGGGGGGGAHGGGYGGGQGAGAGGGYGGG 162
Query: 160 --GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G +GGGG +GGGGGGG+ G
Sbjct: 163 GAGGHGGGGGG----------------------------GNGGGGGGGSGEG 186
Score = 44.3 bits (103), Expect = 5e-05
Identities = 39/127 (30%), Positives = 45/127 (34%), Gaps = 38/127 (29%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG + GGG+G GGG G + GGG GGG
Sbjct: 49 GVGIGGGPGGGSGYGGGSGEGGGAGGHGEGHI-----------------GGGGGGGHGGG 91
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGG G G+GGG + + GG G GGG
Sbjct: 92 AG---------------------GGGGGGPGGGYGGGSGEGGGAGYGGGEAGGHGGGGGG 130
Query: 160 GDYGGGG 166
G GGGG
Sbjct: 131 GAGGGGG 137
Score = 40.8 bits (94), Expect = 6e-04
Identities = 45/169 (26%), Positives = 49/169 (28%), Gaps = 78/169 (46%)
Query: 41 SGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVV 100
S G G + GG GGG G GGG GG +
Sbjct: 45 SAGLGVGIGGGPGGGSGYGGGSGE------------------------GGGAGGHGEGHI 80
Query: 101 GVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGG 160
G G GGG+ GGG GGG+ GGG G GGG
Sbjct: 81 GGGGGG----------------GHGGGAGGGG---GGGPGGGY-------GGGSGEGGGA 114
Query: 161 DYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
YGGG GGGGGGGA G
Sbjct: 115 GYGGG----------------------------EAGGHGGGGGGGAGGG 135
Score = 30.8 bits (68), Expect = 0.60
Identities = 23/67 (34%), Positives = 25/67 (36%), Gaps = 30/67 (44%)
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATV-EGGGNGGG 60
GGGGG+GG G GGGG A A+ GGG GGG
Sbjct: 217 GGGGGSGGG-----------------------------GGGGGGYAAASGYGHGGGAGGG 247
Query: 61 GGGGDRV 67
G G V
Sbjct: 248 EGSGGYV 254
>At4g22020 glycine-rich protein
Length = 396
Score = 54.7 bits (130), Expect = 4e-08
Identities = 57/176 (32%), Positives = 65/176 (36%), Gaps = 45/176 (25%)
Query: 40 GSGGG-GKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAA 98
GSG G G V G G GGGGGGG+ GGG GG
Sbjct: 198 GSGAGAGAGVGGAAGGVGGGGGGGGGE-----------------------GGGANGGSGH 234
Query: 99 VVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDG-----GGWWWWWWQ*GGG 153
G A V+ A G GGG + + GGG G GG + GGG
Sbjct: 235 GSGSGAGGGVSGAAG------GGGGGGGGGGSGGSKVGGGYGHGSGFGGGVGFGNSGGGG 288
Query: 154 CGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G GGGG GGGG+ + G + S GGGGGGG SG
Sbjct: 289 GGGGGGGGGGGGGNGSGYGS----------GSGYGSGMGKGSGSGGGGGGGGGGSG 334
Score = 53.1 bits (126), Expect = 1e-07
Identities = 56/173 (32%), Positives = 60/173 (34%), Gaps = 71/173 (41%)
Query: 40 GSGGG-GKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAA 98
GSG G G V + G G GGGGGGG+ GGG GG
Sbjct: 161 GSGAGAGAGVGGSSGGAGGGGGGGGGE-----------------------GGGANGGSGH 197
Query: 99 VVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGG 158
G A + V A V G GGG GGG+GGG GG G G
Sbjct: 198 GSGAGAGAGVGGAAGGV-------GGGGGG-------GGGEGGGA-------NGGSGHGS 236
Query: 159 GGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGVS 211
G GGG V AA GGGGGGG SG S
Sbjct: 237 GSGAGGG---VSGAA-----------------------GGGGGGGGGGGSGGS 263
Score = 51.2 bits (121), Expect = 4e-07
Identities = 56/171 (32%), Positives = 59/171 (33%), Gaps = 60/171 (35%)
Query: 40 GSGGG-GKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAA 98
GSG G G V T G G GGGGGG GGG GGG +
Sbjct: 120 GSGSGAGAGVGGTTGGVGGGGGGGG-------------------------GGGEGGGSSG 154
Query: 99 VVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGG 158
G + S A G GG S GG GGG GGG G GG
Sbjct: 155 GSGHGSGSGAG----------AGAGVGGSS-------GGAGGGG--------GGGGGEGG 189
Query: 159 GGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G + G G A V A V G GGG GGGAN G
Sbjct: 190 GANGGSGHGSGAGAGAGVGGAAGGVGGGG---------GGGGGEGGGANGG 231
Score = 47.8 bits (112), Expect = 5e-06
Identities = 49/172 (28%), Positives = 52/172 (29%), Gaps = 65/172 (37%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+G G GGG GGG GGG G G G G A
Sbjct: 86 GAGAGMSGYGGVGGGGGRGGGEGGGSSA-------------------NGGSGHGSGSGAG 126
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGG- 158
GV T+ +G GGG GG GG G G GVGG
Sbjct: 127 AGVGGTT-------------GGVGGGGGGGGGGGEGGGSSGGSGHGSGSGAGAGAGVGGS 173
Query: 159 -GGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GG GGGG GGG GGGAN G
Sbjct: 174 SGGAGGGGG-------------------------------GGGGEGGGANGG 194
Score = 44.7 bits (104), Expect = 4e-05
Identities = 45/137 (32%), Positives = 51/137 (36%), Gaps = 45/137 (32%)
Query: 40 GSG-GGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAA 98
GSG GGG + AA GGG GGGG GG +V GGG G G
Sbjct: 237 GSGAGGGVSGAAGGGGGGGGGGGSGGSKV---------------------GGGYGHGSGF 275
Query: 99 VVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*-------- 150
GV + + G GGG GGG+G G+
Sbjct: 276 GGGVGFGN--------------SGGGGGGGGGGGGGGGGGNGSGYGSGSGYGSGMGKGSG 321
Query: 151 -GGGCGVGGGGDYGGGG 166
GGG G GGGG GG G
Sbjct: 322 SGGGGGGGGGGSGGGNG 338
Score = 44.3 bits (103), Expect = 5e-05
Identities = 48/165 (29%), Positives = 56/165 (33%), Gaps = 60/165 (36%)
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGG 61
GGGGG+GG ++G + GSG GG GGG GGGG
Sbjct: 255 GGGGGSGGS----------KVGGGYGH-----------GSGFGGGVGFGNSGGGGGGGGG 293
Query: 62 GGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLAT 121
GGG GGG G G G S + +
Sbjct: 294 GGG------------------------GGGGGNGSGYGSGSGYGSGMGKGS--------- 320
Query: 122 IGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGG 166
G GGG GGG+G G G G G+GGG G GG
Sbjct: 321 -GSGGGGGGGGGGSGGGNGSGSGS-----GEGYGMGGGAGTGNGG 359
Score = 41.2 bits (95), Expect = 4e-04
Identities = 53/174 (30%), Positives = 56/174 (31%), Gaps = 79/174 (45%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG GGGG GGD G G G G A
Sbjct: 60 GGGGGG--------GGGGGGGGEGGD-----------------------GYGHGEGYGAG 88
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGG- 158
G++ V G GGGS A G G G G G G GVGG
Sbjct: 89 AGMSGYGGVGGGGGRGG------GEGGGSSANG---GSGHGSGS-------GAGAGVGGT 132
Query: 159 -GGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGVS 211
GG GGGG GGGG GG +SG S
Sbjct: 133 TGGVGGGGG------------------------------GGGGGGEGGGSSGGS 156
Score = 33.5 bits (75), Expect = 0.092
Identities = 38/131 (29%), Positives = 42/131 (32%), Gaps = 60/131 (45%)
Query: 40 GSGGG-----GKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGG 94
GSG G GK + GGG GGG GGG+ G G G
Sbjct: 307 GSGSGYGSGMGKGSGSGGGGGGGGGGSGGGN-----------------------GSGSGS 343
Query: 95 GRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGC 154
G +G A G G G GGG G G G G
Sbjct: 344 GEGYGMGGGA------------------GTGNGG-------GGGVGFGM-------GIGF 371
Query: 155 GVGGGGDYGGG 165
G+G GG GGG
Sbjct: 372 GIGIGGGSGGG 382
Score = 26.9 bits (58), Expect = 8.6
Identities = 11/16 (68%), Positives = 12/16 (74%)
Query: 152 GGCGVGGGGDYGGGGD 167
GG G GGGG GGGG+
Sbjct: 59 GGGGGGGGGGGGGGGE 74
>At1g05135 unknown protein
Length = 384
Score = 54.7 bits (130), Expect = 4e-08
Identities = 50/170 (29%), Positives = 56/170 (32%), Gaps = 62/170 (36%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+GGG A + GGG GGGG GGG GGG
Sbjct: 157 GAGGGVGGGAGGIGGGGGAGGGG--------------------------GGGVGGGSGHG 190
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G A + A +G GGG GG +GG + GGG G G G
Sbjct: 191 SGFGAGGGIGGGAG------GGVGGGGGGGGGGGGGGGANGGSGHGSGFGAGGGVGGGVG 244
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G GGGG GGGGGGGAN G
Sbjct: 245 GGAGGGG------------------------------GGGGGGGGGANGG 264
Score = 53.5 bits (127), Expect = 9e-08
Identities = 56/172 (32%), Positives = 57/172 (32%), Gaps = 67/172 (38%)
Query: 40 GSGGG-GKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAA 98
G+GGG G V GGG GGGGGGG GGG GG
Sbjct: 194 GAGGGIGGGAGGGVGGGGGGGGGGGG------------------------GGGANGGSGH 229
Query: 99 VVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGG 158
G A G GGG GGG GGG GGG G GG
Sbjct: 230 GSGFGAGG----------------GVGGGV-------GGGAGGG--------GGGGGGGG 258
Query: 159 GGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGV 210
GG GG G A V G GGGGGGG GV
Sbjct: 259 GGANGGSGHGSGFGAGGGVGGGVGGGAG-----------GGGGGGGGGGGGV 299
Score = 52.4 bits (124), Expect = 2e-07
Identities = 64/218 (29%), Positives = 70/218 (31%), Gaps = 70/218 (32%)
Query: 1 VGGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGG 60
+GGGGG GG G+GGG A GGG G G
Sbjct: 70 IGGGGGQGGGF----------------------------GAGGGVGGGAGGGLGGGGGAG 101
Query: 61 GGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLA 120
GGG GGG GGG G A V A +
Sbjct: 102 GGG-------------------------GGGIGGGSGHGGGFGAGGGVGGGAGGGIGGGG 136
Query: 121 TIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVV---------V 171
G GGG G G GGG+ GG G+GGGG GGGG V
Sbjct: 137 GAGGGGGGGVGG---GSGHGGGFGAGGGVGGGAGGIGGGGGAGGGGGGGVGGGSGHGSGF 193
Query: 172 AAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
A + VG GGGGGGGAN G
Sbjct: 194 GAGGGIGGGAGGGVGGGGGG-----GGGGGGGGGANGG 226
Score = 50.4 bits (119), Expect = 7e-07
Identities = 54/172 (31%), Positives = 59/172 (33%), Gaps = 48/172 (27%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG + GGG G GGG G + GGG GGG
Sbjct: 139 GGGGGGGVGGGSGHGGGFGAGGGVGGGAGGIGG----------------GGGAGGGGGGG 182
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
VG + + G GGG A GGG GGG GGG G GGG
Sbjct: 183 VGGGSGHG------------SGFGAGGGIGGGA---GGGVGGGGG------GGGGGGGGG 221
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGVS 211
G GG G A V G GGGGGGG G +
Sbjct: 222 GANGGSGHGSGFGAGGGVGGGVGGGAG-----------GGGGGGGGGGGGAN 262
Score = 49.3 bits (116), Expect = 2e-06
Identities = 50/170 (29%), Positives = 55/170 (31%), Gaps = 65/170 (38%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG G G GGGGGGG GGG GGG
Sbjct: 270 GFGAGGGVGGGVGGGAGGGGGGGGGG-----------------------GGGVGGGSGHG 306
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G A G GGG+ GGG GG GGG G+G G
Sbjct: 307 GGFGAGG----------------GLGGGAGGGLG--GGGGAGG--------GGGGGLGHG 340
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G GGG V I + + V GGG G G+ SG
Sbjct: 341 GGVGGGHGGGVGIGIGIGIGVGV----------------GGGSGQGSGSG 374
Score = 47.0 bits (110), Expect = 8e-06
Identities = 46/130 (35%), Positives = 47/130 (35%), Gaps = 36/130 (27%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG GGGGG G V GGG GGG
Sbjct: 53 GRGGGGGFGGGRGSGGGIGGGGGQGGGFGAGGGV---------------GGGAGGGLGGG 97
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G + G GGG A GGG GGG GGG G GGG
Sbjct: 98 GGAGGGGGGGIGGGS--------GHGGGFGA-----GGGVGGGA-------GGGIG-GGG 136
Query: 160 GDYGGGGDKV 169
G GGGG V
Sbjct: 137 GAGGGGGGGV 146
Score = 43.5 bits (101), Expect = 9e-05
Identities = 48/165 (29%), Positives = 50/165 (30%), Gaps = 58/165 (35%)
Query: 45 GKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAA 104
G+ A G GGGG GG R GGG GGG G A
Sbjct: 43 GRRCAGRGRFGRGGGGGFGGGRGS--------------------GGGIGGGGGQGGGFGA 82
Query: 105 TSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGG 164
V A G GGG A GG GG + GGG G G GG GG
Sbjct: 83 GGGVGGGAGG--------GLGGGGGAGGGGGGGIGGGSGHGGGFGAGGGVGGGAGGGIGG 134
Query: 165 GGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GG GGGGGGG G
Sbjct: 135 GG------------------------------GAGGGGGGGVGGG 149
Score = 40.8 bits (94), Expect = 6e-04
Identities = 48/151 (31%), Positives = 50/151 (32%), Gaps = 57/151 (37%)
Query: 40 GSG-GGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAA 98
GSG G G V V GG GGGGGGG GGG GGG
Sbjct: 268 GSGFGAGGGVGGGVGGGAGGGGGGGGGG----------------------GGGVGGGSGH 305
Query: 99 VVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GG------ 152
G A + A G GGG A GGG GGG GG
Sbjct: 306 GGGFGAGGGLGGGAGG--------GLGGGGGA-----GGGGGGGLGHGGGVGGGHGGGVG 352
Query: 153 -------GCGVGGGGDY--------GGGGDK 168
G GVGGG GGGG +
Sbjct: 353 IGIGIGIGVGVGGGSGQGSGSGSGSGGGGGR 383
Score = 26.9 bits (58), Expect = 8.6
Identities = 18/62 (29%), Positives = 25/62 (40%), Gaps = 23/62 (37%)
Query: 3 GGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGGG 62
GGG GG + + + + V +G GG G+ G G+G GGG
Sbjct: 344 GGGHGGGVGIGIGIGIGVGVG------------------GGSGQG-----SGSGSGSGGG 380
Query: 63 GG 64
GG
Sbjct: 381 GG 382
>At3g17050 unknown protein
Length = 349
Score = 53.9 bits (128), Expect = 7e-08
Identities = 51/170 (30%), Positives = 56/170 (32%), Gaps = 85/170 (50%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+GGGG A GGG+GGG GGG GGG GGG
Sbjct: 210 GAGGGGGAGGGGGLGGGHGGGFGGGA-----------------------GGGLGGGAGGG 246
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGG+ A GGG GGG+ GGG G G G
Sbjct: 247 TGG--------------------GFGGGAGGGA---GGGAGGGF-------GGGAGGGAG 276
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G +GGG GGG GGGA G
Sbjct: 277 GGFGGGA--------------------------------GGGAGGGAGGG 294
Score = 52.0 bits (123), Expect = 3e-07
Identities = 44/132 (33%), Positives = 50/132 (37%), Gaps = 33/132 (25%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+GGG A GGG GGG GGG + GGG GGG
Sbjct: 54 GAGGGFGGGAGGGAGGGLGGGAGGGGGI---------------------GGGAGGGAGGG 92
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGG-----SKAVAA**GGGDGGGWWWWWWQ*GGGC 154
+G A + + A G GGG + GGG GGG GGG
Sbjct: 93 LGGGAGGGLGGGHGGGIGGGAGGGAGGGLGGGHGGGIGGGAGGGSGGGL-------GGGI 145
Query: 155 GVGGGGDYGGGG 166
G G GG GGGG
Sbjct: 146 GGGAGGGAGGGG 157
Score = 50.4 bits (119), Expect = 7e-07
Identities = 44/127 (34%), Positives = 46/127 (35%), Gaps = 49/127 (38%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+GGG A GGG GGG GGG GGG GGG
Sbjct: 262 GAGGGFGGGAGGGAGGGFGGGAGGGA-----------------------GGGAGGGFGGG 298
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G V G GGGS GGG GGG GGG G G G
Sbjct: 299 AGGGHGGGVGG------------GFGGGS-------GGGFGGGA-------GGGAGGGAG 332
Query: 160 GDYGGGG 166
G +GGGG
Sbjct: 333 GGFGGGG 339
Score = 48.5 bits (114), Expect = 3e-06
Identities = 53/171 (30%), Positives = 57/171 (32%), Gaps = 59/171 (34%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGG A GGG+GGG GGG GGG GGG
Sbjct: 108 GIGGGAGGGAGGGLGGGHGGGIGGGA-----------------------GGGSGGGLGGG 144
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
+G A + IG G G A GGG GGG GGG G G G
Sbjct: 145 IGGGAGGGAGGGGGLGGGHGGGIGGGAGGGA-----GGGLGGGH-------GGGIGGGAG 192
Query: 160 GDYGGG-GDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G GGG G + A G GGGGGA G
Sbjct: 193 GGSGGGLGGGIGGGA-----------------------GGGAGGGGGAGGG 220
Score = 48.5 bits (114), Expect = 3e-06
Identities = 42/127 (33%), Positives = 43/127 (33%), Gaps = 55/127 (43%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG GGG GGG GGG GGG GGG
Sbjct: 150 GGGAGGGGGLGGGHGGGIGGGAGGGA-----------------------GGGLGGGHGGG 186
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
+G G GGGS GGG GGG G G G GGG
Sbjct: 187 IGG--------------------GAGGGS-------GGGLGGGIGG-----GAGGGAGGG 214
Query: 160 GDYGGGG 166
G GGGG
Sbjct: 215 GGAGGGG 221
Score = 48.1 bits (113), Expect = 4e-06
Identities = 43/127 (33%), Positives = 46/127 (35%), Gaps = 43/127 (33%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG A GGG GGG GGG GGG GGG
Sbjct: 142 GGGIGGGAGGGAGGGGGLGGGHGGGI-----------------------GGGAGGGAGGG 178
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGS-KAVAA**GGGDGGGWWWWWWQ*GGGCGVGG 158
+G + G GGGS + GGG GGG GGG G GG
Sbjct: 179 LGGGHGGGIGG------------GAGGGSGGGLGGGIGGGAGGGA-------GGGGGAGG 219
Query: 159 GGDYGGG 165
GG GGG
Sbjct: 220 GGGLGGG 226
Score = 46.6 bits (109), Expect = 1e-05
Identities = 47/170 (27%), Positives = 54/170 (31%), Gaps = 54/170 (31%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGK + GG GGG GG GGG GGG
Sbjct: 41 GGFGGGKGFGGGIGAGGGFGGGAGGGA----------------------GGGLGGGAGGG 78
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G+ + + +G GG + GGG GGG GGG G G G
Sbjct: 79 GGIGGGAGGGAGGGLGGGAGGGLG-GGHGGGIGGGAGGGAGGGL-------GGGHGGGIG 130
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G GGG + I GGG GGGA G
Sbjct: 131 GGAGGGSGGGLGGGI------------------------GGGAGGGAGGG 156
Score = 45.8 bits (107), Expect = 2e-05
Identities = 44/128 (34%), Positives = 45/128 (34%), Gaps = 58/128 (45%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+GGG GGG GGG GGG GGG GGG
Sbjct: 270 GAGGGAGGGFGGGAGGGAGGGAGGGF-----------------------GGGAGGGHGGG 306
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVG-G 158
VG G GGGS GGG GGG GGG G G G
Sbjct: 307 VGG--------------------GFGGGS-------GGGFGGGA-------GGGAGGGAG 332
Query: 159 GGDYGGGG 166
GG GGGG
Sbjct: 333 GGFGGGGG 340
Score = 42.7 bits (99), Expect = 2e-04
Identities = 42/126 (33%), Positives = 44/126 (34%), Gaps = 37/126 (29%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG G G GGG G G GGG GGG
Sbjct: 32 GGGGGGGLGGGFGGGKGFGGGIGAGGGF---------------------GGGAGGGAGGG 70
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
+G A + A G GGG A GGG GGG GGG G G G
Sbjct: 71 LGGGAGGGGGIGGG------AGGGAGGGLGGGA---GGGLGGGH-------GGGIGGGAG 114
Query: 160 GDYGGG 165
G GGG
Sbjct: 115 GGAGGG 120
Score = 35.0 bits (79), Expect = 0.032
Identities = 21/53 (39%), Positives = 25/53 (46%)
Query: 114 MVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGG 166
+VVVV+ + C K GGG GG + GGG G GGG G GG
Sbjct: 13 LVVVVIGVVECRRFEKETLGGGGGGGLGGGFGGGKGFGGGIGAGGGFGGGAGG 65
Score = 33.5 bits (75), Expect = 0.092
Identities = 43/145 (29%), Positives = 47/145 (31%), Gaps = 66/145 (45%)
Query: 67 VVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLATIGCGG 126
VVVV+ VV + + GGG GGG G GG
Sbjct: 14 VVVVIGVVECRRFEKETLGGGGGGGLGGGFGGGKGF----------------------GG 51
Query: 127 GSKAVAA**GGGDGGGWWWWWWQ*GGGCGVG-GGGDYGGGGDKVVVAAIMVVVATPVVTV 185
G A GGG GGG GGG G G GGG GGGG
Sbjct: 52 GIGA-----GGGFGGGA-------GGGAGGGLGGGAGGGGG------------------- 80
Query: 186 GRWWRRRRRR*SDGGGGGGGANSGV 210
GGG GGGA G+
Sbjct: 81 ------------IGGGAGGGAGGGL 93
>At3g20470 glycine-rich protein, putative
Length = 174
Score = 53.5 bits (127), Expect = 9e-08
Identities = 52/173 (30%), Positives = 58/173 (33%), Gaps = 73/173 (42%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG GGG GGG GGG GGG +
Sbjct: 64 GFGGGGGLGGGAGGGGGLGGGAGGGA-----------------------GGGFGGGAGSG 100
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GG---GCGV 156
G+ G GGG A GGG GGG+ GG G G
Sbjct: 101 GGLGGGG----------------GAGGGFGGGA---GGGSGGGFGGGAGAGGGLGGGGGA 141
Query: 157 GGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GGGG +GGGG + GGG GGGA +G
Sbjct: 142 GGGGGFGGGGGSGI----------------------------GGGFGGGAGAG 166
Score = 45.4 bits (106), Expect = 2e-05
Identities = 42/134 (31%), Positives = 45/134 (33%), Gaps = 55/134 (41%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG GGG GGGGG G GGG GGG
Sbjct: 86 GGGAGGGFGGGAGSGGGLGGGGGAGGGF---------------------GGGAGGGSGGG 124
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G A G GGG GGG GGG + GGG G+GGG
Sbjct: 125 FGGGA------------------GAGGGLGG-----GGGAGGGGGFGG---GGGSGIGGG 158
Query: 160 --------GDYGGG 165
G +GGG
Sbjct: 159 FGGGAGAGGGFGGG 172
Score = 36.6 bits (83), Expect = 0.011
Identities = 18/31 (58%), Positives = 20/31 (64%), Gaps = 5/31 (16%)
Query: 136 GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGG 166
GGG GGG GGG G+GGGG +GGGG
Sbjct: 44 GGGLGGGGGI-----GGGSGLGGGGGFGGGG 69
>At2g32690 unknown protein
Length = 201
Score = 51.2 bits (121), Expect = 4e-07
Identities = 44/127 (34%), Positives = 49/127 (37%), Gaps = 43/127 (33%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG+G GGGGG + L GGG GGG
Sbjct: 86 GLGGGGGLGGGGGLGGGSGLGGGGG-----------LGGGSGLGGGGGLGGGGGGGLGGG 134
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G+ G GGG GGG GGG GGG G+GGG
Sbjct: 135 GGLGG------------------GAGGGY-------GGGAGGGL-------GGGGGIGGG 162
Query: 160 GDYGGGG 166
G +GGGG
Sbjct: 163 GGFGGGG 169
Score = 47.0 bits (110), Expect = 8e-06
Identities = 43/127 (33%), Positives = 47/127 (36%), Gaps = 41/127 (32%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG + GGG G GGGGG + GGG GGG
Sbjct: 104 GLGGGGGLGGGSGLGGGGGLGGGGGGGLGG-------------------GGGLGGGAGGG 144
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G A + IG GGG GGG GGG+ GGG G G G
Sbjct: 145 YGGGAGGGLGGGGG--------IGGGGGF-------GGGGGGGF-------GGGAGGGFG 182
Query: 160 GDYGGGG 166
GGGG
Sbjct: 183 KGIGGGG 189
Score = 36.6 bits (83), Expect = 0.011
Identities = 37/114 (32%), Positives = 41/114 (35%), Gaps = 47/114 (41%)
Query: 53 EGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVA 112
+G G GGG GGG + + GGG GGG G
Sbjct: 61 KGLGGGGGLGGGGGLGGGGGLGG-------------GGGLGGGGGLGGGG---------- 97
Query: 113 AMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGG 166
G GGGS G G GGG GGG G+GGGG GGGG
Sbjct: 98 ----------GLGGGS-------GLGGGGGL-------GGGSGLGGGGGLGGGG 127
Score = 27.3 bits (59), Expect = 6.6
Identities = 11/16 (68%), Positives = 12/16 (74%)
Query: 151 GGGCGVGGGGDYGGGG 166
G G G+GGGG GGGG
Sbjct: 58 GFGKGLGGGGGLGGGG 73
>At5g46730 unknown protein
Length = 268
Score = 48.9 bits (115), Expect = 2e-06
Identities = 55/174 (31%), Positives = 62/174 (35%), Gaps = 35/174 (20%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCG----GG 95
G GGG V++ GG +GGG GGG E + GGG G GG
Sbjct: 40 GGGGGSGGVSSGGYGGESGGGYGGGSGEGAGGGYGGAEGYAS-------GGGSGHGGGGG 92
Query: 96 RAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCG 155
AA G A+ A G GGG A GG GGG GGG G
Sbjct: 93 GAASSGGYASG-------------AGEGGGGGYGGAA---GGHAGGG--------GGGSG 128
Query: 156 VGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GGG YG GG+ G GGGGGGG+ G
Sbjct: 129 GGGGSAYGAGGEHASGYGNGAGEGGGAGASGYGGGAYGGGGGHGGGGGGGSAGG 182
Score = 48.5 bits (114), Expect = 3e-06
Identities = 43/127 (33%), Positives = 47/127 (36%), Gaps = 47/127 (37%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+G GG A A+ GG GGGGG G GGG GG
Sbjct: 148 GAGEGGGAGASGYGGGAYGGGGGHG------------------------GGGGGGSAGGA 183
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G + G GGG A GGG GG + GGG G GGG
Sbjct: 184 HGGS-------------------GYGGGEGGGAG--GGGSHGGAGGYGG--GGGGGSGGG 220
Query: 160 GDYGGGG 166
G YGGGG
Sbjct: 221 GAYGGGG 227
Score = 47.4 bits (111), Expect = 6e-06
Identities = 49/175 (28%), Positives = 55/175 (31%), Gaps = 67/175 (38%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GG G A GGG G GGGGG G GG A+
Sbjct: 108 GGGGYGGAAGGHAGGGGGGSGGGGGSAY-----------------------GAGGEHASG 144
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGG-----WWWWWWQ*GGGC 154
G A A+ G GGG+ GGG GGG + G G
Sbjct: 145 YGNGAGEGGGAGAS---------GYGGGAYGGGGGHGGGGGGGSAGGAHGGSGYGGGEGG 195
Query: 155 GVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G GGGG +GG G GGGGGGG+ G
Sbjct: 196 GAGGGGSHGGAG------------------------------GYGGGGGGGSGGG 220
Score = 45.8 bits (107), Expect = 2e-05
Identities = 42/128 (32%), Positives = 46/128 (35%), Gaps = 37/128 (28%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+G G A GGG+GGGGGGG G G GGG
Sbjct: 154 GAGASGYGGGAYGGGGGHGGGGGGGSAGGA-----------------HGGSGYGGGEGGG 196
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGG- 158
G + A G GGG GG GGG + GGG G GG
Sbjct: 197 AGGGGSHGGA----------GGYGGGGG--------GGSGGGGAYGGGGAHGGGYGSGGG 238
Query: 159 -GGDYGGG 165
GG YGGG
Sbjct: 239 EGGGYGGG 246
Score = 45.1 bits (105), Expect = 3e-05
Identities = 43/136 (31%), Positives = 48/136 (34%), Gaps = 52/136 (38%)
Query: 40 GSGGGGKAVAA---TVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGR 96
G GGGG A A + GGG GGG GGG GGG G G
Sbjct: 173 GGGGGGSAGGAHGGSGYGGGEGGGAGGGGSHGGAGGYGG-------------GGGGGSGG 219
Query: 97 AAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGC-- 154
G GGG+ GGG+GGG+ GGG
Sbjct: 220 GGAYG-----------------------GGGAHGGGYGSGGGEGGGY-------GGGAAG 249
Query: 155 ----GVGGGGDYGGGG 166
G GGGG +GGGG
Sbjct: 250 GYGGGHGGGGGHGGGG 265
Score = 37.0 bits (84), Expect = 0.008
Identities = 37/117 (31%), Positives = 42/117 (35%), Gaps = 23/117 (19%)
Query: 54 GGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAA 113
GGG GGGGG V + GGG GGG G +
Sbjct: 35 GGGGHGGGGGSGGV------------SSGGYGGESGGGYGGGSGEGAGGGYGGAEGYASG 82
Query: 114 MVVVVLATIGCGGGSKAVAA----**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGG 166
G GGG A+ G G+GGG + GG G GGGG GGGG
Sbjct: 83 ------GGSGHGGGGGGAASSGGYASGAGEGGGGGYGGAA-GGHAGGGGGGSGGGGG 132
>At2g18115 putative protein
Length = 296
Score = 48.5 bits (114), Expect = 3e-06
Identities = 42/128 (32%), Positives = 46/128 (35%), Gaps = 55/128 (42%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG+ + GGG GGGGGGG G G GGG
Sbjct: 195 GIGIGGEPSPGSGGGGGGGGGGGGG------------------------GSGPGGGGGGG 230
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGG G GDG G+ W GGG G GGG
Sbjct: 231 GG---------------------GGGGGG-------GVGDGYGYGRGW---GGGYGGGGG 259
Query: 160 GDYGGGGD 167
G +G GGD
Sbjct: 260 GGWGWGGD 267
Score = 47.0 bits (110), Expect = 8e-06
Identities = 39/121 (32%), Positives = 39/121 (32%), Gaps = 49/121 (40%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG G G GGGGGGG GGG GGG
Sbjct: 208 GGGGGGGGGGGGGSGPGGGGGGGGG------------------------GGGGGGGVGDG 243
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGG GGG GGGW W GG G G G
Sbjct: 244 YGYGR------------------GWGGGY-------GGGGGGGWGWGGDSNGGVWGPGNG 278
Query: 160 G 160
G
Sbjct: 279 G 279
Score = 40.0 bits (92), Expect = 0.001
Identities = 35/92 (38%), Positives = 37/92 (40%), Gaps = 22/92 (23%)
Query: 122 IGCG-GGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVAT 180
IG G GG + + GGG GGG GGG G GGGG GGGG V
Sbjct: 194 IGIGIGGEPSPGSGGGGGGGGGGGGGGSGPGGGGGGGGGG--GGGGGGV----------G 241
Query: 181 PVVTVGRWWRRRRRR*SDGGGGGGGANSGVSW 212
GR W GGG GGG G W
Sbjct: 242 DGYGYGRGW---------GGGYGGGGGGGWGW 264
Score = 37.4 bits (85), Expect = 0.006
Identities = 41/127 (32%), Positives = 41/127 (32%), Gaps = 61/127 (48%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
GSGGGG GGG GGGGG G GGG GGG
Sbjct: 205 GSGGGGG-------GGGGGGGGGSGPG----------------------GGGGGGG---- 231
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGV-GG 158
G GGG V G G G G W GGG G G
Sbjct: 232 -----------------------GGGGGGGGV----GDGYGYGRGWGGGYGGGGGGGWGW 264
Query: 159 GGDYGGG 165
GGD GG
Sbjct: 265 GGDSNGG 271
Score = 30.4 bits (67), Expect = 0.78
Identities = 22/63 (34%), Positives = 26/63 (40%), Gaps = 14/63 (22%)
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGG 61
GGGGG GG +G + + R W + G GGGG GG G G
Sbjct: 232 GGGGGGGG------------VGDGYGYGRGWGGGY--GGGGGGGWGWGGDSNGGVWGPGN 277
Query: 62 GGG 64
GGG
Sbjct: 278 GGG 280
>At1g27710 unknown protein
Length = 212
Score = 48.1 bits (113), Expect = 4e-06
Identities = 52/170 (30%), Positives = 55/170 (31%), Gaps = 73/170 (42%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG G GGGGG V+ GGG GGG
Sbjct: 106 GYGGGGPGYG----GGGYGPGGGGGGVVI--------------------GGGFGGG---- 137
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
A G GGG GGG G G GG G+GGG
Sbjct: 138 --------------------AGYGSGGGLGWDGGNGGGGPGYG--------SGGGGIGGG 169
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G GGG V +G S GGGGGGG G
Sbjct: 170 GGIGGG-----------------VIIGGGGGGCGGSCSGGGGGGGGYGHG 202
Score = 38.9 bits (89), Expect = 0.002
Identities = 42/127 (33%), Positives = 42/127 (33%), Gaps = 48/127 (37%)
Query: 42 GGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVG 101
GGGG V GGG G G GGG L W GG GGG G
Sbjct: 123 GGGGGVVIGGGFGGGAGYGSGGG-----------------LGW---DGGNGGGGPGYGSG 162
Query: 102 VAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGC--GVGGG 159
G GGG GG GGG GG C G GGG
Sbjct: 163 GG-------------------GIGGGGGIGGGVIIGGGGGGC-------GGSCSGGGGGG 196
Query: 160 GDYGGGG 166
G YG GG
Sbjct: 197 GGYGHGG 203
Score = 32.7 bits (73), Expect = 0.16
Identities = 20/64 (31%), Positives = 26/64 (40%), Gaps = 26/64 (40%)
Query: 1 VGGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGG 60
+GGGGG GG +++ G GGGG + + GGG GG
Sbjct: 166 IGGGGGIGGGVII--------------------------GGGGGGCGGSCSGGGGGGGGY 199
Query: 61 GGGG 64
G GG
Sbjct: 200 GHGG 203
>At2g37830 hypothetical protein
Length = 106
Score = 47.8 bits (112), Expect = 5e-06
Identities = 43/127 (33%), Positives = 45/127 (34%), Gaps = 50/127 (39%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG +GGG GGGG GGD GG GGG
Sbjct: 29 GKGGGGG------DGGGGGGGGEGGDGG---------------------GGEDGGGEDVE 61
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
+G A G GG GGG GGG GGG G GGG
Sbjct: 62 IGDGANG----------------GGFGGDGGGGGFGGGGGGGGD-------GGGGGGGGG 98
Query: 160 GDYGGGG 166
G GGGG
Sbjct: 99 GGGGGGG 105
Score = 44.7 bits (104), Expect = 4e-05
Identities = 40/126 (31%), Positives = 43/126 (33%), Gaps = 49/126 (38%)
Query: 41 SGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVV 100
+G GGK GGG GGG GG GG GGG +
Sbjct: 25 TGAGGKGGGGGDGGGGGGGGEGGDGG----------------------GGEDGGGEDVEI 62
Query: 101 GVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGG 160
G A GGG GGDGGG + GGG G GGGG
Sbjct: 63 GDGAN-------------------GGGF--------GGDGGGGGFGGGGGGGGDGGGGGG 95
Query: 161 DYGGGG 166
GGGG
Sbjct: 96 GGGGGG 101
Score = 38.5 bits (88), Expect = 0.003
Identities = 33/103 (32%), Positives = 43/103 (41%), Gaps = 19/103 (18%)
Query: 107 VVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGG 166
++A+ + + V++ G G K GGGDGGG GG G GGGG+ GGG
Sbjct: 8 IIAIASFVGVLIFLAALTGAGGKGG----GGGDGGGGGG-----GGEGGDGGGGEDGGGE 58
Query: 167 DKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
D V G + GGGGGGG + G
Sbjct: 59 D----------VEIGDGANGGGFGGDGGGGGFGGGGGGGGDGG 91
>At1g02710 hypothetical protein
Length = 96
Score = 47.4 bits (111), Expect = 6e-06
Identities = 34/98 (34%), Positives = 42/98 (42%), Gaps = 13/98 (13%)
Query: 112 AAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVV 171
A+ V+++ +G G A GGG G G W GG G GGGG+ GGGG K+
Sbjct: 9 ASAAVIMIGGLGLFGTHSLQAGGNGGGSGKGQWLHGGGGEGGGGEGGGGE-GGGGQKISK 67
Query: 172 AAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G +R S GGGGGGG G
Sbjct: 68 GG------------GGGGSGGGQRSSSGGGGGGGEGDG 93
Score = 42.0 bits (97), Expect = 3e-04
Identities = 39/124 (31%), Positives = 42/124 (33%), Gaps = 44/124 (35%)
Query: 42 GGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVG 101
GG G +++ GGNGGG G G W GGG GGG G
Sbjct: 17 GGLGLFGTHSLQAGGNGGGSGKGQ-------------------WLHGGGGEGGGGEGGGG 57
Query: 102 VAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGD 161
GGG K GGG GGG GG G GG GD
Sbjct: 58 EG---------------------GGGQKISKGGGGGGSGGG----QRSSSGGGGGGGEGD 92
Query: 162 YGGG 165
GGG
Sbjct: 93 GGGG 96
Score = 30.8 bits (68), Expect = 0.60
Identities = 13/24 (54%), Positives = 16/24 (66%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGG 63
GSGGG ++ + GGG G GGGG
Sbjct: 73 GSGGGQRSSSGGGGGGGEGDGGGG 96
>At5g49350 unknown protein
Length = 302
Score = 47.0 bits (110), Expect = 8e-06
Identities = 53/170 (31%), Positives = 55/170 (32%), Gaps = 47/170 (27%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGG T GGG G GGGG GG GGG A
Sbjct: 39 GGEGGGIGGGGTGFGGGGTGVGGGG---------------TGFGGGGLGAGGSGGGGAGG 83
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
+G T G GGG A GGG GG GGG GVGGG
Sbjct: 84 LGSGGT-----------------GVGGGGGLGAGGSGGGGAGGLG------GGGTGVGGG 120
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G GG G + T G GG GGGG N G
Sbjct: 121 GTGGGTGFNGGGTGFGPFGGGGLGTGGG---------GAGGLGGGGGNGG 161
Score = 42.4 bits (98), Expect = 2e-04
Identities = 42/135 (31%), Positives = 48/135 (35%), Gaps = 39/135 (28%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG A GGG GG GGGG V GGG GGG
Sbjct: 90 GVGGGGGLGAGGSGGGGAGGLGGGGTGVG--------------------GGGTGGGTGFN 129
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G +G GGG GGG GG+ GG G+GGG
Sbjct: 130 GGGTGFGPFGG---------GGLGTGGGGAGGLG--GGGGNGGF--------GGGGLGGG 170
Query: 160 GDYGGGGDKVVVAAI 174
G G ++V A+
Sbjct: 171 GLGGEDPPEIVAKAL 185
>At2g05580 unknown protein
Length = 302
Score = 47.0 bits (110), Expect = 8e-06
Identities = 52/171 (30%), Positives = 56/171 (32%), Gaps = 78/171 (45%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG +GG GGGGGGG GGG GGG+
Sbjct: 202 GGGGGG-------QGGHKGGGGGGGQ-------------------GGHKGGGGGGGQGGH 235
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCG-VGG 158
G G GGG GG GGG + GGG G VGG
Sbjct: 236 KG---------------------GGGGGQGG-----GGHKGGGGGQGGHKGGGGGGHVGG 269
Query: 159 GGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GG GGGG R GGGGGGG+ G
Sbjct: 270 GGRGGGGGG-------------------------RGGGGSGGGGGGGSGRG 295
Score = 41.6 bits (96), Expect = 3e-04
Identities = 42/130 (32%), Positives = 44/130 (33%), Gaps = 60/130 (46%)
Query: 40 GSGGGGKAVAATVEGGGNGGGG---GGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGR 96
G GGGG+ GGG GGGG GGG + GG GGG
Sbjct: 226 GGGGGGQGGHKGGGGGGQGGGGHKGGGGGQ----------------------GGHKGGGG 263
Query: 97 AAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGV 156
VG G GGG GGG GGG G G G
Sbjct: 264 GGHVGGG-------------------GRGGG--------GGGRGGG--------GSGGGG 288
Query: 157 GGGGDYGGGG 166
GGG GGGG
Sbjct: 289 GGGSGRGGGG 298
Score = 37.4 bits (85), Expect = 0.006
Identities = 39/129 (30%), Positives = 43/129 (33%), Gaps = 48/129 (37%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG+ +GG GGGGG G + GG GGG+
Sbjct: 103 GGGQGGQKGGGGGQGGQKGGGGGQGGQK----------------------GGGGGGQGGQ 140
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGG--GWWWWWWQ*GGGCGVG 157
G G GGG GGG GG G Q GGG G
Sbjct: 141 KG---------------------GGGGGQGGQKGGGGGGQGGQKGGGGQGGQKGGG---G 176
Query: 158 GGGDYGGGG 166
GG GGGG
Sbjct: 177 QGGQKGGGG 185
Score = 35.8 bits (81), Expect = 0.019
Identities = 39/127 (30%), Positives = 40/127 (30%), Gaps = 59/127 (46%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG+ GG GGGGGG GG GGG
Sbjct: 89 GGGGGGQ--------GGQKGGGGGGQ-----------------------GGQKGGG---- 113
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G GG K GG GGG Q GGG G G G
Sbjct: 114 -----------------------GGQGGQKGGGGGQGGQKGGGGGGQGGQKGGGGG-GQG 149
Query: 160 GDYGGGG 166
G GGGG
Sbjct: 150 GQKGGGG 156
>At3g23450 hypothetical protein
Length = 452
Score = 46.6 bits (109), Expect = 1e-05
Identities = 50/167 (29%), Positives = 52/167 (30%), Gaps = 84/167 (50%)
Query: 43 GGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGV 102
GGGK GGG GGG GGG GGG GGG G
Sbjct: 41 GGGK-------GGGFGGGFGGGA-----------------------GGGVGGGAGGGFGG 70
Query: 103 AATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDY 162
A G GGG GGG GGG GGG G GGGG +
Sbjct: 71 GAGG----------------GFGGGGG------GGGGGGG--------GGGGGFGGGGGF 100
Query: 163 GGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GGG V + GGG GGG G
Sbjct: 101 GGGHGGGVGGGV------------------------GGGHGGGVGGG 123
Score = 45.8 bits (107), Expect = 2e-05
Identities = 41/128 (32%), Positives = 47/128 (36%), Gaps = 43/128 (33%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG +GGG GGGGG G GGG GGG
Sbjct: 303 GIGKGGGIGGGIGKGGGIGGGGGFGK-----------------------GGGIGGGIGKG 339
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GG-GCGVGG 158
G+ G GG K G G GGG + + GG G G+GG
Sbjct: 340 GGIG-------------------GGGGFGKGGGIGGGIGKGGGIGGGFGKGGGIGGGIGG 380
Query: 159 GGDYGGGG 166
GG +GGGG
Sbjct: 381 GGGFGGGG 388
Score = 45.1 bits (105), Expect = 3e-05
Identities = 42/127 (33%), Positives = 45/127 (35%), Gaps = 49/127 (38%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG A GGG GGGGGGG GGG GGG
Sbjct: 65 GGGFGGGAGGGFGGGGGGGGGGGGGG-----------------------GGGFGGGGGFG 101
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GG-GCGVGG 158
G G GGG GGG GGG + + GG G G+G
Sbjct: 102 GGHGG------------------GVGGGV-------GGGHGGGVGGGFGKGGGIGGGIGK 136
Query: 159 GGDYGGG 165
GG GGG
Sbjct: 137 GGGVGGG 143
Score = 43.9 bits (102), Expect = 7e-05
Identities = 42/126 (33%), Positives = 46/126 (36%), Gaps = 49/126 (38%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+GGG A GGG GGGGGGG + GGG GGG
Sbjct: 63 GAGGGFGGGAGGGFGGGGGGGGGGGGGG---------------GGGFGGGGGFGGGHGGG 107
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
VG G GGG GGG GGG+ G G G+GGG
Sbjct: 108 VGG--------------------GVGGGH-------GGGVGGGF-------GKGGGIGGG 133
Query: 160 GDYGGG 165
GGG
Sbjct: 134 IGKGGG 139
Score = 42.4 bits (98), Expect = 2e-04
Identities = 41/130 (31%), Positives = 45/130 (34%), Gaps = 48/130 (36%)
Query: 40 GSGGG---GKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGR 96
G GGG G + + GGG GGGGG + GGG GGG
Sbjct: 363 GIGGGFGKGGGIGGGIGGGGGFGGGGGFGK----------------------GGGIGGGI 400
Query: 97 AAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGV 156
G G GGG G G GGG+ GGG G
Sbjct: 401 GKGGGFGGGGGFGKGG----------GIGGGG-------GFGKGGGFG------GGGFGG 437
Query: 157 GGGGDYGGGG 166
GGGG GGGG
Sbjct: 438 GGGGGGGGGG 447
Score = 42.0 bits (97), Expect = 3e-04
Identities = 40/127 (31%), Positives = 44/127 (34%), Gaps = 41/127 (32%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG +GGG GGGGG G GGG GGG
Sbjct: 325 GFGKGGGIGGGIGKGGGIGGGGGFGK-----------------------GGGIGGGIGKG 361
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G+ G GGG GG GGG + GGG G GGG
Sbjct: 362 GGIGGG----------------FGKGGGIGGGIGGGGGFGGGGGFGKGGGIGGGIGKGGG 405
Query: 160 GDYGGGG 166
+GGGG
Sbjct: 406 --FGGGG 410
Score = 42.0 bits (97), Expect = 3e-04
Identities = 42/129 (32%), Positives = 42/129 (32%), Gaps = 49/129 (37%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGG A GGG GGG GGG GGG GGG
Sbjct: 47 GFGGGFGGGAGGGVGGGAGGGFGGGA-----------------------GGGFGGGGGGG 83
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGG GGG GGG GGG G G G
Sbjct: 84 GGGG-------------------GGGGGGFGGGGGFGGGHGGGV-------GGGVGGGHG 117
Query: 160 GDYGGGGDK 168
G GGG K
Sbjct: 118 GGVGGGFGK 126
Score = 39.3 bits (90), Expect = 0.002
Identities = 39/129 (30%), Positives = 45/129 (34%), Gaps = 21/129 (16%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG +GGG GGG G G + GGG G G
Sbjct: 243 GIGKGGGIGGGIGKGGGIGGGIGKGGGI---------------------GGGIGKGGGIG 281
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G+ + + IG GGG GG GGG + GGG G GGG
Sbjct: 282 GGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGGGFGKGGGIGGGIGKGGG 341
Query: 160 GDYGGGGDK 168
GGG K
Sbjct: 342 IGGGGGFGK 350
Score = 38.1 bits (87), Expect = 0.004
Identities = 40/126 (31%), Positives = 45/126 (34%), Gaps = 29/126 (23%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG +GGG GGG G G V GGG G G
Sbjct: 133 GIGKGGGVGGGIGKGGGIGGGIGKGGGV---------------------GGGIGKGGGIG 171
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G+ + + IG GGG GGG GGG+ G G GVGGG
Sbjct: 172 GGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIG-KGGGVGGGF-------GKGGGVGGG 223
Query: 160 GDYGGG 165
GGG
Sbjct: 224 IGKGGG 229
Score = 37.7 bits (86), Expect = 0.005
Identities = 41/133 (30%), Positives = 47/133 (34%), Gaps = 27/133 (20%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG +GGG GGG G G V GGG G G
Sbjct: 213 GFGKGGGVGGGIGKGGGVGGGFGKGGGV---------------------GGGIGKGGGIG 251
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GG-----GDGGGWWWWWWQ*GG-G 153
G+ + + IG GGG GG G GGG + GG G
Sbjct: 252 GGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIG 311
Query: 154 CGVGGGGDYGGGG 166
G+G GG GGGG
Sbjct: 312 GGIGKGGGIGGGG 324
Score = 36.2 bits (82), Expect = 0.014
Identities = 41/126 (32%), Positives = 44/126 (34%), Gaps = 39/126 (30%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG +GGG GGG G G V + GGG GGG
Sbjct: 193 GIGKGGGIGGGIGKGGGVGGGFGKGGGVGGGIGK---------------GGGVGGGFGKG 237
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
GV IG GGG GGG GGG G G G+GGG
Sbjct: 238 GGVG----------------GGIGKGGGIGGGIG-KGGGIGGGI-------GKGGGIGGG 273
Query: 160 GDYGGG 165
GGG
Sbjct: 274 IGKGGG 279
Score = 36.2 bits (82), Expect = 0.014
Identities = 39/129 (30%), Positives = 44/129 (33%), Gaps = 33/129 (25%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG +GGG GGG G G + GGG G G
Sbjct: 233 GFGKGGGVGGGIGKGGGIGGGIGKGGGI---------------------GGGIGKGGGIG 271
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G+ + + IG GGG G G GGG GGG G GGG
Sbjct: 272 GGIGKGGGIGGGIGKGGGIGGGIGKGGGIGG-----GIGKGGGI-------GGGIGKGGG 319
Query: 160 GDYGGGGDK 168
GGG K
Sbjct: 320 IGGGGGFGK 328
Score = 30.4 bits (67), Expect = 0.78
Identities = 15/25 (60%), Positives = 15/25 (60%), Gaps = 5/25 (20%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGG 64
G GGGG GGG GGGGGGG
Sbjct: 429 GFGGGGFG-----GGGGGGGGGGGG 448
>At4g36230 putative glycine-rich cell wall protein
Length = 221
Score = 45.8 bits (107), Expect = 2e-05
Identities = 43/128 (33%), Positives = 45/128 (34%), Gaps = 40/128 (31%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG + GGG G GG G D R GGG GGG +
Sbjct: 96 GGGGGGGGGQGSGGGGGEGNGGNGKDN----------SHKRNKSSGGGGGGGGGGGGGSG 145
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGG GGG GGG GGG G GGG
Sbjct: 146 NGSGR------------------GRGGGGGG-----GGGGGGGG-------GGGGGGGGG 175
Query: 160 GDYGGGGD 167
G GGG D
Sbjct: 176 GGGGGGSD 183
Score = 45.1 bits (105), Expect = 3e-05
Identities = 33/89 (37%), Positives = 37/89 (41%), Gaps = 33/89 (37%)
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPV 182
G GGG GGG GGG GGG G GGGG G GG+
Sbjct: 89 GGGGGG-------GGGGGGG--------GGGQGSGGGGGEGNGGN--------------- 118
Query: 183 VTVGRWWRRRRRR*SDGGGGGGGANSGVS 211
G+ +R + S GGGGGGG G S
Sbjct: 119 ---GKDNSHKRNKSSGGGGGGGGGGGGGS 144
Score = 42.4 bits (98), Expect = 2e-04
Identities = 43/127 (33%), Positives = 47/127 (36%), Gaps = 37/127 (29%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG G G+GGGGG G+ + R GGG GGG +
Sbjct: 90 GGGGGGGGGGGGGGGQGSGGGGGEGNG----GNGKDNSHKRNKSSGGGGGGGGGGGGGSG 145
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGG GGG GGG GGG G GGG
Sbjct: 146 NGSGR------------------GRGGGG-------GGGGGGG--------GGGGGGGGG 172
Query: 160 GDYGGGG 166
G GGGG
Sbjct: 173 GGGGGGG 179
Score = 42.4 bits (98), Expect = 2e-04
Identities = 38/126 (30%), Positives = 39/126 (30%), Gaps = 58/126 (46%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG + G G GGGGGGG GGG GGG
Sbjct: 135 GGGGGGGGGSGNGSGRGRGGGGGGG------------------------GGGGGGG---- 166
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G GGG GG DG G W G G G G
Sbjct: 167 -----------------------GGGGGGGGGGGGGGGSDGKGGGW-------GFGFGWG 196
Query: 160 GDYGGG 165
GD GG
Sbjct: 197 GDGPGG 202
Score = 39.3 bits (90), Expect = 0.002
Identities = 47/169 (27%), Positives = 51/169 (29%), Gaps = 89/169 (52%)
Query: 41 SGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVV 100
+GGGG GGG GGGGGGG + G G GGG
Sbjct: 88 AGGGG--------GGGGGGGGGGGGQ----------------------GSGGGGGEGNGG 117
Query: 101 GVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGG 160
S ++ GGG GGG GGG GGG G G G
Sbjct: 118 NGKDNSHKRNKSS-----------GGG--------GGGGGGG--------GGGSGNGSGR 150
Query: 161 DYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GGGG GGGGGGG G
Sbjct: 151 GRGGGG--------------------------------GGGGGGGGGGG 167
Score = 38.1 bits (87), Expect = 0.004
Identities = 44/157 (28%), Positives = 51/157 (32%), Gaps = 57/157 (36%)
Query: 53 EGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVA 112
E GGGGGGG GGG GGG+ + G +
Sbjct: 85 ESRAGGGGGGGGG-----------------------GGGGGGGQGSGGGGGEGNGGNGKD 121
Query: 113 AMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVA 172
++ G GGG GGG G G + GGG G GGGG GGGG
Sbjct: 122 NSHKRNKSSGGGGGGGG------GGGGGSGNGSGRGRGGGGGGGGGGGGGGGGGGG---- 171
Query: 173 AIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GGGGGGG + G
Sbjct: 172 ------------------------GGGGGGGGGGSDG 184
>At5g59630 glycine-rich protein - like
Length = 253
Score = 44.7 bits (104), Expect = 4e-05
Identities = 39/116 (33%), Positives = 41/116 (34%), Gaps = 61/116 (52%)
Query: 54 GGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAA 113
GG +GGGGGGG GGG GGG G
Sbjct: 68 GGSSGGGGGGG------------------------GGGGGGGGGGGGG------------ 91
Query: 114 MVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKV 169
G GGG GGG GGG GGG G GGGG GGGG K+
Sbjct: 92 ---------GGGGG--------GGGGGGG--------GGGGGGGGGGGGGGGGGKL 122
Score = 39.7 bits (91), Expect = 0.001
Identities = 38/121 (31%), Positives = 38/121 (31%), Gaps = 69/121 (57%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
GS GGG GGG GGGGGGG GGG GGG
Sbjct: 69 GSSGGGGGGGGGGGGGGGGGGGGGG------------------------GGGGGGG---- 100
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
GGG GGG GGG GGG G GGG
Sbjct: 101 -------------------------GGG--------GGGGGGG--------GGGGGGGGG 119
Query: 160 G 160
G
Sbjct: 120 G 120
Score = 37.7 bits (86), Expect = 0.005
Identities = 29/101 (28%), Positives = 33/101 (31%), Gaps = 53/101 (52%)
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGG 61
GGGGG GG G GGGG GGG GGGG
Sbjct: 80 GGGGGGGG------------------------------GGGGGGGGGGGGGGGGGGGGGG 109
Query: 62 GGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGV 102
GGG GGG GGG+ ++G+
Sbjct: 110 GGGG-----------------------GGGGGGGKLQLLGL 127
Score = 34.7 bits (78), Expect = 0.041
Identities = 15/25 (60%), Positives = 15/25 (60%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGG 64
G GGGG GGG GGGGGGG
Sbjct: 75 GGGGGGGGGGGGGGGGGGGGGGGGG 99
Score = 34.7 bits (78), Expect = 0.041
Identities = 15/25 (60%), Positives = 15/25 (60%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGG 64
G GGGG GGG GGGGGGG
Sbjct: 79 GGGGGGGGGGGGGGGGGGGGGGGGG 103
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.338 0.154 0.636
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,813,607
Number of Sequences: 26719
Number of extensions: 339893
Number of successful extensions: 15823
Number of sequences better than 10.0: 226
Number of HSP's better than 10.0 without gapping: 174
Number of HSP's successfully gapped in prelim test: 56
Number of HSP's that attempted gapping in prelim test: 4127
Number of HSP's gapped (non-prelim): 3909
length of query: 224
length of database: 11,318,596
effective HSP length: 96
effective length of query: 128
effective length of database: 8,753,572
effective search space: 1120457216
effective search space used: 1120457216
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0320b.3


