

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0311.7
(381 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
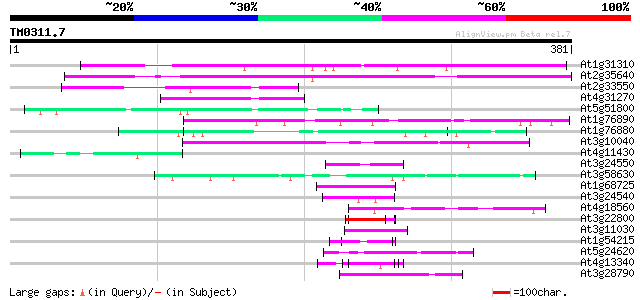
Score E
Sequences producing significant alignments: (bits) Value
At1g31310 unknown protein 233 1e-61
At2g35640 unknown protein 226 1e-59
At2g33550 unknown protein 68 7e-12
At4g31270 unknown protein 63 2e-10
At5g51800 putative protein 56 4e-08
At1g76890 unknown protein 54 2e-07
At1g76880 unknown protein 53 3e-07
At3g10040 unknown protein 51 1e-06
At4g11430 putative proline-rich protein 47 2e-05
At3g24550 protein kinase, putative 45 5e-05
At3g58630 unknown protein 45 7e-05
At1g68725 putative arabinogalactan protein AGP19 45 7e-05
At3g24540 protein kinase, putative 45 9e-05
At4g18560 pherophorin - like protein 44 1e-04
At3g22800 hypothetical protein 44 1e-04
At3g11030 unknown protein 44 1e-04
At1g54215 putative protein 44 1e-04
At5g24620 thaumatin-like protein 44 1e-04
At4g13340 extensin-like protein 44 1e-04
At3g28790 unknown protein 44 1e-04
>At1g31310 unknown protein
Length = 383
Score = 233 bits (594), Expect = 1e-61
Identities = 138/389 (35%), Positives = 203/389 (51%), Gaps = 78/389 (20%)
Query: 49 RDYRKGNWTIQETLILITAKKLDDERRLKASSPPPPPTTTTTPHDPSTTPRPSPAACSAA 108
R+YRKGNWT+ ET++LI AK++DDERR++ S PPP
Sbjct: 12 REYRKGNWTLNETMVLIEAKRMDDERRMRRSIGLPPPEQQQDIR---------------- 55
Query: 109 SSSRTSGELRWKWVENYCWSHGCLRSQNQCNDKWDNLLRDYKKVRDYESK---------- 158
S ELRWKW+E+YCW GC+RSQNQCNDKWDNL+RDYKKVR+YE +
Sbjct: 56 --SNKPAELRWKWIEDYCWRKGCMRSQNQCNDKWDNLMRDYKKVREYERRRVESSITAGE 113
Query: 159 -SESKDHNYNFPSYWTLNKQQRKDHNLPSNMVFEIYQAISDVLQRKN--SQRFITTTT-- 213
S S SYW + K +RK+ +LPSNM+ + YQA+ +V++ K S +T T
Sbjct: 114 SSSSSAPAGETASYWKMEKSERKERSLPSNMLPQTYQALFEVVESKTLPSSTAVTAVTAA 173
Query: 214 -------------------------------PQQQQ--QQQPLVTLVTSTPPPQHPQPLQ 240
P+ Q QQQP++ + PPP QPL
Sbjct: 174 VAAAAAAISSGNGSGGGQIQKVIQQGLGFVVPKVHQIIQQQPVLLPLQPPPPPPPSQPL- 232
Query: 241 AQPLPPPPPPPPQPPATSITPA--------GSEYNHGDQCVKEKTESSGSEQSDQEENNG 292
+PL PPPPPP A I P SEY+ + +T + +
Sbjct: 233 PRPLLLPPPPPPSFHAQPILPTKDSSTDSDTSEYSDTSPAKRRRTMPTTTTAGPSGGGVD 292
Query: 293 SES---SKRRKVRNIGSSIMRSASVLARALRSCEEKKEKRHREMMELEQRRIQMEENRNE 349
E SKR + + +++ RS SV+A A+R EE++++RH+E+M +++RR+++EE+ E
Sbjct: 293 VEEVGRSKRDEETTVAAALSRSVSVIANAIRESEERQDRRHKEVMNVQERRLKIEESNVE 352
Query: 350 VHRQGLATLVTAVSNLSGAIQSFINSQRH 378
++R+G+ LV A++ L+ +I + +S RH
Sbjct: 353 MNREGMNGLVEAINKLASSIFALASSSRH 381
>At2g35640 unknown protein
Length = 340
Score = 226 bits (577), Expect = 1e-59
Identities = 127/369 (34%), Positives = 192/369 (51%), Gaps = 64/369 (17%)
Query: 38 APPLPSSSSAPRDYRKGNWTIQETLILITAKKLDDERRLKASSPPPPPTTTTTPHDPSTT 97
A P R+ RKGNWT+ ETL+LI AKK+DD+RR++ S P
Sbjct: 4 ADPSSGEQIVMRECRKGNWTVSETLVLIEAKKMDDQRRVRRSEKQPEG------------ 51
Query: 98 PRPSPAACSAASSSRTSGELRWKWVENYCWSHGCLRSQNQCNDKWDNLLRDYKKVRDYE- 156
R PA ELRWKW+E YCW GC R+QNQCNDKWDNL+RDYKK+R+YE
Sbjct: 52 -RNKPA------------ELRWKWIEEYCWRRGCYRNQNQCNDKWDNLMRDYKKIREYER 98
Query: 157 SKSESKDHNYNFPSYWTLNKQQRKDHNLPSNMVFEIYQAISDVLQRKN------------ 204
S+ ES + SYW ++K +RK+ NLPSNM+ +IY +S+++ RK
Sbjct: 99 SRVESSFNTVTSSSYWKMDKTERKEKNLPSNMLPQIYDVLSELVDRKTLPSSSSAAAAVG 158
Query: 205 ----------SQRFITTTTPQQQQQQQPLVTLVTSTPPPQHPQPLQ-AQPLPPPPPPPPQ 253
Q+ + P Q + T + + PP PQ L + P PP PPP
Sbjct: 159 NGNGGQILRVCQQSLGFVAPMMAQPMHQIPTTIVLSLPPPPPQSLSLSLPSPPQPPPSSS 218
Query: 254 PPATSITP-AGSEYNHGDQCVKEKTESSGSEQSDQEENNGSESSKRRKVRNIGSSIMRSA 312
A I P G+ + +T + G + +++ +G ++ R
Sbjct: 219 FHAEPIPPTVGTSSTKRRRTTPGETTAGGEREVEEDA--------------VGVALSRCT 264
Query: 313 SVLARALRSCEEKKEKRHREMMELEQRRIQMEENRNEVHRQGLATLVTAVSNLSGAIQSF 372
SV+ + +R EE +E+RH+E++ L++RR+++EE++ E++RQG+ LV A++ L+ +I +
Sbjct: 265 SVITQVIRENEEGQERRHKEVVRLQERRLKIEESKTEINRQGMNGLVDAINQLASSILAL 324
Query: 373 INSQRHGQR 381
+S H R
Sbjct: 325 ASSSCHNNR 333
>At2g33550 unknown protein
Length = 314
Score = 68.2 bits (165), Expect = 7e-12
Identities = 43/163 (26%), Positives = 68/163 (41%), Gaps = 34/163 (20%)
Query: 36 TTAPPLPSSSSAPRDYRKGNWTIQETLILITAKKLDDERRLKASSPPPPPTTTTTPHDPS 95
++AP + R WT QE L+LI K++ + R +
Sbjct: 19 SSAPSNDGGDDGVKTARLPRWTRQEILVLIQGKRVAENRVRRG----------------- 61
Query: 96 TTPRPSPAACSAASSSRTSGELRWKW--VENYCWSHGCLRSQNQCNDKWDNLLRDYKKVR 153
AA + SG++ KW V +YC HG R QC +W NL DYKK++
Sbjct: 62 ----------RAAGMALGSGQMEPKWASVSSYCKRHGVNRGPVQCRKRWSNLAGDYKKIK 111
Query: 154 DYESKSESKDHNYNFPSYWTLNKQQRKDHNLPSNMVFEIYQAI 196
++ES+ + + SYW + R++ LP E+Y +
Sbjct: 112 EWESQIKEETE-----SYWVMRNDVRREKKLPGFFDKEVYDIV 149
>At4g31270 unknown protein
Length = 294
Score = 63.2 bits (152), Expect = 2e-10
Identities = 32/98 (32%), Positives = 54/98 (54%), Gaps = 5/98 (5%)
Query: 103 AACSAASSSRTSGELRWKWVENYCWSHGCLRSQNQCNDKWDNLLRDYKKVRDYESKSESK 162
AA A S+ S +W + C + R+ NQC KWD+L+ DY +++ +ES+
Sbjct: 32 AAVEADCSNALSSFQKWTMITENCNALDVSRNLNQCRRKWDSLMSDYNQIKKWESQYRGT 91
Query: 163 DHNYNFPSYWTLNKQQRKDHNLPSNMVFEIYQAISDVL 200
SYW+L+ +RK NLP ++ E+++AI+ V+
Sbjct: 92 GR-----SYWSLSSDKRKLLNLPGDIDIELFEAINAVV 124
>At5g51800 putative protein
Length = 972
Score = 55.8 bits (133), Expect = 4e-08
Identities = 67/313 (21%), Positives = 103/313 (32%), Gaps = 92/313 (29%)
Query: 11 PPPINIPDP----------------HHHHHHNHHLP--LLHGTTTAPPLPSSSSAPRDYR 52
PPP I P HHHHHH LP + T ++P P PR
Sbjct: 12 PPPNQISSPKDSSLDHQAPNPSLLHHHHHHHQSFLPTPIFIPTVSSPGAPVIPKRPRFST 71
Query: 53 KGNWTIQETLILITAKKLDDERRLKASSPPPPPTTTTTPHDPSTTPRPSPAACSAASSSR 112
+ + L + + + +S P P+T + T SP A +S +
Sbjct: 72 SSGLSPPQWKALPSPSTVPTASTISSS---PTPSTAVVTASSTETAGSSPPGQEATNSEK 128
Query: 113 TS-----------------------GELRW------------------------------ 119
E+ W
Sbjct: 129 QQQPKTESFQHKFRKGKYVSPVWKPNEMLWLARAWRAQYQTQGTGSGSGSVEGRGKTRAE 188
Query: 120 --KWVENYCWSHGCLRSQNQCNDKWDNLLRDYKKVRDYESKSESKDHNYNFPSYWTLNKQ 177
+ V Y HG R KWDN+L +++KV ++E + + SY+ L+
Sbjct: 189 KDREVAEYLNRHGINRDSKIAGTKWDNMLGEFRKVYEWEKCGDQDKYG---KSYFRLSPY 245
Query: 178 QRKDHNLPSNMVFEIYQAISDVLQRKNSQRFITTTTPQQQQQQQPLVTLVTSTPPPQHPQ 237
+RK H LP++ E+YQ ++ + + + T + VT V STPP
Sbjct: 246 ERKQHRLPASFDEEVYQELALFMGPR-----VRAPTINRGGGGGATVT-VASTPP----- 294
Query: 238 PLQAQPLPPPPPP 250
+ LPPP P
Sbjct: 295 --SVEALPPPLYP 305
>At1g76890 unknown protein
Length = 575
Score = 53.5 bits (127), Expect = 2e-07
Identities = 70/286 (24%), Positives = 123/286 (42%), Gaps = 39/286 (13%)
Query: 119 WKWVENYCWSHGCLRSQNQCNDKWDNLLRDYKKVRDYES-KSESKDHNY--NFPSYWTLN 175
W+ + G RS +C +K++N+ + +K+ ++ + KSE K + + ++ TL+
Sbjct: 71 WEEISRKMMELGYKRSSKKCKEKFENVYKYHKRTKEGRTGKSEGKTYRFFEELEAFETLS 130
Query: 176 KQQRKDHNLP--SNMVFEIYQAISDVLQRKNSQRFITTTTPQQQQQQQPL-------VTL 226
Q + + P S+ V A S ++ +I+++ P ++ PL V
Sbjct: 131 SYQPEPESQPAKSSAVITNAPATSSLIP------WISSSNPSTEKSSSPLKHHHQVSVQP 184
Query: 227 VTSTPPPQHPQPLQAQPLP----PPPPPPPQPPATSITPAGSEYNHGDQCVKEKTESSGS 282
+T+ P QP P P QPP ++ N+ + +S S
Sbjct: 185 ITTNPTFLAKQPSSTTPFPFYSSNNTTTVSQPPISN-----DLMNNVSSLNLFSSSTSSS 239
Query: 283 EQSDQEENNGSESSKRRKVRNIGSSIMRSASVLARALRSCEEKKEKRHREMMEL-EQRRI 341
SD+EE++ S R+K R + + L + L +EK +KR E +E E+ RI
Sbjct: 240 TASDEEEDHHQVKSSRKK-RKYWKGLF---TKLTKELMEKQEKMQKRFLETLEYREKERI 295
Query: 342 QMEE--NRNEVHR--QGLATLVTAVSNLS---GAIQSFINSQRHGQ 380
EE E+ R + TL+ SN + AI SF++ GQ
Sbjct: 296 SREEAWRVQEIGRINREHETLIHERSNAAAKDAAIISFLHKISGGQ 341
Score = 31.2 bits (69), Expect = 0.99
Identities = 22/75 (29%), Positives = 36/75 (47%), Gaps = 4/75 (5%)
Query: 113 TSGELRWKWVENYCWSHGCLRSQNQCNDKWDNLLRDYKKVRDYESKS--ESKDHNYNFPS 170
T G L W+ + G RS +C +KW+N+ + +KKV++ K +SK Y F
Sbjct: 422 TKGPL-WEEISAGMRRLGYNRSAKRCKEKWENINKYFKKVKESNKKRPLDSKTCPY-FHQ 479
Query: 171 YWTLNKQQRKDHNLP 185
L ++ K +P
Sbjct: 480 LEALYNERNKSGAMP 494
>At1g76880 unknown protein
Length = 603
Score = 52.8 bits (125), Expect = 3e-07
Identities = 57/265 (21%), Positives = 97/265 (36%), Gaps = 72/265 (27%)
Query: 75 RLKASSPPPPPTTTTTPHDPSTTPRPSPAACSAASSSRT-------------SGELRWKW 121
+L ++ PP + P P+ P+P A S +++T + RW
Sbjct: 352 QLNNNNQQQPPQRSPPPQPPAPLPQPIQAVVSTLDTTKTDNGGDQNMTPAASASSSRWPK 411
Query: 122 VE---------------------NYCWSH--------GCLRSQNQCNDKWDNLLRDYKKV 152
VE W G R+ +C +KW+N+ + +KKV
Sbjct: 412 VEIEALIKLRTNLDSKYQENGPKGPLWEEISAGMRRLGFNRNSKRCKEKWENINKYFKKV 471
Query: 153 RDYESKSESKDHNYNFPSYWTLNKQQRKDHNLPSNMVFEIYQAISDVLQRKNSQRFITTT 212
+ ES + + + P + L+ R+ + SN + S +++ NS +
Sbjct: 472 K--ESNKKRPEDSKTCPYFHQLDALYRERNKFHSNNNIAASSSSSGLVKPDNSVPLMV-- 527
Query: 213 TPQQQQQQQPLVTLVTSTPPPQHPQPLQAQPLPPPPPPPPQPPATSITPAGSEYNHGDQC 272
Q +QQ P VT T+TP P Q+QP SE N D+
Sbjct: 528 --QPEQQWPPAVTTATTTPAAAQPDQ-QSQP--------------------SEQNFDDE- 563
Query: 273 VKEKTESSGSEQSDQEENNGSESSK 297
E T+ ++ ++EEN E +
Sbjct: 564 --EGTDEEYDDEDEEEENEEEEGGE 586
Score = 46.2 bits (108), Expect = 3e-05
Identities = 55/251 (21%), Positives = 93/251 (36%), Gaps = 53/251 (21%)
Query: 119 WKWVENYCWSHGCLRSQNQCNDKWDNLLRDYKKVRDYES-KSESKDHNYNFPSYWTLNKQ 177
W+ V HG +R+ +C +K++N+ + +K+ ++ + KSE K + +
Sbjct: 91 WEEVSRKMAEHGYIRNAKKCKEKFENVYKYHKRTKEGRTGKSEGKTYRF----------- 139
Query: 178 QRKDHNLPSNMVFEIYQAISDVLQRKNSQRFITTTTPQQQQQQQPLVTLVTSTPPPQHPQ 237
D L+ SQ +TT+ QQQ PL + +
Sbjct: 140 -------------------FDQLEALESQ---STTSLHHHQQQTPLRPQQNNNNNNNNNN 177
Query: 238 PLQAQPLPPP-PPPPPQPPATSITPAGSEYN-------HGDQCVKEKTESSG--SEQSDQ 287
PPP P P++SI P + N GD T SS S SD
Sbjct: 178 NSSIFSTPPPVTTVMPTLPSSSIPPYTQQINVPSFPNISGDFLSDNSTSSSSSYSTSSDM 237
Query: 288 EENNGSESSKRRKVR-------NIGSSIMRSASVLARALRSCEEKKEKRHREMMELEQRR 340
E G+ ++++++ R + ++ L R EK+E H ++ E R
Sbjct: 238 EMGGGTATTRKKRKRKWKVFFERLMKQVVDKQEELQRKFLEAVEKRE--HERLVREESWR 295
Query: 341 IQMEENRNEVH 351
+Q N H
Sbjct: 296 VQEIARINREH 306
Score = 35.0 bits (79), Expect = 0.068
Identities = 52/231 (22%), Positives = 84/231 (35%), Gaps = 39/231 (16%)
Query: 83 PPPTTTTTPHDPSTTPRP------------------SPAACSAASSSRTSGELRW----- 119
PPP TT P PS++ P S + S++SS TS ++
Sbjct: 185 PPPVTTVMPTLPSSSIPPYTQQINVPSFPNISGDFLSDNSTSSSSSYSTSSDMEMGGGTA 244
Query: 120 --KWVENYCWSHGCLRSQNQCNDKWDNLLRDY-KKVRDYESKSESKDHNYNFPSYWTLNK 176
+ W R Q DK + L R + + V E + ++ ++ +N+
Sbjct: 245 TTRKKRKRKWKVFFERLMKQVVDKQEELQRKFLEAVEKREHERLVREESWRVQEIARINR 304
Query: 177 QQRKDHNLPS---NMVFEIYQAISDVLQRKNSQRFITTTTPQQQQQQQPLVTLVTSTP-- 231
+ H + + +M A+ LQ+ + ++ Q QQ +P + L +
Sbjct: 305 E----HEILAQERSMSAAKDAAVMAFLQKLSEKQPNQPQPQPQPQQVRPSMQLNNNNQQQ 360
Query: 232 PPQHPQPLQAQPLPPPPPPPPQPPATSITPAGSEYNHGDQCVKEKTESSGS 282
PPQ P P PP P P P S N GDQ + +S S
Sbjct: 361 PPQRSPP----PQPPAPLPQPIQAVVSTLDTTKTDNGGDQNMTPAASASSS 407
>At3g10040 unknown protein
Length = 418
Score = 50.8 bits (120), Expect = 1e-06
Identities = 59/241 (24%), Positives = 99/241 (40%), Gaps = 29/241 (12%)
Query: 118 RWKWVENYCWSHGCLRSQNQCNDKWDNLLRDYKKVRDYESKSESKDHNYNFPSYWTLNKQ 177
+WK V G S QC DK+++L + YK+V D K + N +++
Sbjct: 142 KWKSVSRAMVEKGFSVSPQQCEDKFNDLNKRYKRVNDILGKGIACRVVENQGLLESMDHL 201
Query: 178 QRKDHNLPSNMVFEIYQAISDVLQRKNSQRFITTTTPQQQQQQQPLVTLVTSTPPPQHPQ 237
K + ++ + ++ NS + Q PP Q+P
Sbjct: 202 TPKLKDEVKKLLNSKHLFFREMCAYHNSCGHLGGHDQQ---------------PPQQNPI 246
Query: 238 PLQAQPLPPPPPPPPQPPATSITPAGSEYNHGDQC-VKEKTESSGSEQSDQEENNGSESS 296
+ P P Q AG ++ V+E+ ES +E S+ E SE
Sbjct: 247 SI--------PIPSQQQNCFHAAEAGKMARIAERVEVEEEVESDMAEDSESEMEE-SEEE 297
Query: 297 KRRKVRNIGSSIMR----SASVLARALRSCEEKKEKRHREMMELEQRRIQMEENRNEVHR 352
+ RK R I +++ R +ASV+ +S EKKE R+M+E+E+++I E E+ +
Sbjct: 298 ETRKKRRISTAVKRLREEAASVVEDVGKSVWEKKEWIRRKMLEIEEKKIGYEWEGVEMEK 357
Query: 353 Q 353
Q
Sbjct: 358 Q 358
>At4g11430 putative proline-rich protein
Length = 219
Score = 46.6 bits (109), Expect = 2e-05
Identities = 35/118 (29%), Positives = 43/118 (35%), Gaps = 24/118 (20%)
Query: 8 PIPPPPINIPDPHHHHHHNHHLPLLHGTTTAPPLPSSSSAPRDYRKGNWTIQETLILITA 67
P PPPP++ HHHH P PP P P TI + TA
Sbjct: 99 PPPPPPLSAITTTGHHHHRRSPP--------PPPPPPPPPP--------TITPPVTTTTA 142
Query: 68 KKLDDERRLKASSPPPPP--------TTTTTPHDPSTTPRPSPAACSAASSSRTSGEL 117
R PPPPP TTTTT H P P PA + +++ +L
Sbjct: 143 GHHHHRRSPPPPPPPPPPPPTITPPVTTTTTGHHHHRPPPPPPATTTPITNTSDHHQL 200
Score = 40.0 bits (92), Expect = 0.002
Identities = 19/39 (48%), Positives = 20/39 (50%), Gaps = 8/39 (20%)
Query: 231 PPPQHPQPLQA--------QPLPPPPPPPPQPPATSITP 261
PPP P PL A PPPPPPP PP +ITP
Sbjct: 97 PPPPPPPPLSAITTTGHHHHRRSPPPPPPPPPPPPTITP 135
Score = 39.7 bits (91), Expect = 0.003
Identities = 25/66 (37%), Positives = 28/66 (41%), Gaps = 20/66 (30%)
Query: 209 ITTTTPQQQQQQQPLVTLVTSTPPPQHPQPLQAQPL-------------PPPPPPPPQPP 255
ITTT ++ P PPP P P P+ PPPPPPPP PP
Sbjct: 108 ITTTGHHHHRRSPP------PPPPPPPPPPTITPPVTTTTAGHHHHRRSPPPPPPPPPPP 161
Query: 256 ATSITP 261
T ITP
Sbjct: 162 PT-ITP 166
Score = 37.4 bits (85), Expect = 0.014
Identities = 19/62 (30%), Positives = 30/62 (47%), Gaps = 5/62 (8%)
Query: 211 TTTPQQQQQQQPLVTLVTSTP-----PPQHPQPLQAQPLPPPPPPPPQPPATSITPAGSE 265
TT +Q + P + + T+T PP +P + PPPPP PP ++IT G
Sbjct: 55 TTAGHHRQLRPPSIPVTTNTGHRHCRPPSNPATTNSGHHQLRPPPPPPPPLSAITTTGHH 114
Query: 266 YN 267
++
Sbjct: 115 HH 116
Score = 36.2 bits (82), Expect = 0.031
Identities = 17/42 (40%), Positives = 22/42 (51%), Gaps = 3/42 (7%)
Query: 222 PLVTLVTSTPPPQHPQPLQAQPLPPPPPPPP---QPPATSIT 260
P +T +T H ++ P PPPPPPPP PP T+ T
Sbjct: 131 PTITPPVTTTTAGHHHHRRSPPPPPPPPPPPPTITPPVTTTT 172
Score = 34.7 bits (78), Expect = 0.089
Identities = 15/39 (38%), Positives = 21/39 (53%)
Query: 222 PLVTLVTSTPPPQHPQPLQAQPLPPPPPPPPQPPATSIT 260
P ++ +T+T H + P PPPPPP PP T+ T
Sbjct: 103 PPLSAITTTGHHHHRRSPPPPPPPPPPPPTITPPVTTTT 141
Score = 34.3 bits (77), Expect = 0.12
Identities = 28/94 (29%), Positives = 31/94 (32%), Gaps = 30/94 (31%)
Query: 4 PSTTPIPPPPINIPDP------HHHHHHNHHLPLLHGTTTAPPLPSSSSAPRDYRKGNWT 57
P P PPPP I P HHHH P PP P + + P
Sbjct: 121 PPPPPPPPPPPTITPPVTTTTAGHHHHRRSPPP----PPPPPPPPPTITPP--------- 167
Query: 58 IQETLILITAKKLDDERRLKASSPPPPPTTTTTP 91
+ T R PPPPP TTTP
Sbjct: 168 -----VTTTTTGHHHHR------PPPPPPATTTP 190
Score = 31.6 bits (70), Expect = 0.76
Identities = 25/87 (28%), Positives = 30/87 (33%), Gaps = 29/87 (33%)
Query: 4 PSTTPIPPPPINIPDPHHHHHHNHHLPLLHGTTTAPPLPSSSSAPRDYRKGNWTIQETLI 63
P P PPP I P H+HH P PP P +++ P
Sbjct: 154 PPPPPPPPPTITPPVTTTTTGHHHHRP--------PPPPPATTTP--------------- 190
Query: 64 LITAKKLDDERRLKASSPPPPPTTTTT 90
D +L PPPP TT TT
Sbjct: 191 ---ITNTSDHHQL---HPPPPATTITT 211
Score = 31.2 bits (69), Expect = 0.99
Identities = 23/77 (29%), Positives = 27/77 (34%), Gaps = 31/77 (40%)
Query: 210 TTTTPQQQQQQQPLVTLVTSTPPPQHPQPLQAQPL-----------PPPPPPP------- 251
TTT ++ P PPP P P P+ PPPPPP
Sbjct: 139 TTTAGHHHHRRSP-----PPPPPPPPPPPTITPPVTTTTTGHHHHRPPPPPPATTTPITN 193
Query: 252 --------PQPPATSIT 260
P PPAT+IT
Sbjct: 194 TSDHHQLHPPPPATTIT 210
Score = 30.8 bits (68), Expect = 1.3
Identities = 21/64 (32%), Positives = 25/64 (38%), Gaps = 18/64 (28%)
Query: 222 PLVTLVTSTPPPQHPQPLQAQPLPPPPP-----------------PPPQPPATSITPAGS 264
P VT T+ P P PPPPP PPP PPAT+ TP +
Sbjct: 135 PPVTTTTAGHHHHRRSPPPPPPPPPPPPTITPPVTTTTTGHHHHRPPPPPPATT-TPITN 193
Query: 265 EYNH 268
+H
Sbjct: 194 TSDH 197
Score = 30.0 bits (66), Expect = 2.2
Identities = 28/111 (25%), Positives = 43/111 (38%), Gaps = 21/111 (18%)
Query: 11 PPPINIPDPHHH---------HHHNHHLPLLHGTT-TAPPLPSSSSAPRDYRKGNWTIQE 60
PPP+ HH HHH P + PPLP++++ +R+ ++
Sbjct: 12 PPPLATTAGHHRRSPPPATTGHHHRSPPPAITACHHRRPPLPATTAG--HHRQ----LRP 65
Query: 61 TLILITAKKLDDERRLKASSPPPPPTTTTTPHDPSTTPRPSPAACSAASSS 111
I +T R PP P TT + H P P P SA +++
Sbjct: 66 PSIPVTTNTGHRHCR-----PPSNPATTNSGHHQLRPPPPPPPPLSAITTT 111
>At3g24550 protein kinase, putative
Length = 652
Score = 45.4 bits (106), Expect = 5e-05
Identities = 24/54 (44%), Positives = 30/54 (55%), Gaps = 7/54 (12%)
Query: 215 QQQQQQQPLVT-LVTSTPPPQHPQPLQAQPLPPPPPPPPQPPATSITPAGSEYN 267
QQQ +P +VTS PPP+ P P P PPPPP PPA + GS+Y+
Sbjct: 200 QQQNASRPSDNHVVTSLPPPKPPSP------PRKPPPPPPPPAFMSSSGGSDYS 247
Score = 38.5 bits (88), Expect = 0.006
Identities = 22/54 (40%), Positives = 24/54 (43%), Gaps = 3/54 (5%)
Query: 210 TTTTPQQQQQQQPLVTLVTSTPPPQHPQPLQAQPLPPPPP--PPPQPPATSITP 261
TTTTP P T +S PP P P PP PPP PP S+TP
Sbjct: 23 TTTTPPPAASSPPPTTTPSSPPPSPSTNSTSPPPSSPLPPSLPPPSPPG-SLTP 75
Score = 35.4 bits (80), Expect = 0.052
Identities = 32/114 (28%), Positives = 41/114 (35%), Gaps = 23/114 (20%)
Query: 4 PSTTPIPPPPINIPDPHHHHHHNHHLPLLHGTTTAPPLPSSSSAPRDYRKGNWTIQETLI 63
P TTP P PP P + TTT PP +SS P T
Sbjct: 7 PGTTPSPSPPS---------------PPTNSTTTTPPPAASSPPPTTTPSSPPPSPST-- 49
Query: 64 LITAKKLDDERRLKASSPPPPPTTTTTPHDPSTTP----RPSPAACSAASSSRT 113
+ L S PPP P + TP P +P PSP + + S+ R+
Sbjct: 50 --NSTSPPPSSPLPPSLPPPSPPGSLTPPLPQPSPSAPITPSPPSPTTPSNPRS 101
Score = 33.1 bits (74), Expect = 0.26
Identities = 19/53 (35%), Positives = 21/53 (38%), Gaps = 4/53 (7%)
Query: 212 TTPQQQQQQQPLVTLVTSTPPPQHPQPLQAQPLPPPPPPPPQPPATSITPAGS 264
TTP P T+TPPP P P P PPP P S +P S
Sbjct: 9 TTPSPSPPSPP-TNSTTTTPPPAASSP---PPTTTPSSPPPSPSTNSTSPPPS 57
Score = 32.3 bits (72), Expect = 0.44
Identities = 30/111 (27%), Positives = 41/111 (36%), Gaps = 14/111 (12%)
Query: 5 STTPIPPPPINIPDPHHHHHHNHHLPLLHGTTTAPPLPSSSSAPRDYRKGNWTIQETLIL 64
STT PPP + P P P + T+ P P S P G+ T
Sbjct: 22 STTTTPPPAASSPPPTTTPSSPPPSPSTNSTSPPPSSPLPPSLPPPSPPGSLT------- 74
Query: 65 ITAKKLDDERRLKASSPPPPPTTTTTPHDPSTTPRPS--PAACSAASSSRT 113
L +P PP + TTP +P + P P+ P + S+ RT
Sbjct: 75 ---PPLPQPSPSAPITPSPP--SPTTPSNPRSPPSPNQGPPNTPSGSTPRT 120
Score = 30.8 bits (68), Expect = 1.3
Identities = 29/115 (25%), Positives = 40/115 (34%), Gaps = 6/115 (5%)
Query: 222 PLVTLVTSTPPPQHP---QPLQAQPLPPPP--PPPPQPPATSITPAGSEYNHG-DQCVKE 275
P L S PPP P P QP P P P PP P S + N G
Sbjct: 56 PSSPLPPSLPPPSPPGSLTPPLPQPSPSAPITPSPPSPTTPSNPRSPPSPNQGPPNTPSG 115
Query: 276 KTESSGSEQSDQEENNGSESSKRRKVRNIGSSIMRSASVLARALRSCEEKKEKRH 330
T + S ++ S+ V I + +L C++K+ +RH
Sbjct: 116 STPRTPSNTKPSPPSDSSDGLSTGVVVGIAIGGVAILVILTLICLLCKKKRRRRH 170
Score = 30.8 bits (68), Expect = 1.3
Identities = 16/37 (43%), Positives = 21/37 (56%), Gaps = 2/37 (5%)
Query: 80 SPPPPPT--TTTTPHDPSTTPRPSPAACSAASSSRTS 114
SPP PPT TTTTP +++P P+ S S T+
Sbjct: 14 SPPSPPTNSTTTTPPPAASSPPPTTTPSSPPPSPSTN 50
>At3g58630 unknown protein
Length = 321
Score = 45.1 bits (105), Expect = 7e-05
Identities = 69/289 (23%), Positives = 113/289 (38%), Gaps = 48/289 (16%)
Query: 99 RPSPAACSAAS--SSRTSGELRWKWVENYC-WSHGCLRSQ------NQCNDKWDNLLRDY 149
RPSPA S S + L W Y S G LR + N ND+ N R+
Sbjct: 14 RPSPATLSREDCWSEEATFTLIQAWGNRYVDLSRGNLRQKHWQEVANAVNDRHYNTGRNV 73
Query: 150 K--KVRDYESKSESKDHNYNFPSYWTLNKQQRKDHNLPSNMV--FEIYQAISDVLQRKNS 205
K + Y + + K+ + + K + + N P + + + A+ D+L+
Sbjct: 74 SAAKSQPYRTDVQCKNRIDTLKKKYKVEKARVSESN-PGAYISPWPFFSALDDLLR---- 128
Query: 206 QRFITTTTPQQQQQQQPLVTLVTSTPPPQHPQPLQAQPLPPPPPPP-PQPPATS---ITP 261
+ F T++ P + P + P+ P+P P P+ PATS I
Sbjct: 129 ESFPTSSNPDSTD----------NIPHQRLSLPMSINPVPVAPRSAIPRRPATSPAIIPH 178
Query: 262 AGSEY-------------NHGDQCVKEKTESSGSEQSDQEENNGSESSKRRKVRNIGSSI 308
AG + C + +S GS +S +GS + RK+
Sbjct: 179 AGDDLLGFRGNLNAFAAAAAAAACPASEDDSEGS-RSRSSGRSGSNKKRERKIEK-KQGY 236
Query: 309 MRSASVLARALRSCEEKKEKRHREMMELEQRRIQMEENRNEVHRQGLAT 357
A + R + E +EK+ +EM+ELE++R++ + E HR L T
Sbjct: 237 KEVADAIERLGQIYERVEEKKRKEMVELEKQRMRFAKEL-ECHRMQLFT 284
>At1g68725 putative arabinogalactan protein AGP19
Length = 248
Score = 45.1 bits (105), Expect = 7e-05
Identities = 21/54 (38%), Positives = 25/54 (45%)
Query: 209 ITTTTPQQQQQQQPLVTLVTSTPPPQHPQPLQAQPLPPPPPPPPQPPATSITPA 262
I+ TP Q Q P + T +PPP P P P P P PPP P + PA
Sbjct: 91 ISPATPPPQPPQSPPASAPTVSPPPVSPPPAPTSPPPTPASPPPAPASPPPAPA 144
Score = 38.1 bits (87), Expect = 0.008
Identities = 22/56 (39%), Positives = 25/56 (44%), Gaps = 5/56 (8%)
Query: 210 TTTTPQQQQQQQPLVTL-----VTSTPPPQHPQPLQAQPLPPPPPPPPQPPATSIT 260
TTTTP Q P + VT T PP P PPP PP PPA++ T
Sbjct: 55 TTTTPPVSAAQPPASPVTPPPAVTPTSPPAPKVAPVISPATPPPQPPQSPPASAPT 110
Score = 37.4 bits (85), Expect = 0.014
Identities = 21/53 (39%), Positives = 26/53 (48%), Gaps = 3/53 (5%)
Query: 213 TPQQQQQQQPLVTLVTSTPPPQHPQ--PLQAQPLPPPP-PPPPQPPATSITPA 262
TP + + +TPPPQ PQ P A + PPP PPP P + TPA
Sbjct: 78 TPTSPPAPKVAPVISPATPPPQPPQSPPASAPTVSPPPVSPPPAPTSPPPTPA 130
Score = 35.8 bits (81), Expect = 0.040
Identities = 19/55 (34%), Positives = 22/55 (39%), Gaps = 3/55 (5%)
Query: 210 TTTTPQQQQQQQPLVTL---VTSTPPPQHPQPLQAQPLPPPPPPPPQPPATSITP 261
TTT P P T+TPP QP + PPP P PPA + P
Sbjct: 35 TTTAPPPTTAAPPTTAAPPPTTTTPPVSAAQPPASPVTPPPAVTPTSPPAPKVAP 89
Score = 35.0 bits (79), Expect = 0.068
Identities = 17/50 (34%), Positives = 22/50 (44%), Gaps = 3/50 (6%)
Query: 4 PSTTPIPPPPINIPDPH---HHHHHNHHLPLLHGTTTAPPLPSSSSAPRD 50
PS +PP P P H H H +HH P +PP P + P+D
Sbjct: 159 PSPISLPPAPAPAPTKHKRKHKHKRHHHAPAPAPIPPSPPSPPVLTDPQD 208
Score = 32.7 bits (73), Expect = 0.34
Identities = 16/43 (37%), Positives = 21/43 (48%), Gaps = 3/43 (6%)
Query: 222 PLVTLVTSTPPPQHPQPLQAQPLPPPPPPP---PQPPATSITP 261
P+ + T+ PP P A P P PP QPPA+ +TP
Sbjct: 31 PVTSTTTAPPPTTAAPPTTAAPPPTTTTPPVSAAQPPASPVTP 73
Score = 30.4 bits (67), Expect = 1.7
Identities = 23/99 (23%), Positives = 32/99 (32%), Gaps = 29/99 (29%)
Query: 4 PSTTPIPPPPINIPDPHHHHHHNHHLPLLHGTTTAPPLPSSSSAPRDYRKGNWTIQETLI 63
P P+ PPP+ P P + PP P+ AP +++ +
Sbjct: 146 PPPAPVSPPPVQAPSP----------------ISLPPAPAP--APTKHKRKH-------- 179
Query: 64 LITAKKLDDERRLKASSPPPPPTTTTTPHDPSTTPRPSP 102
K A PP PP+ T P PSP
Sbjct: 180 ---KHKRHHHAPAPAPIPPSPPSPPVLTDPQDTAPAPSP 215
Score = 28.1 bits (61), Expect = 8.4
Identities = 19/74 (25%), Positives = 24/74 (31%), Gaps = 27/74 (36%)
Query: 229 STPPPQHPQPLQAQPLPPP-----------------------PPPPPQPPATS----ITP 261
S PP Q P P+ P P P PP PP PP + P
Sbjct: 152 SPPPVQAPSPISLPPAPAPAPTKHKRKHKHKRHHHAPAPAPIPPSPPSPPVLTDPQDTAP 211
Query: 262 AGSEYNHGDQCVKE 275
A S +G + +
Sbjct: 212 APSPNTNGGNALNQ 225
Score = 28.1 bits (61), Expect = 8.4
Identities = 21/69 (30%), Positives = 29/69 (41%), Gaps = 9/69 (13%)
Query: 35 TTTAPPLPSSSSAPRDYRKGNWTIQETLILITAKKLDDERRLKASSPPPPPTTTTTPHDP 94
TTTAPP P++++ P T T + + + PPP T T+P P
Sbjct: 35 TTTAPP-PTTAAPPTTAAPPPTT--------TTPPVSAAQPPASPVTPPPAVTPTSPPAP 85
Query: 95 STTPRPSPA 103
P SPA
Sbjct: 86 KVAPVISPA 94
Score = 28.1 bits (61), Expect = 8.4
Identities = 14/34 (41%), Positives = 19/34 (55%), Gaps = 2/34 (5%)
Query: 80 SPPP--PPTTTTTPHDPSTTPRPSPAACSAASSS 111
SPPP PP T+P +P P+PA+ A +S
Sbjct: 112 SPPPVSPPPAPTSPPPTPASPPPAPASPPPAPAS 145
>At3g24540 protein kinase, putative
Length = 509
Score = 44.7 bits (104), Expect = 9e-05
Identities = 24/60 (40%), Positives = 34/60 (56%), Gaps = 11/60 (18%)
Query: 213 TPQQQQQQQPLVTLVTSTPPPQH--PQPLQAQPLPPP---------PPPPPQPPATSITP 261
T + + QQ ++ +S PPPQ P+PL ++P PPP PPPPP+ P+TS P
Sbjct: 25 TQLKSRWQQITMSSASSPPPPQVFVPEPLFSEPPPPPKAPVNVSLSPPPPPRSPSTSTPP 84
Score = 28.5 bits (62), Expect = 6.4
Identities = 29/116 (25%), Positives = 42/116 (36%), Gaps = 31/116 (26%)
Query: 1 MTDPSTTPIPPPPINIPDPHHHHHHNHHLPLLHGTTTAPPLPSSSSAPRDYRKGNWTIQE 60
+T S + PPP + +P+P ++ + + PP P S S R GN
Sbjct: 34 ITMSSASSPPPPQVFVPEPLFSEPPPPPKAPVNVSLSPPPPPRSPSTSTPPRLGN----- 88
Query: 61 TLILITAKKLDDERRLKASSPPPP-------PTTTTTPHDPSTTPRPSPAACSAAS 109
+PPPP PTT T S +P PSP+ S +
Sbjct: 89 ------------------RNPPPPASPSGQEPTTPTMTPGFSLSP-PSPSRLSTGA 125
>At4g18560 pherophorin - like protein
Length = 637
Score = 44.3 bits (103), Expect = 1e-04
Identities = 40/143 (27%), Positives = 60/143 (40%), Gaps = 31/143 (21%)
Query: 231 PPPQHPQPLQAQPLPP------PPPPPPQPPATSITPAGSEYNHGDQCVKEKTESSGSEQ 284
PPP P PL QP PP PPPPPP PP S++ A ++ V E E S
Sbjct: 295 PPPPPPPPLLQQPPPPPSVSKAPPPPPPPPPPKSLSIASAKVRR----VPEVVEFYHSLM 350
Query: 285 SDQEENNGSESSKRRKVRNIGSSIMRSASVLARALRSCEEKKEKRHREMM-ELEQRRIQM 343
N+ +S+ G + ++LA + R+M+ E+E R + +
Sbjct: 351 RRDSTNSRRDST--------GGGNAAAEAILANS----------NARDMIGEIENRSVYL 392
Query: 344 EENRNEVHRQG--LATLVTAVSN 364
+ +V QG + L+ V N
Sbjct: 393 LAIKTDVETQGDFIRFLIKEVGN 415
Score = 38.1 bits (87), Expect = 0.008
Identities = 31/130 (23%), Positives = 55/130 (41%), Gaps = 11/130 (8%)
Query: 236 PQPLQAQPLPPPPPPPPQPPATS-------ITPAGSEYNHGDQCVKEKTESSGSEQSDQE 288
P PL PLPPPPPPP +PP++ I P+ + + + +
Sbjct: 25 PSPL---PLPPPPPPPLKPPSSGSATTKPPINPSKPGFTRSFGVYFPRASAQVHATAAAA 81
Query: 289 ENNGSESSKRRKVRNIGS-SIMRSASVLARALRSCEEKKEKRHREMMELEQRRIQMEENR 347
+NG S RR+V + + L A ++ E + ++ + + R++ E +R
Sbjct: 82 SHNGVVSELRRQVEELREREALLKTENLEIADKNGEIDELRKETARLAEDNERLRREFDR 141
Query: 348 NEVHRQGLAT 357
+E R+ T
Sbjct: 142 SEEMRRECET 151
Score = 37.7 bits (86), Expect = 0.011
Identities = 32/126 (25%), Positives = 52/126 (40%), Gaps = 9/126 (7%)
Query: 144 NLLRDYKKVRDYESKSESKDHNYNFPSYWTLNKQQRKDHNLPSNMVFEIYQAISDVLQRK 203
NL+R K+V + E + N +++ D ++ E Y S+ +
Sbjct: 193 NLIRSLKRVGSLRNLPEPITNQENTNK--SISSSGDADGDIYRKDEIESYSRSSNSEELT 250
Query: 204 NSQRFITTTTPQQQQQQQP---LVTLVTSTPPPQHPQPLQAQPLPPPPPPPP----QPPA 256
S T + + + P ++L ST P P ++ P PPPPPPPP PP
Sbjct: 251 ESSSLSTVRSRVPRVPKPPPKRSISLGDSTENRADPPPQKSIPPPPPPPPPPLLQQPPPP 310
Query: 257 TSITPA 262
S++ A
Sbjct: 311 PSVSKA 316
Score = 28.9 bits (63), Expect = 4.9
Identities = 13/30 (43%), Positives = 17/30 (56%), Gaps = 2/30 (6%)
Query: 81 PPPPPTTTTTPHDPSTTPRPSPAACSAASS 110
PPPPP+ + P P P P P + S AS+
Sbjct: 307 PPPPPSVSKAP--PPPPPPPPPKSLSIASA 334
>At3g22800 hypothetical protein
Length = 470
Score = 44.3 bits (103), Expect = 1e-04
Identities = 17/32 (53%), Positives = 19/32 (59%)
Query: 231 PPPQHPQPLQAQPLPPPPPPPPQPPATSITPA 262
PPP P P P PPPPPPPP PP + P+
Sbjct: 381 PPPPPPPPPPPPPPPPPPPPPPPPPPPYVYPS 412
Score = 43.5 bits (101), Expect = 2e-04
Identities = 17/27 (62%), Positives = 17/27 (62%)
Query: 229 STPPPQHPQPLQAQPLPPPPPPPPQPP 255
S PPP P P P PPPPPPPP PP
Sbjct: 378 SPPPPPPPPPPPPPPPPPPPPPPPPPP 404
Score = 42.7 bits (99), Expect = 3e-04
Identities = 17/31 (54%), Positives = 18/31 (57%)
Query: 231 PPPQHPQPLQAQPLPPPPPPPPQPPATSITP 261
PPP P P P PPPPPPPP PP +P
Sbjct: 383 PPPPPPPPPPPPPPPPPPPPPPPPPYVYPSP 413
Score = 40.4 bits (93), Expect = 0.002
Identities = 16/27 (59%), Positives = 16/27 (59%)
Query: 229 STPPPQHPQPLQAQPLPPPPPPPPQPP 255
S P P P P P PPPPPPPP PP
Sbjct: 375 SPPSPPPPPPPPPPPPPPPPPPPPPPP 401
Score = 38.1 bits (87), Expect = 0.008
Identities = 17/34 (50%), Positives = 18/34 (52%), Gaps = 3/34 (8%)
Query: 231 PPPQHPQPLQAQPLPPPPPPP---PQPPATSITP 261
PPP P P P PPPPPPP P PP +P
Sbjct: 387 PPPPPPPPPPPPPPPPPPPPPYVYPSPPPPPPSP 420
Score = 37.7 bits (86), Expect = 0.011
Identities = 16/36 (44%), Positives = 16/36 (44%)
Query: 231 PPPQHPQPLQAQPLPPPPPPPPQPPATSITPAGSEY 266
PPP P P P PPPPPP PP P Y
Sbjct: 397 PPPPPPPPPPPYVYPSPPPPPPSPPPYVYPPPPPPY 432
Score = 37.7 bits (86), Expect = 0.011
Identities = 17/37 (45%), Positives = 18/37 (47%), Gaps = 6/37 (16%)
Query: 230 TPPPQHPQPLQAQPLPPPPPPPPQPPATSITPAGSEY 266
+PPP P P PPPPPPPP PP P Y
Sbjct: 378 SPPPPPPPP------PPPPPPPPPPPPPPPPPPPPPY 408
Score = 37.0 bits (84), Expect = 0.018
Identities = 18/44 (40%), Positives = 18/44 (40%), Gaps = 8/44 (18%)
Query: 231 PPPQHPQPLQAQPLPPPPPP--------PPQPPATSITPAGSEY 266
PPP P P P PPPPPP PP PP P Y
Sbjct: 399 PPPPPPPPPYVYPSPPPPPPSPPPYVYPPPPPPYVYPPPPSPPY 442
Score = 34.7 bits (78), Expect = 0.089
Identities = 16/36 (44%), Positives = 17/36 (46%), Gaps = 5/36 (13%)
Query: 231 PPPQHPQPLQAQPLPPP-----PPPPPQPPATSITP 261
PPP P P P PPP PPPPP P + P
Sbjct: 391 PPPPPPPPPPPPPPPPPYVYPSPPPPPPSPPPYVYP 426
Score = 30.8 bits (68), Expect = 1.3
Identities = 14/28 (50%), Positives = 14/28 (50%), Gaps = 4/28 (14%)
Query: 231 PPPQHPQPLQAQPLPP----PPPPPPQP 254
PPP P P PP PPPP PQP
Sbjct: 426 PPPPPPYVYPPPPSPPYVYPPPPPSPQP 453
Score = 30.0 bits (66), Expect = 2.2
Identities = 13/28 (46%), Positives = 14/28 (49%)
Query: 229 STPPPQHPQPLQAQPLPPPPPPPPQPPA 256
S PPP P P PPPP P PP+
Sbjct: 412 SPPPPPPSPPPYVYPPPPPPYVYPPPPS 439
Score = 29.6 bits (65), Expect = 2.9
Identities = 15/33 (45%), Positives = 16/33 (48%), Gaps = 4/33 (12%)
Query: 227 VTSTPPPQHPQPLQAQPLPPPPPP----PPQPP 255
V +PPP P P PPPPP PP PP
Sbjct: 409 VYPSPPPPPPSPPPYVYPPPPPPYVYPPPPSPP 441
Score = 29.6 bits (65), Expect = 2.9
Identities = 14/30 (46%), Positives = 15/30 (49%), Gaps = 3/30 (10%)
Query: 229 STPPPQHPQPLQAQPLPPPPPPP---PQPP 255
S PP +P P PPPP PP P PP
Sbjct: 419 SPPPYVYPPPPPPYVYPPPPSPPYVYPPPP 448
Score = 28.9 bits (63), Expect = 4.9
Identities = 14/32 (43%), Positives = 15/32 (46%), Gaps = 5/32 (15%)
Query: 231 PPPQHPQPLQAQPLPPPPPPP---PQPPATSI 259
PPP P P P PPP P P P PP +
Sbjct: 435 PPP--PSPPYVYPPPPPSPQPYMYPSPPCNDL 464
>At3g11030 unknown protein
Length = 451
Score = 44.3 bits (103), Expect = 1e-04
Identities = 20/43 (46%), Positives = 22/43 (50%)
Query: 228 TSTPPPQHPQPLQAQPLPPPPPPPPQPPATSITPAGSEYNHGD 270
TS PPP P P P PPPP PPP A TP G + G+
Sbjct: 70 TSPPPPSPPPPSPPPPSPPPPSPPPPAFAVGKTPEGCDVFKGN 112
Score = 41.2 bits (95), Expect = 0.001
Identities = 18/43 (41%), Positives = 21/43 (47%)
Query: 220 QQPLVTLVTSTPPPQHPQPLQAQPLPPPPPPPPQPPATSITPA 262
Q +V +V PPP P P PP PPPP PP + PA
Sbjct: 54 QSAIVVVVDEPPPPPPTSPPPPSPPPPSPPPPSPPPPSPPPPA 96
Score = 32.3 bits (72), Expect = 0.44
Identities = 18/66 (27%), Positives = 28/66 (42%), Gaps = 10/66 (15%)
Query: 81 PPPPPTTTTTPHDPSTT--------PRPSPAACSAASSSRTSGELRWKWVENYCWSHGCL 132
PPPPPT+ P P + P P P A + + + WV++ WS L
Sbjct: 65 PPPPPTSPPPPSPPPPSPPPPSPPPPSPPPPAFAVGKTPEGCDVFKGNWVKD--WSTRPL 122
Query: 133 RSQNQC 138
+++C
Sbjct: 123 YRESEC 128
>At1g54215 putative protein
Length = 169
Score = 44.3 bits (103), Expect = 1e-04
Identities = 20/43 (46%), Positives = 25/43 (57%), Gaps = 4/43 (9%)
Query: 218 QQQQPLVTLVTSTPPPQHPQPLQAQPLPPPPPPPPQPPATSIT 260
Q Q P + + PPP P P P PPPPPPPP PPA +++
Sbjct: 31 QSQDPPLFPQSPPPPPPPPPP----PPPPPPPPPPPPPAVNMS 69
Score = 43.9 bits (102), Expect = 1e-04
Identities = 20/37 (54%), Positives = 20/37 (54%)
Query: 226 LVTSTPPPQHPQPLQAQPLPPPPPPPPQPPATSITPA 262
LV S PP PQ P PPPPPPPP PP PA
Sbjct: 29 LVQSQDPPLFPQSPPPPPPPPPPPPPPPPPPPPPPPA 65
Score = 38.5 bits (88), Expect = 0.006
Identities = 22/60 (36%), Positives = 22/60 (36%), Gaps = 12/60 (20%)
Query: 217 QQQQQPLVTLVTSTPPPQHPQPLQAQPLPPPPPP------------PPQPPATSITPAGS 264
Q Q PL PPP P P P PPPPPP PP P I P S
Sbjct: 31 QSQDPPLFPQSPPPPPPPPPPPPPPPPPPPPPPPAVNMSVETGIPPPPPPVTDMIKPLSS 90
Score = 32.3 bits (72), Expect = 0.44
Identities = 18/55 (32%), Positives = 26/55 (46%), Gaps = 10/55 (18%)
Query: 209 ITTTTPQQQQQQQPLVTLVTSTPPPQHPQPLQAQPLPPPP--------PPPPQPP 255
+ T P ++ ++S PPPQ P ++QP P PP PPPP+ P
Sbjct: 70 VETGIPPPPPPVTDMIKPLSSPPPPQPPP--RSQPPPKPPQKNLPRRHPPPPRSP 122
Score = 28.9 bits (63), Expect = 4.9
Identities = 12/26 (46%), Positives = 15/26 (57%)
Query: 229 STPPPQHPQPLQAQPLPPPPPPPPQP 254
S PPP+ PQ + PPPP P +P
Sbjct: 100 SQPPPKPPQKNLPRRHPPPPRSPEKP 125
Score = 28.9 bits (63), Expect = 4.9
Identities = 12/25 (48%), Positives = 12/25 (48%)
Query: 80 SPPPPPTTTTTPHDPSTTPRPSPAA 104
SPPPPP P P P P P A
Sbjct: 41 SPPPPPPPPPPPPPPPPPPPPPPPA 65
Score = 28.9 bits (63), Expect = 4.9
Identities = 12/25 (48%), Positives = 12/25 (48%)
Query: 79 SSPPPPPTTTTTPHDPSTTPRPSPA 103
S PPPPP P P P P PA
Sbjct: 41 SPPPPPPPPPPPPPPPPPPPPPPPA 65
Score = 28.9 bits (63), Expect = 4.9
Identities = 18/47 (38%), Positives = 22/47 (46%), Gaps = 8/47 (17%)
Query: 229 STPPPQHPQPLQAQPLPPPPPPP------PQPPATSITPAGSEYNHG 269
S+PPP P P ++QP PP PP P PP + P N G
Sbjct: 89 SSPPPPQPPP-RSQP-PPKPPQKNLPRRHPPPPRSPEKPKRDGLNKG 133
>At5g24620 thaumatin-like protein
Length = 420
Score = 43.9 bits (102), Expect = 1e-04
Identities = 32/102 (31%), Positives = 45/102 (43%), Gaps = 12/102 (11%)
Query: 214 PQQQQQQQPLVTLVTSTPPPQHPQPLQAQPLPPPPPPPPQPPATSITPAGSEYNHGDQCV 273
P Q Q QPL PP Q+ QP+ PP PPPP P S+TP N Q +
Sbjct: 315 PTQNQYDQPLA------PPTQNQY---GQPMAPPTPPPPGPNEDSMTPPPQNQNGDGQFM 365
Query: 274 KEKTESSGSEQSDQEENNGSESSKRRKVRNIGSSIMRSASVL 315
T + +DQ N + +R+ ++ S ++R VL
Sbjct: 366 PPPT--LDQDPNDQYMNPPIQDQSQRETQS-SSDLLRPYPVL 404
>At4g13340 extensin-like protein
Length = 760
Score = 43.9 bits (102), Expect = 1e-04
Identities = 18/31 (58%), Positives = 18/31 (58%)
Query: 231 PPPQHPQPLQAQPLPPPPPPPPQPPATSITP 261
PPP P P P PPPPPPPP PP S P
Sbjct: 454 PPPPPPPPPVYSPPPPPPPPPPPPPVYSPPP 484
Score = 43.5 bits (101), Expect = 2e-04
Identities = 24/52 (46%), Positives = 25/52 (47%), Gaps = 5/52 (9%)
Query: 210 TTTTPQQQQQQQPLVTLVTSTPPPQHPQPLQAQPLPPPPPPPPQPPATSITP 261
T T+P P V S PPP P P P PPPPPPPP PP S P
Sbjct: 422 TLTSPPPPSPPPP----VYSPPPPPPPPPPVYSP-PPPPPPPPPPPVYSPPP 468
Score = 42.7 bits (99), Expect = 3e-04
Identities = 20/43 (46%), Positives = 23/43 (52%), Gaps = 5/43 (11%)
Query: 227 VTSTPPPQHPQPLQAQPLPPPPPP-----PPQPPATSITPAGS 264
+TS PPP P P+ + P PPPPPP PP PP P S
Sbjct: 423 LTSPPPPSPPPPVYSPPPPPPPPPPVYSPPPPPPPPPPPPVYS 465
Score = 42.0 bits (97), Expect = 6e-04
Identities = 19/41 (46%), Positives = 20/41 (48%)
Query: 227 VTSTPPPQHPQPLQAQPLPPPPPPPPQPPATSITPAGSEYN 267
V S PPP P P PPPPPPPP PP P Y+
Sbjct: 479 VYSPPPPSPPPPPPPVYSPPPPPPPPPPPPVYSPPPPPVYS 519
Score = 41.6 bits (96), Expect = 7e-04
Identities = 18/35 (51%), Positives = 19/35 (53%)
Query: 227 VTSTPPPQHPQPLQAQPLPPPPPPPPQPPATSITP 261
V S PPP P P PPPPPPPP PP +P
Sbjct: 448 VYSPPPPPPPPPPPPVYSPPPPPPPPPPPPPVYSP 482
Score = 41.2 bits (95), Expect = 0.001
Identities = 18/45 (40%), Positives = 22/45 (48%)
Query: 208 FITTTTPQQQQQQQPLVTLVTSTPPPQHPQPLQAQPLPPPPPPPP 252
F T T P + + PPP P P+ + P PPPPPPPP
Sbjct: 417 FSTPPTLTSPPPPSPPPPVYSPPPPPPPPPPVYSPPPPPPPPPPP 461
Score = 40.4 bits (93), Expect = 0.002
Identities = 17/34 (50%), Positives = 17/34 (50%)
Query: 231 PPPQHPQPLQAQPLPPPPPPPPQPPATSITPAGS 264
PPP P P PPPPPPPP PP P S
Sbjct: 453 PPPPPPPPPPVYSPPPPPPPPPPPPPVYSPPPPS 486
Score = 38.9 bits (89), Expect = 0.005
Identities = 17/44 (38%), Positives = 20/44 (44%)
Query: 222 PLVTLVTSTPPPQHPQPLQAQPLPPPPPPPPQPPATSITPAGSE 265
P + +S PPP P P PPPPPP PP +P E
Sbjct: 513 PPPPVYSSPPPPPSPAPTPVYCTRPPPPPPHSPPPPQFSPPPPE 556
Score = 38.5 bits (88), Expect = 0.006
Identities = 16/31 (51%), Positives = 16/31 (51%)
Query: 231 PPPQHPQPLQAQPLPPPPPPPPQPPATSITP 261
PPP P P PPP PPPP PP S P
Sbjct: 469 PPPPPPPPPPVYSPPPPSPPPPPPPVYSPPP 499
Score = 37.0 bits (84), Expect = 0.018
Identities = 17/43 (39%), Positives = 20/43 (45%), Gaps = 12/43 (27%)
Query: 231 PPPQHPQPLQAQPLPPPPPP------------PPQPPATSITP 261
PPP P P+ + P PPPPPP PP PP +P
Sbjct: 455 PPPPPPPPVYSPPPPPPPPPPPPPVYSPPPPSPPPPPPPVYSP 497
Score = 37.0 bits (84), Expect = 0.018
Identities = 18/45 (40%), Positives = 21/45 (46%), Gaps = 5/45 (11%)
Query: 222 PLVTLVTSTPPPQHPQPLQAQPLPPPPPP-----PPQPPATSITP 261
P+ + +PPP P P PPPPPP PP PP S P
Sbjct: 478 PVYSPPPPSPPPPPPPVYSPPPPPPPPPPPPVYSPPPPPVYSSPP 522
Score = 36.6 bits (83), Expect = 0.023
Identities = 16/34 (47%), Positives = 18/34 (52%), Gaps = 2/34 (5%)
Query: 230 TPPPQHPQPLQAQPL--PPPPPPPPQPPATSITP 261
+PPP P P P+ PPPP PPP PP P
Sbjct: 465 SPPPPPPPPPPPPPVYSPPPPSPPPPPPPVYSPP 498
Score = 36.6 bits (83), Expect = 0.023
Identities = 19/48 (39%), Positives = 20/48 (41%)
Query: 214 PQQQQQQQPLVTLVTSTPPPQHPQPLQAQPLPPPPPPPPQPPATSITP 261
P Q P S+PPP H P P PP PPPP P S P
Sbjct: 546 PPPQFSPPPPEPYYYSSPPPPHSSPPPHSPPPPHSPPPPIYPYLSPPP 593
Score = 35.8 bits (81), Expect = 0.040
Identities = 17/45 (37%), Positives = 21/45 (45%), Gaps = 15/45 (33%)
Query: 232 PPQHPQPLQAQPLPPPPPPPPQ---------------PPATSITP 261
PP P P+ + P PPPPPPPP PP+ + TP
Sbjct: 487 PPPPPPPVYSPPPPPPPPPPPPVYSPPPPPVYSSPPPPPSPAPTP 531
Score = 34.3 bits (77), Expect = 0.12
Identities = 20/58 (34%), Positives = 23/58 (39%), Gaps = 18/58 (31%)
Query: 222 PLVTLVTSTPPPQHPQPLQAQPL---------------PPP---PPPPPQPPATSITP 261
P +VT PPP P P P+ PPP PPPPP PP +P
Sbjct: 394 PRPPVVTPLPPPSLPSPPPPAPIFSTPPTLTSPPPPSPPPPVYSPPPPPPPPPPVYSP 451
Score = 34.3 bits (77), Expect = 0.12
Identities = 18/41 (43%), Positives = 18/41 (43%), Gaps = 7/41 (17%)
Query: 231 PPPQHPQPLQAQPLPP-------PPPPPPQPPATSITPAGS 264
PPP P P Q P PP PPPP PP S P S
Sbjct: 540 PPPHSPPPPQFSPPPPEPYYYSSPPPPHSSPPPHSPPPPHS 580
Score = 34.3 bits (77), Expect = 0.12
Identities = 17/39 (43%), Positives = 20/39 (50%), Gaps = 8/39 (20%)
Query: 229 STPPPQHPQPLQAQPLP------PPPPPPP--QPPATSI 259
S+PPP P P + P P PPPPP P PP T +
Sbjct: 568 SSPPPHSPPPPHSPPPPIYPYLSPPPPPTPVSSPPPTPV 606
Score = 33.5 bits (75), Expect = 0.20
Identities = 18/43 (41%), Positives = 20/43 (45%), Gaps = 3/43 (6%)
Query: 227 VTSTPPPQH---PQPLQAQPLPPPPPPPPQPPATSITPAGSEY 266
V S+PPP P P+ PPPPP P PP S P Y
Sbjct: 517 VYSSPPPPPSPAPTPVYCTRPPPPPPHSPPPPQFSPPPPEPYY 559
Score = 32.7 bits (73), Expect = 0.34
Identities = 17/47 (36%), Positives = 20/47 (42%), Gaps = 12/47 (25%)
Query: 227 VTSTPPPQHPQPLQAQPLPPPP------------PPPPQPPATSITP 261
V+ PP P P + P PPPP PPPP PP +P
Sbjct: 392 VSPRPPVVTPLPPPSLPSPPPPAPIFSTPPTLTSPPPPSPPPPVYSP 438
Score = 32.3 bits (72), Expect = 0.44
Identities = 19/51 (37%), Positives = 23/51 (44%), Gaps = 10/51 (19%)
Query: 213 TPQQQQQQQPLVTLVTSTPPPQHPQPLQAQP-----LPPPPPP---PPQPP 255
+P P + S PPP P P+ + P PPPPPP PP PP
Sbjct: 574 SPPPPHSPPPPIYPYLSPPPP--PTPVSSPPPTPVYSPPPPPPCIEPPPPP 622
Score = 32.3 bits (72), Expect = 0.44
Identities = 14/30 (46%), Positives = 16/30 (52%), Gaps = 5/30 (16%)
Query: 231 PPPQHPQPLQAQPLPPPPPP-----PPQPP 255
PPP P P+ + P P PPPP P PP
Sbjct: 471 PPPPPPPPVYSPPPPSPPPPPPPVYSPPPP 500
Score = 32.0 bits (71), Expect = 0.58
Identities = 27/103 (26%), Positives = 33/103 (31%), Gaps = 36/103 (34%)
Query: 4 PSTTPIPPPPINIPDPHHHHHHNHHLPLLHGTTTAPPLPSSSSAPRDYRKGNWTIQETLI 63
P P PPPP+ P P P + PP P P Y
Sbjct: 469 PPPPPPPPPPVYSPPPPSPP------PPPPPVYSPPPPPPPPPPPPVY------------ 510
Query: 64 LITAKKLDDERRLKASSPPPPPTTTTTPHDPSTTPRPSPAACS 106
SPPPPP ++ P PS P P+P C+
Sbjct: 511 ----------------SPPPPPVYSSPPPPPS--PAPTPVYCT 535
Score = 31.6 bits (70), Expect = 0.76
Identities = 17/47 (36%), Positives = 19/47 (40%), Gaps = 15/47 (31%)
Query: 230 TPPPQH--PQPLQAQPLPPPPP-------------PPPQPPATSITP 261
+PPP H P P+ PPPPP PPP PP P
Sbjct: 574 SPPPPHSPPPPIYPYLSPPPPPTPVSSPPPTPVYSPPPPPPCIEPPP 620
Score = 31.2 bits (69), Expect = 0.99
Identities = 18/54 (33%), Positives = 21/54 (38%), Gaps = 4/54 (7%)
Query: 212 TTPQQQQQQQPLVTLVTSTPPPQHPQPLQAQPLPPPPPP----PPQPPATSITP 261
++P P T PPP P Q PPPP P P PP +S P
Sbjct: 519 SSPPPPPSPAPTPVYCTRPPPPPPHSPPPPQFSPPPPEPYYYSSPPPPHSSPPP 572
Score = 30.8 bits (68), Expect = 1.3
Identities = 21/59 (35%), Positives = 25/59 (41%), Gaps = 7/59 (11%)
Query: 214 PQQQQQQQPLVTLVTSTPPPQHPQPLQAQPLPP-----PPPPPPQPPATSITPAGSEYN 267
P P T V S PPP P ++ P PP PPPPPP +S P Y+
Sbjct: 593 PPPTPVSSPPPTPVYSPPPP--PPCIEPPPPPPCIEYSPPPPPPVVHYSSPPPPPVYYS 649
Score = 30.8 bits (68), Expect = 1.3
Identities = 16/48 (33%), Positives = 21/48 (43%), Gaps = 9/48 (18%)
Query: 214 PQQQQQQQPLVTLVTSTPPPQH-----PQPLQAQPLPPP----PPPPP 252
P P+ ++ PPP P P+ + P PPP PPPPP
Sbjct: 576 PPPHSPPPPIYPYLSPPPPPTPVSSPPPTPVYSPPPPPPCIEPPPPPP 623
Score = 30.4 bits (67), Expect = 1.7
Identities = 24/96 (25%), Positives = 32/96 (33%), Gaps = 26/96 (27%)
Query: 10 PPPPINIPDPHHHHHHNHHLPLLHGTTTAPPLPSSSSAPRDYRKGNWTIQETLILITAKK 69
PPP + P P +++ + P +PP P S P I L
Sbjct: 546 PPPQFSPPPPEPYYYSSPPPPHSSPPPHSPPPPHSPPPP---------IYPYL------- 589
Query: 70 LDDERRLKASSPPPPPTTTTTPHDPSTTPRPSPAAC 105
SPPPPPT ++P P P C
Sbjct: 590 ----------SPPPPPTPVSSPPPTPVYSPPPPPPC 615
Score = 30.0 bits (66), Expect = 2.2
Identities = 17/44 (38%), Positives = 22/44 (49%), Gaps = 7/44 (15%)
Query: 229 STPPPQHPQPLQAQPLPPPP-----PPPPQPPATSITPAGSEYN 267
S+PPP P + P PPPP PPPP+ S P+ Y+
Sbjct: 649 SSPPP--PPVYYSSPPPPPPVHYSSPPPPEVHYHSPPPSPVHYS 690
Score = 30.0 bits (66), Expect = 2.2
Identities = 19/52 (36%), Positives = 22/52 (41%), Gaps = 9/52 (17%)
Query: 222 PLVTLVTSTPPPQHPQPLQAQPLPPPP-----PPPPQPPATSITPAGSEYNH 268
P V +S PPP P+ PPPP PPPP P S P + H
Sbjct: 633 PPVVHYSSPPPP----PVYYSSPPPPPVYYSSPPPPPPVHYSSPPPPEVHYH 680
Score = 30.0 bits (66), Expect = 2.2
Identities = 14/37 (37%), Positives = 16/37 (42%), Gaps = 6/37 (16%)
Query: 231 PPPQHPQPLQAQP------LPPPPPPPPQPPATSITP 261
PPP P P+ + P PPPPP P P P
Sbjct: 501 PPPPPPPPVYSPPPPPVYSSPPPPPSPAPTPVYCTRP 537
Score = 30.0 bits (66), Expect = 2.2
Identities = 18/58 (31%), Positives = 23/58 (39%), Gaps = 10/58 (17%)
Query: 214 PQQQQQQQPLVTLVTSTPPPQHPQPLQAQPLPPP-----PPPP-----PQPPATSITP 261
P+ P + S+PPP P + P P P PPPP P PP +P
Sbjct: 675 PEVHYHSPPPSPVHYSSPPPPPSAPCEESPPPAPVVHHSPPPPMVHHSPPPPVIHQSP 732
Score = 30.0 bits (66), Expect = 2.2
Identities = 22/93 (23%), Positives = 29/93 (30%), Gaps = 33/93 (35%)
Query: 10 PPPPINIPDPHHHHHHNHHLPLLHGTTTAPPLPSSSSAPRDYRKGNWTIQETLILITAKK 69
PPPP + P PH + P ++ + PP P+
Sbjct: 563 PPPPHSSPPPHSPPPPHSPPPPIYPYLSPPPPPTP------------------------- 597
Query: 70 LDDERRLKASSPPPPPTTTTTPHDPSTTPRPSP 102
SSPPP P + P P P P P
Sbjct: 598 --------VSSPPPTPVYSPPPPPPCIEPPPPP 622
Score = 30.0 bits (66), Expect = 2.2
Identities = 24/100 (24%), Positives = 34/100 (34%), Gaps = 36/100 (36%)
Query: 6 TTPIPPPPINI---PDPHHHHHHNHHLPLLHGTTTAPPLPSSSSAPRDYRKGNWTIQETL 62
++P PPPP++ P P H+H P+ + + PP SAP +
Sbjct: 659 SSPPPPPPVHYSSPPPPEVHYHSPPPSPVHYSSPPPPP-----SAPCE------------ 701
Query: 63 ILITAKKLDDERRLKASSPPPPPTTTTTPHDPSTTPRPSP 102
SPPP P +P P P P
Sbjct: 702 ----------------ESPPPAPVVHHSPPPPMVHHSPPP 725
Score = 29.3 bits (64), Expect = 3.8
Identities = 15/39 (38%), Positives = 16/39 (40%), Gaps = 12/39 (30%)
Query: 229 STPPPQ------------HPQPLQAQPLPPPPPPPPQPP 255
S PPPQ P P + P P PPPP PP
Sbjct: 544 SPPPPQFSPPPPEPYYYSSPPPPHSSPPPHSPPPPHSPP 582
Score = 29.3 bits (64), Expect = 3.8
Identities = 17/49 (34%), Positives = 18/49 (36%), Gaps = 11/49 (22%)
Query: 231 PPPQH----PQPLQAQPLPPP-------PPPPPQPPATSITPAGSEYNH 268
PPP H P P PPP PPPPP P P +H
Sbjct: 664 PPPVHYSSPPPPEVHYHSPPPSPVHYSSPPPPPSAPCEESPPPAPVVHH 712
Score = 28.1 bits (61), Expect = 8.4
Identities = 15/38 (39%), Positives = 17/38 (44%), Gaps = 3/38 (7%)
Query: 228 TSTPPPQ---HPQPLQAQPLPPPPPPPPQPPATSITPA 262
+S PPP+ H P PPPPP P S PA
Sbjct: 670 SSPPPPEVHYHSPPPSPVHYSSPPPPPSAPCEESPPPA 707
>At3g28790 unknown protein
Length = 608
Score = 43.9 bits (102), Expect = 1e-04
Identities = 30/84 (35%), Positives = 35/84 (40%), Gaps = 4/84 (4%)
Query: 225 TLVTSTPPPQHPQPLQAQP-LPPPPPPPPQPPATSITPAGSEYNHGDQCVKEKTESSGSE 283
T STP P P P P P P P P PA S AG G + K ES+
Sbjct: 280 TPTPSTPTPSTPTPSTPTPSTPTPSTPTPSTPAPSTPAAGKTSEKGSESASMKKESNSKS 339
Query: 284 QSDQEENNGSESSKRRKVRNIGSS 307
+S E+ S S + K N GSS
Sbjct: 340 ES---ESAASGSVSKTKETNKGSS 360
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.310 0.126 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,606,256
Number of Sequences: 26719
Number of extensions: 581200
Number of successful extensions: 18996
Number of sequences better than 10.0: 671
Number of HSP's better than 10.0 without gapping: 364
Number of HSP's successfully gapped in prelim test: 317
Number of HSP's that attempted gapping in prelim test: 5680
Number of HSP's gapped (non-prelim): 6107
length of query: 381
length of database: 11,318,596
effective HSP length: 101
effective length of query: 280
effective length of database: 8,619,977
effective search space: 2413593560
effective search space used: 2413593560
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0311.7


