

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0309.4
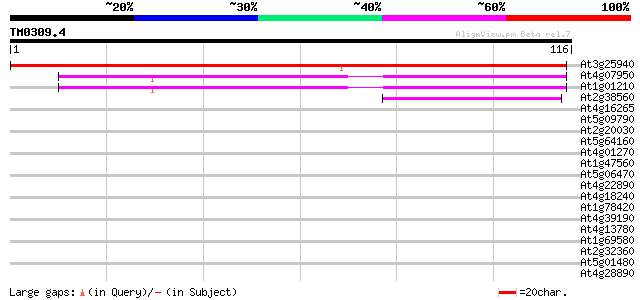
(116 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g25940 unknown protein 140 1e-34
At4g07950 putative DNA-directed RNA polymerase subunit 50 2e-07
At1g01210 unknown protein 47 2e-06
At2g38560 putative elongation factor 41 1e-04
At4g16265 unknown protein 35 0.005
At5g09790 unknown protein 30 0.15
At2g20030 putative RING zinc finger protein 28 0.56
At5g64160 unknown protein 28 0.74
At4g01270 putative RING zinc finger protein 28 0.74
At1g47560 hypothetical protein 28 0.74
At5g06470 putative protein 28 0.96
At4g22890 unknown protein 28 0.96
At4g18240 starch synthase-like protein 28 0.96
At1g78420 unknown protein (At1g78420) 28 0.96
At4g39190 hypothetical protein 27 1.6
At4g13780 methionyl-tRNA synthetase - like protein 27 1.6
At1g69580 transfactor, putative 27 1.6
At2g32360 hypothetical protein 27 2.1
At5g01480 putative protein 26 3.7
At4g28890 unknown protein 26 3.7
>At3g25940 unknown protein
Length = 119
Score = 140 bits (353), Expect = 1e-34
Identities = 67/118 (56%), Positives = 81/118 (67%), Gaps = 3/118 (2%)
Query: 1 MAYSRERDFLFCHYCGTMLTVPSVNYAECPLCKNRSNIKDIEGKEISYTISAEEIRRELG 60
M SRE +FLFC+ CGTML + S YAECP CK N KDI KEI+YT+SAE+IRRELG
Sbjct: 1 MEKSRESEFLFCNLCGTMLVLKSTKYAECPHCKTTRNAKDIIDKEIAYTVSAEDIRRELG 60
Query: 61 IETIEEQ---KVQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRCGHQFQE 115
I E+ + +L K+ K CEKC + E + TRQ RSADEGQTT+Y C C H+F E
Sbjct: 61 ISLFGEKTQAEAELPKIKKACEKCQHPELVYTTRQTRSADEGQTTYYTCPNCAHRFTE 118
>At4g07950 putative DNA-directed RNA polymerase subunit
Length = 106
Score = 49.7 bits (117), Expect = 2e-07
Identities = 31/110 (28%), Positives = 50/110 (45%), Gaps = 12/110 (10%)
Query: 11 FCHYCGTMLTVPSVNYAE-----CPLCKNRSNIKDIEGKEISYTISAEEIRRELGIETIE 65
FC CG +L + CP N +I+ K++ S E + + I T
Sbjct: 3 FCPTCGNLLRYEGGGSSRFFCSTCPYVANIERRVEIKKKQLLVKKSIEPVVTKDDIPTAA 62
Query: 66 EQKVQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRCGHQFQE 115
E + C +CG+ +A F + Q+RSADE ++ FY C +C ++E
Sbjct: 63 ETEAP-------CPRCGHDKAYFKSMQIRSADEPESRFYRCLKCEFTWRE 105
>At1g01210 unknown protein
Length = 106
Score = 46.6 bits (109), Expect = 2e-06
Identities = 30/110 (27%), Positives = 49/110 (44%), Gaps = 12/110 (10%)
Query: 11 FCHYCGTMLTVPSVNYAE-----CPLCKNRSNIKDIEGKEISYTISAEEIRRELGIETIE 65
FC CG +L + CP +I+ K++ S E + + I T
Sbjct: 3 FCPTCGNLLRYEGGGNSRFFCSTCPYVAYIQRQVEIKKKQLLVKKSIEAVVTKDDIPTAA 62
Query: 66 EQKVQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRCGHQFQE 115
E + C +CG+ +A F + Q+RSADE ++ FY C +C ++E
Sbjct: 63 ETEAP-------CPRCGHDKAYFKSMQIRSADEPESRFYRCLKCEFTWRE 105
>At2g38560 putative elongation factor
Length = 378
Score = 40.8 bits (94), Expect = 1e-04
Identities = 15/37 (40%), Positives = 22/37 (58%)
Query: 78 CEKCGNGEATFYTRQMRSADEGQTTFYACTRCGHQFQ 114
C +CG + T+Y Q RSADE TT+ C C + ++
Sbjct: 340 CGRCGQRKCTYYQMQTRSADEPMTTYVTCVNCDNHWK 376
>At4g16265 unknown protein
Length = 114
Score = 35.4 bits (80), Expect = 0.005
Identities = 16/40 (40%), Positives = 23/40 (57%), Gaps = 3/40 (7%)
Query: 78 CEKCGNGEATFYTRQMRSADEGQTTFYAC--TRCGHQFQE 115
C KC +GEA F+ R +EG T F+ C C H+++E
Sbjct: 76 CAKCQHGEAVFFQATAR-GEEGMTLFFVCCNPNCSHRWRE 114
>At5g09790 unknown protein
Length = 352
Score = 30.4 bits (67), Expect = 0.15
Identities = 14/34 (41%), Positives = 22/34 (64%)
Query: 52 AEEIRRELGIETIEEQKVQLSKVNKTCEKCGNGE 85
AE + + + + EE++ + S N TCEKCG+GE
Sbjct: 41 AEIMAKSVPVVEQEEEEDEDSYSNVTCEKCGSGE 74
>At2g20030 putative RING zinc finger protein
Length = 390
Score = 28.5 bits (62), Expect = 0.56
Identities = 10/15 (66%), Positives = 13/15 (86%)
Query: 26 YAECPLCKNRSNIKD 40
+A CPLC+NR NI+D
Sbjct: 159 HATCPLCRNRVNIED 173
>At5g64160 unknown protein
Length = 227
Score = 28.1 bits (61), Expect = 0.74
Identities = 14/43 (32%), Positives = 21/43 (48%), Gaps = 1/43 (2%)
Query: 12 CHYCGTMLTVPSVNYAECPLCKNRSNIKDIEGKEISYTISAEE 54
C CG T S CP+C +R + D++ K+ TI +E
Sbjct: 159 CVQCGVPKTYTSARGMVCPICGDRP-LPDVDAKKKGSTIKDKE 200
>At4g01270 putative RING zinc finger protein
Length = 506
Score = 28.1 bits (61), Expect = 0.74
Identities = 9/19 (47%), Positives = 13/19 (68%)
Query: 22 PSVNYAECPLCKNRSNIKD 40
PS N CP+CK + ++KD
Sbjct: 49 PSTNKRNCPICKQKCSLKD 67
>At1g47560 hypothetical protein
Length = 1564
Score = 28.1 bits (61), Expect = 0.74
Identities = 15/37 (40%), Positives = 25/37 (67%), Gaps = 3/37 (8%)
Query: 44 KEISYTISAEEIRRELGIETIEEQKV---QLSKVNKT 77
K++ YTI+ EEI +LG+ +E +K+ LS V+K+
Sbjct: 721 KDLMYTITPEEIPFQLGLSKVELRKMLKSSLSGVDKS 757
>At5g06470 putative protein
Length = 239
Score = 27.7 bits (60), Expect = 0.96
Identities = 15/45 (33%), Positives = 22/45 (48%), Gaps = 3/45 (6%)
Query: 64 IEEQKVQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTR 108
+EE++ L + + C G FYT +RS + TF AC R
Sbjct: 69 LEEERGILLEFKENCPPGGEDSVVFYTTGLRSV---RKTFEACRR 110
>At4g22890 unknown protein
Length = 324
Score = 27.7 bits (60), Expect = 0.96
Identities = 10/33 (30%), Positives = 15/33 (45%)
Query: 78 CEKCGNGEATFYTRQMRSADEGQTTFYACTRCG 110
C CG +F+ + + G+T CT CG
Sbjct: 272 CPNCGTENTSFFGTILSISSGGKTNTVKCTNCG 304
>At4g18240 starch synthase-like protein
Length = 1071
Score = 27.7 bits (60), Expect = 0.96
Identities = 15/46 (32%), Positives = 26/46 (55%), Gaps = 4/46 (8%)
Query: 23 SVNYAECPLCKNRSNIKDIEGKEISYTISAEEIRRELGIETIEEQK 68
S N A P K + N KD++GKE + +R++LG+ + E ++
Sbjct: 803 SWNPATDPFLKAQFNAKDLQGKE----ENKHALRKQLGLSSAESRR 844
>At1g78420 unknown protein (At1g78420)
Length = 401
Score = 27.7 bits (60), Expect = 0.96
Identities = 18/64 (28%), Positives = 28/64 (43%), Gaps = 10/64 (15%)
Query: 12 CHYCGTMLTVP-SVNYAECPLCKNRSNIKDIEGKEISYTISAEEIRRELGIETIEEQKVQ 70
C C + P S +CP CK + + G + + E GIE +EEQ+V
Sbjct: 80 CTECFLQMKNPNSARPTQCPFCKTPNYAVEYRGVKS---------KEEKGIEQVEEQRVI 130
Query: 71 LSKV 74
+K+
Sbjct: 131 EAKI 134
>At4g39190 hypothetical protein
Length = 277
Score = 26.9 bits (58), Expect = 1.6
Identities = 16/67 (23%), Positives = 30/67 (43%), Gaps = 2/67 (2%)
Query: 34 NRSNIKDIEGKEISYTISAEEIRRELGIETIEEQKVQLSKV--NKTCEKCGNGEATFYTR 91
NR ++E K+I+ TI + + + I++ + K + KV +C + E
Sbjct: 192 NREKQSEVEHKDITMTIEKQNLTEKRQIQSYQRSKSENLKVLEKSSCGRLKRSETDASAE 251
Query: 92 QMRSADE 98
+ S DE
Sbjct: 252 RFDSDDE 258
>At4g13780 methionyl-tRNA synthetase - like protein
Length = 797
Score = 26.9 bits (58), Expect = 1.6
Identities = 9/31 (29%), Positives = 16/31 (51%)
Query: 12 CHYCGTMLTVPSVNYAECPLCKNRSNIKDIE 42
C CG +L + +C +C+N I+D +
Sbjct: 173 CEKCGKLLNPTELKDPKCKVCQNTPRIRDTD 203
>At1g69580 transfactor, putative
Length = 332
Score = 26.9 bits (58), Expect = 1.6
Identities = 15/58 (25%), Positives = 28/58 (47%), Gaps = 2/58 (3%)
Query: 28 ECPLCKNRSNIKDIEGKEISYTISAEE--IRRELGIETIEEQKVQLSKVNKTCEKCGN 83
E PL + S + + + I+ + E+ ++R G++ E K+ L+ K E C N
Sbjct: 269 ELPLMEINSEVMKGKKRSINDVVCVEQPLMKRAFGVDDDEHLKLSLNTYKKDMEACTN 326
>At2g32360 hypothetical protein
Length = 175
Score = 26.6 bits (57), Expect = 2.1
Identities = 17/58 (29%), Positives = 28/58 (47%), Gaps = 10/58 (17%)
Query: 41 IEGKEISYTISAEEIRRELGIETIEEQKVQLSKVNKTCEKCGNGEATFYTRQMRSADE 98
I+G+E + EE R++ +E EE+K EK +GEA + + R +E
Sbjct: 121 IDGEEEDQAMKDEEEDRDVKVEEDEEEK----------EKEKDGEAKYVKEEAREVEE 168
>At5g01480 putative protein
Length = 413
Score = 25.8 bits (55), Expect = 3.7
Identities = 19/82 (23%), Positives = 36/82 (43%), Gaps = 4/82 (4%)
Query: 9 FLFCHYCGTMLTVPSVNYAECPLCKNRSNIKDIEGKEISYTISAEEIRRELGIETIEEQK 68
F FC+ C L + C C N + + +E + + S++ + L + I +
Sbjct: 71 FTFCYVCQIALLGEAYYVYFCSYCPNYYHKECVESPSVFH--SSDHPKHPLQLLWIPDMS 128
Query: 69 VQLSKVNKTCEKCGNG-EATFY 89
++ + +K C C G +A FY
Sbjct: 129 LEEER-DKRCCSCKRGVDALFY 149
>At4g28890 unknown protein
Length = 432
Score = 25.8 bits (55), Expect = 3.7
Identities = 20/94 (21%), Positives = 40/94 (42%), Gaps = 22/94 (23%)
Query: 26 YAECPLCKNRSNIKD-----IEGKEISYTISAEEIRRELGIETI-----EEQKVQLSKVN 75
+A CPLC++R ++++ G + ++ EIR + +E EE+++ +++
Sbjct: 158 HATCPLCRDRVSMEEDSSVLTNGNSFRF-LNQSEIREDSSLELYIEREEEEERIHREELS 216
Query: 76 KTCE-----------KCGNGEATFYTRQMRSADE 98
+ K GN E T + DE
Sbjct: 217 GSSRFSIGESFRKILKLGNKEKTLLDEHVNDKDE 250
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.133 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,472,954
Number of Sequences: 26719
Number of extensions: 86712
Number of successful extensions: 351
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 331
Number of HSP's gapped (non-prelim): 40
length of query: 116
length of database: 11,318,596
effective HSP length: 92
effective length of query: 24
effective length of database: 8,860,448
effective search space: 212650752
effective search space used: 212650752
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0309.4


