

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0288.5
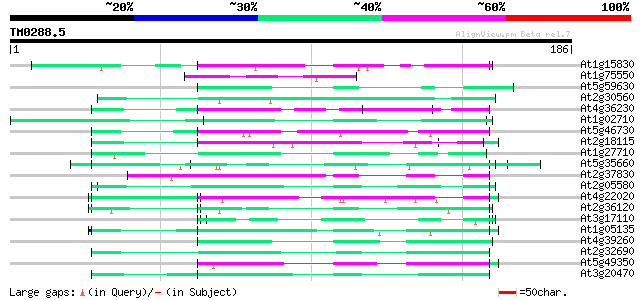
(186 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g15830 hypothetical protein 55 2e-08
At1g75550 hypothetical protein 50 5e-07
At5g59630 glycine-rich protein - like 49 2e-06
At2g30560 putative glycine-rich protein 49 2e-06
At4g36230 putative glycine-rich cell wall protein 49 2e-06
At1g02710 hypothetical protein 49 2e-06
At5g46730 unknown protein 48 3e-06
At2g18115 putative protein 47 5e-06
At1g27710 unknown protein 47 5e-06
At5g35660 unknown protein 47 6e-06
At2g37830 hypothetical protein 47 8e-06
At2g05580 unknown protein 47 8e-06
At4g22020 glycine-rich protein 46 1e-05
At2g36120 unknown protein 45 2e-05
At3g17110 hypothetical protein 44 5e-05
At1g05135 unknown protein 44 5e-05
At4g39260 glycine-rich protein (clone AtGRP8) 44 7e-05
At2g32690 unknown protein 44 7e-05
At5g49350 unknown protein 43 1e-04
At3g20470 glycine-rich protein, putative 42 1e-04
>At1g15830 hypothetical protein
Length = 483
Score = 55.1 bits (131), Expect = 2e-08
Identities = 40/109 (36%), Positives = 48/109 (43%), Gaps = 29/109 (26%)
Query: 63 GGGGGGGGCGRVAATVDG--GGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGS------ 114
G GG GG ++ +G GG GGG + M V+ W GGGGSG+
Sbjct: 317 GNKGGNGGSIKIGVGTNGITGGTGGGEAGAGMQVMQGW---------GGGGSGAATQVMQ 367
Query: 115 ---GGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGGD 160
GGD V M GW GGGG V ++G G GGGGGD
Sbjct: 368 GCGGGDAGAITQV-----MQGW----GGGGAGAVTQVMQGCGGGGGGGD 407
Score = 49.3 bits (116), Expect = 1e-06
Identities = 53/173 (30%), Positives = 61/173 (34%), Gaps = 64/173 (36%)
Query: 8 AAVAMKVVIGWWWGWQWW*VGV--GGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGG 65
A M+V+ GW G V G GGGD+G + M GW
Sbjct: 344 AGAGMQVMQGWGGGGSGAATQVMQGCGGGDAG-----------AITQVMQGW-------- 384
Query: 66 GGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGS----------- 114
GGGG G V + G G GGGG D GGGG G+
Sbjct: 385 -GGGGAGAVTQVMQGCGGGGGGGD------------------GGGGQGTGIGGGGGGEQG 425
Query: 115 -----GGD---RVVAAMVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
GGD +V L MI GGGGG V + G G G GGG
Sbjct: 426 TGVGGGGDTCTQVTHGGGGAPLTMI-----GGGGGEQGVTGSDGGGGRGRGGG 473
Score = 36.6 bits (83), Expect = 0.008
Identities = 37/110 (33%), Positives = 44/110 (39%), Gaps = 26/110 (23%)
Query: 63 GGGGGGGGCGRVAA---TVDGGG---NGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGG 116
GGG G G GR + DGGG N GG + + V G G+GG
Sbjct: 292 GGGRGAEGGGRGSTGEGVTDGGGRTGNKGGNGGSIKIGVGT----------NGITGGTGG 341
Query: 117 DRVVAAMVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGGDRVVVVV 166
A M V M GW GGGG AAT +G GGG + V+
Sbjct: 342 GEAGAGMQV----MQGW----GGGGSG--AATQVMQGCGGGDAGAITQVM 381
Score = 32.3 bits (72), Expect = 0.15
Identities = 27/74 (36%), Positives = 29/74 (38%), Gaps = 22/74 (29%)
Query: 28 GVGGGGGDS--------GDRLVVVVATMVVVVVAMIGWWWWW*GGGG--------GGGGC 71
G+GGGGG GD V + MIG GGGG GGGG
Sbjct: 415 GIGGGGGGEQGTGVGGGGDTCTQVTHGGGGAPLTMIG------GGGGEQGVTGSDGGGGR 468
Query: 72 GRVAATVDGGGNGG 85
GR V GGG G
Sbjct: 469 GRGGGKVAGGGKKG 482
>At1g75550 hypothetical protein
Length = 167
Score = 50.4 bits (119), Expect = 5e-07
Identities = 28/64 (43%), Positives = 29/64 (44%), Gaps = 20/64 (31%)
Query: 59 WWW*GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWW-------LWCQWW*GGGG 111
W W GGGGGGGG G GGG GGGG W W W +W GGGG
Sbjct: 67 WGWGGGGGGGGGGGG-----GGGGGGGGGGG--------WGWGGGGGGGGWYKWGCGGGG 113
Query: 112 SGSG 115
G G
Sbjct: 114 KGKG 117
Score = 35.8 bits (81), Expect = 0.014
Identities = 29/91 (31%), Positives = 29/91 (31%), Gaps = 33/91 (36%)
Query: 67 GGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVV 126
GG G R GGG GGGG GGGG G GG
Sbjct: 60 GGSGSYRWGWGGGGGGGGGGGGG------------------GGGGGGGGG---------- 91
Query: 127 VLAMIGWW*DGGGGGCSMVAATVEGEGNGGG 157
GW GGGGG G G G G
Sbjct: 92 -----GWGWGGGGGGGGWYKWGCGGGGKGKG 117
Score = 33.5 bits (75), Expect = 0.068
Identities = 27/77 (35%), Positives = 27/77 (35%), Gaps = 43/77 (55%)
Query: 83 NGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGC 142
NGG GS R W W GGGG G GG GGGGG
Sbjct: 59 NGGSGSYR-------WGW-------GGGGGGGGG--------------------GGGGG- 83
Query: 143 SMVAATVEGEGNGGGGG 159
G G GGGGG
Sbjct: 84 --------GGGGGGGGG 92
>At5g59630 glycine-rich protein - like
Length = 253
Score = 48.9 bits (115), Expect = 2e-06
Identities = 36/105 (34%), Positives = 40/105 (37%), Gaps = 49/105 (46%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GGGGGGGG G GGG GGGG GGGG G GG
Sbjct: 72 GGGGGGGGGGGGGGGGGGGGGGGGG--------------------GGGGGGGGG------ 105
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGGDRVVVVVL 167
GGGGG G G GGGGG ++ ++ L
Sbjct: 106 --------------GGGGG---------GGGGGGGGGGKLQLLGL 127
Score = 38.5 bits (88), Expect = 0.002
Identities = 23/67 (34%), Positives = 27/67 (39%), Gaps = 25/67 (37%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G GGGGG G GGGGGGGG G GGG GGGG
Sbjct: 84 GGGGGGGGGG-------------------------GGGGGGGGGGGGGGGGGGGGGGGGG 118
Query: 88 SDRVMVV 94
++ ++
Sbjct: 119 GGKLQLL 125
>At2g30560 putative glycine-rich protein
Length = 171
Score = 48.9 bits (115), Expect = 2e-06
Identities = 45/139 (32%), Positives = 54/139 (38%), Gaps = 35/139 (25%)
Query: 30 GGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGG---GGCGRVAATVDGGGNGG- 85
GGGGG G R GGGGGG GGCG + GGG GG
Sbjct: 16 GGGGGSGGGR-----------------------GGGGGGGAKGGCGGGGKSGGGGGGGGY 52
Query: 86 ---GGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGC 142
GS+R + + + GGGG G GG ++ + G GGGGG
Sbjct: 53 MVAPGSNRSSYISRDNFESDPKGGSGGGGKGGGGGGGISGGGAGGKSGCGGGKSGGGGG- 111
Query: 143 SMVAATVEGEGNGGGGGDR 161
G G GGGGG +
Sbjct: 112 ----GGKNGGGCGGGGGGK 126
Score = 36.6 bits (83), Expect = 0.008
Identities = 27/85 (31%), Positives = 31/85 (35%), Gaps = 36/85 (42%)
Query: 80 GGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGG 139
G G+GGGG GGGG GSGG R GGG
Sbjct: 6 GSGSGGGGK-------------------GGGGGGSGGGR-----------------GGGG 29
Query: 140 GGCSMVAATVEGEGNGGGGGDRVVV 164
GG + G+ GGGGG +V
Sbjct: 30 GGGAKGGCGGGGKSGGGGGGGGYMV 54
Score = 34.3 bits (77), Expect = 0.040
Identities = 22/56 (39%), Positives = 23/56 (40%), Gaps = 21/56 (37%)
Query: 63 GGG--GGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGG 116
GGG GGGGG G+ GGG G GG GGGSG GG
Sbjct: 102 GGGKSGGGGGGGKNGGGCGGGGGGKGGK-------------------SGGGSGGGG 138
Score = 33.9 bits (76), Expect = 0.052
Identities = 30/101 (29%), Positives = 31/101 (29%), Gaps = 48/101 (47%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GG GG G GGG GGGG GG G G GG
Sbjct: 2 GGKGGSGS--------GGGGKGGGG--------------------GGSGGGRGG------ 27
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGGDRVV 163
GGGGG G GGGGG +V
Sbjct: 28 --------------GGGGGAKGGCGGGGKSGGGGGGGGYMV 54
>At4g36230 putative glycine-rich cell wall protein
Length = 221
Score = 48.5 bits (114), Expect = 2e-06
Identities = 34/97 (35%), Positives = 37/97 (38%), Gaps = 30/97 (30%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GGGGGGGG G+ + G GNGG G D GGG G GG
Sbjct: 95 GGGGGGGGGGQGSGGGGGEGNGGNGKDNSHKRNKS----------SGGGGGGGG------ 138
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
GGGGG + G G GGGGG
Sbjct: 139 --------------GGGGGSGNGSGRGRGGGGGGGGG 161
Score = 47.8 bits (112), Expect = 3e-06
Identities = 35/97 (36%), Positives = 38/97 (39%), Gaps = 40/97 (41%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GGGGGGGG G + GGG GGGG GGGG G GG
Sbjct: 136 GGGGGGGGSGNGSGRGRGGGGGGGGG-------------------GGGGGGGGG------ 170
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
G GGGGG S +G+G G G G
Sbjct: 171 ---------GGGGGGGGGGGS------DGKGGGWGFG 192
Score = 47.0 bits (110), Expect = 6e-06
Identities = 37/97 (38%), Positives = 41/97 (42%), Gaps = 28/97 (28%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GGGGGGGG G GGG GG GS GGGG G+GG+ +
Sbjct: 89 GGGGGGGGGG-------GGGGGGQGSG------------------GGGGEGNGGNGKDNS 123
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
+ G GGGGG S G G GGGGG
Sbjct: 124 HKRNKSSGGGGGGGGGGGGGS---GNGSGRGRGGGGG 157
Score = 41.6 bits (96), Expect = 2e-04
Identities = 37/113 (32%), Positives = 38/113 (32%), Gaps = 46/113 (40%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G GGGGG SG+ G GGGGGGGG G GGG GGGG
Sbjct: 136 GGGGGGGGSGN-----------------GSGRGRGGGGGGGGGGGGGGGGGGGGGGGGGG 178
Query: 88 SDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGG 140
GGG G GG GW DG GG
Sbjct: 179 --------------------GGGSDGKGGGWGFG---------FGWGGDGPGG 202
Score = 41.6 bits (96), Expect = 2e-04
Identities = 25/55 (45%), Positives = 26/55 (46%), Gaps = 8/55 (14%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGD 117
GGGGGGGG G GGG GGGGSD W W GG G GG+
Sbjct: 157 GGGGGGGGGGGGGGGGGGGGGGGGGSDG----KGGGWGFGFGW----GGDGPGGN 203
Score = 33.5 bits (75), Expect = 0.068
Identities = 21/58 (36%), Positives = 23/58 (39%), Gaps = 20/58 (34%)
Query: 103 CQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGGD 160
C+ GGGG G GG GGGGG + GEGNGG G D
Sbjct: 84 CESRAGGGGGGGGG--------------------GGGGGGGQGSGGGGGEGNGGNGKD 121
>At1g02710 hypothetical protein
Length = 96
Score = 48.5 bits (114), Expect = 2e-06
Identities = 32/96 (33%), Positives = 36/96 (37%), Gaps = 33/96 (34%)
Query: 65 GGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMV 124
GG GGG G+ GGG GGGG GGGG G GG ++
Sbjct: 30 GGNGGGSGKGQWLHGGGGEGGGGE-------------------GGGGEGGGGQKISKG-- 68
Query: 125 VVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGGD 160
GGGGG + G G GGG GD
Sbjct: 69 ------------GGGGGSGGGQRSSSGGGGGGGEGD 92
Score = 47.0 bits (110), Expect = 6e-06
Identities = 47/158 (29%), Positives = 58/158 (35%), Gaps = 62/158 (39%)
Query: 1 MEVIITVAAVAMKVVIGWWWGWQWW*VGVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWW 60
ME I A A ++IG + + GG GG SG W
Sbjct: 1 MEAEIVGEASAAVIMIGGLGLFGTHSLQAGGNGGGSGKGQ------------------WL 42
Query: 61 W*GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVV 120
GGG GGGG +GGG GGG ++ GGGG GSGG +
Sbjct: 43 HGGGGEGGGG--------EGGGGEGGGGQKISK--------------GGGGGGSGGGQRS 80
Query: 121 AAMVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGG 158
++ GGGGG GEG+GGGG
Sbjct: 81 SS-------------GGGGGG---------GEGDGGGG 96
>At5g46730 unknown protein
Length = 268
Score = 48.1 bits (113), Expect = 3e-06
Identities = 38/101 (37%), Positives = 46/101 (44%), Gaps = 12/101 (11%)
Query: 63 GGGGGGGGCGRVAA----TVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDR 118
GG GGGGG G V++ GGG GGG + A + + + GGGSG GG
Sbjct: 37 GGHGGGGGSGGVSSGGYGGESGGGYGGGSGEG-----AGGGYGGAEGYASGGGSGHGGGG 91
Query: 119 VVAAMVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
AA + G +GGGGG A G G GG GG
Sbjct: 92 GGAASSGGYASGAG---EGGGGGYGGAAGGHAGGGGGGSGG 129
Score = 46.2 bits (108), Expect = 1e-05
Identities = 49/143 (34%), Positives = 54/143 (37%), Gaps = 43/143 (30%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATV--------- 78
G GGG G SG G + GGG GGGG G A
Sbjct: 150 GEGGGAGASG-----------------YGGGAYGGGGGHGGGGGGGSAGGAHGGSGYGGG 192
Query: 79 DGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGG 138
+GGG GGGGS + GGGG GSGG A G + GG
Sbjct: 193 EGGGAGGGGSHG-----------GAGGYGGGGGGGSGGGGAYGGGG----AHGGGYGSGG 237
Query: 139 --GGGCSMVAATVEGEGNGGGGG 159
GGG AA G G+GGGGG
Sbjct: 238 GEGGGYGGGAAGGYGGGHGGGGG 260
Score = 44.7 bits (104), Expect = 3e-05
Identities = 33/97 (34%), Positives = 34/97 (35%), Gaps = 8/97 (8%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
G GGGGGG G G GGGG GGGGSG GG A
Sbjct: 86 GHGGGGGGAASSGGYASGAGEGGGGGYGGAAGGHAGG--------GGGGSGGGGGSAYGA 137
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
+GGG G S G G G GGG
Sbjct: 138 GGEHASGYGNGAGEGGGAGASGYGGGAYGGGGGHGGG 174
Score = 41.6 bits (96), Expect = 2e-04
Identities = 34/102 (33%), Positives = 37/102 (35%), Gaps = 16/102 (15%)
Query: 63 GGGGGGGGCGRVAATV-----DGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGD 117
GGGGGGG G +GGG GGGGS + GGGG GSGG
Sbjct: 172 GGGGGGGSAGGAHGGSGYGGGEGGGAGGGGSHG-----------GAGGYGGGGGGGSGGG 220
Query: 118 RVVAAMVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
+GGG G G G GGG G
Sbjct: 221 GAYGGGGAHGGGYGSGGGEGGGYGGGAAGGYGGGHGGGGGHG 262
Score = 40.4 bits (93), Expect = 6e-04
Identities = 34/98 (34%), Positives = 40/98 (40%), Gaps = 9/98 (9%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*G-GGGSGSGGDRVVA 121
GGGGG GG A GGG+GGGG + + G G G+G GG +
Sbjct: 107 GGGGGYGGAAGGHAGGGGGGSGGGGGSA--------YGAGGEHASGYGNGAGEGGGAGAS 158
Query: 122 AMVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
G GGGGG + A G G G GGG
Sbjct: 159 GYGGGAYGGGGGHGGGGGGGSAGGAHGGSGYGGGEGGG 196
Score = 32.7 bits (73), Expect = 0.12
Identities = 29/84 (34%), Positives = 35/84 (41%), Gaps = 5/84 (5%)
Query: 76 ATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW* 135
A+ GG GGGGS V + + GG G G+GG A + G
Sbjct: 33 ASGGGGHGGGGGSGGVS--SGGYGGESGGGYGGGSGEGAGGGYGGAEGYA---SGGGSGH 87
Query: 136 DGGGGGCSMVAATVEGEGNGGGGG 159
GGGGG + G G GGGGG
Sbjct: 88 GGGGGGAASSGGYASGAGEGGGGG 111
Score = 29.6 bits (65), Expect = 0.98
Identities = 20/53 (37%), Positives = 21/53 (38%), Gaps = 23/53 (43%)
Query: 64 GGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGG 116
GGG GGG G AA GGG+GGGG G G GG
Sbjct: 236 GGGEGGGYGGGAAGGYGGGHGGGG-----------------------GHGGGG 265
Score = 29.3 bits (64), Expect = 1.3
Identities = 13/25 (52%), Positives = 13/25 (52%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGG 87
GGG GGG G GGG GGG
Sbjct: 240 GGGYGGGAAGGYGGGHGGGGGHGGG 264
Score = 28.5 bits (62), Expect = 2.2
Identities = 13/25 (52%), Positives = 14/25 (56%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGG 87
GG GGG G GGG+GGGG
Sbjct: 241 GGYGGGAAGGYGGGHGGGGGHGGGG 265
>At2g18115 putative protein
Length = 296
Score = 47.4 bits (111), Expect = 5e-06
Identities = 35/95 (36%), Positives = 39/95 (40%), Gaps = 21/95 (22%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GGGGGGGG G + GGG GGGG + + + W GG G G GG
Sbjct: 207 GGGGGGGGGGGGGGSGPGGGGGGGGGGGGGGGGVGDGYGYGRGWGGGYGGGGGGG----- 261
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGG 157
GW D GG V G GNGGG
Sbjct: 262 --------WGWGGDSNGG--------VWGPGNGGG 280
Score = 45.8 bits (107), Expect = 1e-05
Identities = 32/88 (36%), Positives = 34/88 (38%), Gaps = 22/88 (25%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GGGGGGGG G + GGG GGGG V + W G GG G GG
Sbjct: 208 GGGGGGGGGGGGGSGPGGGGGGGGGGGGGGGGVGDGYGYGRGWGGGYGGGGGGG------ 261
Query: 123 MVVVVLAMIGW--------W*DGGGGGC 142
GW W G GGGC
Sbjct: 262 --------WGWGGDSNGGVWGPGNGGGC 281
Score = 40.8 bits (94), Expect = 4e-04
Identities = 45/155 (29%), Positives = 50/155 (32%), Gaps = 64/155 (41%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGG- 86
G G GG SGD GG GGGG GRV +G G+G G
Sbjct: 130 GCNGAGGSSGD-------------------------GGAGGGGSGRVGRRGNGKGDGKGQ 164
Query: 87 ----------------GSDRVM---VVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVV 127
G DR V + I + + G GG G GG
Sbjct: 165 GDDGGKEDCNDHEKNKGDDRNNGDGVGIGIGIGIGGEPSPGSGGGGGGGG---------- 214
Query: 128 LAMIGWW*DGGGGGCSMVAATVEGEGNGGGGGDRV 162
GGGGG S G G GGGGG V
Sbjct: 215 ---------GGGGGGSGPGGGGGGGGGGGGGGGGV 240
Score = 32.7 bits (73), Expect = 0.12
Identities = 25/64 (39%), Positives = 26/64 (40%), Gaps = 20/64 (31%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGG-----CGRVAATVDGGG 82
G GGGGG GD G+ W GG GGGGG G V G G
Sbjct: 232 GGGGGGGGVGDGY---------------GYGRGWGGGYGGGGGGGWGWGGDSNGGVWGPG 276
Query: 83 NGGG 86
NGGG
Sbjct: 277 NGGG 280
Score = 32.0 bits (71), Expect = 0.20
Identities = 33/124 (26%), Positives = 38/124 (30%), Gaps = 43/124 (34%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSG------- 115
GG G GC + GG GGGGS RV G G+G G
Sbjct: 123 GGEGSCEGCNGAGGSSGDGGAGGGGSGRV----------------GRRGNGKGDGKGQGD 166
Query: 116 --------------------GDRVVAAMVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNG 155
GD V + + + GGGGG G G G
Sbjct: 167 DGGKEDCNDHEKNKGDDRNNGDGVGIGIGIGIGGEPSPGSGGGGGGGGGGGGGGSGPGGG 226
Query: 156 GGGG 159
GGGG
Sbjct: 227 GGGG 230
>At1g27710 unknown protein
Length = 212
Score = 47.4 bits (111), Expect = 5e-06
Identities = 42/132 (31%), Positives = 47/132 (34%), Gaps = 47/132 (35%)
Query: 28 GVGGGG-GDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGG 86
G GGGG G G VV+ GG GGG G G GGNGGG
Sbjct: 113 GYGGGGYGPGGGGGGVVIG-----------------GGFGGGAGYGSGGGLGWDGGNGGG 155
Query: 87 GSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVA 146
G G GG G GG + V++ GGGGGC
Sbjct: 156 GPGY-----------------GSGGGGIGGGGGIGGGVII---------GGGGGGC---G 186
Query: 147 ATVEGEGNGGGG 158
+ G G GGGG
Sbjct: 187 GSCSGGGGGGGG 198
Score = 39.3 bits (90), Expect = 0.001
Identities = 36/102 (35%), Positives = 43/102 (41%), Gaps = 29/102 (28%)
Query: 64 GGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAM 123
GG GGGG G GGG G GG +V+ + GG G GSGG
Sbjct: 105 GGYGGGGPG-----YGGGGYGPGGGGGGVVIGGGFG--------GGAGYGSGGG------ 145
Query: 124 VVVVLAMIGWW*DGG-GGGCSMVAATVEGEGNGGGGGDRVVV 164
+GW DGG GGG + G G GGG G V++
Sbjct: 146 -------LGW--DGGNGGGGPGYGSGGGGIGGGGGIGGGVII 178
>At5g35660 unknown protein
Length = 343
Score = 47.0 bits (110), Expect = 6e-06
Identities = 49/162 (30%), Positives = 57/162 (34%), Gaps = 35/162 (21%)
Query: 21 GWQWW*VGVGGGGGDSGDRLVVVVATMVVVVVAMI----GWWWWW*GGGGGG--GGCGRV 74
GW+ G GGGGG + V + + V G W GG GGG GG G+
Sbjct: 122 GWKGGGGGRGGGGGGGAELETVEIDSASDEDVQNYRGGHGGGWKEGGGQGGGWKGGGGQG 181
Query: 75 AATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW 134
GGG GGG W GGGG G G G W
Sbjct: 182 GGWKGGGGQGGG-------------------WKGGGGQGGGWKGGG--------GQGGGW 214
Query: 135 *DGGGGGCSMVAATVEGEGNGGGGGDRVVVVVLVVAVVTIEW 176
GGG G G G GGGGG + + A I+W
Sbjct: 215 KGGGGQGGGWKGG--GGRGGGGGGGAELENEEINGASEEIQW 254
Score = 46.6 bits (109), Expect = 8e-06
Identities = 44/144 (30%), Positives = 45/144 (30%), Gaps = 69/144 (47%)
Query: 21 GWQWW*VGVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGG-----GGGCGRVA 75
GWQ GGGGG G W GGGGG GG GR
Sbjct: 264 GWQ------GGGGGHGGG----------------------WQGGGGGHGGGWQGGGGRGG 295
Query: 76 ATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW* 135
GGG+GGG W GGGG G GW
Sbjct: 296 GWKGGGGHGGG-------------------WQGGGGRGG-----------------GWKG 319
Query: 136 DGGGGGCSMVAATVEGEGNGGGGG 159
GGGGG G GGGGG
Sbjct: 320 GGGGGGWRGGGGGGGWRGGGGGGG 343
Score = 42.0 bits (97), Expect = 2e-04
Identities = 35/111 (31%), Positives = 39/111 (34%), Gaps = 36/111 (32%)
Query: 61 W*GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVV 120
W GGGG GGG GGG GGG W GGGG G GG
Sbjct: 103 WKGGGGHGGGW-------KGGGGHGGG-----------------WKGGGGGRGGGGGGGA 138
Query: 121 AAMVVVVLAMI------------GWW*DGGGGGCSMVAATVEGEGNGGGGG 159
V + + G W +GGG G +G G GGGG
Sbjct: 139 ELETVEIDSASDEDVQNYRGGHGGGWKEGGGQGGGWKGGGGQGGGWKGGGG 189
Score = 40.8 bits (94), Expect = 4e-04
Identities = 34/105 (32%), Positives = 36/105 (33%), Gaps = 44/105 (41%)
Query: 61 W*GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVV 120
W GGGG GGG GGG+GGG W GGGG G G
Sbjct: 83 WKGGGGHGGGWK------GGGGHGGG-------------------WKGGGGHGGG----- 112
Query: 121 AAMVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGGDRVVVV 165
W GGG G G G GGGGG + V
Sbjct: 113 -------------WKGGGGHGGGWKGGG-GGRGGGGGGGAELETV 143
Score = 40.0 bits (92), Expect = 7e-04
Identities = 44/141 (31%), Positives = 50/141 (35%), Gaps = 51/141 (36%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G GGGGG + + A+ + W GGG GGG G GGG+GGG
Sbjct: 231 GGGGGGGAELENEEINGASEEIQ--------WQGGGGGHGGGWQG------GGGGHGGG- 275
Query: 88 SDRVMVVVAIWWWLWCQWW*GGGGSG----SGGDRVVAAMVVVVLAMIGWW*DGGGGGCS 143
W GGGG G GG R G W GGG G
Sbjct: 276 -----------------WQGGGGGHGGGWQGGGGRG------------GGWKGGGGHGGG 306
Query: 144 MVAATVEG---EGNGGGGGDR 161
G +G GGGGG R
Sbjct: 307 WQGGGGRGGGWKGGGGGGGWR 327
Score = 39.7 bits (91), Expect = 0.001
Identities = 35/110 (31%), Positives = 39/110 (34%), Gaps = 22/110 (20%)
Query: 61 W*GGGGGGGGC----GRVAATVDGGGNGGGG-------SDRVMVVVAIWWWLWCQWW*GG 109
W GGGG GGG G+ GGG GGGG ++ + W GG
Sbjct: 204 WKGGGGQGGGWKGGGGQGGGWKGGGGRGGGGGGGAELENEEINGASEEIQWQGGGGGHGG 263
Query: 110 GGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
G G GG GW GGG G G G GGGG
Sbjct: 264 GWQGGGGGHGG-----------GWQGGGGGHGGGWQGGGGRGGGWKGGGG 302
Score = 38.1 bits (87), Expect = 0.003
Identities = 36/126 (28%), Positives = 38/126 (29%), Gaps = 59/126 (46%)
Query: 56 GWWWWW*GGGGGGGGCGRVAATV----------------------DGGGNGGGGSDRVMV 93
G W GG GGGGG G TV +GGG GGG
Sbjct: 121 GGWKGGGGGRGGGGGGGAELETVEIDSASDEDVQNYRGGHGGGWKEGGGQGGG------- 173
Query: 94 VVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVAATVEGEG 153
W GGGG G G W GGG G +G G
Sbjct: 174 ------------WKGGGGQGGG------------------WKGGGGQGGGWKGGGGQGGG 203
Query: 154 NGGGGG 159
GGGG
Sbjct: 204 WKGGGG 209
Score = 35.8 bits (81), Expect = 0.014
Identities = 36/106 (33%), Positives = 42/106 (38%), Gaps = 25/106 (23%)
Query: 60 WW*GGGGG----GGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSG-- 113
W GGG G GGG GR GG GGGG++ V + Q + GG G G
Sbjct: 113 WKGGGGHGGGWKGGGGGR-------GGGGGGGAELETVEIDSASDEDVQNYRGGHGGGWK 165
Query: 114 SGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
GG + G W GGG G +G G GGGG
Sbjct: 166 EGGGQG------------GGWKGGGGQGGGWKGGGGQGGGWKGGGG 199
>At2g37830 hypothetical protein
Length = 106
Score = 46.6 bits (109), Expect = 8e-06
Identities = 42/123 (34%), Positives = 51/123 (41%), Gaps = 29/123 (23%)
Query: 40 LVVVVATMVVVVV---AMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVA 96
+++ +A+ V V++ A+ G GGG GGGG G GGG GGG D + A
Sbjct: 7 IIIAIASFVGVLIFLAALTGAGGKGGGGGDGGGGGGGGEGGDGGGGEDGGGEDVEIGDGA 66
Query: 97 IWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGG 156
GGGG G GG G DGGGGG G G GG
Sbjct: 67 NGGGFGGDG--GGGGFGGGG---------------GGGGDGGGGG---------GGGGGG 100
Query: 157 GGG 159
GGG
Sbjct: 101 GGG 103
Score = 37.0 bits (84), Expect = 0.006
Identities = 31/89 (34%), Positives = 34/89 (37%), Gaps = 31/89 (34%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G GGGG D G V + G + GGGGG GG G GGG+GGGG
Sbjct: 47 GDGGGGEDGGGEDVEIGD-------GANGGGFGGDGGGGGFGGGG------GGGGDGGGG 93
Query: 88 SDRVMVVVAIWWWLWCQWW*GGGGSGSGG 116
GGGG G GG
Sbjct: 94 GG------------------GGGGGGGGG 104
Score = 36.2 bits (82), Expect = 0.010
Identities = 16/25 (64%), Positives = 16/25 (64%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGG 87
GGGGGGGG G GGG GGGG
Sbjct: 81 GGGGGGGGDGGGGGGGGGGGGGGGG 105
Score = 30.0 bits (66), Expect = 0.75
Identities = 18/44 (40%), Positives = 18/44 (40%), Gaps = 16/44 (36%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGC 71
G GGGGG GD G GGGGGGGGC
Sbjct: 79 GFGGGGGGGGDG----------------GGGGGGGGGGGGGGGC 106
>At2g05580 unknown protein
Length = 302
Score = 46.6 bits (109), Expect = 8e-06
Identities = 42/134 (31%), Positives = 44/134 (32%), Gaps = 67/134 (50%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G GGGGG G + GGGGGGG G GGG GGGG
Sbjct: 213 GGGGGGGQGGHK-----------------------GGGGGGGQGGHKGG--GGGGQGGGG 247
Query: 88 SDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVAA 147
GGG G GG + GGGGG
Sbjct: 248 HK-------------------GGGGGQGGHK------------------GGGGG-----G 265
Query: 148 TVEGEGNGGGGGDR 161
V G G GGGGG R
Sbjct: 266 HVGGGGRGGGGGGR 279
Score = 45.4 bits (106), Expect = 2e-05
Identities = 42/132 (31%), Positives = 44/132 (32%), Gaps = 55/132 (41%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G GGGGG G + GGGG GGG + GG GGGG
Sbjct: 225 GGGGGGGQGGHK---------------------GGGGGGQGGGGHKGGGGGQGGHKGGGG 263
Query: 88 SDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVAA 147
V GGGG G GG G GGGGG S
Sbjct: 264 GGHV----------------GGGGRGGGGG-----------GRGGGGSGGGGGGGS---- 292
Query: 148 TVEGEGNGGGGG 159
G G GGGGG
Sbjct: 293 ---GRGGGGGGG 301
Score = 41.2 bits (95), Expect = 3e-04
Identities = 37/130 (28%), Positives = 41/130 (31%), Gaps = 62/130 (47%)
Query: 30 GGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGGSD 89
GGGGG G + GGGGGG G + GG GGGG
Sbjct: 90 GGGGGQGGQK-----------------------GGGGGGQGGQKGGGGGQGGQKGGGGGQ 126
Query: 90 RVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVAATV 149
GGGG G GG + GGGGG
Sbjct: 127 GGQK--------------GGGGGGQGGQK-----------------GGGGGG-------- 147
Query: 150 EGEGNGGGGG 159
+G GGGGG
Sbjct: 148 QGGQKGGGGG 157
Score = 38.9 bits (89), Expect = 0.002
Identities = 32/100 (32%), Positives = 35/100 (35%), Gaps = 30/100 (30%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GG GGGG G+ GGG G GG GGGG G GG +
Sbjct: 198 GGMKGGGGGGQGGHKGGGGGGGQGGHKG-----------------GGGGGGQGGHK---- 236
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGGDRV 162
G G GGG +G GGGGG V
Sbjct: 237 ---------GGGGGGQGGGGHKGGGGGQGGHKGGGGGGHV 267
Score = 38.5 bits (88), Expect = 0.002
Identities = 39/133 (29%), Positives = 42/133 (31%), Gaps = 61/133 (45%)
Query: 30 GGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGGSD 89
GGGGG G + GGGGGG G+ GG GGGG
Sbjct: 143 GGGGGQGGQK------------------------GGGGGGQGGQKGGGGQGGQKGGGGQG 178
Query: 90 RVMVVVAIWWWLWCQWW*GGGGSG---SGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVA 146
GGGG G GG R M GGGGG
Sbjct: 179 GQK---------------GGGGQGGQKGGGGRGQGGMK-----------GGGGGG----- 207
Query: 147 ATVEGEGNGGGGG 159
+G GGGGG
Sbjct: 208 ---QGGHKGGGGG 217
Score = 36.2 bits (82), Expect = 0.010
Identities = 30/96 (31%), Positives = 34/96 (35%), Gaps = 49/96 (51%)
Query: 64 GGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAM 123
GGGGGG G+ GGG GG G + GGG G GG +
Sbjct: 89 GGGGGGQGGQ-----KGGGGGGQGGQK------------------GGGGGQGGQK----- 120
Query: 124 VVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
GGGGG +G GGGGG
Sbjct: 121 -------------GGGGG--------QGGQKGGGGG 135
Score = 35.8 bits (81), Expect = 0.014
Identities = 28/91 (30%), Positives = 31/91 (33%), Gaps = 43/91 (47%)
Query: 69 GGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVL 128
GG GR GGG GG G + GGGG G GG +
Sbjct: 78 GGIGRGQGGQKGGGGGGQGGQK-----------------GGGGGGQGGQK---------- 110
Query: 129 AMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
GGGGG +G GGGGG
Sbjct: 111 --------GGGGG--------QGGQKGGGGG 125
Score = 35.8 bits (81), Expect = 0.014
Identities = 40/151 (26%), Positives = 45/151 (29%), Gaps = 68/151 (45%)
Query: 30 GGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGG-----GGGCGRVAATVDGGGNG 84
GGGGG G + GGGGG GGG G GGG G
Sbjct: 111 GGGGGQGGQK-----------------------GGGGGQGGQKGGGGGGQGGQKGGGGGG 147
Query: 85 GGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGG----- 139
GG GGGG G GG + G GGG
Sbjct: 148 QGGQK------------------GGGGGGQGGQKGGGGQ--------GGQKGGGGQGGQK 181
Query: 140 ---------GGCSMVAATVEGEGNGGGGGDR 161
GG ++G G GG GG +
Sbjct: 182 GGGGQGGQKGGGGRGQGGMKGGGGGGQGGHK 212
>At4g22020 glycine-rich protein
Length = 396
Score = 45.8 bits (107), Expect = 1e-05
Identities = 37/99 (37%), Positives = 41/99 (41%), Gaps = 18/99 (18%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GGGGGGGG G A G G+G G V GGGG GSGG +V
Sbjct: 216 GGGGGGGGEGGGANGGSGHGSGSGAGGGVSGAAG-------GGGGGGGGGGSGGSKVGGG 268
Query: 123 M--VVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
+G+ GGGGG G G GGGGG
Sbjct: 269 YGHGSGFGGGVGFGNSGGGGG---------GGGGGGGGG 298
Score = 43.9 bits (102), Expect = 5e-05
Identities = 33/99 (33%), Positives = 37/99 (37%), Gaps = 19/99 (19%)
Query: 64 GGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAM 123
G G G G G A V GGG GGGG GGG +G G +
Sbjct: 200 GAGAGAGVGGAAGGVGGGGGGGGGE-------------------GGGANGGSGHGSGSGA 240
Query: 124 VVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGGDRV 162
V G GGGGG S + G G+G G G V
Sbjct: 241 GGGVSGAAGGGGGGGGGGGSGGSKVGGGYGHGSGFGGGV 279
Score = 43.1 bits (100), Expect = 9e-05
Identities = 46/134 (34%), Positives = 51/134 (37%), Gaps = 7/134 (5%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G GGGGG SG V +G+ GGGGGGGG G GGGNG G
Sbjct: 252 GGGGGGGGSGGSKVGGGYGHGSGFGGGVGFGNSGGGGGGGGGGGGG-----GGGGNGSGY 306
Query: 88 SDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGW--W*DGGGGGCSMV 145
+ GGGG GSGG + M G +GGGGG
Sbjct: 307 GSGSGYGSGMGKGSGSGGGGGGGGGGSGGGNGSGSGSGEGYGMGGGAGTGNGGGGGVGFG 366
Query: 146 AATVEGEGNGGGGG 159
G G GGG G
Sbjct: 367 MGIGFGIGIGGGSG 380
Score = 43.1 bits (100), Expect = 9e-05
Identities = 44/145 (30%), Positives = 48/145 (32%), Gaps = 80/145 (55%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGG-------------GCGRV 74
G GGGGG G GGGGGGG G G+
Sbjct: 285 GGGGGGGGGG-------------------------GGGGGGGNGSGYGSGSGYGSGMGKG 319
Query: 75 AATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW 134
+ + GGG GGGGS GG GSGSG G+
Sbjct: 320 SGSGGGGGGGGGGSG------------------GGNGSGSGSGE-------------GY- 347
Query: 135 *DGGGGGCSMVAATVEGEGNGGGGG 159
G GGG G GNGGGGG
Sbjct: 348 --GMGGGA--------GTGNGGGGG 362
Score = 42.7 bits (99), Expect = 1e-04
Identities = 43/132 (32%), Positives = 46/132 (34%), Gaps = 43/132 (32%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G GGGGG+ GD M G+ GG GGGGG GGG GGG
Sbjct: 67 GGGGGGGEGGDG--YGHGEGYGAGAGMSGY-----GGVGGGGG--------RGGGEGGGS 111
Query: 88 SDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVAA 147
S GGSG G A V +G GGGGG
Sbjct: 112 S-------------------ANGGSGHGSGSGAGAGVGGTTGGVGGGGGGGGGG------ 146
Query: 148 TVEGEGNGGGGG 159
GEG G GG
Sbjct: 147 ---GEGGGSSGG 155
Score = 42.7 bits (99), Expect = 1e-04
Identities = 33/97 (34%), Positives = 37/97 (38%), Gaps = 13/97 (13%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GGGGGGGG G + G G+G G V GGGG G GG+ A
Sbjct: 138 GGGGGGGGGGEGGGSSGGSGHGSGSGAGAGAGVG-----GSSGGAGGGGGGGGGEGGGAN 192
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
G G G + V G G GGGGG
Sbjct: 193 G--------GSGHGSGAGAGAGVGGAAGGVGGGGGGG 221
Score = 42.4 bits (98), Expect = 1e-04
Identities = 41/135 (30%), Positives = 47/135 (34%), Gaps = 35/135 (25%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G GGGGG+ G + G G G G G G + GGG GGGG
Sbjct: 141 GGGGGGGEGGG--------------SSGGSGHGSGSGAGAGAGVGGSSGGAGGGGGGGGG 186
Query: 88 SDRVMVVVAIWWWLWCQWW*GG--GGSGSGGDRVVAAMVVVVLAMIGWW*DGGGG-GCSM 144
GG GGSG G A V +G GGGG G
Sbjct: 187 EG------------------GGANGGSGHGSGAGAGAGVGGAAGGVGGGGGGGGGEGGGA 228
Query: 145 VAATVEGEGNGGGGG 159
+ G G+G GGG
Sbjct: 229 NGGSGHGSGSGAGGG 243
Score = 41.6 bits (96), Expect = 2e-04
Identities = 34/97 (35%), Positives = 35/97 (36%), Gaps = 31/97 (31%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GGGGGGGG GGG GGG S GG G GSG A
Sbjct: 137 GGGGGGGG---------GGGEGGGSS-------------------GGSGHGSGSGAGAGA 168
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
V G GGGGG G G+G G G
Sbjct: 169 GVGGSSGGAG---GGGGGGGGEGGGANGGSGHGSGAG 202
Score = 40.8 bits (94), Expect = 4e-04
Identities = 39/133 (29%), Positives = 43/133 (32%), Gaps = 29/133 (21%)
Query: 27 VGVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGG 86
VG GGGGG G + G G G G G G + GGG GGG
Sbjct: 136 VGGGGGGGGGGGE----------GGGSSGGSGHGSGSGAGAGAGVGGSSGGAGGGGGGGG 185
Query: 87 GSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVA 146
G GGG +G G A V G GGGGG
Sbjct: 186 GE-------------------GGGANGGSGHGSGAGAGAGVGGAAGGVGGGGGGGGGEGG 226
Query: 147 ATVEGEGNGGGGG 159
G G+G G G
Sbjct: 227 GANGGSGHGSGSG 239
Score = 40.4 bits (93), Expect = 6e-04
Identities = 30/96 (31%), Positives = 32/96 (33%), Gaps = 33/96 (34%)
Query: 64 GGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAM 123
G G G G G V GGG GGGG G GG SGG
Sbjct: 122 GSGAGAGVGGTTGGVGGGGGGGGGG-------------------GEGGGSSGGS------ 156
Query: 124 VVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
G G G + V + G G GGGGG
Sbjct: 157 --------GHGSGSGAGAGAGVGGSSGGAGGGGGGG 184
Score = 40.0 bits (92), Expect = 7e-04
Identities = 35/98 (35%), Positives = 38/98 (38%), Gaps = 14/98 (14%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*G-GGGSGSGGDRVVA 121
GGGGGGGG GGG GG G D + G GGG G GG
Sbjct: 59 GGGGGGGG--------GGGGGGGEGGDGYGHGEGYGAGAGMSGYGGVGGGGGRGGGEGGG 110
Query: 122 AMVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
+ A G G G + V T G G GGGGG
Sbjct: 111 SS-----ANGGSGHGSGSGAGAGVGGTTGGVGGGGGGG 143
Score = 39.3 bits (90), Expect = 0.001
Identities = 43/135 (31%), Positives = 45/135 (32%), Gaps = 39/135 (28%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
GVGGGGG G A G G G G G G GGG GGGG
Sbjct: 96 GVGGGGGRGGGEGGGSSAN---------GGSGHGSGSGAGAGVGGTTGGVGGGGGGGGGG 146
Query: 88 SDRVMVVVAIWWWLWCQWW*GG---GGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSM 144
+ GG GGSG G A V + G GGGGG
Sbjct: 147 GE------------------GGGSSGGSGHGSGSGAGAGAGVGGSSGGAGGGGGGGG--- 185
Query: 145 VAATVEGEGNGGGGG 159
GEG G GG
Sbjct: 186 ------GEGGGANGG 194
Score = 36.6 bits (83), Expect = 0.008
Identities = 31/97 (31%), Positives = 32/97 (32%), Gaps = 48/97 (49%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
G GGGGGG GGG GGG + GG G GSG
Sbjct: 213 GVGGGGGG---------GGGEGGGAN-------------------GGSGHGSG------- 237
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
G GGG S A G G GGG G
Sbjct: 238 -------------SGAGGGVSGAAGGGGGGGGGGGSG 261
Score = 35.8 bits (81), Expect = 0.014
Identities = 29/95 (30%), Positives = 32/95 (33%), Gaps = 33/95 (34%)
Query: 66 GGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVV 125
G G G+ A GGG GGGG GGGG G GGD
Sbjct: 46 GCGSFYGKGAKRYGGGGGGGGG--------------------GGGGGGEGGDGYGHG--- 82
Query: 126 VVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGGD 160
+G G G M G G G GGG+
Sbjct: 83 ----------EGYGAGAGMSGYGGVGGGGGRGGGE 107
Score = 33.1 bits (74), Expect = 0.088
Identities = 27/88 (30%), Positives = 30/88 (33%), Gaps = 30/88 (34%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G GGGGG SG GG G G G G G G G GG
Sbjct: 325 GGGGGGGGSG-------------------------GGNGSGSGSGEGYGMGGGAGTGNGG 359
Query: 88 SDRVMVVVAIWWWLWCQWW*GGGGSGSG 115
V + I + + GGGSG G
Sbjct: 360 GGGVGFGMGIGFGIGI-----GGGSGGG 382
>At2g36120 unknown protein
Length = 255
Score = 45.4 bits (106), Expect = 2e-05
Identities = 32/98 (32%), Positives = 37/98 (37%), Gaps = 18/98 (18%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GGGGGG G G + +GG +GGG GGGG G G
Sbjct: 169 GGGGGGNGGGGGGGSGEGGAHGGGYGAGGGAGEGYGGGAGAGGHGGGGGGGGGSG----- 223
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGGD 160
GGGGG AA+ G G G GGG+
Sbjct: 224 -------------GGGGGGGGYAAASGYGHGGGAGGGE 248
Score = 44.3 bits (103), Expect = 4e-05
Identities = 39/133 (29%), Positives = 46/133 (34%), Gaps = 54/133 (40%)
Query: 27 VGVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGG 86
+G GGGGG G GGGGGG G G + +GGG G G
Sbjct: 80 IGGGGGGGHGGGAG----------------------GGGGGGPGGGYGGGSGEGGGAGYG 117
Query: 87 GSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVA 146
G + GGGG G+GG GGGGG +
Sbjct: 118 GGE-------------AGGHGGGGGGGAGG-------------------GGGGGGGAHGG 145
Query: 147 ATVEGEGNGGGGG 159
G+G G GGG
Sbjct: 146 GYGGGQGAGAGGG 158
Score = 43.9 bits (102), Expect = 5e-05
Identities = 37/98 (37%), Positives = 38/98 (38%), Gaps = 35/98 (35%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GGG GGGG G GGGNGGG GGGGSG GG
Sbjct: 156 GGGYGGGGAGGHGGG-GGGGNGGG---------------------GGGGSGEGG------ 187
Query: 123 MVVVVLAMIGWW*DGGGGGCSM-VAATVEGEGNGGGGG 159
A G + GGG G A G G GGGGG
Sbjct: 188 ------AHGGGYGAGGGAGEGYGGGAGAGGHGGGGGGG 219
Score = 43.5 bits (101), Expect = 7e-05
Identities = 42/136 (30%), Positives = 45/136 (32%), Gaps = 62/136 (45%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGG---GGCGRVAATVDGGGNG 84
G GGGGG G GGGGGG GG G GGG G
Sbjct: 125 GGGGGGGAGGG------------------------GGGGGGAHGGGYGGGQGAGAGGGYG 160
Query: 85 GGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSM 144
GGG+ GGGG G+GG GGGGG
Sbjct: 161 GGGAG---------------GHGGGGGGGNGG--------------------GGGGGSGE 185
Query: 145 VAATVEGEGNGGGGGD 160
A G G GGG G+
Sbjct: 186 GGAHGGGYGAGGGAGE 201
Score = 43.1 bits (100), Expect = 9e-05
Identities = 31/96 (32%), Positives = 34/96 (35%), Gaps = 33/96 (34%)
Query: 64 GGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAM 123
GGG GGG G + +GGG GG G + GGGG G GG
Sbjct: 53 GGGPGGGSGYGGGSGEGGGAGGHGEGHIG---------------GGGGGGHGGGA----- 92
Query: 124 VVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
GGGGG G G GGG G
Sbjct: 93 -------------GGGGGGGPGGGYGGGSGEGGGAG 115
Score = 42.0 bits (97), Expect = 2e-04
Identities = 40/134 (29%), Positives = 43/134 (31%), Gaps = 53/134 (39%)
Query: 27 VGVGGG-GGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGG 85
VG+GGG GG SG + GG G GGG G GGG GG
Sbjct: 50 VGIGGGPGGGSG-----------------------YGGGSGEGGGAGGHGEGHIGGGGGG 86
Query: 86 GGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMV 145
G GGGG G GG + G GGG
Sbjct: 87 GHGGGAG---------------GGGGGGPGGGYGGGS--------------GEGGGAGYG 117
Query: 146 AATVEGEGNGGGGG 159
G G GGGGG
Sbjct: 118 GGEAGGHGGGGGGG 131
Score = 34.7 bits (78), Expect = 0.030
Identities = 22/62 (35%), Positives = 24/62 (38%), Gaps = 24/62 (38%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G GGGGG G GGGGGGG G AA+ G G G GG
Sbjct: 211 GHGGGGGGGGG------------------------SGGGGGGGGGYAAASGYGHGGGAGG 246
Query: 88 SD 89
+
Sbjct: 247 GE 248
Score = 31.6 bits (70), Expect = 0.26
Identities = 28/89 (31%), Positives = 29/89 (32%), Gaps = 50/89 (56%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G GGGGG SG GGGGGGG A G G+GGG
Sbjct: 214 GGGGGGGGSGG------------------------GGGGGGG-----YAAASGYGHGGG- 243
Query: 88 SDRVMVVVAIWWWLWCQWW*GGGGSGSGG 116
GGG GSGG
Sbjct: 244 --------------------AGGGEGSGG 252
>At3g17110 hypothetical protein
Length = 175
Score = 43.9 bits (102), Expect = 5e-05
Identities = 35/98 (35%), Positives = 37/98 (37%), Gaps = 34/98 (34%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GGG GGGG G GGG GGGG GGGG GSG +
Sbjct: 44 GGGNGGGGGG-------GGGGGGGGG-------------------GGGGDGSGSGSGYGS 77
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGGD 160
G GGGGG G G GGGGG+
Sbjct: 78 GYGSGSGYGGGNNGGGGGG--------GGRGGGGGGGN 107
Score = 42.7 bits (99), Expect = 1e-04
Identities = 35/99 (35%), Positives = 37/99 (37%), Gaps = 42/99 (42%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GGGGGGGG G GGGNGG G+ + G G GSG R
Sbjct: 91 GGGGGGGGRGGGG----GGGNGGSGNG--------------SGYGSGSGYGSGAGR---- 128
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGGDR 161
GGGGG G G GGGGG R
Sbjct: 129 -------------GGGGGGGG-------GGGGGGGGGSR 147
Score = 41.6 bits (96), Expect = 2e-04
Identities = 30/96 (31%), Positives = 35/96 (36%), Gaps = 33/96 (34%)
Query: 64 GGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAM 123
G G G G G + +GGG GGGG GGGG G GGD +
Sbjct: 32 GSGSGSGYGSGSGGGNGGGGGGGGGG------------------GGGGGGGGGDGSGS-- 71
Query: 124 VVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
G G G + + G GN GGGG
Sbjct: 72 -------------GSGYGSGYGSGSGYGGGNNGGGG 94
Score = 40.0 bits (92), Expect = 7e-04
Identities = 32/99 (32%), Positives = 34/99 (34%), Gaps = 38/99 (38%)
Query: 66 GGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVV 125
G G G G + GGGNGGGG GGGG G GG
Sbjct: 30 GRGSGSGSGYGSGSGGGNGGGGGG------------------GGGGGGGGG--------- 62
Query: 126 VVLAMIGWW*DGGGGGCSMVAATVEGEG-----NGGGGG 159
G DG G G + G G NGGGGG
Sbjct: 63 ------GGGGDGSGSGSGYGSGYGSGSGYGGGNNGGGGG 95
Score = 31.2 bits (69), Expect = 0.34
Identities = 38/133 (28%), Positives = 44/133 (32%), Gaps = 65/133 (48%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G GGG G SG+ + G + G G GGG GGG GGGG
Sbjct: 102 GGGGGNGGSGNG-------------SGYGSGSGYGSGAGRGGG---------GGGGGGGG 139
Query: 88 SDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVAA 147
GGGG GS G +G G G +
Sbjct: 140 --------------------GGGGGGSRG-------------------NGSGYG----SG 156
Query: 148 TVEGEGNGGGGGD 160
EG G+G GGGD
Sbjct: 157 YGEGYGSGYGGGD 169
>At1g05135 unknown protein
Length = 384
Score = 43.9 bits (102), Expect = 5e-05
Identities = 42/132 (31%), Positives = 46/132 (34%), Gaps = 33/132 (25%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G GGGGG +G G + GGG GGG G A GGG GGGG
Sbjct: 216 GGGGGGGANGGS----------------GHGSGFGAGGGVGGGVGGGAGGGGGGGGGGGG 259
Query: 88 SDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVAA 147
GG G GSG V G GGGGG +
Sbjct: 260 GAN-----------------GGSGHGSGFGAGGGVGGGVGGGAGGGGGGGGGGGGGVGGG 302
Query: 148 TVEGEGNGGGGG 159
+ G G G GGG
Sbjct: 303 SGHGGGFGAGGG 314
Score = 43.9 bits (102), Expect = 5e-05
Identities = 40/132 (30%), Positives = 43/132 (32%), Gaps = 46/132 (34%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G+GGGGG G +G GG G GGG G A + GGG GGG
Sbjct: 131 GIGGGGGAGGGG------------GGGVGGGSGHGGGFGAGGGVGGGAGGIGGGGGAGGG 178
Query: 88 SDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVAA 147
GGGG G G G GGG A
Sbjct: 179 --------------------GGGGVGGGSGHGSGF--------------GAGGGIGGGAG 204
Query: 148 TVEGEGNGGGGG 159
G G GGGGG
Sbjct: 205 GGVGGGGGGGGG 216
Score = 43.5 bits (101), Expect = 7e-05
Identities = 41/135 (30%), Positives = 44/135 (32%), Gaps = 63/135 (46%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G+GGGGG G GGGG GGG G + GGG GGG
Sbjct: 168 GIGGGGGAGGG------------------------GGGGVGGGSGHGSGFGAGGGIGGGA 203
Query: 88 SDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVAA 147
V GGGG G GG GGGGG A
Sbjct: 204 GGGV----------------GGGGGGGGGG-------------------GGGGG----AN 224
Query: 148 TVEGEGNGGGGGDRV 162
G G+G G G V
Sbjct: 225 GGSGHGSGFGAGGGV 239
Score = 43.1 bits (100), Expect = 9e-05
Identities = 42/133 (31%), Positives = 46/133 (34%), Gaps = 57/133 (42%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G+GGGGG G G+ GGG GGG G GGG GGGG
Sbjct: 69 GIGGGGGQGG------------------GFGAGGGVGGGAGGGLG------GGGGAGGGG 104
Query: 88 SDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGG-GGGCSMVA 146
+ GGGSG GG G+ GG GGG
Sbjct: 105 GGGI-----------------GGGSGHGG---------------GFGAGGGVGGGAGGGI 132
Query: 147 ATVEGEGNGGGGG 159
G G GGGGG
Sbjct: 133 GGGGGAGGGGGGG 145
Score = 42.0 bits (97), Expect = 2e-04
Identities = 38/133 (28%), Positives = 40/133 (29%), Gaps = 63/133 (47%)
Query: 27 VGVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGG 86
VG GGGGG G GGGG GG G + GGG GGG
Sbjct: 207 VGGGGGGGGGGG------------------------GGGGANGGSGHGSGFGAGGGVGGG 242
Query: 87 GSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVA 146
GG G G GG GGGGG
Sbjct: 243 VG-------------------GGAGGGGGG--------------------GGGGGGGANG 263
Query: 147 ATVEGEGNGGGGG 159
+ G G G GGG
Sbjct: 264 GSGHGSGFGAGGG 276
Score = 40.8 bits (94), Expect = 4e-04
Identities = 35/97 (36%), Positives = 37/97 (38%), Gaps = 15/97 (15%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GGGG GGG G GGG GGG V L GGGG+G GG +
Sbjct: 56 GGGGFGGGRGSGGGIGGGGGQGGGFGAGGGVGGGAGGGLG-----GGGGAGGGGGGGIG- 109
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
G GGG G G G GGGGG
Sbjct: 110 ---------GGSGHGGGFGAGGGVGGGAGGGIGGGGG 137
Score = 40.0 bits (92), Expect = 7e-04
Identities = 40/143 (27%), Positives = 48/143 (32%), Gaps = 55/143 (38%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G GGGGG G GGGG GGG G GGG GGG
Sbjct: 284 GAGGGGGGGGG------------------------GGGGVGGGSGHGGGFGAGGGLGGGA 319
Query: 88 SDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVA-----------AMVVVVLAMIGWW*D 136
+ GGGG+G GG + + + + IG
Sbjct: 320 GGGLG---------------GGGGAGGGGGGGLGHGGGVGGGHGGGVGIGIGIGIGVGVG 364
Query: 137 GGGGGCSMVAATVEGEGNGGGGG 159
GG G + G G+GGGGG
Sbjct: 365 GGSG-----QGSGSGSGSGGGGG 382
Score = 39.7 bits (91), Expect = 0.001
Identities = 32/97 (32%), Positives = 35/97 (35%), Gaps = 33/97 (34%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GGGGGGGG G + GGG G GG GGG G GG
Sbjct: 289 GGGGGGGGGGVGGGSGHGGGFGAGGG---------------LGGGAGGGLGGGGGA---- 329
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
GGGGG + G G+GGG G
Sbjct: 330 --------------GGGGGGGLGHGGGVGGGHGGGVG 352
Score = 39.3 bits (90), Expect = 0.001
Identities = 32/96 (33%), Positives = 37/96 (38%), Gaps = 27/96 (28%)
Query: 64 GGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAM 123
GGGGG G GR + GGG G GG GG G G+GG
Sbjct: 55 GGGGGFGGGRGSGGGIGGGGGQGGGFGAG---------------GGVGGGAGG------- 92
Query: 124 VVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
+ G GGGGG + + G G G GGG
Sbjct: 93 -----GLGGGGGAGGGGGGGIGGGSGHGGGFGAGGG 123
Score = 38.1 bits (87), Expect = 0.003
Identities = 33/97 (34%), Positives = 35/97 (36%), Gaps = 32/97 (32%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GGG GGG G V GGG GGGG GGG +G G
Sbjct: 196 GGGIGGGAGGGVGG--GGGGGGGGGG-------------------GGGANGGSGHGSGFG 234
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
V +G GGGGG G GGGGG
Sbjct: 235 AGGGVGGGVGGGAGGGGGG-----------GGGGGGG 260
>At4g39260 glycine-rich protein (clone AtGRP8)
Length = 169
Score = 43.5 bits (101), Expect = 7e-05
Identities = 34/98 (34%), Positives = 36/98 (36%), Gaps = 28/98 (28%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GGGGG GG G + GGG GGG GGG SG GG
Sbjct: 89 GGGGGRGGSGGGYRSGGGGGYSGGG--------------------GGGYSGGGGGGYERR 128
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGGD 160
G + GGGGG EG G GGGD
Sbjct: 129 S--------GGYGSGGGGGGRGYGGGGRREGGGYGGGD 158
Score = 39.3 bits (90), Expect = 0.001
Identities = 34/99 (34%), Positives = 37/99 (37%), Gaps = 43/99 (43%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
G GGGGGG G GGG GG GGG SG GG
Sbjct: 86 GSGGGGGGRGG-----SGGGYRSGG--------------------GGGYSGGGG------ 114
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGGDR 161
G + GGGGG + G G+GGGGG R
Sbjct: 115 ---------GGYSGGGGGGYERRSG---GYGSGGGGGGR 141
>At2g32690 unknown protein
Length = 201
Score = 43.5 bits (101), Expect = 7e-05
Identities = 40/132 (30%), Positives = 44/132 (33%), Gaps = 57/132 (43%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G+GGGGG G GGGGG GG G + GG GG G
Sbjct: 116 GLGGGGGLGG-------------------------GGGGGLGGGGGLGGGAGGGYGGGAG 150
Query: 88 SDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVAA 147
GGG G GG G + GGGGG A
Sbjct: 151 GGL------------------GGGGGIGGG--------------GGFGGGGGGGFGGGAG 178
Query: 148 TVEGEGNGGGGG 159
G+G GGGGG
Sbjct: 179 GGFGKGIGGGGG 190
Score = 41.6 bits (96), Expect = 2e-04
Identities = 42/132 (31%), Positives = 44/132 (32%), Gaps = 45/132 (34%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G+GGGGG G GG GGGGG G GGG GGGG
Sbjct: 62 GLGGGGGLGGG------------------------GGLGGGGGLGGGGGLGGGGGLGGGG 97
Query: 88 SDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVAA 147
GGGSG GG + L G GGGGG
Sbjct: 98 G-------------------LGGGSGLGGGGGLGG--GSGLGGGGGLGGGGGGGLGGGGG 136
Query: 148 TVEGEGNGGGGG 159
G G G GGG
Sbjct: 137 LGGGAGGGYGGG 148
Score = 31.6 bits (70), Expect = 0.26
Identities = 21/60 (35%), Positives = 24/60 (40%), Gaps = 17/60 (28%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G+GGGGG G G + GGG GGG G + GGG GGG
Sbjct: 152 GLGGGGGIGGG-----------------GGFGGGGGGGFGGGAGGGFGKGIGGGGGLGGG 194
>At5g49350 unknown protein
Length = 302
Score = 42.7 bits (99), Expect = 1e-04
Identities = 34/100 (34%), Positives = 40/100 (40%), Gaps = 28/100 (28%)
Query: 63 GGGG---GGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRV 119
GGGG GGGG G + GG+GGGG+ + G GG+G GG
Sbjct: 53 GGGGTGVGGGGTGFGGGGLGAGGSGGGGAGGL----------------GSGGTGVGGGGG 96
Query: 120 VAAMVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
+ A G GG GG V G G GGG G
Sbjct: 97 LGA---------GGSGGGGAGGLGGGGTGVGGGGTGGGTG 127
Score = 40.4 bits (93), Expect = 6e-04
Identities = 34/100 (34%), Positives = 35/100 (35%), Gaps = 33/100 (33%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
G GGGG G G GGG G GGS G GG GSGG V
Sbjct: 51 GFGGGGTGVGGGGTGFGGGGLGAGGSGGG----------------GAGGLGSGGTGV--- 91
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGGDRV 162
GGGGG + G G GGGG V
Sbjct: 92 --------------GGGGGLGAGGSGGGGAGGLGGGGTGV 117
Score = 36.6 bits (83), Expect = 0.008
Identities = 33/97 (34%), Positives = 36/97 (37%), Gaps = 36/97 (37%)
Query: 64 GGGGGGGCGRVAATVDGGGNGGG-GSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GGGG GG G V GGG GGG G + + GGGG G+GG
Sbjct: 103 GGGGAGGLGGGGTGVGGGGTGGGTGFNGGGTGFGPF---------GGGGLGTGG------ 147
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
GG GG G GNGG GG
Sbjct: 148 --------------GGAGGLG------GGGGNGGFGG 164
Score = 35.8 bits (81), Expect = 0.014
Identities = 33/97 (34%), Positives = 37/97 (38%), Gaps = 26/97 (26%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GG GGG G G G G GGGG+ GGGG G+GG A
Sbjct: 39 GGEGGGIGGGGTGFGGGGTGVGGGGTGF-----------------GGGGLGAGGSGGGGA 81
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
+G G GGG + A G G GG GG
Sbjct: 82 ------GGLGSGGTGVGGGGGLGAG---GSGGGGAGG 109
Score = 30.4 bits (67), Expect = 0.57
Identities = 20/54 (37%), Positives = 20/54 (37%), Gaps = 21/54 (38%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGG 116
GGGG G G G GGGNGG GGGG G GG
Sbjct: 139 GGGGLGTGGGGAGGLGGGGGNGGF---------------------GGGGLGGGG 171
>At3g20470 glycine-rich protein, putative
Length = 174
Score = 42.4 bits (98), Expect = 1e-04
Identities = 33/97 (34%), Positives = 35/97 (36%), Gaps = 33/97 (34%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GG GGGGG G + GGG GGGG GGG+G GG A
Sbjct: 45 GGLGGGGGIGGGSGLGGGGGFGGGGG-------------------LGGGAGGGGGLGGGA 85
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
GGG G G G GGGGG
Sbjct: 86 --------------GGGAGGGFGGGAGSGGGLGGGGG 108
Score = 42.0 bits (97), Expect = 2e-04
Identities = 33/97 (34%), Positives = 36/97 (37%), Gaps = 31/97 (31%)
Query: 63 GGGGGGGGCGRVAATVDGGGNGGGGSDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAA 122
GGGG GGG G A GGG GGG + GGGG+G GG
Sbjct: 105 GGGGAGGGFGGGAGGGSGGGFGGGAGAGGGLG-------------GGGGAGGGGG----- 146
Query: 123 MVVVVLAMIGWW*DGGGGGCSMVAATVEGEGNGGGGG 159
GGGGG + G G GGG G
Sbjct: 147 -------------FGGGGGSGIGGGFGGGAGAGGGFG 170
Score = 42.0 bits (97), Expect = 2e-04
Identities = 39/132 (29%), Positives = 45/132 (33%), Gaps = 51/132 (38%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G+GGGGG G + +G + GGGG GGG G G G G GG
Sbjct: 46 GLGGGGGIGGG--------------SGLGGGGGFGGGGGLGGGAGGGGGLGGGAGGGAGG 91
Query: 88 SDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVAA 147
GGG+GSGG GGGGG
Sbjct: 92 G-------------------FGGGAGSGGGL------------------GGGGGAGGGFG 114
Query: 148 TVEGEGNGGGGG 159
G G+GGG G
Sbjct: 115 GGAGGGSGGGFG 126
Score = 39.7 bits (91), Expect = 0.001
Identities = 39/135 (28%), Positives = 43/135 (30%), Gaps = 49/135 (36%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGGG 87
G+GGG G G +G GGG GGG G GGG GGG
Sbjct: 70 GLGGGAGGGGG----------------LGGGAGGGAGGGFGGGAGSGGGLGGGGGAGGG- 112
Query: 88 SDRVMVVVAIWWWLWCQWW*GGGGSGSGGDRVVAAMVVVVLAMIGWW*DGGGGGCSMVAA 147
+ GG G GSGG A G GGG
Sbjct: 113 ------------------FGGGAGGGSGGGFGGGA--------------GAGGGLGGGGG 140
Query: 148 TVEGEGNGGGGGDRV 162
G G GGGGG +
Sbjct: 141 AGGGGGFGGGGGSGI 155
Score = 27.3 bits (59), Expect = 4.9
Identities = 20/59 (33%), Positives = 22/59 (36%), Gaps = 20/59 (33%)
Query: 28 GVGGGGGDSGDRLVVVVATMVVVVVAMIGWWWWW*GGGGGGGGCGRVAATVDGGGNGGG 86
G+GGGGG G + GGGG G G G GGG GGG
Sbjct: 134 GLGGGGGAGGGG--------------------GFGGGGGSGIGGGFGGGAGAGGGFGGG 172
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.332 0.150 0.597
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,603,630
Number of Sequences: 26719
Number of extensions: 264497
Number of successful extensions: 9835
Number of sequences better than 10.0: 210
Number of HSP's better than 10.0 without gapping: 146
Number of HSP's successfully gapped in prelim test: 68
Number of HSP's that attempted gapping in prelim test: 2696
Number of HSP's gapped (non-prelim): 1795
length of query: 186
length of database: 11,318,596
effective HSP length: 93
effective length of query: 93
effective length of database: 8,833,729
effective search space: 821536797
effective search space used: 821536797
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0288.5


