

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0233.21
(309 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
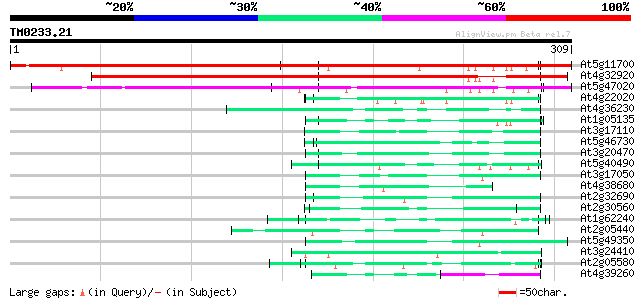
Score E
Sequences producing significant alignments: (bits) Value
At5g11700 putative protein 427 e-120
At4g32920 unknown protein 309 1e-84
At5g47020 putative protein 239 1e-63
At4g22020 glycine-rich protein 53 2e-07
At4g36230 putative glycine-rich cell wall protein 52 4e-07
At1g05135 unknown protein 52 5e-07
At3g17110 hypothetical protein 49 3e-06
At5g46730 unknown protein 47 1e-05
At3g20470 glycine-rich protein, putative 47 1e-05
At5g40490 ribonucleoprotein -like 47 1e-05
At3g17050 unknown protein 46 2e-05
At4g38680 glycine-rich protein 2 (GRP2) 46 3e-05
At2g32690 unknown protein 46 3e-05
At2g30560 putative glycine-rich protein 46 3e-05
At1g62240 unknown protein (At1g62240) 46 3e-05
At2g05440 putative glycine-rich protein 45 4e-05
At5g49350 unknown protein 45 5e-05
At3g24410 hypothetical protein 45 7e-05
At2g05580 unknown protein 44 9e-05
At4g39260 glycine-rich protein (clone AtGRP8) 44 1e-04
>At5g11700 putative protein
Length = 1411
Score = 427 bits (1098), Expect = e-120
Identities = 209/319 (65%), Positives = 248/319 (77%), Gaps = 11/319 (3%)
Query: 1 MARYRFRRLRFLVVAFAVVVFSRQCAC----------DDEFSVTDLDWSVFHQDYSPPAP 50
MAR++F + F++VA AV+ + + D +D + +FHQDYSPPAP
Sbjct: 1 MARFQFCCV-FVLVALAVLAVNPSSSYRFTIVEPKSGSDSDLDSDSESLLFHQDYSPPAP 59
Query: 51 PPPPPHPPSVSCVDDLGGVGTLDTTCKITEDANLTRGVYIAGEGNFNILPGVRFHCEIPG 110
PPPPPH PSVSC +DLGGVG LDTTCKI D NLT VYIAG+GNF ILPGVRFHC IPG
Sbjct: 60 PPPPPHGPSVSCSEDLGGVGFLDTTCKIVADLNLTHDVYIAGKGNFIILPGVRFHCPIPG 119
Query: 111 CWITVNVTGNFSLGSNASIVTGAFELESEYAVFENGSVVNTTCMAGDPPPQTSGTPQGVD 170
C I +NV+GNFSLG+ ++IV G EL + A F NGS VNTT +AG PPPQTSGTPQG+D
Sbjct: 120 CSIAINVSGNFSLGAESTIVAGTLELTAGNASFANGSAVNTTGLAGSPPPQTSGTPQGID 179
Query: 171 GGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGLGGG 230
G GGG+GGRGA CL DTKKLPEDVWGGDAYSWS+LQ P S+GS+G STS+E +YGG GGG
Sbjct: 180 GAGGGHGGRGACCLTDTKKLPEDVWGGDAYSWSTLQKPWSYGSKGGSTSREIDYGGGGGG 239
Query: 231 VVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGGNGFAGGG 290
V++ I +++++N SLLA GG GG KGGGGSGGSIYIKAY+MTG G ISACGG+G+ GGG
Sbjct: 240 KVKMDILQLLDVNGSLLANGGYGGAKGGGGSGGSIYIKAYKMTGIGKISACGGSGYGGGG 299
Query: 291 GGRVSVDVFSRHDEPKISV 309
GGRVSVD+FSRHD+PKI V
Sbjct: 300 GGRVSVDIFSRHDDPKIFV 318
Score = 45.4 bits (106), Expect = 4e-05
Identities = 42/140 (30%), Positives = 63/140 (45%), Gaps = 28/140 (20%)
Query: 171 GGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGLGGG 230
G GGG+GG+G + V GG Y ++L P GS S Y GGG
Sbjct: 615 GSGGGHGGKGGRVCYNNSC----VEGGITYGNANL--PCELGSGSGDFSPG--YSSAGGG 666
Query: 231 VVRLTIHKIVEMNASLLAEGG---DGGN-------------KGGGGSGGSI--YIKAYRM 272
+V I + + + L EG DG + GGGSGG++ +++ +
Sbjct: 667 IV--VIGSMEQPLSGLSLEGSIRVDGESVKRLSRDENGSIVAPGGGSGGTVLLFLRYLIL 724
Query: 273 TGNGIISACGGNGFAGGGGG 292
+ ++S+ GG+G GGGGG
Sbjct: 725 GESSLLSSGGGSGSPGGGGG 744
Score = 42.7 bits (99), Expect = 2e-04
Identities = 49/170 (28%), Positives = 64/170 (36%), Gaps = 37/170 (21%)
Query: 150 NTTCMAGDPPPQTSGTPQGVDGGGG----GYGGRGAGCLVDTKKLPEDVWGGDAYSWSSL 205
N +C+ G + P + G G GY G G +V G S L
Sbjct: 630 NNSCVEGGITYGNANLPCELGSGSGDFSPGYSSAGGGIVVI---------GSMEQPLSGL 680
Query: 206 QNPSSFGSRGASTSKESEY--------GGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKG 257
S G S + S GG GG V L + ++ +SLL+ GG G+ G
Sbjct: 681 SLEGSIRVDGESVKRLSRDENGSIVAPGGGSGGTVLLFLRYLILGESSLLSSGGGSGSPG 740
Query: 258 GGGSGGSIYIKAYRM---TGN---------GIISACGG----NGFAGGGG 291
GGG GG I + TG+ GII A GG +GF G G
Sbjct: 741 GGGGGGGGRIHFHWSNIPTGDIYQPIASVKGIIHARGGAAADDGFYGKNG 790
>At4g32920 unknown protein
Length = 1365
Score = 309 bits (792), Expect = 1e-84
Identities = 150/263 (57%), Positives = 186/263 (70%), Gaps = 16/263 (6%)
Query: 46 SPPAPPPPPPHPPSVSCVDDLGGVGTLDTTCKITEDANLTRGVYIAGEGNFNILPGVRFH 105
SP P P SVSCVDDLGGVG+LD+TCK+ D NLTR + I G+GN ++LPGVR
Sbjct: 44 SPAPEPSPDDDDSSVSCVDDLGGVGSLDSTCKLVADLNLTRDLNITGKGNLHVLPGVRLV 103
Query: 106 CEIPGCWITVNVTGNFSLGSNASIVTGAFELESEYAVFENGSVVNTTCMAGDPPPQTSGT 165
C+ PGC I+VN++GNFSL N+S++ G F L +E A F S V+TT +AG+PPP TSGT
Sbjct: 104 CQFPGCSISVNISGNFSLAENSSVIAGTFRLAAENAEFGLSSAVDTTGLAGEPPPDTSGT 163
Query: 166 PQGVDGGGGGYGGRGAGCLVD-TKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEY 224
P+GV+G GGGYGGRGA CL D T K+PEDV+GGD Y WSSL+ P +GSRG STS E +Y
Sbjct: 164 PEGVEGAGGGYGGRGACCLSDTTTKIPEDVFGGDVYGWSSLEKPEIYGSRGGSTSNEVDY 223
Query: 225 GGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGGN 284
GG GGG V + I + +N S+LA+G GG KGG GNG +SA GG+
Sbjct: 224 GGGGGGTVAIEILGYISLNGSVLADGASGGVKGG---------------GNGRLSASGGD 268
Query: 285 GFAGGGGGRVSVDVFSRHDEPKI 307
G+AGGGGGRVSVD++SRH +PKI
Sbjct: 269 GYAGGGGGRVSVDIYSRHSDPKI 291
Score = 53.5 bits (127), Expect = 1e-07
Identities = 47/141 (33%), Positives = 71/141 (50%), Gaps = 30/141 (21%)
Query: 171 GGGGGYGGRG-AGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGLGG 229
G GGG+GG+G +GC T + GG++Y + L P GS S ++ES GG
Sbjct: 545 GSGGGHGGKGGSGCYNHTC-----IEGGESYGNADL--PCELGS--GSGNEESTDSVAGG 595
Query: 230 GVVRLTIHKIVEMNASLLAEGG---DGGN-----KG--------GGGSGGSI--YIKAYR 271
G++ + + +SL EG DG + KG GGGSGG++ +++
Sbjct: 596 GII--VLGSLEHPLSSLSLEGSITTDGESPRKTLKGLSNSSLGPGGGSGGTVLLFLRTLE 653
Query: 272 MTGNGIISACGGNGFAGGGGG 292
+ + I+S+ GGNG GGGG
Sbjct: 654 IGRSAILSSIGGNGSLKGGGG 674
>At5g47020 putative protein
Length = 1417
Score = 239 bits (610), Expect = 1e-63
Identities = 129/297 (43%), Positives = 174/297 (58%), Gaps = 10/297 (3%)
Query: 13 VVAFAVVVFSRQCACDDEFSVTDLDWSVFHQDYSPPAPPPPPPHPPSVSCVDDLGGVGTL 72
+V V++ S C ++ VT+ + SV + +S A P SV+C DL GVG+L
Sbjct: 4 LVFLCVLLVSTPCFSLSQYGVTEFESSV--RLFSDEASGNSTSSPISVTC-QDLDGVGSL 60
Query: 73 DTTCKITEDANLTRGVYIAGEGNFNILPGVRFHCEIPGCWITVNVTGNFSLGSNASIVTG 132
+TTC + + VY+ G GN NIL V C + GC IT NV+G LG +A IV G
Sbjct: 61 NTTCTLNSNLRFDSDVYVYGTGNLNILAHVLVDCPVEGCMITFNVSGTIHLGQSARIVAG 120
Query: 133 AFELESEYAVFENGSVVNTTCMAGDPPPQTSGTPQGVDGGGGGYGGRGAGCLVDTKKLPE 192
+ + ++ S + TT +AG PP QTSGTP G+DG GGG+GGRGA C+ K
Sbjct: 121 SVVFSAINLTMDSNSSIYTTALAGPPPSQTSGTPYGIDGAGGGHGGRGASCVKSNK---T 177
Query: 193 DVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGLGGGVVRLTIHKIVEMNASLLAEGGD 252
WGGD Y+WSSL +P S+GS G G GGG V+L + V +N ++ A+GGD
Sbjct: 178 TYWGGDVYAWSSLHDPWSYGSEGGVKLSTKNIRGKGGGRVKLILTDTVHVNGTVSADGGD 237
Query: 253 GGNKGGGGSGGSIYIKAYRMTGNGIISACGGNGFAGGGGGRVSVDVFSRHDEPKISV 309
G +GGGGSGGSI I+A G G ISA GG G+ GGGGGR+S+D +S ++ K+ V
Sbjct: 238 AGEEGGGGSGGSICIRA----GYGKISASGGRGWGGGGGGRISLDCYSIQEDVKVFV 290
Score = 44.7 bits (104), Expect = 7e-05
Identities = 46/162 (28%), Positives = 64/162 (39%), Gaps = 17/162 (10%)
Query: 145 NGSVVNTTCMAGDP--PPQTSGTPQGVDGGGGGYGGRGAGCL--VDTKKLPEDVWGGDAY 200
NG V N GDP P + + D G G G + + L ++ G +
Sbjct: 608 NGRVCNGGHTYGDPDFPCELGSGAESPDKSYGNVTGGGMIVIGSIQFPLLTLNLRGSLSS 667
Query: 201 SWSSLQNPSSFGSRGASTSKESEYGGLGGGVVRLTIHKI-VEMNASLLAEGGDGGNKGGG 259
SL P++ G+R S GG GG + L + + + N+SL GG GG GGG
Sbjct: 668 DGQSLWKPTANGNR----SLVGGVGGGSGGTILLFLQMLELSKNSSLSVRGGRGGPLGGG 723
Query: 260 GSGGSIYIKAYRMTGNG--------IISACGGNGFAGGGGGR 293
G GG + M G + + G AG GGR
Sbjct: 724 GGGGGRLHFHWDMLHTGDEYSPVAIVKGSISNRGGAGDNGGR 765
Score = 44.3 bits (103), Expect = 9e-05
Identities = 42/146 (28%), Positives = 66/146 (44%), Gaps = 30/146 (20%)
Query: 171 GGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGL-GG 229
G G G+GGRG + + + GG Y P GS + S + YG + GG
Sbjct: 593 GSGAGHGGRGGSGIFNGRVCN----GGHTYGDPDF--PCELGS--GAESPDKSYGNVTGG 644
Query: 230 GVVRLTIHKI----VEMNASLLAEGGD------GGNKG-----GGGSGGSI--YIKAYRM 272
G++ + + + + SL ++G GN+ GGGSGG+I +++ +
Sbjct: 645 GMIVIGSIQFPLLTLNLRGSLSSDGQSLWKPTANGNRSLVGGVGGGSGGTILLFLQMLEL 704
Query: 273 TGNGIISACGGN----GFAGGGGGRV 294
+ N +S GG G GGGGGR+
Sbjct: 705 SKNSSLSVRGGRGGPLGGGGGGGGRL 730
>At4g22020 glycine-rich protein
Length = 396
Score = 52.8 bits (125), Expect = 2e-07
Identities = 43/130 (33%), Positives = 49/130 (37%), Gaps = 30/130 (23%)
Query: 163 SGTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKES 222
SG GV G GG GG G G GG+ S S GA
Sbjct: 123 SGAGAGVGGTTGGVGGGGGG----------GGGGGEGGGSSGGSGHGSGSGAGAGAGVGG 172
Query: 223 EYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACG 282
GG GGG GG GG +GGG +GGS + + G G+ A G
Sbjct: 173 SSGGAGGG-------------------GGGGGGEGGGANGGSGH-GSGAGAGAGVGGAAG 212
Query: 283 GNGFAGGGGG 292
G G GGGGG
Sbjct: 213 GVGGGGGGGG 222
Score = 51.2 bits (121), Expect = 7e-07
Identities = 44/131 (33%), Positives = 49/131 (36%), Gaps = 29/131 (22%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGS-RGASTSKES 222
G G G GYGG G G GG SS S GS GA
Sbjct: 82 GEGYGAGAGMSGYGGVGGG----------GGRGGGEGGGSSANGGSGHGSGSGAGAGVGG 131
Query: 223 EYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRM-TGNGIISAC 281
GG+GGG GG GG +GGG SGGS + G G+ +
Sbjct: 132 TTGGVGGG-----------------GGGGGGGGEGGGSSGGSGHGSGSGAGAGAGVGGSS 174
Query: 282 GGNGFAGGGGG 292
GG G GGGGG
Sbjct: 175 GGAGGGGGGGG 185
Score = 50.1 bits (118), Expect = 2e-06
Identities = 46/138 (33%), Positives = 54/138 (38%), Gaps = 18/138 (13%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSF--GSRGASTSKE 221
GT GV GGGGG GG G G GG + S + GS G +
Sbjct: 131 GTTGGVGGGGGGGGGGGEG---------GGSSGGSGHGSGSGAGAGAGVGGSSGGAGGGG 181
Query: 222 SEYGGLGGGVVRLTIHKI---VEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGN--- 275
GG GGG + H A G GG GGGG GG + +G+
Sbjct: 182 GGGGGEGGGANGGSGHGSGAGAGAGVGGAAGGVGGGGGGGGGEGGGANGGSGHGSGSGAG 241
Query: 276 -GIISACGGNGFAGGGGG 292
G+ A GG G GGGGG
Sbjct: 242 GGVSGAAGGGGGGGGGGG 259
Score = 47.8 bits (112), Expect = 8e-06
Identities = 44/135 (32%), Positives = 48/135 (34%), Gaps = 14/135 (10%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G GV GGGGG GG G G + GG + G +
Sbjct: 209 GAAGGVGGGGGGGGGEGGGANGGSGHGSGSGAGGGVSGAAGGGGGGGGGGGSGGSKVGGG 268
Query: 224 YG---GLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRM---TGNGI 277
YG G GGGV N+ GG GG GGGG GS Y G G
Sbjct: 269 YGHGSGFGGGVGF--------GNSGGGGGGGGGGGGGGGGGNGSGYGSGSGYGSGMGKGS 320
Query: 278 ISACGGNGFAGGGGG 292
S GG G GG GG
Sbjct: 321 GSGGGGGGGGGGSGG 335
Score = 46.6 bits (109), Expect = 2e-05
Identities = 39/131 (29%), Positives = 47/131 (35%), Gaps = 13/131 (9%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G+ G GGGGG GG G G GG + + G+ G
Sbjct: 172 GSSGGAGGGGGGGGGEGGGAN-----------GGSGHGSGAGAGAGVGGAAGGVGGGGGG 220
Query: 224 YGGLGGGVVRLTIHKIVE-MNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACG 282
GG GGG + H + G GG GGGG G + G+G G
Sbjct: 221 GGGEGGGANGGSGHGSGSGAGGGVSGAAGGGGGGGGGGGSGGSKVGGGYGHGSGFGGGVG 280
Query: 283 -GNGFAGGGGG 292
GN GGGGG
Sbjct: 281 FGNSGGGGGGG 291
Score = 45.4 bits (106), Expect = 4e-05
Identities = 43/135 (31%), Positives = 47/135 (33%), Gaps = 44/135 (32%)
Query: 163 SGTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKES 222
+G GV G GG GG G G GG+ G S
Sbjct: 201 AGAGAGVGGAAGGVGGGGGG------------GGGE-------------GGGANGGSGHG 235
Query: 223 EYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRM-----TGNGI 277
G GGGV A GG GG GGGGSGGS Y G G
Sbjct: 236 SGSGAGGGVSG--------------AAGGGGGGGGGGGSGGSKVGGGYGHGSGFGGGVGF 281
Query: 278 ISACGGNGFAGGGGG 292
++ GG G GGGGG
Sbjct: 282 GNSGGGGGGGGGGGG 296
Score = 44.3 bits (103), Expect = 9e-05
Identities = 39/133 (29%), Positives = 50/133 (37%), Gaps = 20/133 (15%)
Query: 168 GVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYS----WSSLQNPSSFGSRGASTSKESE 223
G GGGGG GG G G GGD Y + + S +G G +
Sbjct: 59 GGGGGGGGGGGGGGG------------EGGDGYGHGEGYGAGAGMSGYGGVGGGGGRGGG 106
Query: 224 YGG----LGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIIS 279
GG GG + + GG GG GGGG GG + +G+G +
Sbjct: 107 EGGGSSANGGSGHGSGSGAGAGVGGTTGGVGGGGGGGGGGGEGGGSSGGSGHGSGSGAGA 166
Query: 280 ACGGNGFAGGGGG 292
G G +GG GG
Sbjct: 167 GAGVGGSSGGAGG 179
Score = 43.5 bits (101), Expect = 1e-04
Identities = 41/130 (31%), Positives = 45/130 (34%), Gaps = 36/130 (27%)
Query: 163 SGTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKES 222
SG GV G GG GG G G K+ GG S FG+ G
Sbjct: 238 SGAGGGVSGAAGGGGGGGGGGGSGGSKV-----GGGYGHGSGFGGGVGFGNSGGGGGG-- 290
Query: 223 EYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACG 282
GG GGG GG GGN G GSG G+G+ G
Sbjct: 291 --GGGGGG-------------------GGGGGNGSGYGSGSGY--------GSGMGKGSG 321
Query: 283 GNGFAGGGGG 292
G GGGGG
Sbjct: 322 SGGGGGGGGG 331
Score = 42.0 bits (97), Expect = 4e-04
Identities = 40/128 (31%), Positives = 43/128 (33%), Gaps = 43/128 (33%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G G GGGGG GG G G GG N S +GS S +
Sbjct: 278 GVGFGNSGGGGGGGGGGGG-------------GGGG------GNGSGYGSGSGYGSGMGK 318
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGG 283
G GGG GG GG GGG GS + Y M G G
Sbjct: 319 GSGSGGG------------------GGGGGGGSGGGNGSGSGSGEGYGMGGG------AG 354
Query: 284 NGFAGGGG 291
G GGGG
Sbjct: 355 TGNGGGGG 362
Score = 38.1 bits (87), Expect = 0.006
Identities = 21/44 (47%), Positives = 22/44 (49%), Gaps = 3/44 (6%)
Query: 250 GGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGGNGFAGGGGGR 293
GG GG GGGG GG Y G G + G G GGGGGR
Sbjct: 63 GGGGGGGGGGGEGGDGYGHG---EGYGAGAGMSGYGGVGGGGGR 103
Score = 36.6 bits (83), Expect = 0.018
Identities = 36/106 (33%), Positives = 42/106 (38%), Gaps = 24/106 (22%)
Query: 198 DAYSWSSLQNPSSFGSRGASTSKESEYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKG 257
D S S + SF +GA YGG GGG GG GG +G
Sbjct: 37 DDCSSRSWRGCGSFYGKGAK-----RYGGGGGGG----------------GGGGGGGGEG 75
Query: 258 GGGSG-GSIYIKAYRMTGNGIISACGGNGFAGGGGGRVSVDVFSRH 302
G G G G Y M+G G + GG G GG GG S + S H
Sbjct: 76 GDGYGHGEGYGAGAGMSGYGGVGGGGGRG--GGEGGGSSANGGSGH 119
>At4g36230 putative glycine-rich cell wall protein
Length = 221
Score = 52.0 bits (123), Expect = 4e-07
Identities = 52/173 (30%), Positives = 62/173 (35%), Gaps = 37/173 (21%)
Query: 120 NFSLGSNASIVTGAFELESEYAVFENGSVVNTTCMAGDPPPQTSGTPQGVDGGGGGYGGR 179
NF +++S++ FEN + G G GGGGG GG+
Sbjct: 46 NFVNATSSSVLDSKSTANISTVEFENALDDQNVNKTEKCESRAGGGGGGGGGGGGGGGGQ 105
Query: 180 GAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGLGGGVVRLTIHKI 239
G+G GG+ S +N SS G G GG GGG
Sbjct: 106 GSGGGGGEGN------GGNGKDNSHKRNKSSGGGGGGG-------GGGGGG--------- 143
Query: 240 VEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGGNGFAGGGGG 292
N S GG GG GGGG GG G G GG G GGGGG
Sbjct: 144 -SGNGSGRGRGGGGGGGGGGGGGGG---------GGG-----GGGGGGGGGGG 181
Score = 34.3 bits (77), Expect = 0.088
Identities = 17/43 (39%), Positives = 22/43 (50%)
Query: 251 GDGGNKGGGGSGGSIYIKAYRMTGNGIISACGGNGFAGGGGGR 293
G GG +G GG+G K + +G G GG G +G G GR
Sbjct: 108 GGGGGEGNGGNGKDNSHKRNKSSGGGGGGGGGGGGGSGNGSGR 150
>At1g05135 unknown protein
Length = 384
Score = 51.6 bits (122), Expect = 5e-07
Identities = 43/132 (32%), Positives = 51/132 (38%), Gaps = 40/132 (30%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G GV GGGGG GG G G + GG + S FG+ G
Sbjct: 202 GAGGGVGGGGGGGGGGGGGGGAN---------GGSGHG-------SGFGAGGGVG----- 240
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRM---TGNGIISA 280
GG+GGG GG GG GGG +GGS + + G G+
Sbjct: 241 -GGVGGGAG---------------GGGGGGGGGGGGANGGSGHGSGFGAGGGVGGGVGGG 284
Query: 281 CGGNGFAGGGGG 292
GG G GGGGG
Sbjct: 285 AGGGGGGGGGGG 296
Score = 51.6 bits (122), Expect = 5e-07
Identities = 42/132 (31%), Positives = 50/132 (37%), Gaps = 40/132 (30%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G G+ GGGG GG G G V GG + S FG+ G
Sbjct: 164 GGAGGIGGGGGAGGGGGGG-----------VGGGSGHG-------SGFGAGGGIGGGAG- 204
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRM---TGNGIISA 280
GG+GGG GG GG GGG +GGS + + G G+
Sbjct: 205 -GGVGGG-----------------GGGGGGGGGGGGANGGSGHGSGFGAGGGVGGGVGGG 246
Query: 281 CGGNGFAGGGGG 292
GG G GGGGG
Sbjct: 247 AGGGGGGGGGGG 258
Score = 46.2 bits (108), Expect = 2e-05
Identities = 39/131 (29%), Positives = 45/131 (33%), Gaps = 36/131 (27%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G G+ GGGG GG G G + GG + G G
Sbjct: 89 GAGGGLGGGGGAGGGGGGG-----------IGGGSGHGGGFGAGGGVGGGAGGGIGGGGG 137
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGG 283
GG GGG V GG G+ GG G+GG + G G GG
Sbjct: 138 AGGGGGGGV-----------------GGGSGHGGGFGAGGGV--------GGGAGGIGGG 172
Query: 284 NGFAGGGGGRV 294
G GGGGG V
Sbjct: 173 GGAGGGGGGGV 183
Score = 45.8 bits (107), Expect = 3e-05
Identities = 43/131 (32%), Positives = 49/131 (36%), Gaps = 17/131 (12%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G+ G+ GGGG GG GAG V GG A G G +
Sbjct: 65 GSGGGIGGGGGQGGGFGAGGGVGGGAGGGLGGGGGAGGGGGGGIGGGSGHGGGFGAGGGV 124
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTG--NGIISAC 281
GG GGG+ GG GG GGG GGS + + G G
Sbjct: 125 GGGAGGGIG---------------GGGGAGGGGGGGVGGGSGHGGGFGAGGGVGGGAGGI 169
Query: 282 GGNGFAGGGGG 292
GG G AGGGGG
Sbjct: 170 GGGGGAGGGGG 180
Score = 45.4 bits (106), Expect = 4e-05
Identities = 42/123 (34%), Positives = 46/123 (37%), Gaps = 35/123 (28%)
Query: 171 GGGGGY-GGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGLGG 229
GGGGG+ GGRG+G + GG FG+ G GGLGG
Sbjct: 55 GGGGGFGGGRGSG---------GGIGGGGGQG-------GGFGAGGGVGGGAG--GGLGG 96
Query: 230 GVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGGNGFAGG 289
G GG GG GGGSG A G G GG G AGG
Sbjct: 97 GG----------------GAGGGGGGGIGGGSGHGGGFGAGGGVGGGAGGGIGGGGGAGG 140
Query: 290 GGG 292
GGG
Sbjct: 141 GGG 143
Score = 44.7 bits (104), Expect = 7e-05
Identities = 45/139 (32%), Positives = 49/139 (34%), Gaps = 52/139 (37%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G GV GG GG GG G G GG A S + S FG+ G
Sbjct: 238 GVGGGVGGGAGGGGGGGGG------------GGGGANGGSG--HGSGFGAGG-------- 275
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYI----------KAYRMT 273
G+GGGV GG G GGGG GG + A
Sbjct: 276 --GVGGGV------------------GGGAGGGGGGGGGGGGGVGGGSGHGGGFGAGGGL 315
Query: 274 GNGIISACGGNGFAGGGGG 292
G G GG G AGGGGG
Sbjct: 316 GGGAGGGLGGGGGAGGGGG 334
Score = 43.9 bits (102), Expect = 1e-04
Identities = 40/130 (30%), Positives = 42/130 (31%), Gaps = 26/130 (20%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G G GGGGG GG G G V GG + G G
Sbjct: 280 GVGGGAGGGGGGGGGGGGG-----------VGGGSGHGGGFGAGGGLGGGAGGGLGGGGG 328
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGG 283
GG GGG L GG G GGG G I I G G G
Sbjct: 329 AGGGGGGG---------------LGHGGGVGGGHGGGVGIGIGIGIGVGVGGGSGQGSGS 373
Query: 284 NGFAGGGGGR 293
+GGGGGR
Sbjct: 374 GSGSGGGGGR 383
Score = 42.0 bits (97), Expect = 4e-04
Identities = 39/129 (30%), Positives = 44/129 (33%), Gaps = 28/129 (21%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G G GGGGG GG G G GG + G G
Sbjct: 242 GVGGGAGGGGGGGGGGGGGAN-----------GGSGHGSGFGAGGGVGGGVGGGAGGGGG 290
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGG 283
GG GGG V GG G+ GG G+GG + A G G + GG
Sbjct: 291 GGGGGGGGV-----------------GGGSGHGGGFGAGGGLGGGAGGGLGGGGGAGGGG 333
Query: 284 NGFAGGGGG 292
G G GGG
Sbjct: 334 GGGLGHGGG 342
Score = 28.9 bits (63), Expect = 3.7
Identities = 18/42 (42%), Positives = 19/42 (44%), Gaps = 6/42 (14%)
Query: 251 GDGGNKGGGGSGGSIYIKAYRMTGNGIISACGGNGFAGGGGG 292
G G GGGG GG R +G GI G G G GGG
Sbjct: 50 GRFGRGGGGGFGGG------RGSGGGIGGGGGQGGGFGAGGG 85
Score = 28.5 bits (62), Expect = 4.8
Identities = 22/53 (41%), Positives = 22/53 (41%), Gaps = 10/53 (18%)
Query: 250 GGDGGNKGGG--GSGGSI--------YIKAYRMTGNGIISACGGNGFAGGGGG 292
G GG GG GSGG I A G G GG G AGGGGG
Sbjct: 53 GRGGGGGFGGGRGSGGGIGGGGGQGGGFGAGGGVGGGAGGGLGGGGGAGGGGG 105
>At3g17110 hypothetical protein
Length = 175
Score = 48.9 bits (115), Expect = 3e-06
Identities = 42/130 (32%), Positives = 47/130 (35%), Gaps = 29/130 (22%)
Query: 163 SGTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKES 222
SG G GGGGG GG G G GG S S S +GS +
Sbjct: 43 SGGGNGGGGGGGGGGGGGGG------------GGGGDGSGSGSGYGSGYGSGSGYGGGNN 90
Query: 223 EYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACG 282
GG GGG GG GG GG G+G + +G G G
Sbjct: 91 GGGGGGGG-----------------RGGGGGGGNGGSGNGSGYGSGSGYGSGAGRGGGGG 133
Query: 283 GNGFAGGGGG 292
G G GGGGG
Sbjct: 134 GGGGGGGGGG 143
Score = 48.5 bits (114), Expect = 5e-06
Identities = 44/130 (33%), Positives = 50/130 (37%), Gaps = 33/130 (25%)
Query: 163 SGTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKES 222
SG+ G GGGGG GG G G GG S S S +GS G+ +
Sbjct: 41 SGSGGGNGGGGGGGGGGGGG----------GGGGGGDGSGSGSGYGSGYGS-GSGYGGGN 89
Query: 223 EYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACG 282
GG GGG GG GG GG GS Y +G+G S G
Sbjct: 90 NGGGGGGG-----------------GRGGGGGGGNGGSGNGSGY-----GSGSGYGSGAG 127
Query: 283 GNGFAGGGGG 292
G GGGGG
Sbjct: 128 RGGGGGGGGG 137
>At5g46730 unknown protein
Length = 268
Score = 47.4 bits (111), Expect = 1e-05
Identities = 42/130 (32%), Positives = 45/130 (34%), Gaps = 27/130 (20%)
Query: 163 SGTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKES 222
SG G GGGGG+GG G G GG Y GS G +
Sbjct: 158 SGYGGGAYGGGGGHGGGGGGGSAG------GAHGGSGYGGGEGGGAGGGGSHGGA----G 207
Query: 223 EYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACG 282
YGG GGG GG G GGG GG G G +A G
Sbjct: 208 GYGGGGGG-----------------GSGGGGAYGGGGAHGGGYGSGGGEGGGYGGGAAGG 250
Query: 283 GNGFAGGGGG 292
G GGGGG
Sbjct: 251 YGGGHGGGGG 260
Score = 47.0 bits (110), Expect = 1e-05
Identities = 41/123 (33%), Positives = 49/123 (39%), Gaps = 20/123 (16%)
Query: 170 DGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGLGG 229
+GGGGGYGG AG GG AY ++ S +G+ GA + G GG
Sbjct: 106 EGGGGGYGG-AAGGHAGGGGGGSGGGGGSAYGAGG-EHASGYGN-GAGEGGGAGASGYGG 162
Query: 230 GVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGGNGFAGG 289
G GG GG GGG +GG+ Y G G GG G GG
Sbjct: 163 GAYG--------------GGGGHGGGGGGGSAGGAHGGSGY---GGGEGGGAGGGGSHGG 205
Query: 290 GGG 292
GG
Sbjct: 206 AGG 208
Score = 41.2 bits (95), Expect = 7e-04
Identities = 39/125 (31%), Positives = 49/125 (39%), Gaps = 16/125 (12%)
Query: 168 GVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGL 227
G GGGG+GG G V + + GG G+ G ++ S +GG
Sbjct: 31 GQASGGGGHGGGGGSGGVSSGGYGGESGGGYGGGSGEGAGGGYGGAEGYASGGGSGHGGG 90
Query: 228 GGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGGNGFA 287
GGG + AS EGG GG GG +GG G G + GG G A
Sbjct: 91 GGGAASSGGY------ASGAGEGGGGGY--GGAAGGH--------AGGGGGGSGGGGGSA 134
Query: 288 GGGGG 292
G GG
Sbjct: 135 YGAGG 139
Score = 40.8 bits (94), Expect = 0.001
Identities = 34/99 (34%), Positives = 36/99 (36%), Gaps = 33/99 (33%)
Query: 168 GVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGL 227
G GG GGYGG G G GG AY +GS G E GG
Sbjct: 201 GSHGGAGGYGGGGGG----------GSGGGGAYGGGGAHG-GGYGSGGG------EGGGY 243
Query: 228 GGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIY 266
GGG A G GG+ GGGG GG Y
Sbjct: 244 GGGA----------------AGGYGGGHGGGGGHGGGGY 266
Score = 28.9 bits (63), Expect = 3.7
Identities = 24/84 (28%), Positives = 29/84 (33%), Gaps = 22/84 (26%)
Query: 212 GSRGASTSKESEYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYR 271
G G+ YGG GG GG G GGG GG+ +
Sbjct: 41 GGGGSGGVSSGGYGGESGG-----------------GYGGGSGEGAGGGYGGAEGYASGG 83
Query: 272 MTGNGIISACGGNGFAGGGGGRVS 295
+G+G GG G A GG S
Sbjct: 84 GSGHG-----GGGGGAASSGGYAS 102
>At3g20470 glycine-rich protein, putative
Length = 174
Score = 47.4 bits (111), Expect = 1e-05
Identities = 40/122 (32%), Positives = 43/122 (34%), Gaps = 29/122 (23%)
Query: 171 GGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGLGGG 230
GGGGG+GG G GG A L + G+ G GGLGGG
Sbjct: 60 GGGGGFGGGGG-------------LGGGAGGGGGLGGGAGGGAGGGFGGGAGSGGGLGGG 106
Query: 231 VVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGGNGFAGGG 290
GG G GGGSGG A G G GG G GGG
Sbjct: 107 G----------------GAGGGFGGGAGGGSGGGFGGGAGAGGGLGGGGGAGGGGGFGGG 150
Query: 291 GG 292
GG
Sbjct: 151 GG 152
Score = 43.5 bits (101), Expect = 1e-04
Identities = 41/129 (31%), Positives = 45/129 (34%), Gaps = 28/129 (21%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G G GGGG GG G G +GG A S L G +
Sbjct: 70 GLGGGAGGGGGLGGGAGGGA--------GGGFGGGAGSGGGLGGGGGAGGGFGGGAGGGS 121
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGG 283
GG GGG + GG GG GGGG GG G+GI GG
Sbjct: 122 GGGFGGGA------------GAGGGLGGGGGAGGGGGFGGG--------GGSGIGGGFGG 161
Query: 284 NGFAGGGGG 292
AGGG G
Sbjct: 162 GAGAGGGFG 170
Score = 38.5 bits (88), Expect = 0.005
Identities = 37/127 (29%), Positives = 39/127 (30%), Gaps = 38/127 (29%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G G GG GG G G G GG + + GS G
Sbjct: 84 GAGGGAGGGFGGGAGSGGGL------------GGGGGAGGGFGGGAGGGSGGGFGGGAGA 131
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGG 283
GGLGGG GG GG GGGGSG G G G
Sbjct: 132 GGGLGGGG----------------GAGGGGGFGGGGGSG----------IGGGFGGGAGA 165
Query: 284 NGFAGGG 290
G GGG
Sbjct: 166 GGGFGGG 172
>At5g40490 ribonucleoprotein -like
Length = 423
Score = 47.0 bits (110), Expect = 1e-05
Identities = 41/125 (32%), Positives = 47/125 (36%), Gaps = 23/125 (18%)
Query: 171 GGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGLGGG 230
GGGGGYGG + P +GG + S+ S G G GG GGG
Sbjct: 314 GGGGGYGGGPGDMYGGSYGEPGGGYGGPSGSYGGGYGSSGIGGYGGGM------GGAGGG 367
Query: 231 VVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGGN--GFAG 288
R G D G GGGG+GG Y G G GG G+ G
Sbjct: 368 GYRG-------------GGGYDMGGVGGGGAGG--YGAGGGGNGGGSFYGGGGGRGGYGG 412
Query: 289 GGGGR 293
GG GR
Sbjct: 413 GGSGR 417
Score = 41.2 bits (95), Expect = 7e-04
Identities = 46/154 (29%), Positives = 53/154 (33%), Gaps = 45/154 (29%)
Query: 156 GDPPPQTSGTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSW--SSLQNPSSFGS 213
G P G G GG GGYGG G +GG Y + + G
Sbjct: 248 GGPYKSGGGYGGGRSGGYGGYGGEFGG------------YGGGGYGGGVGPYRGEPALGY 295
Query: 214 RGASTSKESEYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKG----------GGGSGG 263
G YGG GGG R S+ GG GG G GGG GG
Sbjct: 296 SG-------RYGGGGGGYNR--------GGYSMGGGGGYGGGPGDMYGGSYGEPGGGYGG 340
Query: 264 --SIYIKAYRMTG----NGIISACGGNGFAGGGG 291
Y Y +G G + GG G+ GGGG
Sbjct: 341 PSGSYGGGYGSSGIGGYGGGMGGAGGGGYRGGGG 374
Score = 30.0 bits (66), Expect = 1.7
Identities = 27/94 (28%), Positives = 31/94 (32%), Gaps = 25/94 (26%)
Query: 212 GSRGASTSKESEYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSI------ 265
G G GG GGG + GG+ G GGGG GG +
Sbjct: 242 GGYGGPGGPYKSGGGYGGG-----------RSGGYGGYGGEFGGYGGGGYGGGVGPYRGE 290
Query: 266 ----YIKAYRMTGNGI----ISACGGNGFAGGGG 291
Y Y G G S GG G+ GG G
Sbjct: 291 PALGYSGRYGGGGGGYNRGGYSMGGGGGYGGGPG 324
>At3g17050 unknown protein
Length = 349
Score = 46.2 bits (108), Expect = 2e-05
Identities = 44/132 (33%), Positives = 47/132 (35%), Gaps = 34/132 (25%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
GT G GG GG G GAG +GG A + FG +
Sbjct: 246 GTGGGFGGGAGGGAGGGAG----------GGFGGGAGGGAG----GGFGGGAGGGAGGGA 291
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGG---GSGGSIYIKAYRMTGNGIISA 280
GG GGG A GG GG GGG GSGG A G G
Sbjct: 292 GGGFGGG-----------------AGGGHGGGVGGGFGGGSGGGFGGGAGGGAGGGAGGG 334
Query: 281 CGGNGFAGGGGG 292
GG G AGGG G
Sbjct: 335 FGGGGGAGGGFG 346
Score = 42.0 bits (97), Expect = 4e-04
Identities = 38/129 (29%), Positives = 43/129 (32%), Gaps = 38/129 (29%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G G GGGGG GG G + GG A + G +
Sbjct: 147 GGAGGGAGGGGGLGGGHGGGI-----------GGGAGGGAGGGLGGGHGGGIGGGAGGGS 195
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGG 283
GGLGGG+ GG GG GGGG G G G + G
Sbjct: 196 GGGLGGGI-----------------GGGAGGGAGGGGGAG----------GGGGLGGGHG 228
Query: 284 NGFAGGGGG 292
GF GG GG
Sbjct: 229 GGFGGGAGG 237
Score = 40.4 bits (93), Expect = 0.001
Identities = 41/129 (31%), Positives = 49/129 (37%), Gaps = 26/129 (20%)
Query: 168 GVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGL 227
G+ GGG GG G G GG A + + G+ G GGL
Sbjct: 52 GIGAGGGFGGGAGGGA--------GGGLGGGAGGGGGIGGGAGGGAGGGLGGGAG--GGL 101
Query: 228 GGGVVRLTIHKIVEMNASLLAEGGDGGNKG--GGGSGGSIYIKAYRMTGNGIISACGG-- 283
GGG + + G GG G GGG GG I A +G G+ GG
Sbjct: 102 GGG------------HGGGIGGGAGGGAGGGLGGGHGGGIGGGAGGGSGGGLGGGIGGGA 149
Query: 284 NGFAGGGGG 292
G AGGGGG
Sbjct: 150 GGGAGGGGG 158
Score = 40.0 bits (92), Expect = 0.002
Identities = 42/136 (30%), Positives = 47/136 (33%), Gaps = 27/136 (19%)
Query: 163 SGTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKES 222
+G G GGGG GG G G GG + FG +
Sbjct: 211 AGGGGGAGGGGGLGGGHGGGFGGGA--------GGGLGGGAGGGTGGGFGGGAGGGAGGG 262
Query: 223 EYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKG---GGGSGGSIYIKAYRMTGNGIIS 279
GG GGG A A GG GG G GGG+GG A G G+
Sbjct: 263 AGGGFGGG-------------AGGGAGGGFGGGAGGGAGGGAGGGFGGGAGGGHGGGVGG 309
Query: 280 ACG---GNGFAGGGGG 292
G G GF GG GG
Sbjct: 310 GFGGGSGGGFGGGAGG 325
Score = 40.0 bits (92), Expect = 0.002
Identities = 40/138 (28%), Positives = 44/138 (30%), Gaps = 28/138 (20%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G G GGGG GG G G GG A + G S
Sbjct: 148 GAGGGAGGGGGLGGGHGGGIGGGA--------GGGAGGGLGGGHGGGIGGGAGGGSGGGL 199
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKG---------GGGSGGSIYIKAYRMTG 274
GG+GGG GG GG G GGG+GG + A TG
Sbjct: 200 GGGIGGGA-----------GGGAGGGGGAGGGGGLGGGHGGGFGGGAGGGLGGGAGGGTG 248
Query: 275 NGIISACGGNGFAGGGGG 292
G GG G GGG
Sbjct: 249 GGFGGGAGGGAGGGAGGG 266
Score = 39.7 bits (91), Expect = 0.002
Identities = 37/129 (28%), Positives = 40/129 (30%), Gaps = 38/129 (29%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G G+ GG GG GG G G GG A G
Sbjct: 66 GAGGGLGGGAGGGGGIGGGA------------GGGAGGGLGGGAGGGLGGGHGGGIGGGA 113
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGG 283
GG GGG+ GG GG GGG GGS G G+ GG
Sbjct: 114 GGGAGGGL-----------------GGGHGGGIGGGAGGGS---------GGGLGGGIGG 147
Query: 284 NGFAGGGGG 292
G GGG
Sbjct: 148 GAGGGAGGG 156
Score = 39.3 bits (90), Expect = 0.003
Identities = 40/125 (32%), Positives = 46/125 (36%), Gaps = 26/125 (20%)
Query: 171 GGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGLGGG 230
GGGGG GG G +GG + + G+ G GG+GGG
Sbjct: 34 GGGGGLGGGFGG---------GKGFGGGIGAGGGFGGGAGGGAGGGLGGGAGGGGGIGGG 84
Query: 231 VVRLTIHKIVEMNASLLAEGGDGGNKG---GGGSGGSIYIKAYRMTGNGIISACGGNGFA 287
A A GG GG G GGG GG I A G G+ GG G
Sbjct: 85 -------------AGGGAGGGLGGGAGGGLGGGHGGGIGGGAGGGAGGGLGGGHGG-GIG 130
Query: 288 GGGGG 292
GG GG
Sbjct: 131 GGAGG 135
Score = 37.4 bits (85), Expect = 0.010
Identities = 37/129 (28%), Positives = 40/129 (30%), Gaps = 34/129 (26%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G G GGGG GG G G GG A + G +
Sbjct: 70 GLGGGAGGGGGIGGGAGGGA--------GGGLGGGAGGGLGGGHGGGIGGGAGGGAGGGL 121
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGG 283
GG GGG+ GG G GGG GG I G G GG
Sbjct: 122 GGGHGGGI------------------GGGAGGGSGGGLGGGI--------GGGAGGGAGG 155
Query: 284 NGFAGGGGG 292
G GGG G
Sbjct: 156 GGGLGGGHG 164
Score = 35.8 bits (81), Expect = 0.030
Identities = 40/132 (30%), Positives = 43/132 (32%), Gaps = 43/132 (32%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G+ G+ GG GG G GAG GG A L G G
Sbjct: 194 GSGGGLGGGIGGGAGGGAG------------GGGGAGGGGGLG-----GGHGGGFG---- 232
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACG- 282
GG GGG+ GG G GGG GG A G G G
Sbjct: 233 -GGAGGGL------------------GGGAGGGTGGGFGGGAGGGAGGGAGGGFGGGAGG 273
Query: 283 --GNGFAGGGGG 292
G GF GG GG
Sbjct: 274 GAGGGFGGGAGG 285
Score = 35.4 bits (80), Expect = 0.040
Identities = 36/125 (28%), Positives = 41/125 (32%), Gaps = 37/125 (29%)
Query: 168 GVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGL 227
G+ GG GG G G G + GG A + G+ G GG
Sbjct: 186 GIGGGAGGGSGGGLGGGI----------GGGAGGGAG----GGGGAGGGGGLGGGHGGGF 231
Query: 228 GGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGGNGFA 287
GGG GG G GGG+GG A G G G GF
Sbjct: 232 GGGA------------------GGGLGGGAGGGTGGGFGGGAGGGAGGG-----AGGGFG 268
Query: 288 GGGGG 292
GG GG
Sbjct: 269 GGAGG 273
Score = 34.3 bits (77), Expect = 0.088
Identities = 27/76 (35%), Positives = 31/76 (40%), Gaps = 20/76 (26%)
Query: 220 KESEYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGG---SGGSIYIKAYRMTGNG 276
KE+ GG GGG+ GG GG KG GG +GG A G G
Sbjct: 28 KETLGGGGGGGL-----------------GGGFGGGKGFGGGIGAGGGFGGGAGGGAGGG 70
Query: 277 IISACGGNGFAGGGGG 292
+ GG G GGG G
Sbjct: 71 LGGGAGGGGGIGGGAG 86
>At4g38680 glycine-rich protein 2 (GRP2)
Length = 203
Score = 45.8 bits (107), Expect = 3e-05
Identities = 34/111 (30%), Positives = 38/111 (33%), Gaps = 38/111 (34%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSS--------LQNPSSFGSRG 215
G+ G GGGGGYGGRG G G D Y + +G G
Sbjct: 106 GSGGGYGGGGGGYGGRGGG----------GRGGSDCYKCGEPGHMARDCSEGGGGYGGGG 155
Query: 216 ASTSKESEYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIY 266
YGG GGG GG +GGGG GGS Y
Sbjct: 156 GGYGGGGGYGGGGGGY--------------------GGGGRGGGGGGGSCY 186
Score = 40.0 bits (92), Expect = 0.002
Identities = 45/139 (32%), Positives = 50/139 (35%), Gaps = 37/139 (26%)
Query: 157 DPPPQTSGTPQGVDGGGGGY-GGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRG 215
D P + G GG GG+ GGRG G G Y +G RG
Sbjct: 78 DGAPVQGNSGGGSSGGRGGFGGGRGGG-----------RGSGGGYGGGG----GGYGGRG 122
Query: 216 ASTSKESEYGGLGGGVVRLTIHKIVEMN--ASLLAEGGDGGNKGGGGSGGSIYIKAYRMT 273
GG GG +K E A +EGG G GGGG GG Y
Sbjct: 123 G--------GGRGGS----DCYKCGEPGHMARDCSEGGGGYGGGGGGYGGG---GGYGGG 167
Query: 274 GNGIISACGGNGFAGGGGG 292
G G GG G GGGGG
Sbjct: 168 GGGY----GGGGRGGGGGG 182
Score = 37.4 bits (85), Expect = 0.010
Identities = 33/113 (29%), Positives = 40/113 (35%), Gaps = 23/113 (20%)
Query: 185 VDTKKLPE--DVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGLGGGVVRLTIHKIVEM 242
+D P+ DV G D S G RG G GGG
Sbjct: 63 IDNNNRPKAIDVSGPDGAPVQGNSGGGSSGGRGGFGGGRGGGRGSGGGY----------- 111
Query: 243 NASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIIS---ACGGNGFAGGGGG 292
GG GG GG G GG Y+ G ++ + GG G+ GGGGG
Sbjct: 112 -------GGGGGGYGGRGGGGRGGSDCYKCGEPGHMARDCSEGGGGYGGGGGG 157
Score = 35.4 bits (80), Expect = 0.040
Identities = 38/128 (29%), Positives = 50/128 (38%), Gaps = 24/128 (18%)
Query: 173 GGGYGGRGAGCLVDTKK----LPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGLG 228
GGG +G+ DT+K + D G D + S F S A + E E
Sbjct: 6 GGGERRKGSVKWFDTQKGFGFITPDDGGDDLFVHQSSIRSEGFRSLAAEEAVEFEVE--- 62
Query: 229 GGVVRLTIHKIVEMNASLLAEGGDG----GNKGGGGSGGSIYIKAYRMTGNGIISACGGN 284
+ K ++++ G DG GN GGG SGG R G G G
Sbjct: 63 --IDNNNRPKAIDVS------GPDGAPVQGNSGGGSSGGRGGFGGGRGGGRG-----SGG 109
Query: 285 GFAGGGGG 292
G+ GGGGG
Sbjct: 110 GYGGGGGG 117
>At2g32690 unknown protein
Length = 201
Score = 45.8 bits (107), Expect = 3e-05
Identities = 43/126 (34%), Positives = 44/126 (34%), Gaps = 39/126 (30%)
Query: 168 GVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGL 227
G+ GGGG GG G G GG S L G G GGL
Sbjct: 92 GLGGGGGLGGGSGLG------------GGGGLGGGSGLGGGGGLGGGGGGGLGGG--GGL 137
Query: 228 GGGVVRLTIHKIVEMNASLLAEGGDGGNKGGG-GSGGSIYIKAYRMTGNGIISACGGNGF 286
GGG A GG GG GGG G GG I G G GG GF
Sbjct: 138 GGG-----------------AGGGYGGGAGGGLGGGGGI-------GGGGGFGGGGGGGF 173
Query: 287 AGGGGG 292
GG GG
Sbjct: 174 GGGAGG 179
Score = 44.7 bits (104), Expect = 7e-05
Identities = 44/132 (33%), Positives = 48/132 (36%), Gaps = 28/132 (21%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGA--STSKE 221
G +G+ GGGG GG G G GG L G G S
Sbjct: 58 GFGKGLGGGGGLGGGGGLG------------GGGGLGGGGGLGGGGGLGGGGGLGGGSGL 105
Query: 222 SEYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGG-GSGGSIYIKAYRMTGNGIISA 280
GGLGGG + L GG GG GGG G GG + A G G
Sbjct: 106 GGGGGLGGG-------------SGLGGGGGLGGGGGGGLGGGGGLGGGAGGGYGGGAGGG 152
Query: 281 CGGNGFAGGGGG 292
GG G GGGGG
Sbjct: 153 LGGGGGIGGGGG 164
Score = 37.0 bits (84), Expect = 0.014
Identities = 41/122 (33%), Positives = 42/122 (33%), Gaps = 42/122 (34%)
Query: 171 GGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGLGGG 230
GGGGG GG G G L L GG G G GGLGGG
Sbjct: 118 GGGGGLGGGGGGGLGGGGGLGGGAGGGYG------------GGAG---------GGLGGG 156
Query: 231 VVRLTIHKIVEMNASLLAEGGDGGNKG-GGGSGGSIYIKAYRMTGNGIISACG-GNGFAG 288
GG GG G GGG GG A G GI G G G+ G
Sbjct: 157 -------------------GGIGGGGGFGGGGGGGFGGGAGGGFGKGIGGGGGLGGGYVG 197
Query: 289 GG 290
GG
Sbjct: 198 GG 199
>At2g30560 putative glycine-rich protein
Length = 171
Score = 45.8 bits (107), Expect = 3e-05
Identities = 43/130 (33%), Positives = 47/130 (36%), Gaps = 38/130 (29%)
Query: 163 SGTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKES 222
SG +G GGGG GG G G GG Y + N SS+ SR S
Sbjct: 21 SGGGRGGGGGGGAKGGCGGG------GKSGGGGGGGGYMVAPGSNRSSYISRDNFESDPK 74
Query: 223 EYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACG 282
GG GGG GG GG GGG+GG S CG
Sbjct: 75 --GGSGGGG----------------KGGGGGGGISGGGAGGK--------------SGCG 102
Query: 283 GNGFAGGGGG 292
G GGGGG
Sbjct: 103 GGKSGGGGGG 112
Score = 42.7 bits (99), Expect = 2e-04
Identities = 37/114 (32%), Positives = 42/114 (36%), Gaps = 34/114 (29%)
Query: 166 PQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYG 225
P+G GGGG GG G G + GG A S S G G + G
Sbjct: 73 PKGGSGGGGKGGGGGGG-----------ISGGGAGGKSGCGGGKSGGGGGGGKNG----G 117
Query: 226 GLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIIS 279
G GGG GG G K GGGSGG Y+ A G+ IS
Sbjct: 118 GCGGG-------------------GGGKGGKSGGGSGGGGYMVAPGSNGSSTIS 152
Score = 39.7 bits (91), Expect = 0.002
Identities = 43/139 (30%), Positives = 48/139 (33%), Gaps = 61/139 (43%)
Query: 168 GVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGL 227
G G G G GG+G G GG S G RG GG
Sbjct: 3 GKGGSGSGGGGKGGG-------------GG-----------GSGGGRG---------GGG 29
Query: 228 GGGVVRLTIHKIVEMNASLLAEGG-DGGNKGGGGSGGSIYIKAYRMTGNGIIS------- 279
GGG A+GG GG K GGG GG Y+ A + IS
Sbjct: 30 GGG-----------------AKGGCGGGGKSGGGGGGGGYMVAPGSNRSSYISRDNFESD 72
Query: 280 ---ACGGNGFAGGGGGRVS 295
GG G GGGGG +S
Sbjct: 73 PKGGSGGGGKGGGGGGGIS 91
Score = 37.4 bits (85), Expect = 0.010
Identities = 41/133 (30%), Positives = 47/133 (34%), Gaps = 36/133 (27%)
Query: 163 SGTPQGVDGGGGGY--GGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSK 220
SG+ G GGGGG GGRG G GG A G
Sbjct: 7 SGSGGGGKGGGGGGSGGGRGGG------------GGGGAKG-------------GCGGGG 41
Query: 221 ESEYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISA 280
+S GG GGG + + E G GGGG GG G G IS
Sbjct: 42 KSGGGGGGGGYMVAPGSNRSSYISRDNFESDPKGGSGGGGKGGG---------GGGGISG 92
Query: 281 CGGNGFAGGGGGR 293
G G +G GGG+
Sbjct: 93 GGAGGKSGCGGGK 105
Score = 28.1 bits (61), Expect = 6.3
Identities = 12/21 (57%), Positives = 12/21 (57%)
Query: 163 SGTPQGVDGGGGGYGGRGAGC 183
SG G GGGGG G G GC
Sbjct: 99 SGCGGGKSGGGGGGGKNGGGC 119
>At1g62240 unknown protein (At1g62240)
Length = 227
Score = 45.8 bits (107), Expect = 3e-05
Identities = 40/141 (28%), Positives = 48/141 (33%), Gaps = 30/141 (21%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G G GGGGG GG G G G + ++ + + +GS S E
Sbjct: 91 GIVVGGGGGGGGGGGGGGGS-----------GGSNGSFFNGSGSGTGYGSGDGRVSSSGE 139
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGI------ 277
Y GG S GG GG G GSG + TG
Sbjct: 140 YSASAGG------------GGSGEGSGGGGGGDGSSGSGSGSGSGSGSGTGTASGPDVYM 187
Query: 278 -ISACGGNGFAGGGGGRVSVD 297
+ GG G GGGGG VD
Sbjct: 188 HVEGGGGGGGGGGGGGGGGVD 208
Score = 45.1 bits (105), Expect = 5e-05
Identities = 43/149 (28%), Positives = 51/149 (33%), Gaps = 44/149 (29%)
Query: 143 FENGSVVNTTCMAGDPPPQTSGTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSW 202
F NGS T +GD +SG GGGG G G G GGD S
Sbjct: 117 FFNGSGSGTGYGSGDGRVSSSGEYSASAGGGGSGEGSGGG------------GGGDGSSG 164
Query: 203 SSLQNPSSFGSRGASTSKESEYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSG 262
S + S GS + S Y + EGG GG GGGG G
Sbjct: 165 SGSGSGSGSGSGTGTASGPDVY---------------------MHVEGGGGGGGGGGGGG 203
Query: 263 GSIYIKAYRMTGNGIISACGGNGFAGGGG 291
G G+ + G+G GGG
Sbjct: 204 -----------GGGVDGSGSGSGSGSGGG 221
Score = 42.7 bits (99), Expect = 2e-04
Identities = 42/136 (30%), Positives = 52/136 (37%), Gaps = 36/136 (26%)
Query: 160 PQTSGTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTS 219
P+ G P GGGGG GG GA T+K +D N + +G
Sbjct: 36 PKFDGEPICAAGGGGG-GGGGASVEGGTEKGIDD-------------NANGYGD------ 75
Query: 220 KESEYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIIS 279
G G G H + ++ GG GG GGGG GGS G+G
Sbjct: 76 ------GNGNG----NAHGRADCPGGIVVGGGGGGGGGGGGGGGSGGSNGSFFNGSG--- 122
Query: 280 ACGGNGFAGGGGGRVS 295
G G+ G G GRVS
Sbjct: 123 --SGTGY-GSGDGRVS 135
>At2g05440 putative glycine-rich protein
Length = 154
Score = 45.4 bits (106), Expect = 4e-05
Identities = 51/175 (29%), Positives = 63/175 (35%), Gaps = 46/175 (26%)
Query: 123 LGSNASIVTGAFELESEYAVFENGSVVNTTCMAGDPPPQTSGT--PQGVDGGGGGYGGRG 180
+ S A I+ G F +V S V+ +G P++ T P+G GG GG+GG G
Sbjct: 1 MASKALILLGLF------SVLLVVSEVSAARQSGMVKPESEETVQPEGYGGGHGGHGGHG 54
Query: 181 AGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGLGGGVVRLTIHKIV 240
G GG + G G YGG GG H
Sbjct: 55 GG-------------GGHGHG----------GHNGGGGHGLDGYGGGGGHYGGGGGH--- 88
Query: 241 EMNASLLAEGGDGGNKGGG----GSGGSIYIKAYRMTGNGIISACGGNGFAGGGG 291
GG GG+ GGG G GG Y G G GG G+ GGGG
Sbjct: 89 --------YGGGGGHYGGGGGHYGGGGGHYGGGGGGHGGGGHYGGGGGGYGGGGG 135
Score = 36.2 bits (82), Expect = 0.023
Identities = 19/43 (44%), Positives = 23/43 (53%)
Query: 250 GGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGGNGFAGGGGG 292
GG GG+ G GG GG + G+G+ GG G GGGGG
Sbjct: 45 GGHGGHGGHGGGGGHGHGGHNGGGGHGLDGYGGGGGHYGGGGG 87
Score = 34.3 bits (77), Expect = 0.088
Identities = 27/92 (29%), Positives = 31/92 (33%), Gaps = 32/92 (34%)
Query: 171 GGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGLGGG 230
GGGG YGG G +GG + +G G YGG GGG
Sbjct: 84 GGGGHYGGGGGH------------YGGGGGHYGG--GGGHYGGGGGGHGGGGHYGGGGGG 129
Query: 231 VVRLTIHKIVEMNASLLAEGGDGGNKGGGGSG 262
GG GG+ GGGG G
Sbjct: 130 Y------------------GGGGGHHGGGGHG 143
Score = 30.0 bits (66), Expect = 1.7
Identities = 20/63 (31%), Positives = 27/63 (42%), Gaps = 4/63 (6%)
Query: 230 GVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGGNGFAGG 289
G+V+ + V+ GG GG+ GGGG G + G + GG G G
Sbjct: 28 GMVKPESEETVQPEGYGGGHGGHGGHGGGGGHGHG----GHNGGGGHGLDGYGGGGGHYG 83
Query: 290 GGG 292
GGG
Sbjct: 84 GGG 86
>At5g49350 unknown protein
Length = 302
Score = 45.1 bits (105), Expect = 5e-05
Identities = 46/147 (31%), Positives = 53/147 (35%), Gaps = 23/147 (15%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G GV GGG G+GG G G GG S G GA S
Sbjct: 54 GGGTGVGGGGTGFGGGGLGA-------GGSGGGGAGGLGSGGTGVGGGGGLGAGGSGGGG 106
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKG--GGGSGGSIYIKAYRMTGNGIISAC 281
GGLGGG + + GG GG G GGG+G + TG G
Sbjct: 107 AGGLGGG-------------GTGVGGGGTGGGTGFNGGGTGFGPFGGGGLGTGGGGAGGL 153
Query: 282 GGNGF-AGGGGGRVSVDVFSRHDEPKI 307
GG G G GGG + D P+I
Sbjct: 154 GGGGGNGGFGGGGLGGGGLGGEDPPEI 180
Score = 38.1 bits (87), Expect = 0.006
Identities = 41/133 (30%), Positives = 46/133 (33%), Gaps = 54/133 (40%)
Query: 161 QTSGTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSK 220
+ G G+ GGG G+GG G G V GG + FG G
Sbjct: 37 EVGGEGGGIGGGGTGFGGGGTG-----------VGGGG----------TGFGGGGLGA-- 73
Query: 221 ESEYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGG-GSGGSIYIKAYRMTGNGIIS 279
GG GGG A L GG G GGG G+GGS G G
Sbjct: 74 ----GGSGGG------------GAGGLGSGGTGVGGGGGLGAGGS---------GGG--- 105
Query: 280 ACGGNGFAGGGGG 292
G G GGG G
Sbjct: 106 --GAGGLGGGGTG 116
Score = 34.7 bits (78), Expect = 0.067
Identities = 29/84 (34%), Positives = 33/84 (38%), Gaps = 13/84 (15%)
Query: 210 SFGSRGASTSKESEYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSG-GSIYIK 268
SF S +SE GG GGG+ GG G GGGG+G G +
Sbjct: 24 SFPGNTYSEEDDSEVGGEGGGI-----------GGGGTGFGGGGTGVGGGGTGFGGGGLG 72
Query: 269 AYRMTGNGIIS-ACGGNGFAGGGG 291
A G G GG G GGGG
Sbjct: 73 AGGSGGGGAGGLGSGGTGVGGGGG 96
>At3g24410 hypothetical protein
Length = 240
Score = 44.7 bits (104), Expect = 7e-05
Identities = 48/164 (29%), Positives = 61/164 (36%), Gaps = 27/164 (16%)
Query: 156 GDPPPQ-----------TSGTPQGVDGGGG---------GYGGRGAGCLVDTKKLPEDVW 195
GD PP+ T G GV+GGGG G GG G L E V
Sbjct: 62 GDLPPEGLAGGGGLRLPTRGGGVGVEGGGGDLGGVEGVAGEGGGGEEGLKGEDGGGEGVK 121
Query: 196 GGDAYSWSSLQNPSSFGSRGASTSKESEYGGLGGGVVRL-----TIHKIVEMNASLLAEG 250
GG+ G G + G + GG + + + V+ ++ +G
Sbjct: 122 GGEGMLGDGGGGEGVVGDGGRGDGVKGGEGVMAGGGEGVKGGEGVVGEGVKGGEGVVGDG 181
Query: 251 GDG-GNKGGGGSGGSIYIKAYRMTGNGIISACGGNGFAGGGGGR 293
G G G KGGGG G I + G G GG GGGGGR
Sbjct: 182 GRGEGVKGGGGEGVEGGIGG-TVVGGGGGGRAGGRAVGGGGGGR 224
Score = 39.7 bits (91), Expect = 0.002
Identities = 41/128 (32%), Positives = 43/128 (33%), Gaps = 33/128 (25%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G GV GG G G G G + E V GG+ G RG E
Sbjct: 141 GRGDGVKGGEGVMAGGGEGVKGGEGVVGEGVKGGEGVVGD--------GGRG-----EGV 187
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGG 283
GG G GV EGG GG GGG GG R G G GG
Sbjct: 188 KGGGGEGV-----------------EGGIGGTVVGGGGGGRA---GGRAVGGGGGGRAGG 227
Query: 284 NGFAGGGG 291
GGGG
Sbjct: 228 RAVGGGGG 235
>At2g05580 unknown protein
Length = 302
Score = 44.3 bits (103), Expect = 9e-05
Identities = 41/129 (31%), Positives = 47/129 (35%), Gaps = 33/129 (25%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G QG GGGG GG+G K G + G +G +
Sbjct: 205 GGGQGGHKGGGGGGGQGG------HKGGGGGGGQGGHKGGGGGGQGGGGHKGGGGGQGGH 258
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGG 283
GG GGG V GG GG +GGGGSGG G G GG
Sbjct: 259 KGGGGGGHVGGG------------GRGGGGGGRGGGGSGG----------GGG-----GG 291
Query: 284 NGFAGGGGG 292
+G GGGGG
Sbjct: 292 SGRGGGGGG 300
Score = 43.9 bits (102), Expect = 1e-04
Identities = 42/131 (32%), Positives = 46/131 (35%), Gaps = 20/131 (15%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G G GGGGG GG+ G GG Q G G ++
Sbjct: 104 GGQGGQKGGGGGQGGQKGG---------GGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGG 154
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGG 283
GG GG K GG GG KGGGG GG K G G + GG
Sbjct: 155 GGGGQGG------QKGGGGQGGQKGGGGQGGQKGGGGQGGQ---KGGGGRGQGGMKGGGG 205
Query: 284 NGFAG--GGGG 292
G G GGGG
Sbjct: 206 GGQGGHKGGGG 216
Score = 42.4 bits (98), Expect = 3e-04
Identities = 43/138 (31%), Positives = 50/138 (36%), Gaps = 25/138 (18%)
Query: 161 QTSGTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRG----- 215
Q G QG GGGG GG+ G +K GG ++ G G
Sbjct: 162 QKGGGGQGGQKGGGGQGGQKGGGGQGGQK------GGGGRGQGGMKGGGGGGQGGHKGGG 215
Query: 216 ASTSKESEYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGN 275
+ GG GGG HK GG GG GGG GG ++ G
Sbjct: 216 GGGGQGGHKGGGGGG--GQGGHK----------GGGGGGQGGGGHKGGGGGQGGHKGGGG 263
Query: 276 GIISACGGNGFAGGGGGR 293
G GG G GGGGGR
Sbjct: 264 G--GHVGGGGRGGGGGGR 279
Score = 42.0 bits (97), Expect = 4e-04
Identities = 43/137 (31%), Positives = 50/137 (36%), Gaps = 22/137 (16%)
Query: 161 QTSGTPQGVDGGGGGYGG------RGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSR 214
Q G QG GGGG GG RG G + + G + G
Sbjct: 171 QKGGGGQGGQKGGGGQGGQKGGGGRGQGGMKGGGGGGQGGHKGGGGGGGQGGHKGGGGGG 230
Query: 215 GASTSKESEYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTG 274
G K GG GGG HK GG GG+KGGGG G ++ G
Sbjct: 231 GQGGHKGGGGGGQGGGG-----HKG--------GGGGQGGHKGGGGGG---HVGGGGRGG 274
Query: 275 NGIISACGGNGFAGGGG 291
G GG+G GGGG
Sbjct: 275 GGGGRGGGGSGGGGGGG 291
Score = 41.2 bits (95), Expect = 7e-04
Identities = 41/152 (26%), Positives = 53/152 (33%), Gaps = 22/152 (14%)
Query: 144 ENGSVVNTTCMAGDPPPQTSGTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWS 203
EN V + G+ ++ G+ G GG G G G K GG
Sbjct: 55 ENHFVETDEVLDGEEEEESKNNQGGIGRGQGGQKGGGGGGQGGQKG-----GGGGGQGGQ 109
Query: 204 SLQNPSSFGSRGASTSKESEYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGG 263
G +G + + GG GGG + GG GG KGGGG G
Sbjct: 110 KGGGGGQGGQKGGGGGQGGQKGGGGGG----------QGGQKGGGGGGQGGQKGGGGGG- 158
Query: 264 SIYIKAYRMTGNGIISACGGNGFAG--GGGGR 293
+ + G G GG G G GGGG+
Sbjct: 159 ----QGGQKGGGGQGGQKGGGGQGGQKGGGGQ 186
Score = 30.0 bits (66), Expect = 1.7
Identities = 27/96 (28%), Positives = 30/96 (31%), Gaps = 41/96 (42%)
Query: 168 GVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGGL 227
G GGGGG GG G GG + + G G GG
Sbjct: 247 GHKGGGGGQGGHKGG-------------GGGGHVGGGGRGGGGGGRGG---------GGS 284
Query: 228 GGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGG 263
GGG GG G +GGGG GG
Sbjct: 285 GGG-------------------GGGGSGRGGGGGGG 301
Score = 28.1 bits (61), Expect = 6.3
Identities = 29/102 (28%), Positives = 33/102 (31%), Gaps = 16/102 (15%)
Query: 164 GTPQGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESE 223
G QG GGGG GG+G K GG + G G
Sbjct: 217 GGGQGGHKGGGGGGGQGGH-----KGGGGGGQGGGGHKGGGGGQGGHKGGGGGGHVGGGG 271
Query: 224 YGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSI 265
GG GGG GG GG+ GGG GG +
Sbjct: 272 RGGGGGG-----------RGGGGSGGGGGGGSGRGGGGGGGM 302
>At4g39260 glycine-rich protein (clone AtGRP8)
Length = 169
Score = 43.9 bits (102), Expect = 1e-04
Identities = 23/56 (41%), Positives = 30/56 (53%), Gaps = 8/56 (14%)
Query: 238 KIVEMN-ASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGGNGFAGGGGG 292
+++ +N A GG GG +GG G G YR G G S GG G++GGGGG
Sbjct: 75 RVITVNEAQSRGSGGGGGGRGGSGGG-------YRSGGGGGYSGGGGGGYSGGGGG 123
Score = 41.6 bits (96), Expect = 6e-04
Identities = 41/126 (32%), Positives = 45/126 (35%), Gaps = 42/126 (33%)
Query: 167 QGVDGGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKESEYGG 226
+G GGGGG GG G G GG YS G G S GG
Sbjct: 85 RGSGGGGGGRGGSGGGYRSG---------GGGGYS----------GGGGGGYS-----GG 120
Query: 227 LGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGGNGF 286
GGG R + GG GGG GG Y R G G GG+ +
Sbjct: 121 GGGGYERRS-----------------GGYGSGGGGGGRGYGGGGRREGGGYGGGDGGS-Y 162
Query: 287 AGGGGG 292
GGGGG
Sbjct: 163 GGGGGG 168
Score = 35.8 bits (81), Expect = 0.030
Identities = 20/44 (45%), Positives = 20/44 (45%), Gaps = 7/44 (15%)
Query: 250 GGDGGNKGGGGSGGSIYIKAYRMTGNGIISACGGNGFAGGGGGR 293
GG GG GGGG G Y G S GG G GGGGR
Sbjct: 112 GGGGGYSGGGGGG-------YERRSGGYGSGGGGGGRGYGGGGR 148
Score = 34.3 bits (77), Expect = 0.088
Identities = 22/45 (48%), Positives = 24/45 (52%), Gaps = 12/45 (26%)
Query: 251 GDGGNKGGGGSGGSIYIKAYRMTGNGIISACGGNGFAGGGGGRVS 295
G GG GGGG GGS G G S GG G++GGGGG S
Sbjct: 86 GSGG--GGGGRGGS---------GGGYRSG-GGGGYSGGGGGGYS 118
Score = 31.6 bits (70), Expect = 0.57
Identities = 30/101 (29%), Positives = 32/101 (30%), Gaps = 36/101 (35%)
Query: 163 SGTPQGVD-GGGGGYGGRGAGCLVDTKKLPEDVWGGDAYSWSSLQNPSSFGSRGASTSKE 221
SG G GGGGGY G G GG + G RG
Sbjct: 103 SGGGGGYSGGGGGGYSGGGG--------------GGYERRSGGYGSGGGGGGRGYGGGGR 148
Query: 222 SEYGGLGGGVVRLTIHKIVEMNASLLAEGGDGGNKGGGGSG 262
E GG G GGDGG+ GGGG G
Sbjct: 149 REGGGYG---------------------GGDGGSYGGGGGG 168
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.139 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,930,916
Number of Sequences: 26719
Number of extensions: 513762
Number of successful extensions: 11472
Number of sequences better than 10.0: 308
Number of HSP's better than 10.0 without gapping: 168
Number of HSP's successfully gapped in prelim test: 146
Number of HSP's that attempted gapping in prelim test: 4724
Number of HSP's gapped (non-prelim): 2326
length of query: 309
length of database: 11,318,596
effective HSP length: 99
effective length of query: 210
effective length of database: 8,673,415
effective search space: 1821417150
effective search space used: 1821417150
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0233.21


