

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0230.6
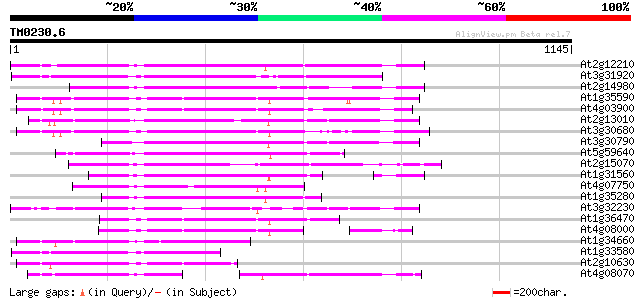
(1145 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g12210 putative TNP2-like transposon protein 640 0.0
At3g31920 hypothetical protein 557 e-158
At2g14980 pseudogene 513 e-145
At1g35590 CACTA-element, putative 499 e-141
At4g03900 putative transposon protein 489 e-138
At2g13010 pseudogene 476 e-134
At3g30680 hypothetical protein 449 e-126
At3g30790 hypothetical protein 449 e-126
At5g59640 serine/threonine-specific protein kinase - like 424 e-118
At2g15070 En/Spm-like transposon protein 422 e-118
At1g31560 putative protein 403 e-112
At4g07750 putative transposon protein 389 e-108
At1g35280 hypothetical protein 382 e-106
At3g32230 hypothetical protein 374 e-103
At1g36470 hypothetical protein 359 4e-99
At4g08000 putative transposon protein 324 2e-88
At1g34660 hypothetical protein 319 7e-87
At1g33580 En/Spm-like transposon protein, putative 306 6e-83
At2g10630 En/Spm transposon hypothetical protein 1 286 6e-77
At4g08070 262 9e-70
>At2g12210 putative TNP2-like transposon protein
Length = 889
Score = 640 bits (1652), Expect = 0.0
Identities = 360/885 (40%), Positives = 487/885 (54%), Gaps = 101/885 (11%)
Query: 2 MESREWIRDPSRRSEEYIRGVNEFLEFAFQNSQVSEKIWCPCTTCANCKSLSRTSVYEHL 61
M + W+R R Y +G +F+ + I CPC C N + S + V +HL
Sbjct: 1 MLDKAWVR-LCRADPGYEKGAQKFVTDVAVALGDIDMIVCPCKDCRNVERKSGSDVVDHL 59
Query: 62 TNPRRGFLRGYK---QWIFHGEKPVASSSTIEEPEHTEMEHDMDGLIHDVLASHTAEDPL 118
RRG YK W HG+ + + E + ++L + A
Sbjct: 60 V--RRGMDEAYKLRADWYHHGDVDSVA----------DCESNFSRWNAEILELYQAAQGF 107
Query: 119 FDD-GERNPQVEENANKFYRLVKDNEQILYPNCKKYSKLSFMVHLYHLKCLHGWSDKSFS 177
D GE + ++F + D E LYP+C +SKLS +V L+ +K +GWSDKSF+
Sbjct: 108 DADLGEIAEDEDIREDEFLAKLADAEIPLYPSCLNHSKLSAIVTLFRIKTKNGWSDKSFN 167
Query: 178 MLLDLLRDAFPEGNVLPKSFYETKKMISELGLGYEKIHACPNDCILYWDKYTNDEVCPKC 237
LL L + P NVL S YE KK + +GYEKIHAC NDC L+ +Y N E CPKC
Sbjct: 168 ELLGTLPEMLPADNVLHTSLYEVKKFLRSFDMGYEKIHACVNDCCLFRKRYKNLENCPKC 227
Query: 238 GASRWKTVTEEVQPNGTKTSEKQKNTPLPKSVPLPGIPAKILRWFPLKPRLQRLFMSSKI 297
ASRWK+ +T E +K G+P K+LR+FP+ PR +R+F S ++
Sbjct: 228 NASRWKS--------NMQTGEAKK-----------GVPQKVLRYFPIIPRFKRMFRSEEM 268
Query: 298 AEAMRWHRDGRLDDGFLRHPADSLAWKKFDSKYQTFALDPHNVRLGLASDGFNPFKTMSI 357
A+ +RWH + DG LRHP DS+ W K + KY FA++ N+RLGL++DGFNPF +
Sbjct: 269 AKDLRWHYSNKSTDGKLRHPVDSVTWDKMNDKYPEFAIEERNLRLGLSTDGFNPFNMKNT 328
Query: 358 THST*PVILVHFNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTYLQPLVKELQELWENGL 417
+S PV+LV++NLPP +CMK+ MLSLLIPGP+ PGN+ID YL+PL+++L LW+ G
Sbjct: 329 RYSCWPVLLVNYNLPPDLCMKKENIMLSLLIPGPQQPGNSIDVYLEPLIEDLNHLWKKGE 388
Query: 418 ETFDAYKKESFQLHAALMWTINDFPAYANLSGWSTKGKYACPCCGFETSSQWLRNGRKFC 477
T+DA+ K +F L A L+WTI+DFPAY NL+G KGK CP CG T S WL+ RK
Sbjct: 389 LTYDAFSKTTFTLKAMLLWTISDFPAYGNLAGCKVKGKMGCPLCGKGTDSMWLKYSRKHV 448
Query: 478 YMSHRRWLTPNHKWRLNGKDFDGTQEFRDPPIRLDGTAQLRQL----------------- 520
+M HR+ L P H +R FDG E L G + L
Sbjct: 449 FMCHRKGLAPTHSYRNKKAWFDGKIEHGRKARILTGREVSQTLKNFKNDFGNAKESGRKR 508
Query: 521 -----------------DEIEDECL---GDQAQRWKKKSILFSLPYWKYNELRHNLDVMH 560
+ EDE L D+ RWKK+SI F L YW+ +RHNLDVMH
Sbjct: 509 KRNDCIESVTDSDDESSESEEDEELEVDEDELSRWKKRSIFFKLSYWEDLPVRHNLDVMH 568
Query: 561 IEKNVCDNIIGTLLNQEGKSKDNYKARADLVDMGIRSILHPQPSPNTNTMLLPRACYQMT 620
+E+NV +I+ TLL+ GKSKD AR DL D+G+R LH P+ LP A + ++
Sbjct: 569 VERNVGASIVSTLLHC-GKSKDGLNARKDLQDLGVRKDLH--PNAQGKRTYLPAAPWSLS 625
Query: 621 TKEKDDFLSVLKNVKAPDECSSNISRCVQVQQRKIFGLKSYDCHVLMQELLPIALRGSLP 680
EK F L K PD SNISR V ++ KI GLKS+D HVLMQ+LLP+AL G LP
Sbjct: 626 KSEKKTFCKRLFEFKGPDGYCSNISRGVSLEDCKIMGLKSHDYHVLMQQLLPVALMGLLP 685
Query: 681 DKVTSVLIDLCNFFKQICSKVLNVKFIEQLESQIAITLCQLETIFPPSFFTIMVHLVVHL 740
+ +I LC+FF +C +V++++ I +E++I TLC E FPP+FF IMVHL VHL
Sbjct: 686 KGPRTAIIRLCSFFNHLCQRVIDIEVISVMEAEIVETLCMFERFFPPTFFDIMVHLTVHL 745
Query: 741 AHEAKVGGPVQYRWMYPIERY*LCIFKLIYFNVRIIYI*NYFFPRFLLTLKSFVRNKAYP 800
EA++GGPV +RWMYP ERY + LK FVRN A P
Sbjct: 746 GREARLGGPVHFRWMYPFERY-------------------------MKVLKDFVRNPARP 780
Query: 801 EGSIAESFLANECLTFCSQYLSGVETRFSRPTRNDDEFDSTLDDG 845
EG IAES+LA EC+ FCS +L + +P RN + ++++ +G
Sbjct: 781 EGCIAESYLAEECMRFCSDFLKKTTSVEEKPERNTEYENNSILEG 825
>At3g31920 hypothetical protein
Length = 725
Score = 557 bits (1435), Expect = e-158
Identities = 314/765 (41%), Positives = 429/765 (56%), Gaps = 52/765 (6%)
Query: 5 REWIRDPSRRSEEYIRGVNEFLEFAFQNSQVSEKIWCPCTTCANCKSLSRTSVYEHLTNP 64
+ W+ P R + EY +G F+ + ++ +++CPC C N ++ EHL
Sbjct: 3 KSWVWLP-RNNIEYEKGATTFVYASSRSLGDPAEMFCPCIDCRNLCHQPIETILEHLV-- 59
Query: 65 RRGFLRGYKQ---WIFHGEKPVASSSTIEEPEHTEMEHDMDGLIHDVLAS---HTAEDPL 118
+G YK+ W HGE + + EP T E + LI H+
Sbjct: 60 MKGMDHKYKRNRCWSKHGE--IRADKLEVEPSSTTSECEAYELIRTAYFDGDVHSDSQNQ 117
Query: 119 FDDGERNPQVEENANKFYRLVKDNEQILYPNCKKYSKLSFMVHLYHLKCLHGWSDKSFSM 178
+D + P+ +E A+ F +K+ E LY C KY+K+S ++ LY +K G S+ F
Sbjct: 118 NEDDSKEPETKEEAD-FREKLKEAETPLYHGCPKYTKVSAIMGLYRIKVKSGMSENYFDQ 176
Query: 179 LLDLLRDAFPEGNVLPKSFYETKKMISELGLGYEKIHACPNDCILYWDKYTNDEVCPKCG 238
LL L+ D P NVLP S E KK + G GY+ IHAC NDCILY +Y + CP+C
Sbjct: 177 LLSLVHDILPGENVLPTSTNEIKKFLKMFGFGYDIIHACQNDCILYRKEYELRDTCPRCN 236
Query: 239 ASRWKTVTEEVQPNGTKTSEKQKNTPLPKSVPLPGIPAKILRWFPLKPRLQRLFMSSKIA 298
ASRW E+ K+T K GIPAK+L +FP+K R +R+F S K+A
Sbjct: 237 ASRW---------------ERDKHTGEEKK----GIPAKVLGYFPIKDRFKRMFRSKKMA 277
Query: 299 EAMRWHRDGRLDDGFLRHPADSLAWKKFDSKYQTFALDPHNVRLGLASDGFNPFKTMSIT 358
E RWH DDG +RH DSLAW + + K+ FA +P N+RLGL++DG NPF +
Sbjct: 278 EDQRWHFSNASDDGTMRHHVDSLAWAQVNDKWPEFAAEPRNLRLGLSADGMNPFSIQNTK 337
Query: 359 HST*PVILVHFNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTYLQPLVKELQELWENGLE 418
+ST P++LV++N+ P MCMK ML+LLI GP P NNID YL PL+ +L++LW G+
Sbjct: 338 YSTWPILLVNYNMAPTMCMKAENIMLTLLIHGPTAPSNNIDVYLAPLIDDLKDLWSEGIH 397
Query: 419 TFDAYKKESFQLHAALMWTINDFPAYANLSGWSTKGKYACPCCGFETSSQWLRNGRKFCY 478
+D++KKESF L A +W+I+D+PA L+G KGK AC CG +T +WL+ RK Y
Sbjct: 398 VYDSFKKESFTLRAMFLWSISDYPALGTLAGCKVKGKQACNVCGKDTPFRWLKFSRKHVY 457
Query: 479 MSHRRWLTPNHKWRLNGKDFDGTQEFRDPPIRLDGTAQLRQLDEIEDECLGDQAQRWKKK 538
+ +RR L P H +R FD T E +GT Q E E L K
Sbjct: 458 LRNRRRLRPGHPYRRRRGWFDNTVE--------EGTTSRIQTGEEIFEALK------ALK 503
Query: 539 SILFSLPYWKYNEL--RHNLDVMHIEKNVCDNIIGTLLNQEGKSKDNYKARADLVDMGIR 596
IL L W+ ++ RHN+DVMH+EKNV D I+ ++L Q KSKD AR DL DMG R
Sbjct: 504 MILGDL--WRETDMPVRHNIDVMHVEKNVSDAIV-SMLMQSVKSKDGLNARKDLEDMGTR 560
Query: 597 SILHPQPSPNTNTMLLPRACYQMTTKEKDDFLSVLKNVKAPDECSSNISRCVQVQQRKIF 656
S LHPQ LP A Y ++ EK F L + PD +NIS CV + I
Sbjct: 561 SNLHPQ--LRVKRTYLPPAAYWLSKDEKRRFCKRLARFRGPDGYCANISNCVSMDPPVIG 618
Query: 657 GLKSYDCHVLMQELLPIALRGSLPDKVTSVLIDLCNFFKQICSKVLNVKFIEQLESQIAI 716
GLKS+D HVL+Q LLP+ALRG LP + LC++F ++C +V++ + + LES++
Sbjct: 619 GLKSHDHHVLLQNLLPVALRGLLPRVPRVAISRLCHYFNRLCQRVIDPEKLMSLESELVE 678
Query: 717 TLCQLETIFPPSFFTIMVHLVVHLAHEAKVGGPVQYRWMYPIERY 761
T+CQLE FPPS F IM HL +HLA E ++GGPV +RWMYP ERY
Sbjct: 679 TMCQLERYFPPSLFDIMFHLPIHLARETRLGGPVHFRWMYPFERY 723
>At2g14980 pseudogene
Length = 665
Score = 513 bits (1321), Expect = e-145
Identities = 289/724 (39%), Positives = 395/724 (53%), Gaps = 98/724 (13%)
Query: 122 GERNPQVEENANKFYRLVKDNEQILYPNCKKYSKLSFMVHLYHLKCLHGWSDKSFSMLLD 181
GE + ++ ++ + D E LYP+C +SKLS +V L+ LK +GWSDKSF+ LL+
Sbjct: 5 GEIAERDDKKEDELLAKLVDAETPLYPSCVNHSKLSAIVSLFRLKTKNGWSDKSFNDLLE 64
Query: 182 LLRDAFPEGNVLPKSFYETKKMISELGLGYEKIHACPNDCILYWDKYTNDEVCPKCGASR 241
L + PE NVL S YE KK + +GY+KIHAC NDC L+ KY + C KC ASR
Sbjct: 65 TLPEMLPEDNVLHTSLYEVKKFLKSFDMGYQKIHACVNDCCLFRKKYKKLQNCLKCNASR 124
Query: 242 WKTVTEEVQPNGTKTSEKQKNTPLPKSVPLPGIPAKILRWFPLKPRLQRLFMSSKIAEAM 301
WKT T E +K G+P K+LR+F + PRL+R+F S ++A +
Sbjct: 125 WKT--------NMHTGEVKK-----------GVPQKVLRYFSIVPRLKRMFRSEEMARDL 165
Query: 302 RWHRDGRLDDGFLRHPADSLAWKKFDSKYQTFALDPHNVRLGLASDGFNPFKTMSITHST 361
RWH + DG L HP DS+ W + ++KY FA + N+RLGL+++GFNPF + +S
Sbjct: 166 RWHFHNKSTDGKLCHPVDSVTWDQMNAKYPLFASEERNLRLGLSTNGFNPFNMKNTRYSC 225
Query: 362 *PVILVHFNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTYLQPLVKELQELWENGLETFD 421
PV+LV++NLPP +CMK+ ML+LLIPG + PGN+ID YL+PL+ +L LW+NG T+D
Sbjct: 226 WPVLLVNYNLPPDLCMKKENIMLTLLIPGSQQPGNSIDVYLEPLIDDLCHLWDNGEWTYD 285
Query: 422 AYKKESFQLHAALMWTINDFPAYANLSGWSTKGKYACPCCGFETSSQWLRNGRKFCYMSH 481
A+ K SF L A L+WTI+DFPAY NL+G KGK AC CG T S WL+ RK YM H
Sbjct: 286 AFSKSSFTLKAMLLWTISDFPAYGNLAGCKVKGKMACLLCGKHTESMWLKFSRKHVYMGH 345
Query: 482 RRWLTPNHKWRLNGKDFDGTQEFRDPPIRLDGTAQLRQLDEIEDECLGDQAQRWKKKSIL 541
R L P+H++R FDG E R L G L ++ + K+K
Sbjct: 346 RMGLPPSHRFRNKKSWFDGKAEHRRKSRILSGLEISHNLKNFQNNFGNFKQSTRKRKQPE 405
Query: 542 FSLPYWKYNELRHNLDVMHIEKNVCDNIIGTLLNQEGKSKDNYKARADLVDMGIRSILHP 601
LP +RHNLDVMH+E+NV +I+ TL++ GKSKD AR DL +GIR LHP
Sbjct: 406 SDLP------VRHNLDVMHVERNVVASIVSTLMHC-GKSKDGVNARKDLEVLGIRKDLHP 458
Query: 602 QPSPNTNTMLLPRACYQMTTKEKDDFLSVLKNVKAPDECSSNISRCVQVQQRKIFGLKSY 661
Q LP A + ++ EK F L + K PD S+I+R V ++
Sbjct: 459 QTKGKRT--YLPAALWSLSKSEKKLFCKRLFDFKGPDGYCSDIARGVSLED--------- 507
Query: 662 DCHVLMQELLPIALRGSLPDKVTSVLIDLCNFFKQICSKVLNVKFIEQLESQIAITLCQL 721
C +V++++ + ++E++I TLC
Sbjct: 508 ------------------------------------CKRVIDMEQLLEIEAEIVETLCLF 531
Query: 722 ETIFPPSFFTIMVHLVVHLAHEAKVGGPVQYRWMYPIERY*LCIFKLIYFNVRIIYI*NY 781
E+ FPPSFF IMVHL VHL EA +GGPV +RWMYP ERY
Sbjct: 532 ESFFPPSFFDIMVHLTVHLGREALLGGPVHFRWMYPFERY-------------------- 571
Query: 782 FFPRFLLTLKSFVRNKAYPEGSIAESFLANECLTFCSQYLSGVETRFSRPTRNDDEFDST 841
+ LK FVRN A PEG IAE +LA EC+ FCS++L + RN + +++
Sbjct: 572 -----MKVLKDFVRNTARPEGCIAECYLAEECIQFCSEFLKKTTKVQEKADRNTEYENNS 626
Query: 842 LDDG 845
+ G
Sbjct: 627 ILGG 630
>At1g35590 CACTA-element, putative
Length = 1180
Score = 499 bits (1286), Expect = e-141
Identities = 315/897 (35%), Positives = 450/897 (50%), Gaps = 137/897 (15%)
Query: 15 SEEYIRGVNEFLEFAFQNSQVSE---KIWCPCTTCANCKSL-SRTSVYEHLTNPRRGFLR 70
S EY+ GV EF+ FA V K CPC+ C N K + S V HL + +GF+
Sbjct: 30 SAEYVAGVEEFMTFANSQPIVQSCRGKFHCPCSVCKNEKHIISGRRVSSHLFS--QGFMP 87
Query: 71 GYKQWIFHGEK---PVASSST---------------IEEPEHTEMEHDM--------DGL 104
Y W HGE+ + +S T +E+P + +M +D D +
Sbjct: 88 DYYVWYKHGEELNMDIGTSYTDRTYFSENHEEVGNVVEDP-YVDMVNDAFNFNVGYDDNV 146
Query: 105 IHDVLASHTAEDPLFDDGERNPQVEENANKFYRLVKDNEQILYPNCKK-YSKLSFMVHLY 163
HD H ++ RN ++NKFY L++ +LY C++ S+LS L
Sbjct: 147 GHDDCYHHDGSYQNVEEPVRN-----HSNKFYDLLEGANNLLYDGCREGQSQLSLASRLM 201
Query: 164 HLKCLHGWSDKSFSMLLDLLRDAFPEGNVLPKSFYETKKMISELGLGYEKIHACPNDCIL 223
H K + S+K + ++ D PEGN S Y+T+K++ LGL Y I C N+C+L
Sbjct: 202 HNKAEYNMSEKLVDSVCEMFTDFLPEGNQATTSHYQTEKLMHNLGLPYHTIDVCKNNCML 261
Query: 224 YWDKYTNDEVCPKCGASRWKTVTEEVQPNGTKTSEKQKNTPLPKSVPLPGIPAKILRWFP 283
+W + ++ C CGA RWK + ++ T +P S + + P
Sbjct: 262 FWKEDEKEDQCRFCGAQRWKP------------KDDRRRTKVPYSR---------MWYLP 300
Query: 284 LKPRLQRLFMSSKIAEAMRWHRDGRLDDGFLRHPADSLAWKKFDSKYQTFALDPHNVRLG 343
+ RL+R++ K A AMRWH + + +G + HP+D+ W+ F + FA +P NV LG
Sbjct: 301 IADRLKRMYQCHKTAAAMRWHAEHQSKEGEMNHPSDAAEWRYFQELHPRFAEEPRNVYLG 360
Query: 344 LASDGFNPFKTMSITHST*PVILVHFNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTYLQ 403
L +DGFNPF MS HS PVIL +NLPP MCM + L++L GP P ++D +LQ
Sbjct: 361 LCTDGFNPFG-MSRNHSLWPVILTPYNLPPGMCMNTEYLFLTILNYGPNHPRASLDVFLQ 419
Query: 404 PLVKELQELWENGLETFDAYKKESFQLHAALMWTINDFPAYANLSGWSTKGKYACPCCGF 463
PL++EL+ELW G++ ++ ++F L A L+WTI+DFPAY LSGW+T GK +CP C
Sbjct: 420 PLIEELKELWCTGVDAYEVSLSQNFNLKAVLLWTISDFPAYNMLSGWTTHGKLSCPVCME 479
Query: 464 ETSSQWLRNGRKFCYMS-HRRWLTPNHKWRLNGKDF----DGTQEFRDPPIRLDGT---- 514
T S +L GRK C+ HRR+L H R N KDF D + E+ PP L G
Sbjct: 480 STKSFYLPIGRKTCWFDCHRRFLPHGHPSRRNKKDFLKGRDASSEY--PPESLTGEQVYY 537
Query: 515 AQLRQLDEIEDECLG-----------DQAQRWKKKSILFSLPYWKYNELRHNLDVMHIEK 563
+L ++ + + +G + W K+SIL+ L YWK LRHN+DVMH EK
Sbjct: 538 ERLASVNPPKTKDVGGNGHEKKMRGYGKEHNWHKESILWELSYWKDLNLRHNIDVMHTEK 597
Query: 564 NVCDNIIGTLLNQEGKSKDNYKARADLVDMGIRSILHPQPSPNTNTMLLPRACYQMTTKE 623
N DNI+ TLL +GKSKDN +R D+ R LH +T P Y +T +
Sbjct: 598 NFLDNIMNTLLGVKGKSKDNIMSRLDIEKYCSRPGLHID-----STGKAPFPPYTLTEQA 652
Query: 624 KDDFLSVLK-NVKAPDECSSNISRCVQVQQRKIFGLKSYDCHVLMQELLPIALRGSLPDK 682
K +K +V+ PD SS+++ CV ++ K G+KS+DCH+ M+ LLP L
Sbjct: 653 KQSLFQCVKHDVRFPDGYSSDLASCVDLENGKFSGMKSHDCHIFMERLLPFIFAELLDRN 712
Query: 683 VTSVL-----------IDLCN------------FFKQICSKVLNVKFIEQLESQIAITLC 719
V L I LCN FF+ +CS+ L ++ L+ I + +C
Sbjct: 713 VHLALSGTILTIFYGEIFLCNYLTSTYILGIGAFFRDLCSRTLQTSRVQILKQNIVLIIC 772
Query: 720 QLETIFPPSFFTIMVHLVVHLAHEAKVGGPVQYRWMYPIERY*LCIFKLIYFNVRIIYI* 779
LE IFPPSFF +M HL +H +EA++GGPVQYRWMYP
Sbjct: 773 NLEKIFPPSFFDVMEHLPIHRPYEAELGGPVQYRWMYP---------------------- 810
Query: 780 NYFFPRFLLTLKSFVRNKAYPEGSIAESFLANECLTFCSQYLSGVETRFSRPTRNDD 836
F RF LK +NK Y SI +S++ +E F Y + SR TR D+
Sbjct: 811 ---FERFFKKLKGKAKNKRYAASSIVKSYINDEIAYFSEHYFTDHIQTKSRLTRFDE 864
>At4g03900 putative transposon protein
Length = 817
Score = 489 bits (1260), Expect = e-138
Identities = 305/859 (35%), Positives = 434/859 (50%), Gaps = 130/859 (15%)
Query: 15 SEEYIRGVNEFLEFAFQNSQVSE---KIWCPCTTCANCKSL-SRTSVYEHLTNPRRGFLR 70
S EY+ GV EF+ FA V K +CPC+ C N K + S V HL + F+
Sbjct: 30 SAEYVAGVEEFMTFANSQPIVQSCRGKFYCPCSVCKNEKHIISGRRVSSHLFSHE--FMP 87
Query: 71 GYKQWIFHGEK---PVASSST---------------IEEPEHTEMEHDM--------DGL 104
Y W HGE+ + +S T +E+P + +M +D D +
Sbjct: 88 DYYVWYKHGEELNMDIGTSYTDRTYFSENHEEVGNVVEDP-YVDMVNDAFNFNVGYDDNV 146
Query: 105 IHDVLASHTAEDPLFDDGERNPQVEENANKFYRLVKDNEQILYPNCKK-YSKLSFMVHLY 163
HD H ++ RN ++NKFY L++ LY C++ S+LS L
Sbjct: 147 GHDDNYHHDGSYQNVEEPVRN-----HSNKFYDLLEGANNPLYDGCREGQSQLSLASRLM 201
Query: 164 HLKCLHGWSDKSFSMLLDLLRDAFPEGNVLPKSFYETKKMISELGLGYEKIHACPNDCIL 223
H K + S+K + ++ PEGN S Y+T+K++ LGL Y I N+C+L
Sbjct: 202 HNKAEYNMSEKLVDSVCEMFTAFLPEGNQATTSHYQTEKLMRNLGLPYHTIDVYKNNCML 261
Query: 224 YWDKYTNDEVCPKCGASRWKTVTEEVQPNGTKTSEKQKNTPLPKSVPLPGIPAKILRWFP 283
+W + ++ C CGA RWK + ++ T +P S + + P
Sbjct: 262 FWKEDEKEDQCRFCGAQRWKP------------KDDRRRTKVPYSR---------MWYLP 300
Query: 284 LKPRLQRLFMSSKIAEAMRWHRDGRLDDGFLRHPADSLAWKKFDSKYQTFALDPHNVRLG 343
+ RL+R++ S K A AMRWH + + +G + HP+D+ W+ F + FA +PHNV LG
Sbjct: 301 IADRLKRMYQSHKTAAAMRWHAEHQSKEGEMNHPSDAAEWRYFQELHPRFAEEPHNVYLG 360
Query: 344 LASDGFNPFKTMSITHST*PVILVHFNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTYLQ 403
L +DGFNPF MS HS PVIL +NLPP MCM + L++L GP P ++D +LQ
Sbjct: 361 LYTDGFNPFG-MSRNHSLWPVILTPYNLPPGMCMNTEYLFLTILNSGPNHPRASLDVFLQ 419
Query: 404 PLVKELQELWENGLETFDAYKKESFQLHAALMWTINDFPAYANLSGWSTKGKYACPCCGF 463
PL++EL+ELW G++ +D ++F L A L+WTI+DFPAY+ LSGW+T GK +CP C
Sbjct: 420 PLIEELKELWCTGVDAYDVSLSQNFNLKAVLLWTISDFPAYSMLSGWTTHGKLSCPVCME 479
Query: 464 ETSSQWLRNGRKFCYMS-HRRWLTPNHKWRLNGKDF----DGTQEFRDPPIRLDGT---- 514
T S +L NGRK C+ HRR+L H R N KDF D + E+ PP L G
Sbjct: 480 STKSFYLPNGRKTCWFDCHRRFLPHGHPSRRNKKDFLKGRDASSEY--PPESLTGEQVYY 537
Query: 515 AQLRQLDEIEDECLG-----------DQAQRWKKKSILFSLPYWKYNELRHNLDVMHIEK 563
+L ++ + + +G + W K+SIL+ L YWK LRHN+DVMH EK
Sbjct: 538 ERLASVNPPKTKDVGGNGHEKKMRGYGKEHNWHKESILWELSYWKDLNLRHNIDVMHTEK 597
Query: 564 NVCDNIIGTLLNQEGKSKDNYKARADLVDMGIRSILHPQPSPNTNTMLLPRACYQMTTKE 623
N DNI+ TLL +GKSKDN +R D+ R LH +T P Y++T
Sbjct: 598 NFLDNIMNTLLGVKGKSKDNIMSRLDIEKYCSRPGLHID-----STGKAPFPPYKLT--- 649
Query: 624 KDDFLSVLKNVKAPDECSSNISRCVQVQQRKIFGLKSYDCHVLMQELLPIALRGSLPDKV 683
+E ++ +CV+ R G+KS+DCHV M+ LLP L V
Sbjct: 650 --------------EEAKQSLFQCVKHDVRFPDGMKSHDCHVFMERLLPFIFAELLDRNV 695
Query: 684 TSVLIDLCNFFKQICSKVLNVKFIEQLESQIAITLCQLETIFPPSFFTIMVHLVVHLAHE 743
L + FF+ +CS+ L ++ L+ I + +C LE IFPPSFF +M HL +HL +E
Sbjct: 696 HLALSGIRAFFRDLCSRTLQTSRVQILKQNIVLIICNLEKIFPPSFFDVMEHLPIHLPYE 755
Query: 744 AKVGGPVQYRWMYPIERY*LCIFKLIYFNVRIIYI*NYFFPRFLLTLKSFVRNKAYPEGS 803
A++GGPVQYRWMYP F RF LK +NK Y GS
Sbjct: 756 AELGGPVQYRWMYP-------------------------FERFFKKLKGKAKNKRYAAGS 790
Query: 804 IAESFLANECLTFCSQYLS 822
I ES++ +E F Y +
Sbjct: 791 IVESYINDEIAYFSEHYFA 809
>At2g13010 pseudogene
Length = 1040
Score = 476 bits (1226), Expect = e-134
Identities = 300/839 (35%), Positives = 427/839 (50%), Gaps = 110/839 (13%)
Query: 39 IWCPCTTCANCKSLSRTSVYEHLTNPRRGFLRGYKQWIFHGE----------KPVASSST 88
I CPC C N K L V +HL N RGF+ Y W+ HG+ + + SS+
Sbjct: 211 ILCPCRKCLNEKYLEFNVVKKHLYN--RGFMPNYYVWLRHGDVVEIAHELVIQIMVSSNL 268
Query: 89 I--------EEPEHTEMEHDMDGLIHDVL--ASHTAEDPLFDDGERNPQVEENANKFYRL 138
+ + P+ ++ + HD++ A H ++ P V+ A FY +
Sbjct: 269 VILIMMRIMQVPDQFGYGYNQENRFHDMVTNAFHETIASFPENISEEPNVD--AQHFYDM 326
Query: 139 VKDNEQILYPNCKK-YSKLSFMVHLYHLKCLHGWSDKSFSMLLDLLRDAFPEGNVLPKSF 197
+ Q +Y C++ SKLS + +K + S+K +L+++ P N+ +S+
Sbjct: 327 LDAANQPIYEGCREGNSKLSLASRMMTIKADNNLSEKCMDSWAELIKEYLPPDNISAESY 386
Query: 198 YETKKMISELGLGYEKIHACPNDCILYWDKYTNDEVCPKCGASRWKTVTEEVQPNGTKTS 257
YE +K++S GL E I C + C+++W N + C CG R++T G +T
Sbjct: 387 YEIQKLVSSHGLPSEMIDVCIDYCMIFWGDDVNLQECRFCGKPRYQTT-------GGRTR 439
Query: 258 EKQKNTPLPKSVPLPGIPAKILRWFPLKPRLQRLFMSSKIAEAMRWHRDGRLDDGFLRHP 317
+P K + + P+ RL+RL+ S + A MRWH DG + HP
Sbjct: 440 ----------------VPYKRMWYLPITDRLKRLYQSERTAAKMRWHAQHTTADGEITHP 483
Query: 318 ADSLAWKKFDSKYQTFALDPHNVRLGLASDGFNPFKTMSITHST*PVILVHFNLPPWMCM 377
+D+ AWK F + Y FA + NV LGL +DGF+PF +S PVIL + LPP MCM
Sbjct: 484 SDAKAWKHFQTVYPDFANECRNVYLGLCTDGFSPFGLCGRQYSLWPVILTPYKLPPDMCM 543
Query: 378 KQSFFMLSLLIPGPKGPGNNIDTYLQPLVKELQELWENGLETFDAYKKESFQLHAALMWT 437
+ F LS+L+PGPK P +D +LQPL+ EL++LW G+ TFD K++F + A LMWT
Sbjct: 544 QSEFMFLSILVPGPKHPKRALDVFLQPLIHELKKLWYEGVHTFDYSSKQNFNMRAMLMWT 603
Query: 438 INDFPAYANLSGWSTKGKYACPCCGFETSSQWLRNGRKFCYMS-HRRWLTPNHKWRLNGK 496
I+DFPAY LSGW+ G+ P RK C+ + HRR+L +H +R N K
Sbjct: 604 ISDFPAYRMLSGWTMHGRLYVP-----------TKDRKPCWFNCHRRFLPLHHPYRRNKK 652
Query: 497 DFDGTQEFR-DPPIRLDGTAQLRQLD--EIEDEC-----------LGDQ---AQRWKKKS 539
F + R PP + GT Q+D ++ C + D+ W K+S
Sbjct: 653 LFRKNKIVRLLPPSYVSGTDLFEQIDYYGAQETCKRGGNWHTTANMPDEYGSTHNWYKQS 712
Query: 540 ILFSLPYWKYNELRHNLDVMHIEKNVCDNIIGTLLNQEGKSKDNYKARADLVDMGIRSIL 599
I + LPYWK LRHNL VMHIEKN DNI+ TLLN +GK+KD K+R DL ++ + L
Sbjct: 713 IFWQLPYWKDLLLRHNLYVMHIEKNFFDNIMNTLLNLQGKTKDTLKSRLDLAEICSKPEL 772
Query: 600 HPQPSPNTNTMLLPRACYQMTTKEKDD-FLSVLKNVKAPDECSSNISRCVQVQQRKIFGL 658
H T LP + ++++ + K F V+ VK PD S SRCV+ Q +K G+
Sbjct: 773 HV-----TRDGKLPVSKFRLSNEAKKALFEWVVAEVKFPDGYVSKFSRCVE-QGQKFSGM 826
Query: 659 KSYDCHVLMQELLPIALRGSLPDKVTSVLIDLCNFFKQICSKVLNVKFIEQLESQIAITL 718
KS+DCHV MQ LLP L LP + + + FFK +C++ L IEQL+ I + L
Sbjct: 827 KSHDCHVFMQRLLPFVLVELLPTNIHEAIAGIGVFFKDLCTRTLTTDTIEQLDKNIPVLL 886
Query: 719 CQLETIFPPSFFTIMVHLVVHLAHEAKVGGPVQYRWMYPIERY*LCIFKLIYFNVRIIYI 778
C LE +F P+FF +M HL +HL HE +GGPVQ+RWMYP
Sbjct: 887 CNLEKLFSPAFFDVMEHLPIHLPHEVALGGPVQFRWMYP--------------------- 925
Query: 779 *NYFFPRFLLTLKSFVRNKAYPEGSIAESFLANECLTFCSQYLS-GVETRFSRPTRNDD 836
F RF+ +LK +N A EGSI L E F S Y S V T+ +RP DD
Sbjct: 926 ----FERFMKSLKGKAKNLARVEGSIVAGSLTEETSHFTSYYFSPSVRTKKTRPRCYDD 980
>At3g30680 hypothetical protein
Length = 1116
Score = 449 bits (1155), Expect = e-126
Identities = 304/896 (33%), Positives = 434/896 (47%), Gaps = 163/896 (18%)
Query: 15 SEEYIRGVNEFLEFAFQNSQVSE---KIWCPCTTCANCKSL-SRTSVYEHLTNPRRGFLR 70
S EY+ GV EF+ FA V K CPC+ C N K + S V HL + +GF+
Sbjct: 30 SAEYVAGVEEFMTFANSQPIVQSCRGKFHCPCSVCKNEKHIISGRRVSSHLFS--QGFMP 87
Query: 71 GYKQWIFHGEK---PVASSST---------------IEEPEHTEMEHDM--------DGL 104
Y W HGE+ + +S T +E+P + +M +D D +
Sbjct: 88 DYYVWYKHGEELNMDIGTSYTDRTYFSENHEEVGNVVEDP-YVDMVNDAFNFNVGYDDNV 146
Query: 105 IHDVLASHTAEDPLFDDGERNPQVEENANKFYRLVKDNEQILYPNCKK-YSKLSFMVHLY 163
HD H ++ RN ++NKFY L++ LY C++ S+LS L
Sbjct: 147 GHDDCYHHDGSYQNVEEPVRN-----HSNKFYDLLEGANNPLYDGCREGQSQLSLASRLM 201
Query: 164 HLKCLHGWSDKSFSMLLDLLRDAFPEGNVLPKSFYETKKMISELGLGYEKIHACPNDCIL 223
H K + S+K + ++ D PEGN S Y+T+K++ LGL Y I C N+C+L
Sbjct: 202 HNKAEYNMSEKLVDSVCEMFTDFLPEGNQATTSHYQTEKLMRNLGLPYHTIDVCKNNCML 261
Query: 224 YWDKYTNDEVCPKCGASRWKTVTEEVQPNGTKTSEKQKNTPLPKSVPLPGIPAKILRWFP 283
+W + ++ C CGA RWK + ++ T +P S + + P
Sbjct: 262 FWKEDEKEDQCRFCGAQRWKP------------KDDRRRTKVPYSR---------MWYLP 300
Query: 284 LKPRLQRLFMSSKIAEAMRWHRDGRLDDGFLRHPADSLAWKKFDSKYQTFALDPHNVRLG 343
+ +L+R++ S K A AMRWH + + +G + HP+D+ W+ F + FA +P NV LG
Sbjct: 301 IADQLKRMYQSHKTAAAMRWHAEHQSKEGEMNHPSDAAEWRYFQELHPRFAEEPRNVYLG 360
Query: 344 LASDGFNPFKTMSITHST*PVILVHFNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTYLQ 403
L +DGFNPF +MS HS PVIL +NLPP MCM + L++L GP P ++D +LQ
Sbjct: 361 LCTDGFNPF-SMSRNHSLWPVILTPYNLPPRMCMNTEYLFLTILNSGPNHPRASLDVFLQ 419
Query: 404 PLVKELQELWENGLETFDAYKKESFQLHAALMWTINDFPAYANLSGWSTKGKYACPCCGF 463
PL++EL+ELW G++ +D ++F L A L+WTI+DFPAY+ LSGW+T GK +CP C
Sbjct: 420 PLIEELKELWCTGVDAYDVSLSQNFNLKAVLLWTISDFPAYSMLSGWTTHGKLSCPVCME 479
Query: 464 ETSSQWLRNGRKFCYMS-HRRWLTPNHKWRLNGKDF----DGTQEFRDPPIRLDGT---- 514
T S +L NGRK C+ HRR+L H R N KDF D + E+ PP L G
Sbjct: 480 STKSFYLPNGRKTCWFDCHRRFLPHGHPSRRNKKDFLKGRDASSEY--PPESLTGEQVYY 537
Query: 515 AQLRQLDEIEDECLG-----------DQAQRWKKKSILFSLPYWKYNELRHNLDVMHIEK 563
+L ++ + + +G + W K+SIL+ L YWK LRHN+DVMH EK
Sbjct: 538 ERLASVNPPKTKDVGGNGHEKKMRGYGKEHNWHKESILWELSYWKDLNLRHNIDVMHTEK 597
Query: 564 NVCDNIIGTLLNQEGKSKDNYKARADLVDMGIRSILHPQPSPNTNTMLLPRACYQMTTKE 623
N DNI+ TLL +GKSKDN +R D+ R LH +
Sbjct: 598 NFLDNIMNTLLGVKGKSKDNIMSRLDIEKYCSRPGLHIDST------------------- 638
Query: 624 KDDFLSVLKNVKAPDECSSNISRCVQVQQRKIFGLKSYDCHVLMQELLPIALRGSLPDKV 683
+K+ D C +R L + L+ + +AL G +
Sbjct: 639 ---------GMKSHD--------CHVFMER----LLPFIFAELLDRNVHLALSG-----I 672
Query: 684 TSVLIDLCNFFKQICSKVLNVKFIEQLESQIAITLCQLETIFPPSFFTIMVHLVVHLAHE 743
+ DLC S+ L ++ L+ I + +C LE IFPPSFF +M HL +HL +E
Sbjct: 673 GAFFRDLC-------SRTLQTSRVQILKQNIVLIICNLEKIFPPSFFDVMEHLPIHLPYE 725
Query: 744 AKVGGPVQYRWMYPIERY*LCIFKLIYFNVRIIYI*NYFFPRFLLTLKSFVRNKAYPEGS 803
A++GGPVQYRWMYP F RF LK +NK Y GS
Sbjct: 726 AELGGPVQYRWMYP-------------------------FERFFKKLKGKAKNKRYAAGS 760
Query: 804 IAESFLANECLTFCSQYLSGVETRFSRPTR-NDDEFDSTLDDG--SFLHPMGRPLG 856
I ES++ +E F Y + SR TR N+ E G + +GRP G
Sbjct: 761 IVESYINDEIAYFSEHYFADHIQTKSRLTRFNEGEVPVYHVPGVPNIFMQVGRPSG 816
>At3g30790 hypothetical protein
Length = 953
Score = 449 bits (1154), Expect = e-126
Identities = 259/669 (38%), Positives = 362/669 (53%), Gaps = 74/669 (11%)
Query: 188 PEGNVLPKSFYETKKMISELGLGYEKIHACPNDCILYWDKYTNDEVCPKCGASRWKTVTE 247
P N+ +S++E +K++ LGL E I C + C+++W N + C CG R++T +
Sbjct: 127 PPDNISAESYFEIQKLVLSLGLPSEMIDVCIDYCMIFWGDDVNLQECRFCGKPRYQTTSG 186
Query: 248 EVQPNGTKTSEKQKNTPLPKSVPLPGIPAKILRWFPLKPRLQRLFMSSKIAEAMRWHRDG 307
+ +P K + + P+ RL+RL+ S + A MRWH
Sbjct: 187 RTR-----------------------VPYKRMWYLPITDRLKRLYQSERTAAKMRWHAQH 223
Query: 308 RLDDGFLRHPADSLAWKKFDSKYQTFALDPHNVRLGLASDGFNPFKTMSITHST*PVILV 367
DG ++HP+D+ AWK F + Y FA + NV LGL +DGF+PF +S +IL
Sbjct: 224 TTADGEIKHPSDTKAWKHFQTVYPDFASECRNVYLGLCTDGFSPFVLSGRQYSLWHIILT 283
Query: 368 HFNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTYLQPLVKELQELWENGLETFDAYKKES 427
+NLPP M M+ F LS+L+PGPK P +D +LQPL+ EL++LW G+ TFD K++
Sbjct: 284 PYNLPPDMYMQSEFLFLSILVPGPKHPKRALDVFLQPLIHELKKLWYEGVHTFDYSSKQN 343
Query: 428 FQLHAALMWTINDFPAYANLSGWSTKGKYACPCCGFETSSQWLRNGRKFCYMS-HRRWLT 486
F + A LMWTI+DFPAY LSGW+T G+ +CP C T + L+ GRK C+ HRR+L
Sbjct: 344 FNMRAVLMWTISDFPAYGMLSGWTTHGRLSCPYCMGRTDAFQLKKGRKPCWFDYHRRFLP 403
Query: 487 PNHKWRLNGKDFDGTQEFR-DPPIRLDGTAQLRQLD--EIEDEC-----------LGDQ- 531
+H +R N K F + R PP + GT +Q+D ++ C + D+
Sbjct: 404 LHHPYRRNKKLFRKNKIVRLPPPSYVSGTDLFKQIDYYGAQETCKRGGNWLTTANMPDEY 463
Query: 532 --AQRWKKKSILFSLPYWKYNELRHNLDVMHIEKNVCDNIIGTLLNQEGKSKDNYKARAD 589
W K+SI + LPYWK LRH LDVMHIEKN DNI+ TLLN +GK+KD+ K+R D
Sbjct: 464 GSTHNWHKQSIFWQLPYWKDLFLRHCLDVMHIEKNFFDNIMNTLLNVQGKTKDSLKSRLD 523
Query: 590 LVDMGIRSILHPQPSPNTNTMLLPRACYQMTTKEKDD-FLSVLKNVKAPDECSSNISRCV 648
L ++ R LH T LP + ++++ + K F V+ VK PD S SRCV
Sbjct: 524 LEEICSRPELHV-----TRDGKLPVSKFRLSNEAKKALFEWVVAEVKFPDGYVSKFSRCV 578
Query: 649 QVQQRKIFGLKSYDCHVLMQELLPIALRGSLPDKVTSVLIDLCNFFKQICSKVLNVKFIE 708
+ Q +K G+KS+DCHV MQ LLP L LP + + + FFK +C++ L IE
Sbjct: 579 E-QGQKFSGMKSHDCHVFMQRLLPFVLVELLPTNIHEAIAGIGVFFKDLCTRTLTTYTIE 637
Query: 709 QLESQIAITLCQLETIFPPSFFTIMVHLVVHLAHEAKVGGPVQYRWMYPIERY*LCIFKL 768
QL+ I + LC LE +F P FF +M HL +HL HE +GGPVQ+RWM
Sbjct: 638 QLDKNIPVLLCNLEKLFLPVFFDVMEHLPIHLLHEVALGGPVQFRWM------------- 684
Query: 769 IYFNVRIIYI*NYFFPRFLLTLKSFVRNKAYPEGSIAESFLANECLTFCSQYLS-GVETR 827
Y F +F+ +LK +N A EGSI L E F S Y S V T+
Sbjct: 685 ------------YLFEQFMKSLKGKAKNLARVEGSIVAGSLTEETSHFTSYYFSPSVRTK 732
Query: 828 FSRPTRNDD 836
+RP R DD
Sbjct: 733 KTRPRRYDD 741
Score = 38.1 bits (87), Expect = 0.029
Identities = 26/77 (33%), Positives = 37/77 (47%), Gaps = 6/77 (7%)
Query: 4 SREW-IRDPSRR-SEEYIRGVNEFLEFAFQNSQVSEK--IWCPCTTCANCKSLSRTSVYE 59
S W +R R+ + EY G+ F+ A E I+CPC C N K L V +
Sbjct: 52 SESWRVRSNDRKVTREYYDGLRGFMYQAINEPFARENGTIFCPCRKCLNEKYLEFNVVKK 111
Query: 60 HLTNPRRGFLRGYKQWI 76
HL N RGF+ Y +++
Sbjct: 112 HLYN--RGFMPNYYEYL 126
>At5g59640 serine/threonine-specific protein kinase - like
Length = 986
Score = 424 bits (1089), Expect = e-118
Identities = 246/604 (40%), Positives = 338/604 (55%), Gaps = 47/604 (7%)
Query: 94 HTEMEHDMDGLIHDVLASHTAEDPLFDD-GERNPQVEENANKFYRLVKDNEQILYPNCKK 152
HTE ++ D S +E FD E +P E A KFY ++ +Q +Y CK+
Sbjct: 20 HTEPVDPYVEMVSDAFGSTESE---FDQMREEDPNFE--AKKFYDILDAAKQPIYDGCKE 74
Query: 153 -YSKLSFMVHLYHLKCLHGWSDKSFSMLLDLLRDAFPEGNVLPKSFYETKKMISELGLGY 211
SKLS L LK + S + ++++ PEGN PKS+YE KK++ LGL Y
Sbjct: 75 GLSKLSLAARLMSLKTDNNLSQNCMDSIAQIMQEYLPEGNNSPKSYYEIKKLMRSLGLPY 134
Query: 212 EKIHACPNDCILYWDKYTNDEVCPKCGASRWKTVTEEVQPNGTKTSEKQKNTPLPKSVPL 271
+KI C ++C+++W + +E C C R++ Q G K+
Sbjct: 135 QKIDVCQDNCMIFWKETEKEEYCLFCKKDRYRPT----QKIGQKS--------------- 175
Query: 272 PGIPAKILRWFPLKPRLQRLFMSSKIAEAMRWHRDGRLDDGFLRHPADSLAWKKFDSKYQ 331
IP + + + P+ RL+RL+ S A+ MRWH + DG + HP+D AWK F +
Sbjct: 176 --IPYRQMFYLPIADRLKRLYQSHNTAKHMRWHAEHLASDGEMGHPSDGEAWKHFHKVHP 233
Query: 332 TFALDPHNVRLGLASDGFNPFKTMSITHST*PVILVHFNLPPWMCMKQSFFMLSLLIPGP 391
FA P NV LGL +DGFNPF +S PVIL +NLPP MCMKQ F L++L+PGP
Sbjct: 234 DFASKPRNVYLGLCTDGFNPFGMSGHNYSLWPVILTPYNLPPEMCMKQEFMFLTILVPGP 293
Query: 392 KGPGNNIDTYLQPLVKELQELWENGLETFDAYKKESFQLHAALMWTINDFPAYANLSGWS 451
P ++D +LQPL++EL++LW NG+E +D K++F L LMWTI+DFPAY LSGW+
Sbjct: 294 NHPKRSLDIFLQPLIEELKDLWVNGVEAYDISTKQNFLLKVVLMWTISDFPAYGMLSGWT 353
Query: 452 TKGKYACPCCGFETSSQWLRNGRKFCYMS-HRRWLTPNHKWRLNGKDFDGTQEFRD-PPI 509
T G+ ACP C +T + WL+NGRK C+ HR +L NH +R N KDF + D P
Sbjct: 354 THGRLACPYCSDQTGAFWLKNGRKTCWFDCHRCFLPVNHSYRGNKKDFKKGKVVEDSKPE 413
Query: 510 RLDGTAQLRQLDEIED--ECLGD--------QAQRWKKKSILFSLPYWKYNELRHNLDVM 559
L G ++ + +C G+ + W K+SIL+ L YWK +LRHNLDVM
Sbjct: 414 ILTGEELYNEVCCLPKTVDCGGNHGRLEGYGKTHNWHKQSILWELSYWKDLKLRHNLDVM 473
Query: 560 HIEKNVCDNIIGTLLNQEGKSKDNYKARADLVDMGIRSILHPQPSPNTNTMLLPRACYQM 619
HIEKNV DN I TLLN +GK+KDN K+R DL + R LH P P +++
Sbjct: 474 HIEKNVLDNFIKTLLNVQGKTKDNIKSRLDLQENCNRKDLHLTPEGKA-----PIPKFRL 528
Query: 620 TTKEKDDFLSVL-KNVKAPDECSSNISRCVQVQQRKIFGLKSYDCHVLMQELLPIALRGS 678
K+ FL L K+VK D SS+++ CV + RK+ G+KS+DCHVLMQ L PIA
Sbjct: 529 KPDAKEIFLRWLEKDVKFSDGYSSSLANCVDLPGRKLTGMKSHDCHVLMQRLFPIAF-AE 587
Query: 679 LPDK 682
L DK
Sbjct: 588 LMDK 591
>At2g15070 En/Spm-like transposon protein
Length = 672
Score = 422 bits (1084), Expect = e-118
Identities = 271/765 (35%), Positives = 381/765 (49%), Gaps = 144/765 (18%)
Query: 121 DGERNPQ--VEENANKFYRLVKDNEQILYPNCKKYSKLSFMVHLYHLKCLHGWSDKSFSM 178
DGE + + V + + F +KD E LY C KY+K+S ++ LY +K G S+ F
Sbjct: 42 DGEEDSKEPVTKEESYFREKLKDVETPLYYGCPKYTKVSAIMGLYRIKVKSGMSENYFDQ 101
Query: 179 LLDLLRDAFPEGNVLPKSFYETKKMISELGLGYEKIHACPNDCILYWDKYTNDEVCPKCG 238
LL L+ D P NVLP S E KK + G GY+ IHACPNDCILY +Y + C C
Sbjct: 102 LLSLVHDILPGENVLPTSTNEIKKFLKMFGFGYDIIHACPNDCILYKKEYELRDTCLGCS 161
Query: 239 ASRWKTVTEEVQPNGTKTSEKQKNTPLPKSVPLPGIPAKILRWFPLKPRLQRLFMSSKIA 298
ASRWK T E++K GIPAK+L +F +K R +R+F S K+
Sbjct: 162 ASRWK--------RDKHTGEEKK-----------GIPAKVLWYFSIKDRFKRMFRSKKMV 202
Query: 299 EAMRWHRDGRLDDGFLRHPADSLAWKKFDSKYQTFALDPHNVRLGLASDGFNPFKTMSIT 358
E ++WH DG +RHP DSLAW + + K+ FA + N+RLGL++D NPF +
Sbjct: 203 EDLQWHFSNASVDGTMRHPIDSLAWAQVNDKWPQFAAESRNLRLGLSTDKMNPFSIQNTK 262
Query: 359 HST*PVILVHFNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTYLQPLVKELQELWENGLE 418
+ST PV+LV++N+ P MCMK ML+LLI GPK P NNID YL PL+ +L++L +G++
Sbjct: 263 YSTWPVLLVNYNMAPTMCMKAKNIMLTLLIHGPKAPSNNIDVYLAPLINDLKDLSSDGIQ 322
Query: 419 TFDAYKKESFQLHAALMWTINDFPAYANLSGWSTKGKYACPCCGFETSSQWLRNGRKFCY 478
+D++ KE+F L A L+W+IND+P L+G
Sbjct: 323 VYDSFFKENFTLRAMLLWSINDYPTLGTLAGC---------------------------- 354
Query: 479 MSHRRWLTPNHKWRLNGKDFDGTQEFRDPPIRLDGTAQLRQLDEIEDECLGDQAQ--RWK 536
KDF +F P L+ + ++ D ED+ G+ RWK
Sbjct: 355 -----------------KDF--KNDFGRP---LERNNKRKRSDLCEDDEYGEDTHQWRWK 392
Query: 537 KKSILFSLPYWKYNELRHNLDVMHIEKNVCDNIIGTLLNQEGKSKDNYKARADLVDMGIR 596
K+SILF LPYWK +RHN+DVMH+EKNV D I+ TL+ Q KSKD AR +L DMGIR
Sbjct: 393 KRSILFELPYWKDLPVRHNIDVMHVEKNVSDVIVSTLM-QSAKSKDGLNARKNLEDMGIR 451
Query: 597 SILHPQPSPNTNTMLLPRACYQMTTKEKDDFLSVLKNVKAPDECSSNISRCVQVQQRKIF 656
S LH Q +L A + ++ EK F L + PD +NIS CV V I
Sbjct: 452 SNLHIQLRGKRTYLL--HAAFWLSKDEKKRFCKRLARFRGPDGYCANISNCVSVDPPVIG 509
Query: 657 GLKSYDCHVLMQELLPIALRGSLPDKVTSVLIDLCNFFKQICSKVLNVKFIEQLESQIAI 716
GLKS+D HVL+Q LLP+ALRG LP + + L ++F ++C +V++ + + LE ++
Sbjct: 510 GLKSHDHHVLLQNLLPVALRGLLPKRPRVAISRLGHYFNRLCQRVIDPEKLISLEYELVE 569
Query: 717 TLCQLETIFPPSFFTIMVHLVVHLAHEAKVGGPVQYRWMYPIERY*LCIFKLIYFNVRII 776
T+CQLE R+M ++ Y
Sbjct: 570 TMCQLE------------------------------RYMKTLKAY--------------- 584
Query: 777 YI*NYFFPRFLLTLKSFVRNKAYPEGSIAESFLANECLTFCSQYLSGVETRFSRPTRNDD 836
+K+F +A +AE +LA EC+ FC ++L + RN+
Sbjct: 585 -------------VKNFATLEA----CMAEGYLAGECMAFCLKFLQNSLSVPEVVNRNEG 627
Query: 837 EFDSTLDDGSFLHPMGRPLGRKRQQKFQLKKRKRVSRAKLEKKSV 881
D + L GRPL + Q K+R R L +V
Sbjct: 628 V------DSNSLVLEGRPLHKATQVTLTSKERDIAHRYVLMNMAV 666
>At1g31560 putative protein
Length = 1431
Score = 403 bits (1036), Expect = e-112
Identities = 218/515 (42%), Positives = 286/515 (55%), Gaps = 61/515 (11%)
Query: 162 LYHLKCLHGWSDKSFSMLLDLLRDAFPEGNVLPKSFYETKKMISELGLGYEKIHACPNDC 221
L +K +GWSDKSF+ LL L + P NVL S YE KK + +GYEKIHAC NDC
Sbjct: 83 LAKIKTKNGWSDKSFNELLGTLPEMLPADNVLRTSLYEVKKFLKSFDMGYEKIHACVNDC 142
Query: 222 ILYWDKYTNDEVCPKCGASRWKTVTEEVQPNGTKTSEKQKNTPLPKSVPLPGIPAKILRW 281
L+ +Y N E CPKC ASRWK N +T E +K G+P K+LR+
Sbjct: 143 CLFRKRYKNLENCPKCNASRWK--------NYMQTGEAKK-----------GVPQKVLRY 183
Query: 282 FPLKPRLQRLFMSSKIAEAMRWHRDGRLDDGFLRHPADSLAWKKFDSKYQTFALDPHNVR 341
F + PR +++F S ++A+ +RWH + DG RHP DS+ W K KY FA + N+R
Sbjct: 184 FLIIPRFKKMFRSKEMAKDLRWHYSNKSTDGKHRHPVDSVTWDKMIDKYPEFAAEERNMR 243
Query: 342 LGLASDGFNPFKTMSITHST*PVILVHFNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTY 401
LGL++DGFNPF + +S PV+LV++NLPP CMK MLSLLIPGP+ PGN+ID Y
Sbjct: 244 LGLSTDGFNPFNMKNTRYSCWPVLLVNYNLPPDQCMKNENIMLSLLIPGPQQPGNSIDVY 303
Query: 402 LQPLVKELQELWENGLETFDAYKKESFQLHAALMWTINDFPAYANLSGWSTKGKYACPCC 461
L+P +++L LW+ G T+DA+ K +F L A L WTI+DFPAY NL+G KGK CP C
Sbjct: 304 LEPFIEDLNHLWKKGELTYDAFSKTTFTLKAMLFWTISDFPAYGNLAGCKVKGKMGCPLC 363
Query: 462 GFETSSQWLRNGRKFCYMSHRRWLTPNHKWRLNGKDFDGTQEFRDPPIRLDGTAQLRQLD 521
G T S WL+ RK +M HR+ L P H ++ FDG E L G L
Sbjct: 364 GKGTDSMWLKYSRKHVFMCHRKGLAPTHSYKNKKAWFDGKVEHGRKARILTGREVSHNLK 423
Query: 522 EIEDE---------------CLG----------------------DQAQRWKKKSILFSL 544
+++ C+ D+ RWKK+SI F L
Sbjct: 424 NFKNDFGNAKESGRKRKRNDCIESVTDSDNESSESEEDEEVEVDEDELSRWKKRSIFFKL 483
Query: 545 PYWKYNELRHNLDVMHIEKNVCDNIIGTLLNQEGKSKDNYKARADLVDMGIRSILHPQ-P 603
YW+ +RHNLDVMH+E+NV +I+ TLL+ GKSKD +R DL D+G+R LHP
Sbjct: 484 SYWENLPVRHNLDVMHVERNVGASIVSTLLHC-GKSKDGLNSRKDLQDLGVRKDLHPNAQ 542
Query: 604 SPNTNTMLLPRACYQMTTKEKDDFLSVLKNVKAPD 638
TN LP A + ++ EK F L K PD
Sbjct: 543 GKRTN---LPAAPWSLSKSEKKTFCKHLFEFKGPD 574
Score = 70.5 bits (171), Expect = 5e-12
Identities = 36/103 (34%), Positives = 53/103 (50%), Gaps = 25/103 (24%)
Query: 743 EAKVGGPVQYRWMYPIERY*LCIFKLIYFNVRIIYI*NYFFPRFLLTLKSFVRNKAYPEG 802
EA++GGP+ +RWMYP ERY + LK FVRN P+
Sbjct: 576 EARLGGPIHFRWMYPFERY-------------------------MKVLKDFVRNSVKPDV 610
Query: 803 SIAESFLANECLTFCSQYLSGVETRFSRPTRNDDEFDSTLDDG 845
IAES+LA EC+ FCS +L + +P RN + ++++ +G
Sbjct: 611 CIAESYLAEECMQFCSDFLKKTTSVEEKPERNTEYENNSILEG 653
>At4g07750 putative transposon protein
Length = 569
Score = 389 bits (999), Expect = e-108
Identities = 210/511 (41%), Positives = 290/511 (56%), Gaps = 67/511 (13%)
Query: 129 EENANKFYRLVKDNEQILYPNCKKYSKLSFMVHLYHLKCLHGWSDKSFSMLLDLLRDAFP 188
++ ++F + D E LYP+C +SKLS +V L+ L +GWSDKSF+ LL++L + P
Sbjct: 60 DKKEDEFLAKLADAETPLYPSCVNHSKLSAIVSLFRLNTKNGWSDKSFNDLLEILPEMLP 119
Query: 189 EGNVLPKSFYETKKMISELGLGYEKIHACPNDCILYWDKYTNDEVCPKCGASRWKTVTEE 248
+ NVL S Y+ KK + +GY+KIHAC NDC L+ Y + CPKC ASRWKT
Sbjct: 120 KDNVLHTSLYKVKKFLKSFDMGYQKIHACVNDCCLFRKNYKKLDSCPKCNASRWKT---- 175
Query: 249 VQPNGTKTSEKQKNTPLPKSVPLPGIPAKILRWFPLKPRLQRLFMSSKIAEAMRWHRDGR 308
E +K G+P K+LR+FP+ PRL+ +F S ++ + +RWH +
Sbjct: 176 ----NLHIGEIKK-----------GVPQKVLRYFPIIPRLKGMFRSKEMDKDLRWHFSNK 220
Query: 309 LDDGFLRHPADSLAWKKFDSKYQTFALDPHNVRLGLASDGFNPFKTMSITHST*PVILVH 368
D L HP D + W + ++KY +FA + N+R GL++DGFNPF + +S P +
Sbjct: 221 SSDEKLCHPVDLVTWDQMNAKYPSFAGEERNLRHGLSTDGFNPFNMKNTRYSCWPDL--- 277
Query: 369 FNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTYLQPLVKELQELWENGLETFDAYKKESF 428
CMK+ ML+LLIPGP+ PGNNID YL+PL+++L LW+NG T+DAY K +F
Sbjct: 278 -------CMKKENIMLTLLIPGPQQPGNNIDVYLEPLIEDLCHLWDNGELTYDAYSKSTF 330
Query: 429 QLHAALMWTINDFPAYANLSGWSTKGKYACPCCGFETSSQWLRNGRKFCYMSHRRWLTPN 488
L A L+WTI+DFPAY NL+G KGK CP CG T S WL+ RK YM H + L P
Sbjct: 331 DLKAMLLWTISDFPAYGNLAGCKVKGKMGCPLCGKNTDSMWLKFSRKHVYMCHIKGLPPT 390
Query: 489 HKWRLNGKDFDGTQE-------------------FRDPPIRLDGTAQLRQL--------- 520
H +R+ FDG E F++ L +A R+
Sbjct: 391 HHFRMKKSWFDGKAENGRKRRILSGQEIYQNLKNFKNNFGTLKQSASKRKRTVNIEADSD 450
Query: 521 ---------DEIEDECLGDQAQRWKKKSILFSLPYWKYNELRHNLDVMHIEKNVCDNIIG 571
++ +D+ ++ RWKKKSI F LPYW+ +RHNLDVMH+E+NV +II
Sbjct: 451 SDDQSSESEEDEQDQVDEEELSRWKKKSIFFKLPYWQELLVRHNLDVMHVERNVAASIIS 510
Query: 572 TLLNQEGKSKDNYKARADLVDMGIRSILHPQ 602
TLL+ KSKD AR DL +GIR LHP+
Sbjct: 511 TLLHC-WKSKDGLNARKDLEVLGIRKDLHPK 540
>At1g35280 hypothetical protein
Length = 557
Score = 382 bits (981), Expect = e-106
Identities = 207/485 (42%), Positives = 275/485 (56%), Gaps = 59/485 (12%)
Query: 188 PEGNVLPKSFYETKKMISELGLGYEKIHACPNDCILYWDKYTNDEVCPKCGASRWKTVTE 247
P NVL S YE KK + +GYEKIHAC NDC L+ +Y N E CPKC ASRWK+
Sbjct: 3 PADNVLHTSLYEVKKFLKSFDMGYEKIHACVNDCCLFRKRYKNLENCPKCNASRWKS--- 59
Query: 248 EVQPNGTKTSEKQKNTPLPKSVPLPGIPAKILRWFPLKPRLQRLFMSSKIAEAMRWHRDG 307
+T E +K G+P K+LR+FP+ PR +R+F S ++A+ +RWH
Sbjct: 60 -----NMQTGEAKK-----------GVPQKVLRYFPIIPRFKRMFRSEEMAKDLRWHYSN 103
Query: 308 RLDDGFLRHPADSLAWKKFDSKYQTFALDPHNVRLGLASDGFNPFKTMSITHST*PVILV 367
+ DG LRHP DS+ W K + KY FA + N+RLGL++DGFNPF + +S PV+LV
Sbjct: 104 KSIDGKLRHPVDSVTWDKMNDKYPEFAAEERNLRLGLSTDGFNPFNMKNTRYSCWPVLLV 163
Query: 368 HFNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTYLQPLVKELQELWENGLETFDAYKKES 427
++NLP +CMK+ MLSLLIPGP+ PGN+ID YL+PL+++L LW+ G T+DA+ K +
Sbjct: 164 NYNLPLDLCMKKENIMLSLLIPGPQQPGNSIDVYLEPLIEDLNHLWKKGELTYDAFSKTT 223
Query: 428 FQLHAALMWTINDFPAYANLSGWSTKGKYACPCCGFETSSQWLRNGRKFCYMSHRRWLTP 487
F L A L+WTI+DF AY NL+G KGK CP CG T S WL+ RK +M HR+ L P
Sbjct: 224 FTLKAMLLWTISDFSAYGNLAGCKVKGKMGCPLCGKGTDSMWLKYSRKHVFMCHRKGLAP 283
Query: 488 NHKWRLNGKDFDGTQEFRDPPIRLDGTAQLRQL--------------------------- 520
H +R FDG E L G + L
Sbjct: 284 THSYRNKKAWFDGKIEHGRKARILTGREVSQTLKNFKNDFRNAKESGRKRKRNDCIESVT 343
Query: 521 -------DEIEDECL---GDQAQRWKKKSILFSLPYWKYNELRHNLDVMHIEKNVCDNII 570
+ EDE L D+ RWKK+SI F L YW+ +RHNLDVMH+E+NV +I+
Sbjct: 344 DSDDESSESEEDEELEVDKDELSRWKKRSIFFKLSYWEDLPVRHNLDVMHVERNVGASIV 403
Query: 571 GTLLNQEGKSKDNYKARADLVDMGIRSILHPQPSPNTNTMLLPRACYQMTTKEKDDFLSV 630
TLL+ GKSKD AR DL D+G+R LH P+ LP A + ++ E+ SV
Sbjct: 404 STLLHC-GKSKDGLNARKDLQDLGVRKDLH--PNAQGKRTYLPAAPWSLSKSERKHSASV 460
Query: 631 LKNVK 635
++K
Sbjct: 461 CLSLK 465
>At3g32230 hypothetical protein
Length = 1169
Score = 374 bits (961), Expect = e-103
Identities = 264/849 (31%), Positives = 393/849 (46%), Gaps = 168/849 (19%)
Query: 3 ESREWIRDPSRRSEEYIRGVNEFLEFAFQNS--QVSEKIWCPCTTCANCKSLSRTSVYEH 60
+S E + + + + I G+ F+ A S + + I+CPC C+ + +S
Sbjct: 147 KSEEIYNEVAPKKKGRIYGLRGFMYQATNESFARANGSIFCPC----RCRDSAGSS---- 198
Query: 61 LTNPRRGFLRGYKQWIFHGEKPVASSSTIEEPEHTEMEHDMDGLIHDVL--ASHTAEDPL 118
N G + + +G + ++ ++ + HD++ A H
Sbjct: 199 --NMNYGVVEPTNYYENYGHH-----------QESDQGYNHENRFHDMVTDAFHETTAAF 245
Query: 119 FDDGERNPQVEENANKFYRLVKDNEQILYPNCKK-YSKLSFMVHLYHLKCLHGWSDKSFS 177
++ P V+ A +FY ++ + +Y C++ SKLS + +K + S+
Sbjct: 246 HENIIEEPNVD--AQRFYDMLDAANKPIYEECREGQSKLSLASRMMTIKADNNLSENCMD 303
Query: 178 MLLDLLRDAFPEGNVLPKSFYETKKMISELGLGYEKIHACPNDCILYWDKYTNDEVCPKC 237
+L+++ N+ +S+YE +K++S LGL E I C + C+++W N + C C
Sbjct: 304 SWAELIKEYLIADNISAESYYEIQKLVSSLGLPSEMIDVCIDYCMIFWKDDENLQECRFC 363
Query: 238 GASRWKTVTEEVQPNGTKTSEKQKNTPLPKSVPLPGIPAKILRWFPLKPRLQRLFMSSKI 297
G R+ QP G +T +P K + + P+ RL+RL+ S +
Sbjct: 364 GKPRY-------QPTGRRTR----------------VPYKRMWYLPITDRLKRLYQSERT 400
Query: 298 AEAMRWHRDGRLDDGFLRHPADSLAWKKFDSKYQTFALDPHNVRLGLASDGFNPFKTMSI 357
A MRWH + +G + HP+D+ AWK F + Y FA + NV LGL +DGF+PF
Sbjct: 401 AAKMRWHAEHSTVNGEVTHPSDAKAWKHFQTVYPDFASECRNVYLGLCTDGFSPF----- 455
Query: 358 THST*PVILVHFNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTYLQPLVKELQELWENGL 417
GPK P +D +LQPL+ EL+ LW G+
Sbjct: 456 --------------------------------GPKHPKRALDVFLQPLIHELKNLWYEGV 483
Query: 418 ETFDAYKKESFQLHAALMWTINDFPAYANLSGWSTKGKYACPCCGFETSSQWLRNGRK-F 476
+TFD K++F + A LMWTI+DFPAY LSGW+T G+ +CP C T + L++GRK F
Sbjct: 484 QTFDYSSKQNFNMRAMLMWTISDFPAYGMLSGWTTHGRLSCPYCMDRTDAFQLKHGRKSF 543
Query: 477 CYMSHRRWLTPNHKWRLNGKDFDGTQE-------FRDPPIRLDGTAQLRQLDEIEDECLG 529
+ +L+ N + D+ G+QE + P DG
Sbjct: 544 VRLPPPVYLSGNDLF--ENIDYYGSQETCKRGGNWHTPANMPDGYG-------------- 587
Query: 530 DQAQRWKKKSILFSLPYWKYNELRHNLDVMHIEKNVCDNIIGTLLNQEGKSKDNYKARAD 589
A W K+SI + LPYWK LRHNLD GK+KD+ K+R D
Sbjct: 588 -SAHNWHKQSIFWQLPYWKDLLLRHNLD--------------------GKTKDSMKSRLD 626
Query: 590 LVDMGIRSILHPQPSPNTNTMLLPRACYQMTTKEKDDFLSVLK-NVKAPDECSSNISRCV 648
L ++ R LH T LP +++TT+ K +K VK PD S SRCV
Sbjct: 627 LAEICNRPDLH-----ITRDGKLPVPKFRLTTEGKKALFEWVKAEVKFPDGYLSKFSRCV 681
Query: 649 QVQQRKIFGLKSYDCHVLMQELLPIALRGSLPDKVTSVLIDLCNFFKQICSKVLNVKFIE 708
+ Q +K G+KS+DCHV +Q LLP LP V + FF+ +C++ L I+
Sbjct: 682 E-QGQKFSGMKSHDCHVFIQRLLPFVFLELLPANVHDAIEGA--FFRDLCTRTLTTDGIQ 738
Query: 709 QLESQIAITLCQLETIFPPSFFTIMVHLVVHLAHEAKVGGPVQYRWMYPIERY*LCIFKL 768
QL+ I + LC LE +FPP+FF +M HL +HL HEA +GGPVQ+RWMYP
Sbjct: 739 QLDENIPMLLCNLEKVFPPTFFDVMEHLSIHLPHEAALGGPVQFRWMYP----------- 787
Query: 769 IYFNVRIIYI*NYFFPRFLLTLKSFVRNKAYPEGSIAESFLANECLTFCSQYLS-GVETR 827
F RF+ TLK +N A EGSI L E F S Y S V T+
Sbjct: 788 --------------FERFMKTLKGKAKNLARVEGSIVAGSLTEETTHFTSYYFSPTVRTK 833
Query: 828 FSRPTRNDD 836
+RP R DD
Sbjct: 834 KTRPRRYDD 842
>At1g36470 hypothetical protein
Length = 753
Score = 359 bits (922), Expect = 4e-99
Identities = 200/510 (39%), Positives = 281/510 (54%), Gaps = 50/510 (9%)
Query: 184 RDAFPEGNVLPKSFYETKKMISELGLGYEKIHACPNDCILYWDKYTNDEVCPKCGASRWK 243
RD PEGN S Y+T+K++ LGL Y I C N+C+L W + ++ C CGA RWK
Sbjct: 54 RDFLPEGNQATTSHYQTEKLMRNLGLPYHTIDVCQNNCMLSWKEDEKEDQCRFCGAKRWK 113
Query: 244 TVTEEVQPNGTKTSEKQKNTPLPKSVPLPGIPAKILRWFPLKPRLQRLFMSSKIAEAMRW 303
+ ++ T +P S + + P+ RL+R++ S K A AMRW
Sbjct: 114 P------------KDDRRRTKVPYSR---------MWYLPIGDRLKRMYQSHKTAAAMRW 152
Query: 304 HRDGRLDDGFLRHPADSLAWKKFDSKYQTFALDPHNVRLGLASDGFNPFKTMSITHST*P 363
H + + +G + H +D+ W+ F + FA +P NV LGL +DGFNPF MS HS P
Sbjct: 153 HAEHQSKEGEMNHLSDAAEWRYFQGLHPEFAEEPRNVYLGLCTDGFNPFG-MSRNHSLWP 211
Query: 364 VILVHFNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTYLQPLVKELQELWENGLETFDAY 423
VIL +NLPP MCM + L++L GP P ++D +LQPL++EL+E W G++ +D
Sbjct: 212 VILTPYNLPPGMCMNTEYLFLTILNSGPNHPRASLDVFLQPLIEELKEFWSTGVDAYDVS 271
Query: 424 KKESFQLHAALMWTINDFPAYANLSGWSTKGKYACPCCGFETSSQWLRNGRKFCYMS-HR 482
++F L A L+WTI+DFPAY L GW+T GK +CP C T S +L NGRK C+ HR
Sbjct: 272 LSQNFNLKAVLLWTISDFPAYNMLLGWTTHGKLSCPVCMESTKSFYLPNGRKTCWFDCHR 331
Query: 483 RWLTPNHKWRLNGKDF----DGTQEFRDPPIRLDGT----AQLRQLDEIEDECLG----- 529
R+L H R N KDF D + E+ PP L G +L ++ + + +G
Sbjct: 332 RFLPHGHPSRRNRKDFLKGRDASSEY--PPESLTGEQVYYERLASVNPPKTKDVGGNGHE 389
Query: 530 ------DQAQRWKKKSILFSLPYWKYNELRHNLDVMHIEKNVCDNIIGTLLNQEGKSKDN 583
+ W K+SIL+ L YWK LRHN+DVMH EKN DNI+ TL+ +GKSKDN
Sbjct: 390 KKMRGYGKEHNWHKESILWELSYWKDLNLRHNIDVMHTEKNFLDNIMNTLMRVKGKSKDN 449
Query: 584 YKARADLVDMGIRSILHPQPSPNTNTMLLPRACYQMTTKEKDDFLSVLK-NVKAPDECSS 642
+R D+ R LH S P Y +T + K L +K +++ PD SS
Sbjct: 450 IMSRLDIEKFCSRPGLHIDSSGKA-----PFPAYTLTEEAKQSLLQCVKYDIRFPDGYSS 504
Query: 643 NISRCVQVQQRKIFGLKSYDCHVLMQELLP 672
+++ CV + K G+KS+DCHV M+ LLP
Sbjct: 505 DLASCVDLDNGKFSGMKSHDCHVFMERLLP 534
>At4g08000 putative transposon protein
Length = 609
Score = 324 bits (830), Expect = 2e-88
Identities = 176/440 (40%), Positives = 249/440 (56%), Gaps = 44/440 (10%)
Query: 181 DLLRDAFPEGNVLPKSFYETKKMISELGLGYEKIHACPNDCILYWDKYTNDEVCPKCGAS 240
DLL + PEGN S Y+T+K++ LGL Y I C N+C+L+W + ++ C CGA
Sbjct: 73 DLLEEFLPEGNQATSSHYQTEKLMRNLGLPYHTIDVCSNNCMLFWKEDEQEDHCRFCGAQ 132
Query: 241 RWKTVTEEVQPNGTKTSEKQKNTPLPKSVPLPGIPAKILRWFPLKPRLQRLFMSSKIAEA 300
RWK + ++ T +P S + + P+ RL+R++ S K A A
Sbjct: 133 RWKP------------KDDRRRTKVPYSR---------MWYLPIGDRLKRMYQSHKTAAA 171
Query: 301 MRWHRDGRLDDGFLRHPADSLAWKKFDSKYQTFALDPHNVRLGLASDGFNPFKTMSITHS 360
MRWH + + +G + HP+D+ W+ F + FA +P NV LGL +DGFNPF MS HS
Sbjct: 172 MRWHAEHQSKEGEMNHPSDAAEWRYFQELHPRFAEEPRNVYLGLCTDGFNPFG-MSRNHS 230
Query: 361 T*PVILVHFNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTYLQPLVKELQELWENGLETF 420
PVIL +NLPP MCM + L++L P P ++D +LQPL++EL+ELW G++ +
Sbjct: 231 LWPVILTPYNLPPSMCMNTKYLFLTILNVEPNHPRASLDVFLQPLIEELKELWSTGVDAY 290
Query: 421 DAYKKESFQLHAALMWTINDFPAYANLSGWSTKGKYACPCCGFETSSQWLRNGRKFCYMS 480
D ++F L A L+WTI+DFPAY+ LSGW T GK++CP C T S +L NGRK C+
Sbjct: 291 DVSLSQNFNLKAVLLWTISDFPAYSMLSGWITHGKFSCPICMESTKSFYLPNGRKTCWFD 350
Query: 481 -HRRWLTPNHKWRLNGKDF----DGTQEFRDPPIRLDGT----AQLRQLDEIEDECLG-- 529
HRR+L H R N KDF D + E+ PP L G +L ++ + + +G
Sbjct: 351 CHRRFLPHGHPSRRNKKDFLKGRDSSSEY--PPKSLTGEQVYYERLASVNPTKTKDVGGN 408
Query: 530 ---------DQAQRWKKKSILFSLPYWKYNELRHNLDVMHIEKNVCDNIIGTLLNQEGKS 580
+ W K+SIL+ L YW LRHN+DVMH EKN D+I+ TL++ + KS
Sbjct: 409 GHEKKMPGYGKEHNWHKESILWELSYWNDLNLRHNIDVMHTEKNFLDDIMNTLMSVKDKS 468
Query: 581 KDNYKARADLVDMGIRSILH 600
KDN +R D+ RS LH
Sbjct: 469 KDNIMSRLDIEIFCSRSDLH 488
Score = 100 bits (249), Expect = 5e-21
Identities = 51/130 (39%), Positives = 70/130 (53%), Gaps = 25/130 (19%)
Query: 693 FFKQICSKVLNVKFIEQLESQIAITLCQLETIFPPSFFTIMVHLVVHLAHEAKVGGPVQY 752
FF+ +CS+ L ++ L+ I + +C LE IFPPSFF +M HL +HL +E ++GGPVQY
Sbjct: 497 FFRDLCSRTLQKSRVQILKQNIVLIICNLEKIFPPSFFDVMEHLPIHLPYEPELGGPVQY 556
Query: 753 RWMYPIERY*LCIFKLIYFNVRIIYI*NYFFPRFLLTLKSFVRNKAYPEGSIAESFLANE 812
RWMYP ER FF +F K +NK Y GSI ES++ +E
Sbjct: 557 RWMYPFER---------------------FFKKF----KGKAKNKRYAAGSIVESYINDE 591
Query: 813 CLTFCSQYLS 822
F Y +
Sbjct: 592 IAYFSEYYFA 601
>At1g34660 hypothetical protein
Length = 501
Score = 319 bits (817), Expect = 7e-87
Identities = 176/492 (35%), Positives = 274/492 (54%), Gaps = 43/492 (8%)
Query: 15 SEEYIRGVNEFLEFAFQNSQVSE--KIWCPCTTCANCKSLSRTSVYEHLTNPRRGFLRGY 72
++E+ G+ +F+ FA E K++CPC+TC N K L S+++HL + RGF+ GY
Sbjct: 13 TKEFKEGLVDFMRFAANQEIPLECGKMYCPCSTCGNEKFLPVDSIWKHLYS--RGFMPGY 70
Query: 73 KQWIFHGE--KPVASSSTIEE-----PEHTEMEHDMDGLIHD----VLASHTAEDPLFDD 121
W HG+ + +A+S+ +E PE + + +I D + A +
Sbjct: 71 YIWFSHGKDFETLANSNEDDEIHGLEPEDNNSNEEEENIIRDPTGVIQMVTNAFRETTES 130
Query: 122 GERNPQVEENANKFYRLVKDNEQILYPNCKK-YSKLSFMVHLYHLKCLHGWSDKSFSMLL 180
G P +E A KF+ ++ ++ +Y CK+ +S+LS L ++K + S+ +
Sbjct: 131 GIEEPNLE--AKKFFDMLDAAKKPIYDGCKEGHSRLSAATRLMNIKTEYNLSEDCVDAIA 188
Query: 181 DLLRDAFPEGNVLPKSFYETKKMISELGLGYEKIHACPNDCILYWDKYTNDEVCPKCGAS 240
D ++D PE N+ P S+YE +K++S LGL Y+ I C ++C+++W N + C C
Sbjct: 189 DFVKDLLPEPNLAPGSYYEIQKLVSSLGLPYQMIDVCVDNCMIFWRDTVNLDACTFCQKP 248
Query: 241 RWKTVTEEVQPNGTKTSEKQKNTPLPKSVPLPGIPAKILRWFPLKPRLQRLFMSSKIAEA 300
R++ TS K + +P K + + P+ RL+RL+ S + A
Sbjct: 249 RFQV-----------TSGKSR------------VPFKRMWYLPITDRLKRLYQSERTAGP 285
Query: 301 MRWHRDGRLDDGFLRHPADSLAWKKFDSKYQTFALDPHNVRLGLASDGFNPFKTMSITHS 360
MRWH + DG + HP+D+ AWK F+S Y+ FA + NV LGL +DGFNPF +S
Sbjct: 286 MRWHAE-HSSDGNISHPSDAEAWKHFNSMYKDFANEHRNVYLGLCTDGFNPFGKSGRKYS 344
Query: 361 T*PVILVHFNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTYLQPLVKELQELWENGLETF 420
PVIL +NLPP +CMK+ F LS+L+PGP+ P ++D +LQPL+ ELQ LW G+ +
Sbjct: 345 LWPVILTPYNLPPSLCMKREFLFLSILVPGPEHPRRSLDVFLQPLIHELQLLWSEGVVAY 404
Query: 421 DAYKKESFQLHAALMWTINDFPAYANLSGWSTKGKYACPCCGFETSSQWLRNGRKFCYMS 480
D K +F + A LMWTI+DFPAY LSGW+T G+ +CP C + L++G+K C+
Sbjct: 405 DVSLKNNFVMRAVLMWTISDFPAYGMLSGWTTHGRLSCPYCQDTIDAFQLKHGKKSCWFD 464
Query: 481 -HRRWLTPNHKW 491
HRR+L NH +
Sbjct: 465 CHRRFLPKNHSY 476
>At1g33580 En/Spm-like transposon protein, putative
Length = 428
Score = 306 bits (783), Expect = 6e-83
Identities = 168/430 (39%), Positives = 242/430 (56%), Gaps = 30/430 (6%)
Query: 5 REWIRDPSRRSEEYIRGVNEFLEFAFQNSQVSEKIWCPCTTCANCKSLSRTSVYEHLTNP 64
+ W+ P R S EY +G +F+ + + +++CPCT C N +V EHL
Sbjct: 13 KSWVWLP-RNSIEYEKGATKFVYASSRRLGDPSEMFCPCTDCRNLCHQPIDTVLEHLVI- 70
Query: 65 RRGFLRGYKQ---WIFHGEKPVASSSTIEEPEHTEM-EHDMDGLIHDVLASHTAEDPLFD 120
+G YK+ W HGE + +E +E +++ + H+ +
Sbjct: 71 -KGMNHKYKRNGCWSKHGE---IRADKLEAESSSEFGAYELIRTAYFDGEDHSDSQNQNE 126
Query: 121 DGERNPQVEENANKFYRLVKDNEQILYPNCKKYSKLSFMVHLYHLKCLHGWSDKSFSMLL 180
+ + P +E ++ F +KD E LY C KY+K+S ++ LY +K G S+ F LL
Sbjct: 127 NDSKEPSTKEESD-FREKLKDAETPLYYGCPKYTKVSAIMRLYRIKVKSGMSENYFDQLL 185
Query: 181 DLLRDAFPEGNVLPKSFYETKKMISELGLGYEKIHACPNDCILYWDKYTNDEVCPKCGAS 240
L+ D P NVL S E KK + G GY+ IHACPNDCILY +Y + CP+C A
Sbjct: 186 SLVHDILPGENVLLTSTNEIKKFLKMFGFGYDIIHACPNDCILYRKEYELRDTCPRCSAF 245
Query: 241 RWKTVTEEVQPNGTKTSEKQKNTPLPKSVPLPGIPAKILRWFPLKPRLQRLFMSSKIAEA 300
RW E+ K+T K GIPAK+LR+FP+K R +R+F S K+AE
Sbjct: 246 RW---------------ERDKHTGEEKK----GIPAKVLRYFPIKDRFKRMFRSKKMAED 286
Query: 301 MRWHRDGRLDDGFLRHPADSLAWKKFDSKYQTFALDPHNVRLGLASDGFNPFKTMSITHS 360
+RWH DDG +RHP DSLAW + + K+ FA +P N+RLGL++DG NPF + +S
Sbjct: 287 LRWHFSNASDDGTMRHPVDSLAWAQVNYKWPQFAAEPRNLRLGLSTDGMNPFSIQNTKYS 346
Query: 361 T*PVILVHFNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTYLQPLVKELQELWENGLETF 420
T PV+LV++N+ P MCMK ML+LLIPGP P NNID YL PL+ +L++LW +G++ +
Sbjct: 347 TWPVLLVNYNMAPTMCMKAENIMLTLLIPGPTTPNNNIDVYLAPLIDDLKDLWSDGIQVY 406
Query: 421 DAYKKESFQL 430
D++ KESF L
Sbjct: 407 DSFLKESFTL 416
>At2g10630 En/Spm transposon hypothetical protein 1
Length = 515
Score = 286 bits (731), Expect = 6e-77
Identities = 165/469 (35%), Positives = 247/469 (52%), Gaps = 46/469 (9%)
Query: 15 SEEYIRGVNEFLEFAFQNSQVSE---KIWCPCTTCANCKSL-SRTSVYEHLTNPRRGFLR 70
S EY+ GV +F+ FA V K CPC+ C N K + S V HL + GF+
Sbjct: 30 SAEYVAGVEQFMTFANSQPIVQSSRGKFHCPCSVCKNEKHIISGRRVSSHLFS--HGFMH 87
Query: 71 GYKQWIFHGEK------PVASSSTIEEPEHTEMEHDMDG----LIHDVLASHTAEDPLF- 119
Y W HGE+ ++ T H E+ + ++ +++D + D +
Sbjct: 88 DYYVWYKHGEEMNMDLGTSYTNRTYFSENHEEVGNIVEDPYVDMVNDAFNFNVGYDDNYR 147
Query: 120 -DDGERNPQ--VEENANKFYRLVKDNEQILYPNCK-KYSKLSFMVHLYHLKCLHGWSDKS 175
DD +N + V ++NKFY L++ LY C+ + S+LS L H K + ++K
Sbjct: 148 HDDSYQNVEEPVRNHSNKFYDLLEGANNPLYDGCRERQSQLSLASRLMHNKAEYNMNEKF 207
Query: 176 FSMLLDLLRDAFPEGNVLPKSFYETKKMISELGLGYEKIHACPNDCILYWDKYTNDEVCP 235
+ + D PE N S Y+T+K++ LGL Y I C N+C+L+W + ++ C
Sbjct: 208 VDSVCETFTDFLPEENQATTSHYQTEKLMRNLGLPYHTIDVCQNNCMLFWKEEEKEDQCR 267
Query: 236 KCGASRWKTVTEEVQPNGTKTSEKQKNTPLPKSVPLPGIPAKILRWFPLKPRLQRLFMSS 295
CGA RWK + ++ T +P S + + P+ RL+R++ S
Sbjct: 268 FCGAKRWKP------------KDDRRRTKVPYSC---------MWYLPIGDRLKRMYQSH 306
Query: 296 KIAEAMRWHRDGRLDDGFLRHPADSLAWKKFDSKYQTFALDPHNVRLGLASDGFNPFKTM 355
K A AMRWH + + +G + HP+D+ W+ F + FA DP NV LGL +DGFNPF M
Sbjct: 307 KTAAAMRWHAEHQSKEGEMNHPSDAAEWRYFQGLHPQFAEDPRNVYLGLCTDGFNPFG-M 365
Query: 356 SITHST*PVILVHFNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTYLQPLVKELQELWEN 415
S HS PVIL +NLPP MCM + L++L GP P ++D +LQPL++EL+ELW
Sbjct: 366 SRNHSLWPVILTPYNLPPGMCMNTEYLFLTILNSGPNHPRASLDVFLQPLIEELKELWST 425
Query: 416 GLETFDAYKKESFQLHAALMWTINDFPAYANLSGWSTKGKYACPCCGFE 464
G++ +D ++F L A L+WTI+DFPAY+ LSGW+T+ C C FE
Sbjct: 426 GVDAYDVSLSQNFNLKAVLLWTISDFPAYSMLSGWTTQ---FCKCSMFE 471
>At4g08070
Length = 767
Score = 262 bits (669), Expect = 9e-70
Identities = 157/410 (38%), Positives = 212/410 (51%), Gaps = 68/410 (16%)
Query: 470 LRNGRKFCYMSHRRWLTPNHKWRLNGKDFDGTQEFRDPPIRLDG---------------- 513
++N RK YM HR L P+H++R FDG E R L G
Sbjct: 346 MKNTRKHVYMGHRMGLPPSHRFRNKKSWFDGKAEHRRKSRILSGLEISHNLKNFQNNFGN 405
Query: 514 -----------------------TAQLRQLDEIEDECLGDQAQRWKKKSILFSLPYWKYN 550
++ + +E E E ++ RWKK+SI F L YW+
Sbjct: 406 FKQSTRKRKQPESVKSGSDSDDLSSDSEEDEEEEAEVDEEELSRWKKRSIFFRLSYWQDL 465
Query: 551 ELRHNLDVMHIEKNVCDNIIGTLLNQEGKSKDNYKARADLVDMGIRSILHPQPSPNTNTM 610
+RHNLDVMH+E+NV +I+ TLL+ GKSKD AR DL +GIR LHPQ
Sbjct: 466 PVRHNLDVMHVERNVAASIVSTLLHC-GKSKDGLNARKDLEVLGIRKDLHPQTKGKRT-- 522
Query: 611 LLPRACYQMTTKEKDDFLSVLKNVKAPDECSSNISRCVQVQQRKIFGLKSYDCHVLMQEL 670
LP A + ++ EK F L + K PD S+I+R V ++ K+ GLKS+D HVLMQ+L
Sbjct: 523 YLPAALWSLSKSEKKLFCKRLFDFKGPDGYCSDIARGVSLEDCKVTGLKSHDYHVLMQQL 582
Query: 671 LPIALRGSLPDKVTSVLIDLCNFFKQICSKVLNVKFIEQLESQIAITLCQLETIFPPSFF 730
LPIALRG LP + LC FF +C +V++++ + ++E++I TLC E FPPSFF
Sbjct: 583 LPIALRGLLPKGPRVAIGRLCKFFNHLCQRVIDMEQLLEMEAEIVETLCLFERFFPPSFF 642
Query: 731 TIMVHLVVHLAHEAKVGGPVQYRWMYPIERY*LCIFKLIYFNVRIIYI*NYFFPRFLLTL 790
IM HL VHL EA++GGPV +RWMYP ERY + L
Sbjct: 643 DIMEHLTVHLGREARLGGPVHFRWMYPFERY-------------------------MKVL 677
Query: 791 KSFVRNKAYPEGSIAESFLANECLTFCSQYLSGVETRFSRPTRNDDEFDS 840
K FVRN PEG IAE +LA EC+ F S++ T+ + EF++
Sbjct: 678 KDFVRNTTRPEGCIAECYLAEECIQFYSEFFKKT-TKVQEKADRNTEFEN 726
Score = 226 bits (576), Expect = 6e-59
Identities = 126/319 (39%), Positives = 173/319 (53%), Gaps = 27/319 (8%)
Query: 37 EKIWCPCTTCANCKSLSRTSVYEHLTNPRRGFLRGYKQ---WIFHGEKPVASSSTIEEPE 93
+ I CPC C N S V +HL RG YK+ W HGE S E +
Sbjct: 50 DMIVCPCIDCRNIDRHSGCVVVDHLVT--RGMDEAYKRRCDWYHHGELVSGGES---ESK 104
Query: 94 HTEMEHDMDGLIHDVLASHTAEDPLFDDGERNPQVEENANKFYRLVKDNEQILYPNCKKY 153
++ ++ L D + GE + ++ ++F + D E LYP+C +
Sbjct: 105 VSQWNDEVVELYQAAEYLDEEFDAMVQLGEIAERDDKKEDEFLAKLADAETPLYPSCVNH 164
Query: 154 SKLSFMVHLYHLKCLHGWSDKSFSMLLDLLRDAFPEGNVLPKSFYETKKMISELGLGYEK 213
SKLS +V L+ LK +GWSDKSF+ LL+ L + PE NVL S YE KK + +GY+K
Sbjct: 165 SKLSAIVSLFRLKTKNGWSDKSFNDLLETLSEMLPEDNVLHTSLYEVKKFLKSFDMGYQK 224
Query: 214 IHACPNDCILYWDKYTNDEVCPKCGASRWKTVTEEVQPNGTKTSEKQKNTPLPKSVPLPG 273
IHAC NDC L+ KY + CPKC ASRWKT T E +K G
Sbjct: 225 IHACVNDCCLFRKKYKKLQNCPKCNASRWKT--------NMHTGEVKK-----------G 265
Query: 274 IPAKILRWFPLKPRLQRLFMSSKIAEAMRWHRDGRLDDGFLRHPADSLAWKKFDSKYQTF 333
+P K+LR+FP+ PRL+R+F S ++A +RWH + DG LRHP DS+ W + ++KY F
Sbjct: 266 VPQKVLRYFPIIPRLKRMFRSEEMARDLRWHFHNKSTDGKLRHPVDSVTWDQMNAKYPLF 325
Query: 334 ALDPHNVRLGLASDGFNPF 352
A + N+RLGL++DGFNPF
Sbjct: 326 ASEERNLRLGLSTDGFNPF 344
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.336 0.147 0.497
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,301,125
Number of Sequences: 26719
Number of extensions: 1143939
Number of successful extensions: 4561
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 4367
Number of HSP's gapped (non-prelim): 78
length of query: 1145
length of database: 11,318,596
effective HSP length: 110
effective length of query: 1035
effective length of database: 8,379,506
effective search space: 8672788710
effective search space used: 8672788710
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0230.6


