

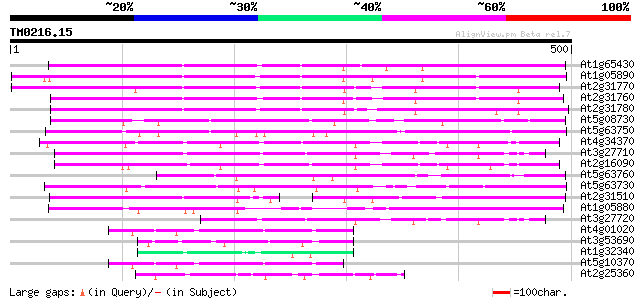
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0216.15
(500 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g65430 unknown protein 394 e-110
At1g05890 unknown protein 387 e-107
At2g31770 putative ARI-like RING zinc finger protein 363 e-101
At2g31760 putative ARI-like RING zinc finger protein 358 5e-99
At2g31780 similar to Ariadne protein from Drosophila 353 1e-97
At5g08730 putative protein 187 9e-48
At5g63750 ARI-like RING zinc finger protein-like 184 1e-46
At4g34370 unknown protein 167 2e-41
At3g27710 putative protein 167 2e-41
At2g16090 unknown protein 167 2e-41
At5g63760 unknown protein 157 1e-38
At5g63730 ARI-like RING zinc finger protein-like 156 3e-38
At2g31510 putative RING zinc finger protein 154 1e-37
At1g05880 unknown protein 141 7e-34
At3g27720 hypothetical protein 117 1e-26
At4g01020 unknown protein 89 4e-18
At3g53690 unknown protein 78 1e-14
At1g32340 RING finger protein, putative 78 1e-14
At5g10370 putative protein 77 2e-14
At2g25360 hypothetical protein 76 4e-14
>At1g65430 unknown protein
Length = 567
Score = 394 bits (1011), Expect = e-110
Identities = 206/472 (43%), Positives = 284/472 (59%), Gaps = 17/472 (3%)
Query: 35 KRENFTILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCEDEEKV 94
+++N+++LSE +I +LQED I+++++VL I S+ LL H W V++V + W DEEKV
Sbjct: 47 RQQNYSVLSEADICKLQEDDISRISTVLSISRNSSAILLRHYNWCVSRVHDEWFADEEKV 106
Query: 95 QKAAGLLNQPKVEIDWLKDL-CQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNINKGP 153
+ A GLL +P V+ +L C +CF + K+ +A C HP C++CW YI T IN GP
Sbjct: 107 RDAVGLLEKPVVDFPTDGELDCGICFETFLSDKLHAAACGHPFCDSCWEGYITTAINDGP 166
Query: 154 SKCLTLRCPDPSCDVAVDRDMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYV 213
CLTLRCPDPSC AV +DMI LA + K Y + + SYV+ K +WCPAPGCDY
Sbjct: 167 G-CLTLRCPDPSCRAAVGQDMINLLAPDKDKQKYTSYFVRSYVEDNRKTKWCPAPGCDYA 225
Query: 214 VCYQGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDCETAGRWMKKIDKSTSENGKWILA 273
V + G + DV C C +SFCW C EE+H PVDC+T +W+ K + + SEN WILA
Sbjct: 226 VNFVVGSG---NYDVNCRCCYSFCWNCAEEAHRPVDCDTVSKWVLK-NSAESENMNWILA 281
Query: 274 HTKPCPRCKSLIEKNQGFKYMTC--KCGFQFCWVCFHEQLKCESRSCSPFIRMHIMEFQG 331
++KPCP+CK IEKNQG ++TC C F+FCW+C + F + E
Sbjct: 282 NSKPCPKCKRPIEKNQGCMHITCTPPCKFEFCWLCLGAWTE-HGEKTGGFYACNRYEAAK 340
Query: 332 TDG----EEVEINMEKDNPKKCNHYYECWARNDFLSKSI---LQKLIDLDRKYLSMSLQK 384
DG E M K++ ++ HYYE WA N + L+K+ D + LS +
Sbjct: 341 QDGIYDETEKRREMAKNSLERYTHYYERWATNQSSRQKALLDLKKMQTDDIEKLSDIQCQ 400
Query: 385 RETYFDFVKKAWQQIAECRRILKWTNVYGYFLPKNEKAKKQFSEYIQVEAEVALEKLQQC 444
E+ F+ +AW QI ECRR+LKWT YG+++P E K+ F EY+Q EAE LE+L QC
Sbjct: 401 PESQLKFIIEAWLQIVECRRVLKWTYAYGFYIPDQEHGKRVFFEYLQGEAESGLERLHQC 460
Query: 445 AKVEETMFQD-NYPEESLNKFQEKVIHLTDVTKIYFENLLRAFEDGLDKVEA 495
A+ E + D P E N+F+ K+ LT VTK YFENL+RA E+GL V +
Sbjct: 461 AEKELLPYLDAKGPSEDFNEFRTKLAGLTSVTKNYFENLVRALENGLSDVNS 512
>At1g05890 unknown protein
Length = 552
Score = 387 bits (993), Expect = e-107
Identities = 209/518 (40%), Positives = 296/518 (56%), Gaps = 30/518 (5%)
Query: 2 DSIFDMKSLSHGLVCGLRERDKNLPNTE----EEEED---------KRENFTILSELEIK 48
D + DM S+ + G D + T+ E + D + N+ +L E +I+
Sbjct: 5 DDMHDMDSVDYDYYSGGTYDDNDSDETDFGFGEADTDDAAIIASYRSKSNYVVLKEEDIR 64
Query: 49 RLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCEDEEKVQKAAGLLNQPKVEI 108
R Q D + +V++VL I D A LL H W+V+KV + W DEE+V++ G+L P V
Sbjct: 65 RHQNDDVGRVSAVLSITDVEASTLLLHYHWSVSKVNDEWFADEERVRRTVGILEGPVVTT 124
Query: 109 -DWLKDLCQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNINKGPSKCLTLRCPDPSCD 167
D + C +CF S +I+S C HP C CW YI T IN GP CL L+CPDPSC
Sbjct: 125 PDGREFTCGICFDSYTLEEIVSVSCGHPFCATCWTGYISTTINDGPG-CLMLKCPDPSCP 183
Query: 168 VAVDRDMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDLD 227
A+ RDMI +LAS+ K Y ++ L SYV+ +M+WCPAPGC++ + + G D
Sbjct: 184 AAIGRDMIDKLASKEDKEKYYRYFLRSYVEVNREMKWCPAPGCEHAIDFAGG---TESYD 240
Query: 228 VTCLCYHSFCWRCGEESHSPVDCETAGRWMKKIDKSTSENGKWILAHTKPCPRCKSLIEK 287
V+CLC HSFCW C EE+H PVDC+T G+W+ K + + SEN WILA++KPCP+CK IEK
Sbjct: 241 VSCLCSHSFCWNCTEEAHRPVDCDTVGKWILK-NSAESENMNWILANSKPCPKCKRPIEK 299
Query: 288 NQGFKYMTC--KCGFQFCWVCFHEQLKCESRSCSPFI--RMHIMEFQGT-DGEEVEINME 342
N G +MTC C F+FCW+C + + R+ + R + +G D E M
Sbjct: 300 NHGCMHMTCTPPCKFEFCWLCLNAWTEHGERTGGFYACNRYEAAKQEGLYDEAERRREMA 359
Query: 343 KDNPKKCNHYYECWARNDFLSKSI---LQKLIDLDRKYLSMSLQKRETYFDFVKKAWQQI 399
K++ ++ HYYE WA N + LQK+ LS E+ F+ +AW QI
Sbjct: 360 KNSLERYTHYYERWASNQVSRQKAMGDLQKMQSEKLGKLSDIQCTPESQLKFIAEAWLQI 419
Query: 400 AECRRILKWTNVYGYFLPKNEKAKKQFSEYIQVEAEVALEKLQQCAKVEETMFQ-DNYPE 458
ECRR+LKWT YGY+L + AKK F EY+Q EAE LE+L +C + + +F+ P
Sbjct: 420 IECRRVLKWTYAYGYYL--QDHAKKPFFEYLQGEAESGLERLHKCVEKDIEVFELAEGPS 477
Query: 459 ESLNKFQEKVIHLTDVTKIYFENLLRAFEDGLDKVEAR 496
E N F+ K+ LT +TK +FENL++A E+GL V+++
Sbjct: 478 EEFNHFRTKLTGLTSITKTFFENLVKALENGLADVDSQ 515
>At2g31770 putative ARI-like RING zinc finger protein
Length = 543
Score = 363 bits (933), Expect = e-101
Identities = 192/505 (38%), Positives = 278/505 (55%), Gaps = 32/505 (6%)
Query: 2 DSIFDMKSLSHGLVCGLRERDKNLPNTEEEEEDKRENFTILSELEIKRLQEDAINKVASV 61
D + D KS G + + + +++ IL E +I +LQ D I +V+S+
Sbjct: 6 DDMIDNKSGEENYSYGGGNESDDYNDVVDTIIPSEKSYVILKEEDILKLQRDDIERVSSI 65
Query: 62 LCIPDASACFLLCHNLWNVTKVLESWCEDEEKVQKAAGLLNQPKVEIDW---------LK 112
L + LL H W V+KV + W DEE+++KA GLL +P V+ + +
Sbjct: 66 LSLSQVEVIVLLLHYNWCVSKVEDEWFTDEERIRKAVGLLKEPVVDFNGGEKDKKCRKVN 125
Query: 113 DLCQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNINKGPSKCLTLRCPDPSCDVAVDR 172
C +CF S + +I C HP C+ CW YI T I GP CL ++CP+PSC AV +
Sbjct: 126 IQCGICFESYTREEIARVSCGHPYCKTCWAGYITTKIEDGPG-CLRVKCPEPSCSAAVGK 184
Query: 173 DMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDLDVTCLC 232
DMI ++ Y +++L SYV+ K++WCP+PGC Y V + G + DV+CLC
Sbjct: 185 DMIEDVTETKVNEKYSRYILRSYVEDGKKIKWCPSPGCGYAVEFGGSE--SSSYDVSCLC 242
Query: 233 YHSFCWRCGEESHSPVDCETAGRWMKKIDKSTSENGKWILAHTKPCPRCKSLIEKNQGFK 292
+ FCW C E++HSPVDC+T +W+ K ++ SEN W+LA++KPCP CK IEKN G
Sbjct: 243 SYRFCWNCSEDAHSPVDCDTVSKWIFK-NQDESENKNWMLANSKPCPECKRPIEKNDGCN 301
Query: 293 YMTCK--CGFQFCWVCFHEQLKCESRSCSPFIRMHIMEFQGTDGEEVEINMEKDNPKKCN 350
+MTC CG +FCW+C + + S +C+ F+ + E + + + K+
Sbjct: 302 HMTCSAPCGHEFCWICL-KAYRRHSGACNRFV---------VEQAESKRALLQSEIKRYT 351
Query: 351 HYYECWARND---FLSKSILQKLIDLDRKYLSMSLQKRETYFDFVKKAWQQIAECRRILK 407
HYY WA N + L+KL + K LS + ET F AW QI ECRR+LK
Sbjct: 352 HYYVRWAENQSSRLKAMRDLEKLQSVQLKELSDNQCTSETQLQFTVDAWLQIIECRRVLK 411
Query: 408 WTNVYGYFLPKNEKAKKQFSEYIQVEAEVALEKLQQCAKVEETMF--QDNYPEESLNKFQ 465
WT YGY+L + K++F EY+Q EAE LE+L CA+ E F + P ++ N F+
Sbjct: 412 WTYAYGYYL--QDLPKRKFFEYLQGEAESGLERLHHCAENELKQFFIKSEDPSDTFNAFR 469
Query: 466 EKVIHLTDVTKIYFENLLRAFEDGL 490
K+ LT VTK YFENL++A E+GL
Sbjct: 470 MKLTGLTTVTKTYFENLVKALENGL 494
>At2g31760 putative ARI-like RING zinc finger protein
Length = 514
Score = 358 bits (918), Expect = 5e-99
Identities = 184/466 (39%), Positives = 271/466 (57%), Gaps = 28/466 (6%)
Query: 37 ENFTILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCEDEEKVQK 96
+++ I+ E EI +LQ D I +V+++L + A LL H W V+K+ + W DEE+++K
Sbjct: 41 KSYVIIKEEEILKLQRDDIERVSTILFLSQVEAIVLLLHYHWCVSKLEDEWFTDEERIRK 100
Query: 97 AAGLLNQPKVEIDWLK-DL-CQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNINKGPS 154
G+L +P V+++ + D+ C +CF S + +I S C HP C+ CW YI T I GP
Sbjct: 101 TVGILKEPVVDVNGTEVDIQCGICFESYTRKEIASVSCGHPYCKTCWTGYITTKIEDGPG 160
Query: 155 KCLTLRCPDPSCDVAVDRDMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVV 214
CL ++CP+PSC V +DMI E+ + K Y ++ L SYV+ KM+WCP+PGC+ V
Sbjct: 161 -CLRVKCPEPSCYAVVGQDMIDEVTEKKDKDKYYRYFLRSYVEDGKKMKWCPSPGCECAV 219
Query: 215 CYQGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDCETAGRWMKKIDKSTSENGKWILAH 274
+ G DV CLC + FCW C E++HSPVDCET +W+ K ++ SEN WILA+
Sbjct: 220 EFGESSG----YDVACLCSYRFCWNCSEDAHSPVDCETVSKWIFK-NQDESENKNWILAN 274
Query: 275 TKPCPRCKSLIEKNQGFKYMTC--KCGFQFCWVCFHEQLKCESRSCSPFIRMHIMEFQGT 332
+KPCP+CK IEK+ G +MTC CG +FCW+C + + +C+ ++
Sbjct: 275 SKPCPKCKRPIEKSHGCNHMTCSASCGHRFCWIC--GKSYSDHYACNNYVE--------- 323
Query: 333 DGEEVEINMEKDNPKKCNHYYECWARND---FLSKSILQKLIDLDRKYLSMSLQKRETYF 389
D + + + + K+ HYY W N + S L+K + K LS + K +
Sbjct: 324 DADHDKRTLLQSEIKRYTHYYVRWVENQSSRLKAMSDLEKFQSVQLKQLSDNQCKPKIDL 383
Query: 390 DFVKKAWQQIAECRRILKWTNVYGYFLPKNEKAKKQFSEYIQVEAEVALEKLQQCAKVEE 449
F+ AW QI ECRR+LKWT YGY+L + AK+ EY+Q EAE LE+L CA+ E
Sbjct: 384 QFIVDAWLQIIECRRVLKWTYAYGYYL--DNLAKRPLFEYLQGEAETGLERLHHCAENEL 441
Query: 450 TMF--QDNYPEESLNKFQEKVIHLTDVTKIYFENLLRAFEDGLDKV 493
F + P ++ N F+ K+ LT VTK YF+NL++A E+GL V
Sbjct: 442 KQFFIKSEDPSDTFNAFRMKLTGLTKVTKTYFDNLVKALENGLADV 487
>At2g31780 similar to Ariadne protein from Drosophila
Length = 542
Score = 353 bits (906), Expect = 1e-97
Identities = 185/476 (38%), Positives = 271/476 (56%), Gaps = 28/476 (5%)
Query: 37 ENFTILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCEDEEKVQK 96
+++ ++ E +I +LQ D I +V++VL + + LL H W V+K+ + W DEE+++K
Sbjct: 59 KSYVVVKEEDILKLQRDDIEQVSTVLSVSQVESIVLLLHYHWCVSKLEDEWFTDEERIRK 118
Query: 97 AAGLLNQPKVEIDWLK-DL-CQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNINKGPS 154
G+L +P V+++ + D+ C +CF S + +I C HP C+ CW YI T I GP
Sbjct: 119 TVGILKEPVVDVNGTEVDIQCGICFESYTRKEIARVSCGHPYCKTCWTGYITTKIEDGPG 178
Query: 155 KCLTLRCPDPSCDVAVDRDMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVV 214
CL ++CP+PSC V +DMI E+ + K Y ++ L SYV+ KM+WCP+PGC+Y V
Sbjct: 179 -CLRVKCPEPSCYAVVGQDMIDEVTEKKDKDKYYRYFLRSYVEDGKKMKWCPSPGCEYAV 237
Query: 215 CYQGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDCETAGRWMKKIDKSTSENGKWILAH 274
+ +G + DV+CLC + FCW C E++HSPVDCET +W+ K +K SEN WILA
Sbjct: 238 EFGVNG--SSSYDVSCLCSYKFCWNCCEDAHSPVDCETVSKWLLK-NKDESENMNWILAK 294
Query: 275 TKPCPRCKSLIEKNQGFKYMTCK--CGFQFCWVCFHEQLKCESRSCSPFIRMHIMEFQGT 332
TKPCP+CK IEKN G +M+C C FCW C Q + ++C+ F+
Sbjct: 295 TKPCPKCKRPIEKNTGCNHMSCSAPCRHYFCWACL--QPLSDHKACN--------AFKAD 344
Query: 333 DGEEVEINMEKDNPKKCNHYYECWARND---FLSKSILQKLIDLDRKYLSMSLQKRETYF 389
+ +E + KD + H+YE WA N + S L+K ++ K LS ET
Sbjct: 345 NEDETKRKRAKDAIDRYTHFYERWAFNQSSRLKAMSDLEKWQSVELKQLSDIQSTPETQL 404
Query: 390 DFVKKAWQQIAECRRILKWTNVYGYFLPKNEKAKKQFSEYIQV-----EAEVALEKLQQC 444
F AW QI ECRR+LKWT YGY++ E+ K+ F+ + EAE LE+L C
Sbjct: 405 SFTVDAWLQIIECRRVLKWTYAYGYYILSQERNKRVFARTFSLSCCSAEAENGLERLHHC 464
Query: 445 AKVEETMF--QDNYPEESLNKFQEKVIHLTDVTKIYFENLLRAFEDGLDKVEARRS 498
A+ E F + P ++ + + K+I LT TK YFENL++A E+GL V S
Sbjct: 465 AEEELKQFIGKIEDPSKNFGELRAKLIDLTKATKTYFENLVKALENGLVDVAYNES 520
>At5g08730 putative protein
Length = 500
Score = 187 bits (476), Expect = 9e-48
Identities = 127/472 (26%), Positives = 218/472 (45%), Gaps = 43/472 (9%)
Query: 37 ENFTILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCEDEEKVQK 96
+ +++L++ +++ I +++ V + + A +L WN K + +++EK
Sbjct: 6 QKYSVLAKTQVREKMMKEIEQISEVFLVSKSDATVILIRLGWNSFKASDLLGDNKEKFLA 65
Query: 97 AAGL---LNQPKVEIDWLKDLCQVCFRSSEKGKILSAG--CAHPLCEACWMDYIDTNINK 151
GL LN D R + G L + C+H CW +Y+ + K
Sbjct: 66 KLGLARVLNSNSSSAD----------RETGDGDYLVSTPFCSHKFSTTCWSEYLSDALKK 115
Query: 152 GPSKCLTLRCPDPSCDVAVDRDMIRELASESSKVMYEQFLLWSYVDS-KAKMRWCPAPGC 210
+ + C C +V D I +L +E K MYE ++L S+++ KA ++WCPA GC
Sbjct: 116 NKEQRGLISCLSQDCVASVGPDTIEQL-TEPVKEMYENYILESFMECHKATIKWCPASGC 174
Query: 211 DYVVCYQGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDCETAGRWMK-KIDKSTSENGK 269
+Y V Q DG + V CLC H+FCW CG ESH PV C+ A W +D+S S +
Sbjct: 175 EYAVELQEDGNEDNVISVVCLCGHTFCWTCGLESHRPVSCKKASIWWTYLLDQSRSIS-- 232
Query: 270 WILAHTKPCPRCKSLIEKN--QGFKYMTCKCGFQFCWVCFHEQLKCESR-SCSPFIRMHI 326
WI +TK CP+CK +++N ++ + C C FCW+C + + + +CSP
Sbjct: 233 WIHTNTKSCPKCKIPVQQNGDPNYRLINCICSNNFCWICLRTEEQHQGNWNCSP------ 286
Query: 327 MEFQGTDGEEVEINMEKDNPKKCNHYYECWARNDFLSKSILQKLIDLDRKYLSMSLQK-- 384
+ VE + + W K KL L+ K + ++
Sbjct: 287 VAVPAAGPSTVEFSQIL--------HLNLWEAGHEALKKAKSKLRALEEKIIPKLIENCG 338
Query: 385 -RETYFDFVKKAWQQIAECRRILKWTNVYGYFLPKNEKAKKQFSEYIQVEAEVALEKLQQ 443
E V++A +CR++LKW+ V+ Y + + E KKQ+ ++++ A L
Sbjct: 339 ATELDIRTVREAGMLSVQCRQVLKWSCVFDYSIIEYESTKKQYLKHLRALASTML--CMH 396
Query: 444 CAKVEETMFQDNYPEESLNKFQEKVIHLTDVTKIYFENLLRAFEDGLDKVEA 495
K++E + PE+ N ++ K+ T T +F+ ++ EDG +V+A
Sbjct: 397 EGKLDELIHLALSPEDFTN-YKHKLEISTTCTGNHFDGFIKELEDGKPEVKA 447
>At5g63750 ARI-like RING zinc finger protein-like
Length = 536
Score = 184 bits (466), Expect = 1e-46
Identities = 130/500 (26%), Positives = 228/500 (45%), Gaps = 46/500 (9%)
Query: 33 EDKREN-FTILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCEDE 91
E+ RE +++L+ ++K + I +++ + + A LL W+ +V E E+
Sbjct: 2 ENNREGPYSVLTRDQLKGNMKKQIAEISEIFSLSKPDATVLLMFLRWDSHEVSEFLVENN 61
Query: 92 EKVQKAAGLLNQPKVEIDWLKDL-----CQVCFRSSEKGKILSAG--CAHPLCEACWMDY 144
EKV +GL V +D +DL C +CF++ + G L + C+H C++CW Y
Sbjct: 62 EKVLSESGL---KPVVVDPNQDLYKISSCGICFKTCDDGDYLISTPFCSHMFCKSCWRKY 118
Query: 145 IDTNINKGPSKCLTLRCPDPSCDVAVDRDMIRELASESSKVMYEQFLLWSYVDSKA--KM 202
++ N + CP +C AV D I++L + + MY +++L SY++ ++
Sbjct: 119 LEKNFYLVEKTQTRISCPHGACQAAVGPDTIQKL-TVCDQEMYVEYILRSYIEGNKVLEI 177
Query: 203 RWCPAPGCDYVVCYQG---DGGCARD--LDVTCLCYHSFCWRCGEESHSPVDCETAGRWM 257
++CPA C+YV+ + DG D +V CLC H FCWRC ESH PV C A W+
Sbjct: 178 KYCPAQDCNYVIEFHQKNHDGADQEDYGFNVVCLCGHIFCWRCMLESHKPVTCNNASDWL 237
Query: 258 -KKIDKSTSENGK------------------WILAHTKPCPRC--KSLIEKNQGFKYMTC 296
+ ++ + E+G+ I A K CP C + + Q +++TC
Sbjct: 238 FRDLNSLSKESGEKPLSLSSFETREKTYPLSSIKATKKVCPHCLRPADLGTKQYLRFLTC 297
Query: 297 KCGFQFCWVCFHEQLKCESRSCSPFIRMHIMEFQGTDGEEVEINMEKDNPKKCNHYYECW 356
C +FCW C + ++ S M F+G + +E E +N C W
Sbjct: 298 ACNGRFCWKCMQPEEAHKTESGFYKFCNVSMTFEGRAPKTLEGRAEPEN--SC---VGLW 352
Query: 357 ARNDFLSKSILQKLIDLDRKYLSMSLQKRETYFDFVKKAWQQIAECRRILKWTNVYGYFL 416
++ K L + + E F ++K I +CR++LKW+ VY Y
Sbjct: 353 KASEVSLKQAKSDLQAFEESNIKNPSDLTEKDFTIIRKGLMLIVQCRQVLKWSCVYDYLH 412
Query: 417 PKNEKAKKQFSEYIQVEAEVALEKLQQCAKVEETMFQDNYPEESLNKFQEKVIHLTDVTK 476
+ E +K+++ ++Q +A +E + EE + E+ + KV T
Sbjct: 413 AEYEMSKREYLRFLQADATSLVESFSKTLN-EEIGRASSATYENFCCVKHKVTIETSNIG 471
Query: 477 IYFENLLRAFEDGLDKVEAR 496
YF + ++ ++GLD V+ +
Sbjct: 472 NYFYHFIKTLQEGLDDVKVK 491
>At4g34370 unknown protein
Length = 597
Score = 167 bits (422), Expect = 2e-41
Identities = 128/500 (25%), Positives = 216/500 (42%), Gaps = 61/500 (12%)
Query: 27 NTEEEE----EDKRENFTILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTK 82
+ EE E KR N ++++ + Q + + +V +L I + A LL H W+V K
Sbjct: 25 DNEESELQPLSSKRSNTQVITQESLLAAQREDLLRVMELLSIKEHHARTLLIHYQWDVEK 84
Query: 83 VLESWCED-EEKVQKAAGLL-------NQPKVEIDWLKDLCQVCFRSSEKGKILSAGCAH 134
+ + E ++ + AG+ N + + C VC + C H
Sbjct: 85 LFAVFVEKGKDSLFSGAGVTVFDYQYGNSSFPQSSQMS--CDVCMEDLPGDHMTRMDCGH 142
Query: 135 PLCEACWMDYIDTNINKGPSKCLTLRCPDPSCDVAVDRDMIRELASESSKVM---YEQFL 191
C CW ++ IN+G SK +RC C+ D D++R L S+ + ++++L
Sbjct: 143 CFCNNCWTEHFTVQINEGQSK--RIRCMAHQCNAICDEDIVRSLVSKKRPDLAAKFDRYL 200
Query: 192 LWSYVDSKAKMRWCPA-PGCDYVVCYQGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDC 250
L SY++ ++WCP+ P C + + D C +V C C FC+ C ++HSP C
Sbjct: 201 LESYIEDNRMVKWCPSTPHCGNAIRAEDDKLC----EVECSCGLQFCFSCLCQAHSPCSC 256
Query: 251 ETAGRWMKKIDKSTSENGKWILAHTKPCPRCKSLIEKNQGFKYMTCKCGFQFCWVC---- 306
W KK + SE WI HTK CP+C +EKN G + C CG FCW+C
Sbjct: 257 LMWELWRKKC-RDESETINWITVHTKLCPKCYKPVEKNGGCNLVRCICGQCFCWLCGGAT 315
Query: 307 --FHEQLKCESRSCSPFIRMHIMEFQGTDGEEVEINMEKDNPKKCNHYYECWARNDFLSK 364
H SC + D +E ++ K + + HY+ + + SK
Sbjct: 316 GSDHTYRSIAGHSCGRY----------QDDKEKQMERAKRDLNRYTHYHHRYKAHTDSSK 365
Query: 365 SILQKLIDLDRKYLSMSLQKRETY---FDFVKKAWQQIAECRRILKWTNVYGYFL----- 416
+ KL D + +S S +KRE F +V ++ RR+L ++ + Y++
Sbjct: 366 -LEDKLRDTIHEKVSKS-EKRELKLKDFSWVTNGLDRLFRSRRVLSYSYAFAYYMFGEEM 423
Query: 417 ------PKNEKAKKQFSEYIQVEAEVALEKLQQCAKVEETMFQDNYPEESLNKFQEKVIH 470
P+ + KK E Q + E +EKL Q +EE D + + + + ++I+
Sbjct: 424 FKDEMTPEEREIKKNLFEDQQQQLESNVEKLSQF--LEEPF--DEFSNDKVMAIRIQIIN 479
Query: 471 LTDVTKIYFENLLRAFEDGL 490
L+ + + E+ L
Sbjct: 480 LSVAVDTLCKKMYECIENDL 499
>At3g27710 putative protein
Length = 537
Score = 167 bits (422), Expect = 2e-41
Identities = 126/464 (27%), Positives = 203/464 (43%), Gaps = 52/464 (11%)
Query: 41 ILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCED-EEKVQKAAG 99
++ + + Q++ + +V +L + + A LL + WNV K+ + + ++++ AG
Sbjct: 45 VIKKESLVAAQKEILVRVMELLSVKENQARTLLIYYQWNVEKLFSVFADQGKDRMFSCAG 104
Query: 100 L-LNQPKVEIDWLKDLCQVCFRSSEKGKILSA-GCAHPLCEACWMDYIDTNINKGPSKCL 157
L + P + C VC +++ C H C CW+ + IN+G SK
Sbjct: 105 LTVFVPSLVTSKKTMKCDVCMEDDLPSNVMTRMECGHRFCNDCWIGHFTVKINEGESK-- 162
Query: 158 TLRCPDPSCDVAVDRDMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPA-PGCDYVVCY 216
+ C C D D++R+L S Y++FL+ SYV+ ++WCP+ P C +
Sbjct: 163 RILCMAHECKAICDEDVVRKLVSPELADRYDRFLIESYVEDNNMVKWCPSKPHCGSAIRK 222
Query: 217 QGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDCETAGRWMKKIDKSTSENGKWILAHTK 276
DG ++V C C FC+ C ESHSP C W KK + SE WI +TK
Sbjct: 223 IEDGHDV--VEVGCSCGLQFCFSCLSESHSPCSCLMWKLWKKKCEDE-SETVNWITVNTK 279
Query: 277 PCPRCKSLIEKNQGFKYMTCKCGFQFCWVC------FHEQLKCESRSCSPFIRMHIMEFQ 330
CP+C I+K G MTCKCG FCW+C H SC + + + +
Sbjct: 280 LCPKCSKPIQKRDGCNLMTCKCGQHFCWLCGQATGRDHTYTSIAGHSCGRYKDEKVRQLE 339
Query: 331 GTDGEEVEINMEKDNPKKCNHYYECWAR------NDFLSKSILQKLIDLDRKYLSMSLQK 384
++D + +++Y A D L KSIL+K + QK
Sbjct: 340 ---------RAQRDLDRYTHYHYRYKAHIDSLKLEDKLRKSILEKAVSNS----ETKDQK 386
Query: 385 RETYFDFVKKAWQQIAECRRILKWTNVYGYFL-----------PKNEKAKKQFSEYIQVE 433
+ +V A ++ RRIL + + +++ K + KK E Q +
Sbjct: 387 VFKEYSWVTDAVNRLFISRRILSQSYPFAFYMFGEELFKDEMSEKEREIKKNLFEDQQQQ 446
Query: 434 AEVALEKLQQCAKVEETMFQDNYPEESLNKFQEKVIHLTDVTKI 477
E +EKL +K+ E F D Y E K E + HLT++T +
Sbjct: 447 LEGNVEKL---SKILEEPF-DEYDHE---KVVEMMRHLTNLTAV 483
>At2g16090 unknown protein
Length = 593
Score = 167 bits (422), Expect = 2e-41
Identities = 128/482 (26%), Positives = 216/482 (44%), Gaps = 56/482 (11%)
Query: 41 ILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCED-EEKVQKAAG 99
++++ + Q + + +V +L + + A LL H W+V K+ E ++ + AG
Sbjct: 43 VITKESLLAAQREDLRRVMELLSVKEHHARTLLIHYRWDVEKLFAVLVEKGKDSLFSGAG 102
Query: 100 ---LLNQP---KVEIDWLKDLCQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNINKGP 153
L NQ V C +C ++ C H C CW + IN+G
Sbjct: 103 VTLLENQSCDSSVSGSSSMMSCDICVEDVPGYQLTRMDCGHSFCNNCWTGHFTVKINEGQ 162
Query: 154 SKCLTLRCPDPSCDVAVDRDMIRELASESSKVM---YEQFLLWSYVDSKAKMRWCPA-PG 209
SK + C C+ D D++R L S+S + +++FLL SY++ ++WCP+ P
Sbjct: 163 SKRII--CMAHKCNAICDEDVVRALVSKSQPDLAEKFDRFLLESYIEDNKMVKWCPSTPH 220
Query: 210 CDYVVCYQGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDCETAGRWMKK-IDKSTSENG 268
C + + D C +V C C FC+ C ++HSP C W KK D+S + N
Sbjct: 221 CGNAIRVEDDELC----EVECSCGLQFCFSCSSQAHSPCSCVMWELWRKKCFDESETVN- 275
Query: 269 KWILAHTKPCPRCKSLIEKNQGFKYMTCKCGFQFCWVC------FHEQLKCESRSCSPFI 322
WI HTKPCP+C +EKN G +TC C FCW+C H + SC F
Sbjct: 276 -WITVHTKPCPKCHKPVEKNGGCNLVTCLCRQSFCWLCGEATGRDHTWARISGHSCGRF- 333
Query: 323 RMHIMEFQGTDGEEVEINMEKDNPKKCNHYYECWARNDFLSKSILQKLIDLDRKYLSMSL 382
+ +E ++ K + K+ HY+ + + SK + KL + K +S+S
Sbjct: 334 ---------QEDKEKQMERAKRDLKRYMHYHNRYKAHIDSSK-LEAKLSNNISKKVSIS- 382
Query: 383 QKRETY---FDFVKKAWQQIAECRRILKWTNVYGYFLPKNEKAKKQFS-----------E 428
+KRE F + ++ RR+L ++ + +++ +E K + S E
Sbjct: 383 EKRELQLKDFSWATNGLHRLFRSRRVLSYSYPFAFYMFGDELFKDEMSSEEREIKQNLFE 442
Query: 429 YIQVEAEVALEKLQQCAKVEETMFQDNYPEESLNKFQEKVIHLTDVTKIYFENLLRAFED 488
Q + E +EKL +K E F D + ++ + + + +VI+L+ EN+ E+
Sbjct: 443 DQQQQLEANVEKL---SKFLEEPF-DQFADDKVMQIRIQVINLSVAVDTLCENMYECIEN 498
Query: 489 GL 490
L
Sbjct: 499 DL 500
>At5g63760 unknown protein
Length = 452
Score = 157 bits (397), Expect = 1e-38
Identities = 108/378 (28%), Positives = 174/378 (45%), Gaps = 36/378 (9%)
Query: 132 CAHPLCEACWMDYIDTNINKGPSKCLTLRCPDPSCDVAVDRDMIRELASESSKVMYEQFL 191
C+H C+ACW Y+ N + CPD C AV + + +L + MYE ++
Sbjct: 54 CSHKFCKACWSKYLKKNFFSVEKNHTAISCPDRDCRAAVGPETVEKLTVRD-QAMYELYI 112
Query: 192 LWSYVDSKA--KMRWCPAPGCDYVVCYQ-GDGGCARDLDVTCLCYHSFCWRCGEESHSPV 248
L SY + K++ CPA GC+YV+ + L++ CLC H FCWRC ESH PV
Sbjct: 113 LKSYREKYLGWKLKLCPARGCNYVIEFHLASEDEEHSLNIVCLCGHIFCWRCMLESHRPV 172
Query: 249 DCETAGRWMKK-IDKSTSENGK-----WILAHTKPCPRCKSLIE---KNQGFKYMTCKCG 299
C A W+ + ++K E K WI A+TKPCP C +E + +++TC C
Sbjct: 173 TCNNASDWLSRDLEKLIEEVDKPSTVSWIDANTKPCPHCFIPVEIDGERPWAQFLTCVCS 232
Query: 300 FQFCWVCFHE-QLKCESRSCSPFIRMHIMEFQGTDGEEVEIN-MEKDNPKKCNHYYECWA 357
+FCW CF + S SC R + F + + I+ ++ N + N +
Sbjct: 233 GRFCWKCFRSPETHGTSGSCLAPARSSNVGFNHWNRAKPGISCLDLWNASQVNLVNAKYE 292
Query: 358 RNDFLSKSILQKLIDLDRKYLSMSLQKRETYFDFVKKAWQQIAECRRILKWTNVYGYFLP 417
F +SI++K DL +E +++ I +CR+ LKW+ Y Y
Sbjct: 293 LEAF-EESIIKKPSDL-----------KEQDVKVLREGLMLIVQCRQFLKWSCAYDYIHT 340
Query: 418 KNEKAKKQFSEYIQVEAEVALEKLQQCAKVEETMFQDNYPEESLNKFQEKVIHLTDVTKI 477
+ + AK+++ ++Q A + Q K EET E+ K++ T
Sbjct: 341 EYDMAKREYLRFLQQNASGIVHSFSQSIK-EET--------EAKELTCGKLLSETTNIGN 391
Query: 478 YFENLLRAFEDGLDKVEA 495
+F + ++ +GL +V+A
Sbjct: 392 FFYHFIKTLREGLPEVQA 409
Score = 31.2 bits (69), Expect = 1.4
Identities = 35/142 (24%), Positives = 49/142 (33%), Gaps = 40/142 (28%)
Query: 216 YQGDGGCAR-DLDVTCLCYHSFCWRCG-----------EESHSPVDCE----TAGRWMKK 259
Y GD DL T C H FC C E++H+ + C A +
Sbjct: 37 YDGDAVVVDGDLISTPFCSHKFCKACWSKYLKKNFFSVEKNHTAISCPDRDCRAAVGPET 96
Query: 260 IDKST-------------SENGKWILAHTKPCPR--CKSLI------EKNQGFKYMTCKC 298
++K T S K++ K CP C +I E + + C C
Sbjct: 97 VEKLTVRDQAMYELYILKSYREKYLGWKLKLCPARGCNYVIEFHLASEDEEHSLNIVCLC 156
Query: 299 GFQFCWVCF---HEQLKCESRS 317
G FCW C H + C + S
Sbjct: 157 GHIFCWRCMLESHRPVTCNNAS 178
>At5g63730 ARI-like RING zinc finger protein-like
Length = 506
Score = 156 bits (394), Expect = 3e-38
Identities = 125/482 (25%), Positives = 208/482 (42%), Gaps = 43/482 (8%)
Query: 32 EEDKRENFTILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCEDE 91
E D R +++L+ EI + IN+++ + I ++ A LL + W+ +V E E++
Sbjct: 2 EYDGRRPYSVLTRNEITVKMKKQINEISDIFFISNSDATVLLMYLRWDSLRVSERLGENK 61
Query: 92 EKVQKAAGLLN-------QPKVEIDWLKDLCQVCFRSSEKGKILSAGCAHPLCEACWMDY 144
EK+ +GL + EI D+ + + I C+H W +Y
Sbjct: 62 EKLLMDSGLKSVMIDPSPDSSSEISLETDVYEF---DGDNDLISMPFCSHKFDSKYWREY 118
Query: 145 IDTNINKGPSKCLTLRCPDPSCDVAVDRDMIRELASESSKVMYEQFLLWSYVDSKAKM-- 202
++ N T+ CPD C AV D I +L + MYE+++ SY++ +
Sbjct: 119 LEKNFYYVEKIQTTISCPDQDCRSAVGPDTIEKLTVRDQE-MYERYIWRSYIEGNKVLMI 177
Query: 203 RWCPAPGCDYVVCY--QGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDCETAGRWMKKI 260
+ CPA CDYV+ + + D L+V C+C H FCWRC ESH PV C A W+
Sbjct: 178 KQCPARNCDYVIEFHQENDDDDEYSLNVVCICGHIFCWRCRLESHRPVSCNKASDWLCSA 237
Query: 261 DKSTSENGKWIL---AHTKPCPRCKSLIEKNQGF-KYMTCKCGFQFCWVCFH--EQLKCE 314
S+ + T CP C +E + +++TC C +FC C E K E
Sbjct: 238 TMKISDESFSLYPTKTKTVTCPHCLCSLESDTKMPQFLTCVCRLRFCSRCLRSEEAHKIE 297
Query: 315 SRSCSPFIRMHIMEFQGTDGEEVEINMEKDNPKKCNHYYECWARNDFLSKSILQKLIDLD 374
+ I+ EV I E D C E +KS L+ + +
Sbjct: 298 AVDSGFCIKT-----------EVGILCE-DRWNVCQKLLE-------QAKSDLEAFEETN 338
Query: 375 RKYLSMSLQKRETYFDFVKKAWQQIAECRRILKWTNVYGYFLPKNEKAKKQFSEYIQVEA 434
K S L++++ +++ I +CRR+LKW VY YF + E + K++ Y+Q A
Sbjct: 339 IKKPSDLLREQDIMI--IREGLMLIVQCRRVLKWCCVYDYFHTEYENS-KEYLRYLQGNA 395
Query: 435 EVALEKLQQCAKVEETMFQDNYPEESLNKFQEKVIHLTDVTKIYFENLLRAFEDGLDKVE 494
L+ + ++ + E F+ + T YF + ++ +DGL V+
Sbjct: 396 IATLQSYSNTLQEQKDIVLAAATYEECTFFRHTIPTATSNIGNYFYDFMKTLQDGLVDVK 455
Query: 495 AR 496
+
Sbjct: 456 VK 457
>At2g31510 putative RING zinc finger protein
Length = 565
Score = 154 bits (389), Expect = 1e-37
Identities = 89/230 (38%), Positives = 133/230 (57%), Gaps = 12/230 (5%)
Query: 271 ILAHTKPCPRCKSLIEKNQGFKYMTCK--CGFQFCWVCFHEQLKCESRSCSPFI--RMHI 326
ILA++KPCPRCK IEKNQG +MTC C ++FCW+C + R+ + R +
Sbjct: 241 ILANSKPCPRCKRPIEKNQGCMHMTCTPPCKYEFCWLCLGAWMDHGERTGGFYACNRYEV 300
Query: 327 MEFQGTDGEEVEINMEKDNPKKCNHYYECWARNDFLSKSILQKLIDLDRKYLSMSLQKRE 386
+ +G ++ ++++ + N + R LS+S+ + L K + E
Sbjct: 301 AKQEGQSRQKAMADLQQAQMQ--NVRLVMFFRILLLSESLRLFISKLSDKQCTP-----E 353
Query: 387 TYFDFVKKAWQQIAECRRILKWTNVYGYFLPKNEKAKKQFSEYIQVEAEVALEKLQQCAK 446
+ F+ +AW QI ECRR+LKWT YGY+LP++E AK+QF EY+Q EAE LE+L QC +
Sbjct: 354 SQLKFILEAWLQIIECRRVLKWTYAYGYYLPEHEHAKRQFFEYLQGEAESGLERLHQCVE 413
Query: 447 VEETMFQ-DNYPEESLNKFQEKVIHLTDVTKIYFENLLRAFEDGLDKVEA 495
+ F P + N F+ K+ LT VTK YFENL++A E+GL V++
Sbjct: 414 KDLVQFLIAEGPSKDFNDFRTKLAGLTSVTKNYFENLVKALENGLADVDS 463
Score = 146 bits (369), Expect = 2e-35
Identities = 86/228 (37%), Positives = 114/228 (49%), Gaps = 29/228 (12%)
Query: 36 RENFTILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCEDEEKVQ 95
++NF +L E +I+R Q D I +V+ VL I + A LL H W+V +V + W DEE+V+
Sbjct: 57 QKNFCVLREEDIRRHQMDNIERVSVVLSITEVEASILLRHFHWSVGRVHDEWFADEERVR 116
Query: 96 KAAGLLNQPKVEIDWLKDL-CQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNINKGPS 154
K G+L V +L C +CF S KI S C HP C CW YI T IN GP
Sbjct: 117 KTVGILESHVVPPSDDSELTCGICFDSYPPEKIASVSCGHPFCTTCWTGYISTTINDGPG 176
Query: 155 KCLTLRCPDPSCDVAVDRDMIRELASESSKVMYEQFLLWSYVDSKAK------------- 201
CL LRCPDPSC AV DM+ +LASE K Y ++ L SY++ K
Sbjct: 177 -CLMLRCPDPSCLAAVGHDMVDKLASEDEKEKYNRYFLRSYIEDNRKGLEIMMFPACARL 235
Query: 202 -------MRWCPAPGCDYVVCYQGDGGCARDLDVTCL--CYHSFCWRC 240
P P C + + + GC + +TC C + FCW C
Sbjct: 236 AFAGMILANSKPCPRCKRPI--EKNQGC---MHMTCTPPCKYEFCWLC 278
>At1g05880 unknown protein
Length = 426
Score = 141 bits (356), Expect = 7e-34
Identities = 117/473 (24%), Positives = 194/473 (40%), Gaps = 88/473 (18%)
Query: 35 KRENFTILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCEDEEKV 94
+R T+L+E +I+ L E + V+ + A A LL H WNV + + W + V
Sbjct: 15 QRNYATVLTEEDIRALMEIDVQSVSDFTSLSKAEATLLLSHLRWNVDCICKQWSAGAQSV 74
Query: 95 QKAAGLLNQPKVEIDWLKD----LCQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNIN 150
+ + GLL E+D D C C S + S C H +C CW +I+ I+
Sbjct: 75 RDSVGLL-----ELDPPSDDNEYFCGACGESHPHKNLASVSCGHRICTRCWTSHINKIIS 129
Query: 151 KGPSK--CLTLRCP-----DPSCDVAVDRDMIRELASESSKVMYEQFLLWSYVDSKAK-- 201
+ P+ L L+CP SC +V D I AS+ K + +L ++
Sbjct: 130 EKPAAEWNLWLKCPVRVGLHASCPASVGLDTIERFASKREKGLDAPLILVQVLEMPVSPV 189
Query: 202 MRWCPAPGCDYVVCYQGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDCETAGRWMKKID 261
W + G E++HSPVDC+TA +W+ I
Sbjct: 190 TVWLDSVGI-------------------------------EDAHSPVDCKTAAKWLA-IH 217
Query: 262 KSTSENGKWILAHTKPCPRCKSLIEKNQGFKYMTCKCGFQFCWVCFHEQLKCESRSCSPF 321
S +N + +FC V + E ++
Sbjct: 218 VSDIQNPER------------------------------RFCHVDWIEDMEGTGGD---- 243
Query: 322 IRMHIMEFQGTDGEEVEINMEKDNPKKCNHYYECWARNDFLSKSILQKLIDLDRKYLSMS 381
+H F ++ M + + + YE W N+ L + L LD LS +
Sbjct: 244 --LHFCTFDAVLSDQ-RGKMSESDSNRYEDCYENWDSNELLMQKEQANLPKLDTIELSNT 300
Query: 382 LQKRETYFDFVKKAWQQIAECRRILKWTNVYGYFLPKNEKAKKQFSEYIQVEAEVALEKL 441
+ + F+ +A QI ECRR+L+WT VYGY+L ++E K+ + Q + +E L
Sbjct: 301 QLENVSQLKFILEAGLQIIECRRVLEWTYVYGYYLREDEVGKQNLLKDTQERLKKFVENL 360
Query: 442 QQCAKVEETMFQ-DNYPEESLNKFQEKVIHLTDVTKIYFENLLRAFEDGLDKV 493
+ C + F+ + P + N F+ K+ LT +T+ ++EN+++ E+GL V
Sbjct: 361 KHCLETNLQPFRYEEEPSKDFNAFRIKLTELTSLTRNHYENVVKDVENGLASV 413
>At3g27720 hypothetical protein
Length = 493
Score = 117 bits (293), Expect = 1e-26
Identities = 92/332 (27%), Positives = 149/332 (44%), Gaps = 49/332 (14%)
Query: 171 DRDMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPA-PGCDYVVCYQGDGGCARDLD-V 228
++D +L S +++FL+ SYV+ ++WCP+ P C + D G D+D V
Sbjct: 136 EKDDEVQLVSTELAEKFDRFLIESYVEDNNMVKWCPSTPHCGNAIRNIKDDG---DVDEV 192
Query: 229 TCLCYHSFCWRCGEESHSPVDCETAGRWMKKIDKSTSENGKWILAHTKPCPRCKSLIEKN 288
C C FC+ C ESHSP C W KK + SE W+ +TK CP+C I+K
Sbjct: 193 ECSCGLQFCFSCLSESHSPCSCLMWKLWKKKCEDE-SETVNWMTVNTKLCPKCSKPIQKR 251
Query: 289 QGFKYMTCKCGFQFCWVC------FHEQLKCESRSCSPFIRMHIMEFQGTDGEEVEINME 342
G +MTCKCG FCW+C H SC + + + + +
Sbjct: 252 DGCNHMTCKCGQHFCWLCGQATGRDHSYSSIAGHSCGRYKEEKVRQLE---------RAQ 302
Query: 343 KDNPKKCNHYYECWAR------NDFLSKSILQKLIDLDRKYLSMSLQKRETYFDFVKKAW 396
+D + +++Y A D L KSIL+K + L+ + + K + ++ A
Sbjct: 303 RDLDRYTHYHYRYKAHIDSLKLEDKLKKSILKKAV-LNSETKDQKVFKE---YSWIIDAV 358
Query: 397 QQIAECRRILKWTNVYGYFL-----------PKNEKAKKQFSEYIQVEAEVALEKLQQCA 445
++ RRIL ++ + +++ + KK E Q + E +E+L +
Sbjct: 359 NRLFRSRRILSYSYPFVFYMFGKELFKDDMSDEERNIKKNLFEDQQQQLEGNVERL---S 415
Query: 446 KVEETMFQDNYPEESLNKFQEKVIHLTDVTKI 477
K+ E F D Y E K E + HLT++T +
Sbjct: 416 KILEEPF-DEYDHE---KVVEMMRHLTNLTAV 443
>At4g01020 unknown protein
Length = 1787
Score = 89.4 bits (220), Expect = 4e-18
Identities = 62/225 (27%), Positives = 92/225 (40%), Gaps = 15/225 (6%)
Query: 89 EDEEKVQKAAGLLNQPKVEI----DWLKDLCQVCFRSSEKGKILSAGCAHPLCEACWMDY 144
E ++VQK L + K + D ++ C +C + G L GC+H C+AC ++
Sbjct: 1531 EMRQEVQKMVNELAREKSALGEKPDEIELECPICLSEVDDGYSLE-GCSHLFCKACLLEQ 1589
Query: 145 IDT---NINKGPSKCLTLRCPDPSCDVAVDRDMIRELASESSKVMYEQFLLWSYVDSKAK 201
+ N + P C + C P V DM L+ E + L S K
Sbjct: 1590 FEASMRNFDAFPILCSHIDCGAP----IVVADMRALLSQEKLDELISASLSAFVTSSDGK 1645
Query: 202 MRWCPAPGCDYVVCYQGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDCETAGRWMKKID 261
+R+C P C + G + C+ C RC E H + CE ++ + D
Sbjct: 1646 LRFCSTPDCPSIYRVAGPQESGEPF-ICGACHSETCTRCHLEYHPLITCERYKKFKENPD 1704
Query: 262 KSTSENGKWILAHTKPCPRCKSLIEKNQGFKYMTCKCGFQFCWVC 306
S + K K CP CKS IEK G ++ C+CG CW C
Sbjct: 1705 LSLKDWAKG--KDVKECPICKSTIEKTDGCNHLQCRCGKHICWTC 1747
>At3g53690 unknown protein
Length = 320
Score = 77.8 bits (190), Expect = 1e-14
Identities = 54/204 (26%), Positives = 88/204 (42%), Gaps = 26/204 (12%)
Query: 115 CQVCFRSS---EKGKILSAGCAHPLCEACWMDYIDTNINKGPSKCLTLRCPDPSCDVAVD 171
C++C S E +I GC+H C C YI + L++ CP C ++
Sbjct: 115 CEICVDSKSIIESFRI--GGCSHFYCNDCVSKYIAAKLQ---DNILSIECPVSGCSGRLE 169
Query: 172 RDMIRELASESSKVMYEQF--LLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDLDVT 229
D R++ K +++++ L V ++K +CP C +V + +D +
Sbjct: 170 PDQCRQILP---KEVFDRWGDALCEAVVMRSKKFYCPYKDCSALVFLEESEVKMKDSECP 226
Query: 230 CLCYHSFCWRCGEESHSPVDCETAGRWMKK-------IDKSTSENGKWILAHTKPCPRCK 282
C+ C CG + H + CE + + + ++ KW K CP CK
Sbjct: 227 H-CHRMVCVECGTQWHPEMTCEEFQKLAANERGRDDILLATMAKQKKW-----KRCPSCK 280
Query: 283 SLIEKNQGFKYMTCKCGFQFCWVC 306
IEK+QG YM C+CG FC+ C
Sbjct: 281 FYIEKSQGCLYMKCRCGLAFCYNC 304
>At1g32340 RING finger protein, putative
Length = 688
Score = 77.8 bits (190), Expect = 1e-14
Identities = 54/212 (25%), Positives = 83/212 (38%), Gaps = 29/212 (13%)
Query: 115 CQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNINKGPSKCLTLRCPDPSCDVAVDRDM 174
C +CF S + C H C C Y D ++ +G L+CPD C V +
Sbjct: 378 CCICFSESAGIDFVKLPCQHFFCLKCMKTYTDIHVTEGTVN--KLKCPDSKCGETVPPGI 435
Query: 175 IRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDLDVTCLCYH 234
++ L + + +E +L ++S + +CP C+ C + + L + CY
Sbjct: 436 LKRLLGDEAYERWETLMLQKTLESMTDVAYCPR--CE-TPCIEDE----EQLALCFKCYF 488
Query: 235 SFCWRCGEESHSPVDC----------------ETAGRWMKKIDKSTSE---NGKWILAHT 275
SFC C E+ H V C G ++ +K + K I+
Sbjct: 489 SFCTLCKEKRHVGVACMSPELRLQILQERQDSSRLGEEQRRKEKEMINEIMSVKVIMKSA 548
Query: 276 KPCPRCKSLIEKNQGFKYMTC-KCGFQFCWVC 306
K CP CK I + G M C CG FC+ C
Sbjct: 549 KQCPSCKIAISRTGGCNKMVCNNCGQYFCYRC 580
>At5g10370 putative protein
Length = 1751
Score = 77.4 bits (189), Expect = 2e-14
Identities = 59/216 (27%), Positives = 88/216 (40%), Gaps = 15/216 (6%)
Query: 89 EDEEKVQKAAGLLNQPKVEI----DWLKDLCQVCFRSSEKGKILSAGCAHPLCEACWMDY 144
E ++VQK L + K + D ++ C +C + G L GC+H C+AC ++
Sbjct: 1534 EMRQEVQKMVNELAREKSALGEKPDEIEVECPICLSEVDDGYSLE-GCSHLFCKACLLEQ 1592
Query: 145 IDT---NINKGPSKCLTLRCPDPSCDVAVDRDMIRELASESSKVMYEQFLLWSYVDSKAK 201
+ N + P C + C P V DM L+ E ++ L S K
Sbjct: 1593 FEASMRNFDAFPILCSHIDCGAP----IVLADMRALLSQEKLDELFSASLSSFVTSSDGK 1648
Query: 202 MRWCPAPGCDYVVCYQGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDCETAGRWMKKID 261
R+C P C V G + C+ C RC E H + CE ++ + D
Sbjct: 1649 FRFCSTPDCPSVYRVAGPQESGEPF-ICGACHSEICTRCHLEYHPLITCERYKKFKENPD 1707
Query: 262 KSTSENGKWILAHTKPCPRCKSLIEKNQGFKYMTCK 297
S + K + K CP CKS IEK G +M C+
Sbjct: 1708 LSLKDWAKG--KNVKECPICKSTIEKTDGCNHMKCR 1741
>At2g25360 hypothetical protein
Length = 373
Score = 76.3 bits (186), Expect = 4e-14
Identities = 62/257 (24%), Positives = 112/257 (43%), Gaps = 37/257 (14%)
Query: 113 DLCQVCFRSSEKGKIL--SAGCAHPLCEACWMDYIDTNINKGPSKCLTLRCPDPSCDVAV 170
++C +CF +E ++ + C H C C Y++ + G C D C +
Sbjct: 88 EICSICFEETEGERMFFTTEKCVHRHCFPCVKQYVEVKLLSGTVPT----CLDDGCKFKL 143
Query: 171 DRDMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDL--DV 228
+ ++ + M++Q + + + ++ +CP P C ++ + + DL D
Sbjct: 144 TLESCSKVLTLELIEMWKQKMKEDSIPAAERI-YCPYPNCSMLMS-KTELSSESDLSNDR 201
Query: 229 TCL-CYHSFCWRCGEESHSPVDCETAGRWMKKID----------KSTSENGKWILAHTKP 277
+C+ C FC C SHS + C KK+ KS +++ KW +
Sbjct: 202 SCVKCCGLFCIDCKVPSHSDLSCAE----YKKLHHDPLVDELKLKSLAKDKKW-----RQ 252
Query: 278 CPRCKSLIEKNQGFKYMTCKCGFQFCWVCFHEQLKCESRSCSP--FIRMHIMEFQGTDGE 335
C C+ +IE + +MTC+CG+QFC+ C + K + ++CS + H + D
Sbjct: 253 CKMCRHMIELSHACNHMTCRCGYQFCYQC-EVEWKNDQKTCSSGCLLTGH-GYYDDYDYN 310
Query: 336 EVEINMEKDNPKKCNHY 352
E E + E D CN+Y
Sbjct: 311 EPEYDFEVDT---CNYY 324
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.136 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,356,657
Number of Sequences: 26719
Number of extensions: 558259
Number of successful extensions: 2179
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 36
Number of HSP's successfully gapped in prelim test: 38
Number of HSP's that attempted gapping in prelim test: 1967
Number of HSP's gapped (non-prelim): 108
length of query: 500
length of database: 11,318,596
effective HSP length: 103
effective length of query: 397
effective length of database: 8,566,539
effective search space: 3400915983
effective search space used: 3400915983
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0216.15


