

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
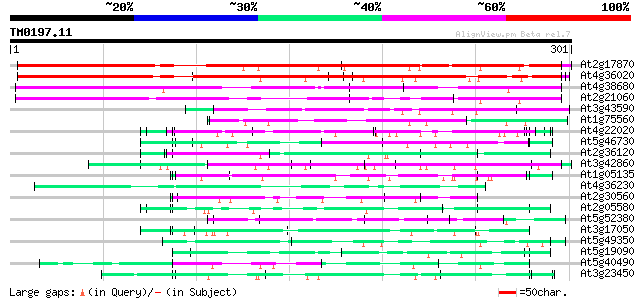
Query= TM0197.11
(301 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g17870 putative glycine-rich, zinc-finger DNA-binding protein 320 6e-88
At4g36020 glycine-rich protein 317 5e-87
At4g38680 glycine-rich protein 2 (GRP2) 202 1e-52
At2g21060 glycine-rich protein (AtGRP2) 171 3e-43
At3g43590 putative protein 110 7e-25
At1g75560 DNA-binding protein 96 2e-20
At4g22020 glycine-rich protein 89 2e-18
At5g46730 unknown protein 86 3e-17
At2g36120 unknown protein 86 3e-17
At3g42860 unknown protein (At3g42860) 84 7e-17
At1g05135 unknown protein 84 7e-17
At4g36230 putative glycine-rich cell wall protein 82 4e-16
At2g30560 putative glycine-rich protein 81 6e-16
At2g05580 unknown protein 78 5e-15
At5g52380 unknown protein 78 7e-15
At3g17050 unknown protein 78 7e-15
At5g49350 unknown protein 74 7e-14
At5g19090 unknown protein 74 1e-13
At5g40490 ribonucleoprotein -like 71 8e-13
At3g23450 hypothetical protein 71 8e-13
>At2g17870 putative glycine-rich, zinc-finger DNA-binding protein
Length = 301
Score = 320 bits (820), Expect = 6e-88
Identities = 164/311 (52%), Positives = 197/311 (62%), Gaps = 42/311 (13%)
Query: 5 RFSGVVQWFNNVKGFGFIKPDDGGEDLFVHQSSIRSDGYRTLLEGDLVEFSIATGDNDKT 64
R G V WF++ KG+GFI PDDGGE+LFVHQSSI SDG+R+L G+ VE+ IA G + KT
Sbjct: 10 RSIGKVSWFSDGKGYGFITPDDGGEELFVHQSSIVSDGFRSLTLGESVEYEIALGSDGKT 69
Query: 65 KAVDVTGPNGAALQPTRKDSAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCH 124
KA++VT P G +L +K+++ RG GG C+NCG+ GH+A+DC
Sbjct: 70 KAIEVTAPGGGSLN--KKENSSRG-----------------SGGNCFNCGEVGHMAKDCD 110
Query: 125 -----RSNNNGGG---GGGAACYNCGDAGHLARDCNRSNN-NSGGGGAG---CYNCGDTG 172
+S GGG GG CY CGD GH ARDC +S NSGGGG G CY+CG+ G
Sbjct: 111 GGSGGKSFGGGGGRRSGGEGECYMCGDVGHFARDCRQSGGGNSGGGGGGGRPCYSCGEVG 170
Query: 173 HLARDC------NRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRC 226
HLA+DC NR GG G GG CY CGG GH ARDC + G G GG GS +C+ C
Sbjct: 171 HLAKDCRGGSGGNRYGGGGGRGSGGDGCYMCGGVGHFARDCRQNGGGNVGGGGS-TCYTC 229
Query: 227 GGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCD-GGVAVSGGGGNAGRNTCFN 285
GG+GH+A+ C T+K PS GG GG C+ CG GHLARDCD G SGGGG G N CF
Sbjct: 230 GGVGHIAKVC-TSKIPSGGGGGGRACYECGGTGHLARDCDRRGSGSSGGGG--GSNKCFI 286
Query: 286 CGKPGHFAREC 296
CGK GHFAREC
Sbjct: 287 CGKEGHFAREC 297
Score = 131 bits (329), Expect = 5e-31
Identities = 65/136 (47%), Positives = 81/136 (58%), Gaps = 15/136 (11%)
Query: 179 NRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNG----GGGGP---GSASCFRCGGIGH 231
N+ NS G GG C+ CG GH+A+DC G G GGGG G C+ CG +GH
Sbjct: 83 NKKENSSRGSGGN--CFNCGEVGHMAKDCDGGSGGKSFGGGGGRRSGGEGECYMCGDVGH 140
Query: 232 MARDCATAKGPSSGGVGGGG--CFRCGEVGHLARDCDGGVAVS----GGGGNAGRNTCFN 285
ARDC + G +SGG GGGG C+ CGEVGHLA+DC GG + GGG +G + C+
Sbjct: 141 FARDCRQSGGGNSGGGGGGGRPCYSCGEVGHLAKDCRGGSGGNRYGGGGGRGSGGDGCYM 200
Query: 286 CGKPGHFARECIEASG 301
CG GHFAR+C + G
Sbjct: 201 CGGVGHFARDCRQNGG 216
>At4g36020 glycine-rich protein
Length = 299
Score = 317 bits (812), Expect = 5e-87
Identities = 160/309 (51%), Positives = 190/309 (60%), Gaps = 40/309 (12%)
Query: 5 RFSGVVQWFNNVKGFGFIKPDDGGEDLFVHQSSIRSDGYRTLLEGDLVEFSIATGDNDKT 64
R +G V WFN KG+GFI PDDG +LFVHQSSI S+GYR+L GD VEF+I G + KT
Sbjct: 10 RSTGKVNWFNASKGYGFITPDDGSVELFVHQSSIVSEGYRSLTVGDAVEFAITQGSDGKT 69
Query: 65 KAVDVTGPNGAALQPTRKDSAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCH 124
KAV+VT P G +L +K++ RG NG GGGG GCYNCG+ GH+++DC
Sbjct: 70 KAVNVTAPGGGSL---KKENNSRG----------NGARRGGGGSGCYNCGELGHISKDCG 116
Query: 125 RSNNNGGGG----GGAACYNCGDAGHLARDCNRSNNN-----SGGGGAGCYNCGDTGHLA 175
GGG GG CYNCGD GH ARDC + N + GG GCY CGD GH+A
Sbjct: 117 IGGGGGGGERRSRGGEGCYNCGDTGHFARDCTSAGNGDQRGATKGGNDGCYTCGDVGHVA 176
Query: 176 RDCNRSNNSGGGG-----GGGGACYTCGGFGHLARDCMR---GGNGGGGGPGSASCFRCG 227
RDC + + G GG CYTCG GH ARDC + GN GG GS +C+ CG
Sbjct: 177 RDCTQKSVGNGDQRGAVKGGNDGCYTCGDVGHFARDCTQKVAAGNVRSGGGGSGTCYSCG 236
Query: 228 GIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAGRNTCFNCG 287
G+GH+ARDCAT + PS G C++CG GHLARDCD SGGGGN N C+ CG
Sbjct: 237 GVGHIARDCATKRQPSRG------CYQCGGSGHLARDCDQ--RGSGGGGND--NACYKCG 286
Query: 288 KPGHFAREC 296
K GHFAREC
Sbjct: 287 KEGHFAREC 295
Score = 126 bits (317), Expect = 1e-29
Identities = 60/136 (44%), Positives = 75/136 (55%), Gaps = 9/136 (6%)
Query: 172 GHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGG---GPGSASCFRCGG 228
G L ++ N N GGGG CY CG GH+++DC GG GGGG G C+ CG
Sbjct: 80 GSLKKENNSRGNGARRGGGGSGCYNCGELGHISKDCGIGGGGGGGERRSRGGEGCYNCGD 139
Query: 229 IGHMARDCATA-KGPSSGGVGGG--GCFRCGEVGHLARDCDGGVAVSG---GGGNAGRNT 282
GH ARDC +A G G GG GC+ CG+VGH+ARDC +G G G +
Sbjct: 140 TGHFARDCTSAGNGDQRGATKGGNDGCYTCGDVGHVARDCTQKSVGNGDQRGAVKGGNDG 199
Query: 283 CFNCGKPGHFARECIE 298
C+ CG GHFAR+C +
Sbjct: 200 CYTCGDVGHFARDCTQ 215
Score = 93.6 bits (231), Expect = 1e-19
Identities = 39/81 (48%), Positives = 47/81 (57%), Gaps = 5/81 (6%)
Query: 99 NGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNS 158
N GGGG G CY+CG GH+ARDC G CY CG +GHLARDC++ +
Sbjct: 221 NVRSGGGGSGTCYSCGGVGHIARDCATKRQPSRG-----CYQCGGSGHLARDCDQRGSGG 275
Query: 159 GGGGAGCYNCGDTGHLARDCN 179
GG CY CG GH AR+C+
Sbjct: 276 GGNDNACYKCGKEGHFARECS 296
Score = 91.3 bits (225), Expect = 6e-19
Identities = 44/120 (36%), Positives = 58/120 (47%), Gaps = 4/120 (3%)
Query: 185 GGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSS 244
G G T G G L ++ GNG G G + C+ CG +GH+++DC G
Sbjct: 64 GSDGKTKAVNVTAPGGGSLKKENNSRGNGARRGGGGSGCYNCGELGHISKDCGIGGGGGG 123
Query: 245 G---GVGGGGCFRCGEVGHLARDC-DGGVAVSGGGGNAGRNTCFNCGKPGHFARECIEAS 300
G GG GC+ CG+ GH ARDC G G G + C+ CG GH AR+C + S
Sbjct: 124 GERRSRGGEGCYNCGDTGHFARDCTSAGNGDQRGATKGGNDGCYTCGDVGHVARDCTQKS 183
>At4g38680 glycine-rich protein 2 (GRP2)
Length = 203
Score = 202 bits (515), Expect = 1e-52
Identities = 104/211 (49%), Positives = 127/211 (59%), Gaps = 20/211 (9%)
Query: 4 ERFSGVVQWFNNVKGFGFIKPDDGGEDLFVHQSSIRSDGYRTLLEGDLVEFSIATGDNDK 63
ER G V+WF+ KGFGFI PDDGG+DLFVHQSSIRS+G+R+L + VEF + +N++
Sbjct: 9 ERRKGSVKWFDTQKGFGFITPDDGGDDLFVHQSSIRSEGFRSLAAEEAVEFEVEIDNNNR 68
Query: 64 TKAVDVTGPNGAALQPTR---KDSAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLA 120
KA+DV+GP+GA +Q GFGG RGG R +GGG GGGGGG G
Sbjct: 69 PKAIDVSGPDGAPVQGNSGGGSSGGRGGFGGGRGGGRGSGGGYGGGGGGYGGRG------ 122
Query: 121 RDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNR 180
GGG GG+ CY CG+ GH+ARDC+ GGGG G Y G G+
Sbjct: 123 ---------GGGRGGSDCYKCGEPGHMARDCSEGGGGYGGGGGG-YG-GGGGYGGGGGGY 171
Query: 181 SNNSGGGGGGGGACYTCGGFGHLARDCMRGG 211
GGGGGGG+CY+CG GH ARDC GG
Sbjct: 172 GGGGRGGGGGGGSCYSCGESGHFARDCTSGG 202
Score = 84.7 bits (208), Expect = 5e-17
Identities = 51/137 (37%), Positives = 59/137 (42%), Gaps = 46/137 (33%)
Query: 183 NSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGP 242
NSGGG GG GGFG GG GGG G G GG G G
Sbjct: 85 NSGGGSSGGR-----GGFG--------GGRGGGRGSGGGYGGGGGGYG----------GR 121
Query: 243 SSGGVGGGGCFRCGEVGHLARDCD-----------------------GGVAVSGGGGNAG 279
GG GG C++CGE GH+ARDC GG G GG G
Sbjct: 122 GGGGRGGSDCYKCGEPGHMARDCSEGGGGYGGGGGGYGGGGGYGGGGGGYGGGGRGGGGG 181
Query: 280 RNTCFNCGKPGHFAREC 296
+C++CG+ GHFAR+C
Sbjct: 182 GGSCYSCGESGHFARDC 198
>At2g21060 glycine-rich protein (AtGRP2)
Length = 201
Score = 171 bits (434), Expect = 3e-43
Identities = 97/235 (41%), Positives = 124/235 (52%), Gaps = 50/235 (21%)
Query: 4 ERFSGVVQWFNNVKGFGFIKPDDGGEDLFVHQSSIRSDGYRTLLEGDLVEFSIATGDNDK 63
+R G V+WF+ KGFGFI P DGG+DLFVHQSSIRS+G+R+L + VEF + ++ +
Sbjct: 13 DRRKGTVKWFDTQKGFGFITPSDGGDDLFVHQSSIRSEGFRSLAAEESVEFDVEVDNSGR 72
Query: 64 TKAVDVTGPNGAALQPTRKDSAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDC 123
KA++V+GP+GA P + +S G G RGG GG GGG GGG Y G G
Sbjct: 73 PKAIEVSGPDGA---PVQGNSGGGGSSGGRGGFGGGGGRGGGRGGGSYGGGYGGR----- 124
Query: 124 HRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNN 183
+GG GGG GG C+ CG+ GH+AR+C++
Sbjct: 125 ----GSGGRGGG------------------------GGDNSCFKCGEPGHMARECSQGGG 156
Query: 184 SGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCAT 238
GGGGGG Y GG G GG GGG SC+ CG GH ARDC +
Sbjct: 157 GYSGGGGGGR-YGSGGGG--------GGGGGG-----LSCYSCGESGHFARDCTS 197
Score = 90.5 bits (223), Expect = 1e-18
Identities = 58/131 (44%), Positives = 66/131 (50%), Gaps = 41/131 (31%)
Query: 183 NSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGP 242
NSGGGG GG GGFG GG G GGG G S GG G G
Sbjct: 89 NSGGGGSSGGR----GGFG--------GGGGRGGGRGGGSYG--GGYG----------GR 124
Query: 243 SSGGVGGGG----CFRCGEVGHLARDC-DGGVAVS------------GGGGNAGRNTCFN 285
SGG GGGG CF+CGE GH+AR+C GG S GGGG G +C++
Sbjct: 125 GSGGRGGGGGDNSCFKCGEPGHMARECSQGGGGYSGGGGGGRYGSGGGGGGGGGGLSCYS 184
Query: 286 CGKPGHFAREC 296
CG+ GHFAR+C
Sbjct: 185 CGESGHFARDC 195
>At3g43590 putative protein
Length = 551
Score = 110 bits (276), Expect = 7e-25
Identities = 66/202 (32%), Positives = 85/202 (41%), Gaps = 28/202 (13%)
Query: 110 CYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCG 169
C+ CG H A+ C + ++ CY C GH A+DC N G GA C CG
Sbjct: 191 CFICGSLEHGAKQCSKGHD---------CYICKKTGHRAKDCPDKYKN-GSKGAVCLRCG 240
Query: 170 DTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGI 229
D GH C + CY C FGHL C+ GN + SC+RCG +
Sbjct: 241 DFGHDMILCKYEYSKEDLKDV--QCYICKSFGHLC--CVEPGNSLSW---AVSCYRCGQL 293
Query: 230 GHMARDCATAKGPSSGGVGG-----------GGCFRCGEVGHLARDCDGGVAVSGGGGNA 278
GH C S+ C+RCGE GH AR+C ++S G
Sbjct: 294 GHSGLACGRHYEESNENDSATPERLFNSREASECYRCGEEGHFARECPNSSSISTSHGRE 353
Query: 279 GRNTCFNCGKPGHFARECIEAS 300
+ C+ C GHFAREC +S
Sbjct: 354 SQTLCYRCNGSGHFARECPNSS 375
Score = 91.7 bits (226), Expect = 4e-19
Identities = 62/191 (32%), Positives = 75/191 (38%), Gaps = 20/191 (10%)
Query: 95 GERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRS 154
G +G G CY C TGH A+DC N G GA C CGD GH C
Sbjct: 195 GSLEHGAKQCSKGHDCYICKKTGHRAKDCPDKYKN--GSKGAVCLRCGDFGHDMILCKYE 252
Query: 155 NNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMR----G 210
+ CY C GHL C NS +CY CG GH C R
Sbjct: 253 YSKEDLKDVQCYICKSFGHLC--CVEPGNS---LSWAVSCYRCGQLGHSGLACGRHYEES 307
Query: 211 GNGGGGGP-------GSASCFRCGGIGHMARDC--ATAKGPSSGGVGGGGCFRCGEVGHL 261
P ++ C+RCG GH AR+C +++ S G C+RC GH
Sbjct: 308 NENDSATPERLFNSREASECYRCGEEGHFARECPNSSSISTSHGRESQTLCYRCNGSGHF 367
Query: 262 ARDCDGGVAVS 272
AR+C VS
Sbjct: 368 ARECPNSSQVS 378
>At1g75560 DNA-binding protein
Length = 257
Score = 95.9 bits (237), Expect = 2e-20
Identities = 76/225 (33%), Positives = 89/225 (38%), Gaps = 63/225 (28%)
Query: 107 GGGCYNCGDTGHLARDCHR---SNNNGGGGGGAA-------CYNCGDAGHLARDCNRSNN 156
G C NC GH ARDC NN G G AA C+NC + GH+A +C +
Sbjct: 54 GNLCNNCKRPGHFARDCSNVSVCNNCGLPGHIAAECTAESRCWNCREPGHVASNC----S 109
Query: 157 NSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGG 216
N G C++CG +GH ARDC +NS G C C GHLA DC
Sbjct: 110 NEG----ICHSCGKSGHRARDC---SNSDSRAGDLRLCNNCFKQGHLAADC--------- 153
Query: 217 GPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDC--------DGG 268
+C C GH+ARDC C C GH+AR C D G
Sbjct: 154 -TNDKACKNCRTSGHIARDCR----------NDPVCNICSISGHVARHCPKGDSNYSDRG 202
Query: 269 VAVSGGG--------------GNAGRNTCFNCGKPGHFARECIEA 299
V GG G + C NCG GH A EC A
Sbjct: 203 SRVRDGGMQRGGLSRMSRDREGVSAMIICHNCGGRGHRAYECPSA 247
Score = 84.0 bits (206), Expect = 9e-17
Identities = 53/162 (32%), Positives = 66/162 (40%), Gaps = 44/162 (27%)
Query: 108 GGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYN 167
G C++CG +GH ARDC SN++ G C NC GHLA DC C N
Sbjct: 112 GICHSCGKSGHRARDC--SNSDSRAGDLRLCNNCFKQGHLAADCTNDK--------ACKN 161
Query: 168 CGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNG----------GGGG 217
C +GH+ARDC C C GH+AR C +G + GG
Sbjct: 162 CRTSGHIARDCRND----------PVCNICSISGHVARHCPKGDSNYSDRGSRVRDGGMQ 211
Query: 218 PGSAS--------------CFRCGGIGHMARDCATAKGPSSG 245
G S C CGG GH A +C +A+ G
Sbjct: 212 RGGLSRMSRDREGVSAMIICHNCGGRGHRAYECPSARVADRG 253
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/52 (38%), Positives = 22/52 (41%), Gaps = 12/52 (23%)
Query: 249 GGGCFRCGEVGHLARDCDGGVAVSGGGGNAGRNTCFNCGKPGHFARECIEAS 300
G C C GH ARDC + + C NCG PGH A EC S
Sbjct: 54 GNLCNNCKRPGHFARDC------------SNVSVCNNCGLPGHIAAECTAES 93
>At4g22020 glycine-rich protein
Length = 396
Score = 89.4 bits (220), Expect = 2e-18
Identities = 66/203 (32%), Positives = 78/203 (37%), Gaps = 18/203 (8%)
Query: 85 APRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDA 144
A G GG GG GGGGGGGG G + G +GH + + GG G A G
Sbjct: 125 AGAGVGGTTGGVGGGGGGGGGGGEGGGSSGGSGHGSGSGAGAGAGVGGSSGGAGGGGGGG 184
Query: 145 GHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLA 204
G N + + G GAG G G + GGG GG + G G
Sbjct: 185 GGEGGGANGGSGHGSGAGAGAGVGGAAGGVGGGGGGGGGEGGGANGGSGHGSGSGAGGGV 244
Query: 205 RDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARD 264
GG GGGGG GS GG GH + +SGG GGGG
Sbjct: 245 SGAAGGGGGGGGGGGSGGSKVGGGYGHGSGFGGGVGFGNSGGGGGGG------------- 291
Query: 265 CDGGVAVSGGGGNAGRNTCFNCG 287
GG GGGG G + + G
Sbjct: 292 --GG---GGGGGGGGNGSGYGSG 309
Score = 82.4 bits (202), Expect = 3e-16
Identities = 70/210 (33%), Positives = 81/210 (38%), Gaps = 32/210 (15%)
Query: 89 FGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLA 148
+GG GG GGGGGGGG G G S G GGGG G+ G
Sbjct: 58 YGGGGGG---GGGGGGGGGEGGDGYGHGEGYGAGAGMSGYGGVGGGGGR--GGGEGG--G 110
Query: 149 RDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARD-- 206
N + + G GAG G TG + GGGGGGG + GG GH +
Sbjct: 111 SSANGGSGHGSGSGAGAGVGGTTGGV------GGGGGGGGGGGEGGGSSGGSGHGSGSGA 164
Query: 207 --------CMRGGNGGGGGPGSASCFRCGGIGH-MARDCATAKGPSSGGVGGGGCFRCGE 257
G GGGGG G GG GH G ++GGVGGGG GE
Sbjct: 165 GAGAGVGGSSGGAGGGGGGGGGEGGGANGGSGHGSGAGAGAGVGGAAGGVGGGGGGGGGE 224
Query: 258 VGHL--------ARDCDGGVAVSGGGGNAG 279
G GGV+ + GGG G
Sbjct: 225 GGGANGGSGHGSGSGAGGGVSGAAGGGGGG 254
Score = 79.7 bits (195), Expect = 2e-15
Identities = 68/201 (33%), Positives = 83/201 (40%), Gaps = 22/201 (10%)
Query: 85 APRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDA 144
A G GG GG GGGGGGGG G G +GH + +G GGG G +
Sbjct: 203 AGAGVGGAAGGV--GGGGGGGGGEGGGANGGSGHGSGSGAGGGVSGAAGGGGGGGGGGGS 260
Query: 145 GHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCG-GFGH- 202
G + + GGG G N G G GGGGGG G+ Y G G+G
Sbjct: 261 GGSKVGGGYGHGSGFGGGVGFGNSGGGGG-----GGGGGGGGGGGGNGSGYGSGSGYGSG 315
Query: 203 LARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSG-GVGGGGCFRCGEVGHL 261
+ + GG GGGGG GS GG G + G G G+GGG G G +
Sbjct: 316 MGKGSGSGGGGGGGGGGSG-----GGNG-------SGSGSGEGYGMGGGAGTGNGGGGGV 363
Query: 262 ARDCDGGVAVSGGGGNAGRNT 282
G + GGG+ G T
Sbjct: 364 GFGMGIGFGIGIGGGSGGGTT 384
Score = 76.6 bits (187), Expect = 1e-14
Identities = 66/204 (32%), Positives = 74/204 (35%), Gaps = 27/204 (13%)
Query: 74 GAALQPTRKDSAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGG 133
GA + T G GG GGE GG G G G G A S GGGG
Sbjct: 126 GAGVGGTTGGVGGGGGGGGGGGEGGGSSGGSGHGSG----SGAGAGAGVGGSSGGAGGGG 181
Query: 134 GGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGA 193
GG G+ G N + + G GAG G G + GGG GG
Sbjct: 182 GGGG----GEGG----GANGGSGHGSGAGAGAGVGGAAGGVGGGGGGGGGEGGGANGGSG 233
Query: 194 CYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCF 253
+ G G GG GGGGG GS GG GH + +SGG GGGG
Sbjct: 234 HGSGSGAGGGVSGAAGGGGGGGGGGGSGGSKVGGGYGHGSGFGGGVGFGNSGGGGGGG-- 291
Query: 254 RCGEVGHLARDCDGGVAVSGGGGN 277
GG GGGGN
Sbjct: 292 -------------GGGGGGGGGGN 302
Score = 75.9 bits (185), Expect = 3e-14
Identities = 73/220 (33%), Positives = 81/220 (36%), Gaps = 49/220 (22%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHL 147
G+GG GG GG GGG GGG G +GH S + G G G G G
Sbjct: 93 GYGGVGGG----GGRGGGEGGGSSANGGSGH------GSGSGAGAGVGGTTGGVGGGG-- 140
Query: 148 ARDCNRSNNNSGGGGAGCYNCGDTGH-------LARDCNRSNNSGGGGGGGGACY---TC 197
GGGG G + G +GH S+ GGGGGGG
Sbjct: 141 --------GGGGGGGEGGGSSGGSGHGSGSGAGAGAGVGGSSGGAGGGGGGGGGEGGGAN 192
Query: 198 GGFGH-----------LARDCMRGGNGGGGGPGSASCFRCG-----GIGHMARDCATAKG 241
GG GH A + GG GGGGG G + G G G A G
Sbjct: 193 GGSGHGSGAGAGAGVGGAAGGVGGGGGGGGGEGGGANGGSGHGSGSGAGGGVSGAAGGGG 252
Query: 242 PSSGGVGGGGCFRCGEVGHLARDCDGGVAV--SGGGGNAG 279
GG G GG G GH GGV SGGGG G
Sbjct: 253 GGGGGGGSGGSKVGGGYGH-GSGFGGGVGFGNSGGGGGGG 291
Score = 69.3 bits (168), Expect = 2e-12
Identities = 66/210 (31%), Positives = 76/210 (35%), Gaps = 56/210 (26%)
Query: 71 GPNGAALQPTRKDSAPRGFGGWRGGERRNGGGGGGGG-GGCYNCGDTGH---LARDCHRS 126
G G A + S GG G GGGGGGGG GG G GH
Sbjct: 223 GEGGGANGGSGHGSGSGAGGGVSGAAGGGGGGGGGGGSGGSKVGGGYGHGSGFGGGVGFG 282
Query: 127 NNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGG 186
N+ GGGGGG G G N SG G Y G + + SGG
Sbjct: 283 NSGGGGGGGGGGGGGGGGG----------NGSGYGSGSGYGSG--------MGKGSGSGG 324
Query: 187 GGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGG 246
GGGGGG GG+GGG G GS S G G+ G +G
Sbjct: 325 GGGGGG-----------------GGSGGGNGSGSGS-----GEGY-----GMGGGAGTGN 357
Query: 247 VGGGGCFRCGEVGHLARDCDGGVAVSGGGG 276
GGGG +G G+ + GG G
Sbjct: 358 GGGGGVGFGMGIGF-------GIGIGGGSG 380
Score = 54.7 bits (130), Expect = 6e-08
Identities = 43/112 (38%), Positives = 48/112 (42%), Gaps = 10/112 (8%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGC---YNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDA 144
GFG GG GGGGGGGGGG Y G +G+ + S + GGGGGG G
Sbjct: 280 GFGNSGGGGGGGGGGGGGGGGGNGSGYGSG-SGYGSGMGKGSGSGGGGGGGGG----GSG 334
Query: 145 GHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSG-GGGGGGGACY 195
G GGGAG N G G + G GGG GGG Y
Sbjct: 335 GGNGSGSGSGEGYGMGGGAGTGN-GGGGGVGFGMGIGFGIGIGGGSGGGTTY 385
Score = 52.4 bits (124), Expect = 3e-07
Identities = 54/165 (32%), Positives = 64/165 (38%), Gaps = 20/165 (12%)
Query: 131 GGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGG 190
GG G + N D + R + G GA Y G G GGGGG
Sbjct: 23 GGSCGDSVENNNDDDDCSSRSWRGCGSFYGKGAKRYGGGGGG--------GGGGGGGGGE 74
Query: 191 GGACYTCG-GFGHLARDCMRGGNGGGGGPG---SASCFRCGGIGHMARDCATA-KGPSSG 245
GG Y G G+G A GG GGGGG G GG GH + A A G ++G
Sbjct: 75 GGDGYGHGEGYGAGAGMSGYGGVGGGGGRGGGEGGGSSANGGSGHGSGSGAGAGVGGTTG 134
Query: 246 GVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAGRNTCFNCGKPG 290
GVGGGG G +GG + G G +G G G
Sbjct: 135 GVGGGG-------GGGGGGGEGGGSSGGSGHGSGSGAGAGAGVGG 172
Score = 45.1 bits (105), Expect = 5e-05
Identities = 37/112 (33%), Positives = 40/112 (35%), Gaps = 17/112 (15%)
Query: 197 CGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCG 256
CG F GG GGGGG G G GH A A GGVGGGG G
Sbjct: 47 CGSFYGKGAKRYGGGGGGGGGGGGGGGEGGDGYGHGEGYGAGAGMSGYGGVGGGGGRGGG 106
Query: 257 EVGHLARD-----------------CDGGVAVSGGGGNAGRNTCFNCGKPGH 291
E G + + GGV GGGG G + G GH
Sbjct: 107 EGGGSSANGGSGHGSGSGAGAGVGGTTGGVGGGGGGGGGGGEGGGSSGGSGH 158
>At5g46730 unknown protein
Length = 268
Score = 85.5 bits (210), Expect = 3e-17
Identities = 70/196 (35%), Positives = 76/196 (38%), Gaps = 20/196 (10%)
Query: 88 GFGGWRGGERRNGG---GGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDA 144
G GG GG +GG G G GGGG Y GH G GGGG + Y G
Sbjct: 86 GHGGGGGGAASSGGYASGAGEGGGGGYGGAAGGHAG-----GGGGGSGGGGGSAYGAG-- 138
Query: 145 GHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLA 204
G A GG GA Y G G GGGGGGG A GG G+
Sbjct: 139 GEHASGYGNGAGEGGGAGASGYGGGAYGG-------GGGHGGGGGGGSAGGAHGGSGYGG 191
Query: 205 RDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPS-SGGVGGGGCFRCGEVGHLAR 263
+ GG GGGG G A + GG G A G + GG G GG G G A
Sbjct: 192 GE--GGGAGGGGSHGGAGGYGGGGGGGSGGGGAYGGGGAHGGGYGSGGGEGGGYGGGAAG 249
Query: 264 DCDGGVAVSGGGGNAG 279
GG GG G G
Sbjct: 250 GYGGGHGGGGGHGGGG 265
Score = 85.5 bits (210), Expect = 3e-17
Identities = 72/213 (33%), Positives = 85/213 (39%), Gaps = 40/213 (18%)
Query: 71 GPNGAALQPTRKDSAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNG 130
G +G + A G+GG G G G GGGGGG + G A + G
Sbjct: 55 GESGGGYGGGSGEGAGGGYGGAEGYASGGGSGHGGGGGGAASSGGYASGAGE-----GGG 109
Query: 131 GGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGG 190
GG GGAA + G G SGGGG Y G N +G GGG
Sbjct: 110 GGYGGAAGGHAGGGG----------GGSGGGGGSAYGAGG----EHASGYGNGAGEGGGA 155
Query: 191 GGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSS----GG 246
G + Y G +G GG+GGGGG GSA GG G+ + A G S GG
Sbjct: 156 GASGYGGGAYGG------GGGHGGGGGGGSAGGAH-GGSGYGGGEGGGAGGGGSHGGAGG 208
Query: 247 VGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAG 279
GGGG G GG A GGG + G
Sbjct: 209 YGGGGGGGSG----------GGGAYGGGGAHGG 231
Score = 75.5 bits (184), Expect = 3e-14
Identities = 65/200 (32%), Positives = 79/200 (39%), Gaps = 26/200 (13%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGD-----TGHLARDCHRS---NNNGGGGGGAACY 139
G GG G + GG GG GG Y G G+ + + S + +GGGGGGAA
Sbjct: 38 GHGGGGGSGGVSSGGYGGESGGGYGGGSGEGAGGGYGGAEGYASGGGSGHGGGGGGAA-- 95
Query: 140 NCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGG 199
+G A + +G GG G Y GH GG GGGGG+ Y GG
Sbjct: 96 ---SSGGYA-------SGAGEGGGGGYGGAAGGH------AGGGGGGSGGGGGSAYGAGG 139
Query: 200 FGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVG 259
G GG G G GG GH + G + GG G GG G G
Sbjct: 140 EHASGYGNGAGEGGGAGASGYGGGAYGGGGGHGGGGGGGSAGGAHGGSGYGGGEGGGAGG 199
Query: 260 HLARDCDGGVAVSGGGGNAG 279
+ GG GGGG+ G
Sbjct: 200 GGSHGGAGGYGGGGGGGSGG 219
Score = 67.0 bits (162), Expect = 1e-11
Identities = 63/189 (33%), Positives = 75/189 (39%), Gaps = 27/189 (14%)
Query: 99 NGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNS 158
N G GGGG G +G ++ + + GG GGG+ G G + +
Sbjct: 29 NYGQASGGGGH-GGGGGSGGVSSGGYGGESGGGYGGGSGEGAGGGYGGAEGYASGGGSGH 87
Query: 159 GGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGP 218
GGGG G + G A GGGGG GGA GG GG GGG G
Sbjct: 88 GGGGGGAASSGGYASGA-------GEGGGGGYGGA---AGGHA--------GGGGGGSGG 129
Query: 219 GSASCFRCG-----GIGHMARD--CATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAV 271
G S + G G G+ A + A A G G GGGG G G A GG
Sbjct: 130 GGGSAYGAGGEHASGYGNGAGEGGGAGASGYGGGAYGGGGGHGGGGGGGSAGGAHGGSGY 189
Query: 272 SGG-GGNAG 279
GG GG AG
Sbjct: 190 GGGEGGGAG 198
Score = 65.1 bits (157), Expect = 4e-11
Identities = 60/193 (31%), Positives = 71/193 (36%), Gaps = 55/193 (28%)
Query: 99 NGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNS 158
+GGGG GGGGG +G ++ + + GG GGG S
Sbjct: 34 SGGGGHGGGGG------SGGVSSGGYGGESGGGYGGG----------------------S 65
Query: 159 GGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGP 218
G G G Y G G+ S G GGGGG + GG+ A G GGGGG
Sbjct: 66 GEGAGGGYG-GAEGYA------SGGGSGHGGGGGGAASSGGYASGA------GEGGGGGY 112
Query: 219 GSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNA 278
G A+ GG G GG GGGG G G A G GG G +
Sbjct: 113 GGAAGGHAGGGG--------------GGSGGGGGSAYGAGGEHASGYGNGAGEGGGAGAS 158
Query: 279 GRNTCFNCGKPGH 291
G G GH
Sbjct: 159 GYGGGAYGGGGGH 171
Score = 52.8 bits (125), Expect = 2e-07
Identities = 38/111 (34%), Positives = 43/111 (38%), Gaps = 25/111 (22%)
Query: 90 GGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLAR 149
GG GG GG GGG GGG + G G+ GGG GG Y G A
Sbjct: 181 GGAHGGSGYGGGEGGGAGGGGSHGGAGGY-------GGGGGGGSGGGGAYGGGGA----- 228
Query: 150 DCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGF 200
+ SGGG G Y G G GGG GGG + GG+
Sbjct: 229 --HGGGYGSGGGEGGGYGGGAAGGY-----------GGGHGGGGGHGGGGY 266
Score = 50.4 bits (119), Expect = 1e-06
Identities = 41/118 (34%), Positives = 56/118 (46%), Gaps = 10/118 (8%)
Query: 168 CGDTGHLARDCNRSNNSGGGGGGGGA-CYTCGGFGHLARDCMRGGN--GGGGGPGSASCF 224
C T + +++ GG GGGGG+ + GG+G + GG+ G GGG G A +
Sbjct: 20 CYATSRNLLNYGQASGGGGHGGGGGSGGVSSGGYGGESGGGYGGGSGEGAGGGYGGAEGY 79
Query: 225 RCG---GIGHMARDCATAKGPSSG-GVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNA 278
G G G A++ G +SG G GGGG + GH GG GGGG+A
Sbjct: 80 ASGGGSGHGGGGGGAASSGGYASGAGEGGGGGYGGAAGGHAG---GGGGGSGGGGGSA 134
>At2g36120 unknown protein
Length = 255
Score = 85.5 bits (210), Expect = 3e-17
Identities = 76/217 (35%), Positives = 84/217 (38%), Gaps = 39/217 (17%)
Query: 88 GFGGWRGGERRNGGGGG-GGGGGCYNCGDTGHLARDCHRSNNNGGGG------------- 133
G GG GG GGG G GGG G + G G H GGGG
Sbjct: 51 GIGGGPGGGSGYGGGSGEGGGAGGHGEGHIGGGGGGGHGGGAGGGGGGGPGGGYGGGSGE 110
Query: 134 GGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGA 193
GG A Y G+AG GGGG G + G G + +GGG GGGGA
Sbjct: 111 GGGAGYGGGEAGGHGGGGGGGAGGGGGGGGGAHGGGYGG------GQGAGAGGGYGGGGA 164
Query: 194 CYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCF 253
GG G GGNGGGGG GS GG G+ A G + G GGG
Sbjct: 165 GGHGGGGG--------GGNGGGGGGGSGEGGAHGG-GY------GAGGGAGEGYGGG--- 206
Query: 254 RCGEVGHLARDCDGGVAVSGGGGNAGRNTCFNCGKPG 290
G GH GG + GGGG G G G
Sbjct: 207 -AGAGGHGGGGGGGGGSGGGGGGGGGYAAASGYGHGG 242
Score = 76.6 bits (187), Expect = 1e-14
Identities = 59/172 (34%), Positives = 66/172 (38%), Gaps = 10/172 (5%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHL 147
G GG GG GGGGG GGG G+ G + +GGGGGG A G G
Sbjct: 83 GGGGGHGGGAGGGGGGGPGGGYGGGSGEGGGAGYGGGEAGGHGGGGGGGAGGGGGGGG-- 140
Query: 148 ARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGG-----GACYTCGGF-- 200
G G G Y G G N GGGGG G G Y GG
Sbjct: 141 GAHGGGYGGGQGAGAGGGYGGGGAGGHGGGGGGGNGGGGGGGSGEGGAHGGGYGAGGGAG 200
Query: 201 -GHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGG 251
G+ G GGGGG G + GG G+ A G + GG G GG
Sbjct: 201 EGYGGGAGAGGHGGGGGGGGGSGGGGGGGGGYAAASGYGHGGGAGGGEGSGG 252
Score = 74.7 bits (182), Expect = 6e-14
Identities = 60/175 (34%), Positives = 66/175 (37%), Gaps = 42/175 (24%)
Query: 84 SAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGD 143
S G G+ GGE GGGGGGG G GGGGGG + G
Sbjct: 108 SGEGGGAGYGGGEAGGHGGGGGGGAG--------------------GGGGGGGGAHGGGY 147
Query: 144 AGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFG-- 201
G + GGGGAG + G G N GGGGG G GG+G
Sbjct: 148 GGGQGAG---AGGGYGGGGAGGHGGGGGG--------GNGGGGGGGSGEGGAHGGGYGAG 196
Query: 202 -----HLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGG 251
GG+GGGGG G S GG G A A G GG GGG
Sbjct: 197 GGAGEGYGGGAGAGGHGGGGGGGGGS----GGGGGGGGGYAAASGYGHGGGAGGG 247
Score = 68.2 bits (165), Expect = 5e-12
Identities = 49/150 (32%), Positives = 55/150 (36%), Gaps = 40/150 (26%)
Query: 71 GPNGAALQPTRKDSAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNG 130
G +G + A G+GG G GGGGG GGGG G+ G H
Sbjct: 141 GAHGGGYGGGQGAGAGGGYGGGGAGGHGGGGGGGNGGGGGGGSGEGG-----AHGGGYGA 195
Query: 131 GGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGG 190
GGG G GG GAG + G G SGGGGGG
Sbjct: 196 GGGAG-------------------EGYGGGAGAGGHGGGGGG--------GGGSGGGGGG 228
Query: 191 GGACYTCGGFGHLARDCMRGGNGGGGGPGS 220
GG G+GH G G GGG GS
Sbjct: 229 GGGYAAASGYGH--------GGGAGGGEGS 250
Score = 44.3 bits (103), Expect = 8e-05
Identities = 22/55 (40%), Positives = 26/55 (47%)
Query: 85 APRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACY 139
A G+GG G GGGGGGGG G G G+ A + GGG G+ Y
Sbjct: 199 AGEGYGGGAGAGGHGGGGGGGGGSGGGGGGGGGYAAASGYGHGGGAGGGEGSGGY 253
Score = 34.7 bits (78), Expect = 0.065
Identities = 27/69 (39%), Positives = 30/69 (43%), Gaps = 16/69 (23%)
Query: 211 GNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVA 270
G G GGGPG S G G + + A G G +GGG G GH GG A
Sbjct: 49 GVGIGGGPGGGS-----GYGGGSGEGGGAGGHGEGHIGGG-----GGGGH------GGGA 92
Query: 271 VSGGGGNAG 279
GGGG G
Sbjct: 93 GGGGGGGPG 101
>At3g42860 unknown protein (At3g42860)
Length = 372
Score = 84.3 bits (207), Expect = 7e-17
Identities = 44/135 (32%), Positives = 58/135 (42%), Gaps = 16/135 (11%)
Query: 162 GAGCYNCGDTGHLARDCNRSNNSG--GGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPG 219
G CY CG GH ARDC +++G G C+ CG GH +RDC PG
Sbjct: 236 GTPCYKCGKEGHWARDCTVQSDTGPVKSTSAAGDCFKCGKPGHWSRDCTAQSGNPKYEPG 295
Query: 220 -------SASCFRCGGIGHMARDCA-------TAKGPSSGGVGGGGCFRCGEVGHLARDC 265
S C++CG GH +RDC G + G C++CG+ GH +RDC
Sbjct: 296 QMKSSSSSGECYKCGKQGHWSRDCTGQSSNQQFQSGQAKSTSSTGDCYKCGKAGHWSRDC 355
Query: 266 DGGVAVSGGGGNAGR 280
+ G R
Sbjct: 356 TSPAQTTNTPGKRQR 370
Score = 83.2 bits (204), Expect = 2e-16
Identities = 44/120 (36%), Positives = 59/120 (48%), Gaps = 14/120 (11%)
Query: 191 GGACYTCGGFGHLARDCMRGGNGGGGGPGSAS--CFRCGGIGHMARDCATAKGPSSGGVG 248
G CY CG GH ARDC + G SA+ CF+CG GH +RDC G G
Sbjct: 236 GTPCYKCGKEGHWARDCTVQSDTGPVKSTSAAGDCFKCGKPGHWSRDCTAQSGNPKYEPG 295
Query: 249 -------GGGCFRCGEVGHLARDCDGGVAVSGGGGNAGRNT-----CFNCGKPGHFAREC 296
G C++CG+ GH +RDC G + ++T C+ CGK GH++R+C
Sbjct: 296 QMKSSSSSGECYKCGKQGHWSRDCTGQSSNQQFQSGQAKSTSSTGDCYKCGKAGHWSRDC 355
Score = 82.8 bits (203), Expect = 2e-16
Identities = 44/120 (36%), Positives = 53/120 (43%), Gaps = 19/120 (15%)
Query: 107 GGGCYNCGDTGHLARDCHRSNNNG---GGGGGAACYNCGDAGHLARDCNRSNNN------ 157
G CY CG GH ARDC ++ G C+ CG GH +RDC + N
Sbjct: 236 GTPCYKCGKEGHWARDCTVQSDTGPVKSTSAAGDCFKCGKPGHWSRDCTAQSGNPKYEPG 295
Query: 158 ---SGGGGAGCYNCGDTGHLARDC-NRSNNSGGGGG------GGGACYTCGGFGHLARDC 207
S CY CG GH +RDC +S+N G G CY CG GH +RDC
Sbjct: 296 QMKSSSSSGECYKCGKQGHWSRDCTGQSSNQQFQSGQAKSTSSTGDCYKCGKAGHWSRDC 355
Score = 55.8 bits (133), Expect = 3e-08
Identities = 67/268 (25%), Positives = 87/268 (32%), Gaps = 57/268 (21%)
Query: 71 GPNGAALQPTRKD----------SAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLA 120
GP AAL+ ++ D S R GG +R+ GGGG G +
Sbjct: 41 GPYMAALKGSKSDQWQQSPLNPASKSRSVAVTTGGFQRSDGGGGVAGEQDFPEKSCPCGV 100
Query: 121 RDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNR 180
C +N G Y C NR N GG G + D +
Sbjct: 101 GICLILTSNTPKNPGRKFYKCP---------NREEN----GGCGFFQWCDAVQSSGTSTT 147
Query: 181 SNNSGGGGG-----------GGGAC---------------YTCGGFGHLARDCMRGGNGG 214
++NS G G G G C Y C F +
Sbjct: 148 TSNSYGNGNDTKFPDHQCPCGAGLCRVLTAKTGENVGRQFYRCPVFEGSCGFFKWCNDNV 207
Query: 215 GGGPGSASCFRCGGIGHM-ARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSG 273
P S S + G R AK G C++CG+ GH ARDC
Sbjct: 208 VSSPTSYSVTKNSNFGDSDTRGYQNAKT-------GTPCYKCGKEGHWARDCTVQSDTGP 260
Query: 274 GGGNAGRNTCFNCGKPGHFARECIEASG 301
+ CF CGKPGH++R+C SG
Sbjct: 261 VKSTSAAGDCFKCGKPGHWSRDCTAQSG 288
Score = 50.4 bits (119), Expect = 1e-06
Identities = 34/120 (28%), Positives = 47/120 (38%), Gaps = 15/120 (12%)
Query: 43 YRTLLEGDLVEFSIATGDNDKTKAVDVTGPNGAALQP---TRKDSAPRGFGGWRGGERRN 99
Y+ EG D K+ G +P +R +A G + G+ ++
Sbjct: 240 YKCGKEGHWARDCTVQSDTGPVKSTSAAGDCFKCGKPGHWSRDCTAQSGNPKYEPGQMKS 299
Query: 100 GGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAA--------CYNCGDAGHLARDC 151
G CY CG GH +RDC ++N G A CY CG AGH +RDC
Sbjct: 300 SSSSGE----CYKCGKQGHWSRDCTGQSSNQQFQSGQAKSTSSTGDCYKCGKAGHWSRDC 355
>At1g05135 unknown protein
Length = 384
Score = 84.3 bits (207), Expect = 7e-17
Identities = 69/188 (36%), Positives = 77/188 (40%), Gaps = 21/188 (11%)
Query: 90 GGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLAR 149
GG GG GGGGGGGGGG G +GH GGG GG A G G
Sbjct: 201 GGAGGGVGGGGGGGGGGGGGGGANGGSGH-GSGFGAGGGVGGGVGGGAGGGGGGGGGGGG 259
Query: 150 DCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMR 209
N + + G GAG G G A GGGGGGGG GG GH
Sbjct: 260 GANGGSGHGSGFGAGGGVGGGVGGGA-------GGGGGGGGGGGGGVGGGSGH------G 306
Query: 210 GGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGV 269
GG G GGG G + GG G A G GG+G GG G G + G+
Sbjct: 307 GGFGAGGGLGGGAGGGLGGGG-------GAGGGGGGGLGHGGGVGGGHGGGVGIGIGIGI 359
Query: 270 AVSGGGGN 277
V GGG+
Sbjct: 360 GVGVGGGS 367
Score = 84.0 bits (206), Expect = 9e-17
Identities = 74/205 (36%), Positives = 80/205 (38%), Gaps = 45/205 (21%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHL 147
G GG GG GG GGGGGGG G +GH + GGG GG A G G
Sbjct: 161 GVGGGAGGIGGGGGAGGGGGGGVG--GGSGHGS-----GFGAGGGIGGGAGGGVGGGG-- 211
Query: 148 ARDCNRSNNNSGGGGAGCYNCGDTGH---------LARDCNRSNNSGGGGGGGGACYTCG 198
GGGG G N G +GH + GGGGGGGG G
Sbjct: 212 -------GGGGGGGGGGGAN-GGSGHGSGFGAGGGVGGGVGGGAGGGGGGGGGGGGGANG 263
Query: 199 GFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGG----GGCFR 254
G GH G G GGG G GG+G A G GGVGG GG F
Sbjct: 264 GSGH------GSGFGAGGGVG-------GGVGGGAGGGGGGGGGGGGGVGGGSGHGGGFG 310
Query: 255 CGEVGHLARDCDGGVAVSGGGGNAG 279
G G L GG+ GG G G
Sbjct: 311 AG--GGLGGGAGGGLGGGGGAGGGG 333
Score = 84.0 bits (206), Expect = 9e-17
Identities = 75/217 (34%), Positives = 76/217 (34%), Gaps = 21/217 (9%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHL 147
GFGG RG GGGGG GGG G G GGGGGG G G
Sbjct: 59 GFGGGRGSGGGIGGGGGQGGGFGAGGGVGGGAGGGLGGGGGAGGGGGGGIGGGSGHGGGF 118
Query: 148 ARDCNRSNNNSGG-GGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARD 206
GG GG G G G + G GGG GG GG G
Sbjct: 119 GAGGGVGGGAGGGIGGGGGAGGGGGGGVGGGSGHGGGFGAGGGVGGGAGGIGGGGGAGGG 178
Query: 207 CMRGGNGGGGGPGSASCFRCGG-IGHMARDCATAKGPSSGGVGGGGCFRCGE-------- 257
GG G GGG G S F GG IG A G GG GGGG G
Sbjct: 179 ---GGGGVGGGSGHGSGFGAGGGIGGGAGGGVGGGGGGGGGGGGGGGANGGSGHGSGFGA 235
Query: 258 ---VGHLARDCDGGVAVSGGGGNAGRNTCFNCGKPGH 291
VG GG GGGG G N G GH
Sbjct: 236 GGGVGGGVGGGAGGGGGGGGGGGGGAN-----GGSGH 267
Score = 82.8 bits (203), Expect = 2e-16
Identities = 73/209 (34%), Positives = 82/209 (38%), Gaps = 24/209 (11%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHL 147
G GG GG GGG GGGGGG G +GH GGG GG G G
Sbjct: 85 GVGGGAGGGLGGGGGAGGGGGGGIG-GGSGH-GGGFGAGGGVGGGAGGGIGGGGGAGGGG 142
Query: 148 ARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGG-------GACYTC-GG 199
+ + GG GAG G G A +GGGGGGG G+ + GG
Sbjct: 143 GGGVGGGSGHGGGFGAG----GGVGGGAGGIGGGGGAGGGGGGGVGGGSGHGSGFGAGGG 198
Query: 200 FGHLARDCMRGGNGGGGGPGSASCFRCGGIGH---------MARDCATAKGPSSGGVGGG 250
G A + GG GGGGG G GG GH + G GG GGG
Sbjct: 199 IGGGAGGGVGGGGGGGGGGGGGGGAN-GGSGHGSGFGAGGGVGGGVGGGAGGGGGGGGGG 257
Query: 251 GCFRCGEVGHLARDCDGGVAVSGGGGNAG 279
G G GH + GG G GG AG
Sbjct: 258 GGGANGGSGHGSGFGAGGGVGGGVGGGAG 286
Score = 81.3 bits (199), Expect = 6e-16
Identities = 72/207 (34%), Positives = 79/207 (37%), Gaps = 37/207 (17%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGG--GAACYNCGDAG 145
G GG GG GGG GGGGGG G +GH GG GG G G G
Sbjct: 123 GVGGGAGGGIGGGGGAGGGGGGGVG-GGSGHGGGFGAGGGVGGGAGGIGGGGGAGGGGGG 181
Query: 146 HLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGH--- 202
+ + GGG G G G GGGGGGGG GG GH
Sbjct: 182 GVGGGSGHGSGFGAGGGIGGGAGGGVG---------GGGGGGGGGGGGGGANGGSGHGSG 232
Query: 203 ----------LARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGC 252
+ GG GGGGG G A+ GG GH + A G GGVGGG
Sbjct: 233 FGAGGGVGGGVGGGAGGGGGGGGGGGGGAN----GGSGHGSG--FGAGGGVGGGVGGGAG 286
Query: 253 FRCGEVGHLARDCDGGVAVSGGGGNAG 279
G G GG V GG G+ G
Sbjct: 287 GGGGGGG------GGGGGVGGGSGHGG 307
Score = 79.3 bits (194), Expect = 2e-15
Identities = 72/210 (34%), Positives = 79/210 (37%), Gaps = 39/210 (18%)
Query: 87 RGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGH 146
RG GG GG R +GGG GGGGG G G + GGG GG G G
Sbjct: 54 RGGGGGFGGGRGSGGGIGGGGGQGGGFGAGGGVGGGAGGGLGGGGGAGG------GGGGG 107
Query: 147 LARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARD 206
+ GGG G G G GGG GGGG GG G
Sbjct: 108 IGGGSGHGGGFGAGGGVGGGAGGGIG-----------GGGGAGGGGG----GGVG----- 147
Query: 207 CMRGGNGGGGGPGSASCFRCGGIGHMARDC---ATAKGPSSGGVGGGGCFRC--GEVGHL 261
GG+G GGG G+ GG+G A A G GGVGGG G G +
Sbjct: 148 ---GGSGHGGGFGAG-----GGVGGGAGGIGGGGGAGGGGGGGVGGGSGHGSGFGAGGGI 199
Query: 262 ARDCDGGVAVSGGGGNAGRNTCFNCGKPGH 291
GGV GGGG G G GH
Sbjct: 200 GGGAGGGVGGGGGGGGGGGGGGGANGGSGH 229
Score = 56.6 bits (135), Expect = 2e-08
Identities = 58/178 (32%), Positives = 62/178 (34%), Gaps = 60/178 (33%)
Query: 88 GFGGWRGGERRNGG-------------GGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGG 134
G GG GG NGG GGG GGG + GGGGG
Sbjct: 251 GGGGGGGGGGANGGSGHGSGFGAGGGVGGGVGGG-----------------AGGGGGGGG 293
Query: 135 GAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGAC 194
G G +GH GG GAG G L GGG GGGG
Sbjct: 294 GGGGGVGGGSGH-----------GGGFGAG-------GGLGGGAGGGLGGGGGAGGGGG- 334
Query: 195 YTCGGFGHLARDCMRGGNGGGGGPGSASCFRCG-GIGHMARDCATAKGPSSGGVGGGG 251
GG GH GG GGG G G G G+G + G SG GGGG
Sbjct: 335 ---GGLGH------GGGVGGGHGGGVGIGIGIGIGVG-VGGGSGQGSGSGSGSGGGGG 382
Score = 53.1 bits (126), Expect = 2e-07
Identities = 53/163 (32%), Positives = 59/163 (35%), Gaps = 25/163 (15%)
Query: 119 LARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDC 178
L C G G A + D R R GGGG G G +
Sbjct: 15 LLNSCVECVLGDGSVVGPARFRDDDCRWGRRCAGRGRFGRGGGGGFGGGRGSGGGI---- 70
Query: 179 NRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCA- 237
GGGGG GG GG G A + GG G GGG G GG GH A
Sbjct: 71 ------GGGGGQGGGFGAGGGVGGGAGGGLGGGGGAGGGGGGGI---GGGSGHGGGFGAG 121
Query: 238 -TAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAG 279
G + GG+GGGG G G GG V GG G+ G
Sbjct: 122 GGVGGGAGGGIGGGG----GAGG------GGGGGVGGGSGHGG 154
>At4g36230 putative glycine-rich cell wall protein
Length = 221
Score = 82.0 bits (201), Expect = 4e-16
Identities = 79/243 (32%), Positives = 92/243 (37%), Gaps = 65/243 (26%)
Query: 14 NNVKGFGFIKPDDGGEDLFVHQSSIRSDGYRTLLEGDLVEFSIATGDNDKTKAVDVTGPN 73
N V G +K D E FV+ +S ++ VEF A D + K
Sbjct: 29 NCVNGLRDLKERDASESNFVNATSSSVLDSKSTANISTVEFENALDDQNVNK-------- 80
Query: 74 GAALQPTRKDSAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGG 133
T K + G GG GG GGGGG G GG G+ G+ + H+ N + GGG
Sbjct: 81 ------TEKCESRAG-GGGGGGGGGGGGGGGQGSGGGGGEGNGGNGKDNSHKRNKSSGGG 133
Query: 134 GGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGA 193
GG GGGG G N G R GGGGGGGG
Sbjct: 134 GG---------------------GGGGGGGGSGNGSGRG-------RGGGGGGGGGGGGG 165
Query: 194 CYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCF 253
GG G GG GGGGG G S + GG G G GG G GG F
Sbjct: 166 ----GGGG--------GGGGGGGGGGGGSDGKGGGWGF---------GFGWGGDGPGGNF 204
Query: 254 -RC 255
RC
Sbjct: 205 GRC 207
Score = 38.9 bits (89), Expect = 0.003
Identities = 27/84 (32%), Positives = 30/84 (35%), Gaps = 15/84 (17%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHL 147
G G RG GGGGGGGGGG GGGGGG + G G
Sbjct: 145 GNGSGRGRGGGGGGGGGGGGGG---------------GGGGGGGGGGGGGGGSDGKGGGW 189
Query: 148 ARDCNRSNNNSGGGGAGCYNCGDT 171
+ GG C+ G T
Sbjct: 190 GFGFGWGGDGPGGNFGRCWYPGCT 213
>At2g30560 putative glycine-rich protein
Length = 171
Score = 81.3 bits (199), Expect = 6e-16
Identities = 60/170 (35%), Positives = 70/170 (40%), Gaps = 42/170 (24%)
Query: 91 GWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARD 150
G +GG GGG GGGGGG GGGGGG A CG G
Sbjct: 2 GGKGGSGSGGGGKGGGGGGS---------------GGGRGGGGGGGAKGGCGGGG----- 41
Query: 151 CNRSNNNSGGGGAGCY----NCGDTGHLARDCNRSNNSGGG-----GGGGGACYTCGGFG 201
+ GGGG G Y + +++RD S+ GG GGGGG + GG G
Sbjct: 42 -----KSGGGGGGGGYMVAPGSNRSSYISRDNFESDPKGGSGGGGKGGGGGGGISGGGAG 96
Query: 202 HLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGG 251
+ GG GGGG G + CGG G G S GG GGGG
Sbjct: 97 --GKSGCGGGKSGGGGGGGKNGGGCGGGG------GGKGGKSGGGSGGGG 138
Score = 68.9 bits (167), Expect = 3e-12
Identities = 52/140 (37%), Positives = 59/140 (42%), Gaps = 30/140 (21%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCY----NCGDTGHLARDCHRSNNNGGGGGGAACYNCGD 143
G GG +GG G GGGGGGG Y + +++RD S+ GG GGG G
Sbjct: 29 GGGGAKGGCGGGGKSGGGGGGGGYMVAPGSNRSSYISRDNFESDPKGGSGGGGK----GG 84
Query: 144 AGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHL 203
G SGGG G CG SGGGGGGG CGG G
Sbjct: 85 GG--------GGGISGGGAGGKSGCG-----------GGKSGGGGGGGKNGGGCGGGGGG 125
Query: 204 ARDCMRGGNGGGG---GPGS 220
GG+GGGG PGS
Sbjct: 126 KGGKSGGGSGGGGYMVAPGS 145
Score = 61.6 bits (148), Expect = 5e-10
Identities = 45/136 (33%), Positives = 52/136 (38%), Gaps = 23/136 (16%)
Query: 87 RGFGGWRGGERRNGGGGGGGGGGCYNCGDTG------------------HLARDCHRSNN 128
+G GG G R GGGGGG GGC G +G +++RD S+
Sbjct: 14 KGGGGGGSGGGRGGGGGGGAKGGCGGGGKSGGGGGGGGYMVAPGSNRSSYISRDNFESDP 73
Query: 129 NGGGGGGAACYNCG---DAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSG 185
GG GGG G G SGGGG G N G G + SG
Sbjct: 74 KGGSGGGGKGGGGGGGISGGGAGGKSGCGGGKSGGGGGGGKNGGGCG--GGGGGKGGKSG 131
Query: 186 GGGGGGGACYTCGGFG 201
GG GGGG G G
Sbjct: 132 GGSGGGGYMVAPGSNG 147
Score = 36.2 bits (82), Expect = 0.022
Identities = 23/63 (36%), Positives = 28/63 (43%), Gaps = 2/63 (3%)
Query: 90 GGWRGGERRNGGG--GGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHL 147
GG GG+ GGG GGGGGGG G G +S GGGG + +
Sbjct: 92 GGGAGGKSGCGGGKSGGGGGGGKNGGGCGGGGGGKGGKSGGGSGGGGYMVAPGSNGSSTI 151
Query: 148 ARD 150
+RD
Sbjct: 152 SRD 154
Score = 35.4 bits (80), Expect = 0.038
Identities = 20/54 (37%), Positives = 23/54 (42%), Gaps = 15/54 (27%)
Query: 82 KDSAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGG 135
K G G GG +NGGG GGGGGG + +GGG GG
Sbjct: 98 KSGCGGGKSGGGGGGGKNGGGCGGGGGG---------------KGGKSGGGSGG 136
Score = 33.5 bits (75), Expect = 0.14
Identities = 24/91 (26%), Positives = 31/91 (33%), Gaps = 23/91 (25%)
Query: 209 RGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGG 268
+GG+G GGG G GG G GG G G C GG
Sbjct: 4 KGGSGSGGG-----------------------GKGGGGGGSGGGRGGGGGGGAKGGCGGG 40
Query: 269 VAVSGGGGNAGRNTCFNCGKPGHFARECIEA 299
GGGG G + + +R+ E+
Sbjct: 41 GKSGGGGGGGGYMVAPGSNRSSYISRDNFES 71
Score = 32.3 bits (72), Expect = 0.32
Identities = 18/45 (40%), Positives = 22/45 (48%), Gaps = 2/45 (4%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGH--LARDCHRSNNNG 130
G GG GG+ GGG GGGG G G ++RD S+ G
Sbjct: 118 GCGGGGGGKGGKSGGGSGGGGYMVAPGSNGSSTISRDKFESDTKG 162
>At2g05580 unknown protein
Length = 302
Score = 78.2 bits (191), Expect = 5e-15
Identities = 68/206 (33%), Positives = 81/206 (39%), Gaps = 21/206 (10%)
Query: 87 RGFGGWRGGERRNGGGGGG--GGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDA 144
+G GG +GG++ GGG GG GGGG G G + GGGGGG G
Sbjct: 110 KGGGGGQGGQKGGGGGQGGQKGGGGGGQGGQKG--GGGGGQGGQKGGGGGGQGGQKGGGG 167
Query: 145 GHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLA 204
+ GGGG G G G R + GGGGGG G GG G
Sbjct: 168 QGGQKGGGGQGGQKGGGGQG----GQKGGGGR--GQGGMKGGGGGGQGGHKGGGGGGG-- 219
Query: 205 RDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARD 264
+GG+ GGGG G + GG G G GG G GG G GH+
Sbjct: 220 ----QGGHKGGGGGGGQGGHKGGGGGGQG-----GGGHKGGGGGQGGHKGGGGGGHVGGG 270
Query: 265 CDGGVAVSGGGGNAGRNTCFNCGKPG 290
GG GGG +G G+ G
Sbjct: 271 GRGGGGGGRGGGGSGGGGGGGSGRGG 296
Score = 77.4 bits (189), Expect = 9e-15
Identities = 66/198 (33%), Positives = 76/198 (38%), Gaps = 45/198 (22%)
Query: 88 GFGGWRGGERRNGGGG------GGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNC 141
G GG +GG++ GGGG GGG GG G G + GGGG G
Sbjct: 143 GGGGGQGGQKGGGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGRGQGGMKG 202
Query: 142 GDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFG 201
G G + GGGG G G GH GGGGGGG + GG G
Sbjct: 203 GGGGG-------QGGHKGGGGGG----GQGGH----------KGGGGGGGQGGHKGGGGG 241
Query: 202 HLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHL 261
+GG GG GG GG GH+ +G GG GGGG G
Sbjct: 242 GQGGGGHKGGGGGQGGHKGG-----GGGGHVG---GGGRGGGGGGRGGGGSGGGG----- 288
Query: 262 ARDCDGGVAVSGGGGNAG 279
GG + GGGG G
Sbjct: 289 -----GGGSGRGGGGGGG 301
Score = 75.9 bits (185), Expect = 3e-14
Identities = 67/206 (32%), Positives = 76/206 (36%), Gaps = 22/206 (10%)
Query: 74 GAALQPTRKDSAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGG 133
G Q +K G GG +GG GG GGGGG G G + GGGG
Sbjct: 91 GGGGQGGQKGGGGGGQGGQKGGGGGQGGQKGGGGGQGGQKGGGGG-----GQGGQKGGGG 145
Query: 134 GGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGA 193
GG G G + GGG G G G + GGGG G G
Sbjct: 146 GGQGGQKGGGGG------GQGGQKGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGRGQGG 199
Query: 194 CYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCF 253
GG G +GG+ GGGG G + GG G KG GG GGGG
Sbjct: 200 MKGGGGGG-------QGGHKGGGGGGGQGGHKGGGGG---GGQGGHKGGGGGGQGGGG-H 248
Query: 254 RCGEVGHLARDCDGGVAVSGGGGNAG 279
+ G G GG GGGG G
Sbjct: 249 KGGGGGQGGHKGGGGGGHVGGGGRGG 274
Score = 74.3 bits (181), Expect = 7e-14
Identities = 64/192 (33%), Positives = 71/192 (36%), Gaps = 25/192 (13%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHL 147
G G +GG++ GGGGGG GG G G GGGGG G G
Sbjct: 79 GIGRGQGGQK---GGGGGGQGGQKGGGGGG--------QGGQKGGGGGQGGQKGGGGG-- 125
Query: 148 ARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDC 207
GGGG G G G + GGGGGG G GG G
Sbjct: 126 ----QGGQKGGGGGGQGGQKGGGGG------GQGGQKGGGGGGQGGQKGGGGQGGQKGGG 175
Query: 208 MRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDG 267
+GG GGGG G G G M +G GG GGGG + G G G
Sbjct: 176 GQGGQKGGGGQGGQKGGGGRGQGGMKGGGGGGQGGHKGGGGGGG--QGGHKGGGGGGGQG 233
Query: 268 GVAVSGGGGNAG 279
G GGGG G
Sbjct: 234 GHKGGGGGGQGG 245
Score = 71.2 bits (173), Expect = 6e-13
Identities = 64/169 (37%), Positives = 68/169 (39%), Gaps = 52/169 (30%)
Query: 71 GPNGAALQPTRKDSAPRGFGGWRGGERRNGGGG-----GGGGGGCYNCGDTGHLARDCHR 125
G G Q +K RG GG +GG GGGG GGGGGG G GH
Sbjct: 179 GQKGGGGQGGQKGGGGRGQGGMKGG----GGGGQGGHKGGGGGG----GQGGH------- 223
Query: 126 SNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSG 185
GGGGGG G GH GGGG G G GH + + G
Sbjct: 224 ---KGGGGGG------GQGGH-----------KGGGGGGQ---GGGGHKGGGGGQGGHKG 260
Query: 186 GGGGG--GGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHM 232
GGGGG GG GG G RGG G GGG G S GG G M
Sbjct: 261 GGGGGHVGGGGRGGGGGG-------RGGGGSGGGGGGGSGRGGGGGGGM 302
Score = 71.2 bits (173), Expect = 6e-13
Identities = 62/180 (34%), Positives = 70/180 (38%), Gaps = 24/180 (13%)
Query: 74 GAALQPTRKDSAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNN--GG 131
G Q +K G GG +GG + G GGGG GG G G R GG
Sbjct: 144 GGGGQGGQKGGGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGRGQGGMKGG 203
Query: 132 GGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGG 191
GGGG + G G + + GGGG G G G + GGGGG G
Sbjct: 204 GGGGQGGHKGGGGGG-----GQGGHKGGGGGGG--QGGHKGGGGGGQGGGGHKGGGGGQG 256
Query: 192 GACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGG 251
G GG GH+ GG GG GG GS GG G G GG GGGG
Sbjct: 257 GH-KGGGGGGHVGGGGRGGGGGGRGGGGSGG----GGGG----------GSGRGGGGGGG 301
>At5g52380 unknown protein
Length = 268
Score = 77.8 bits (190), Expect = 7e-15
Identities = 42/130 (32%), Positives = 57/130 (43%), Gaps = 8/130 (6%)
Query: 107 GGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCY 166
G GC+ C H+A+ C + C C GH ++C NN S CY
Sbjct: 73 GEGCFICHSKTHIAKLCPEKSE---WERNKICLQCRRRGHSLKNCPEKNNESSEKKL-CY 128
Query: 167 NCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRC 226
NCGDTGH C GG +C+ C G GH++++C N G P C C
Sbjct: 129 NCGDTGHSLSHCPYPMED--GGTKFASCFICKGQGHISKNCPE--NKHGIYPMGGCCKVC 184
Query: 227 GGIGHMARDC 236
G + H+ +DC
Sbjct: 185 GSVAHLVKDC 194
Score = 72.4 bits (176), Expect = 3e-13
Identities = 38/115 (33%), Positives = 47/115 (40%), Gaps = 4/115 (3%)
Query: 110 CYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCG 169
C C GH ++C NN CYNCGD GH C + G A C+ C
Sbjct: 101 CLQCRRRGHSLKNCPEKNNESSEK--KLCYNCGDTGHSLSHCPYPMEDGGTKFASCFICK 158
Query: 170 DTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCF 224
GH++++C N G GG C CG HL +DC N P S F
Sbjct: 159 GQGHISKNC--PENKHGIYPMGGCCKVCGSVAHLVKDCPDKFNQESAQPKKTSRF 211
Score = 68.6 bits (166), Expect = 4e-12
Identities = 43/132 (32%), Positives = 55/132 (41%), Gaps = 11/132 (8%)
Query: 135 GAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGAC 194
G C+ C H+A+ C + C C GH ++C NN C
Sbjct: 73 GEGCFICHSKTHIAKLC--PEKSEWERNKICLQCRRRGHSLKNCPEKNNESSEKK---LC 127
Query: 195 YTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFR 254
Y CG GH C GG ASCF C G GH++++C K G GGC +
Sbjct: 128 YNCGDTGHSLSHCPYPMEDGG--TKFASCFICKGQGHISKNCPENK---HGIYPMGGCCK 182
Query: 255 -CGEVGHLARDC 265
CG V HL +DC
Sbjct: 183 VCGSVAHLVKDC 194
Score = 43.5 bits (101), Expect = 1e-04
Identities = 29/113 (25%), Positives = 42/113 (36%), Gaps = 21/113 (18%)
Query: 191 GGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGG 250
G C+ C H+A+ C + C +C GH ++C SS
Sbjct: 73 GEGCFICHSKTHIAKLCPEKSEW----ERNKICLQCRRRGHSLKNCPEKNNESSEKKL-- 126
Query: 251 GCFRCGEVGHLARDC-----DGGVAVSGGGGNAGRNTCFNCGKPGHFARECIE 298
C+ CG+ GH C DGG + +CF C GH ++ C E
Sbjct: 127 -CYNCGDTGHSLSHCPYPMEDGGTKFA---------SCFICKGQGHISKNCPE 169
Score = 38.9 bits (89), Expect = 0.003
Identities = 17/49 (34%), Positives = 24/49 (48%), Gaps = 1/49 (2%)
Query: 103 GGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDC 151
GG C+ C GH++++C N +G G C CG HL +DC
Sbjct: 147 GGTKFASCFICKGQGHISKNC-PENKHGIYPMGGCCKVCGSVAHLVKDC 194
Score = 35.0 bits (79), Expect = 0.050
Identities = 21/82 (25%), Positives = 31/82 (37%), Gaps = 10/82 (12%)
Query: 215 GGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGG 274
G PG CF C H+A+ C + C +C GH ++C
Sbjct: 69 GMKPGEG-CFICHSKTHIAKLCPEKSEWERNKI----CLQCRRRGHSLKNCP-----EKN 118
Query: 275 GGNAGRNTCFNCGKPGHFAREC 296
++ + C+NCG GH C
Sbjct: 119 NESSEKKLCYNCGDTGHSLSHC 140
>At3g17050 unknown protein
Length = 349
Score = 77.8 bits (190), Expect = 7e-15
Identities = 74/200 (37%), Positives = 74/200 (37%), Gaps = 31/200 (15%)
Query: 88 GFGGWRGGERRNGGGGGGGGG--GCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAG 145
G GG GG G GGG GGG G G G L G GGGG A G G
Sbjct: 166 GIGGGAGGGAGGGLGGGHGGGIGGGAGGGSGGGLGGGIGGGAGGGAGGGGGA----GGGG 221
Query: 146 HLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLAR 205
L GGG G G TG GGG GGG GGFG A
Sbjct: 222 GLGGGHGGGFGGGAGGGLGGGAGGGTG---------GGFGGGAGGGAGGGAGGGFGGGAG 272
Query: 206 DCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGG------GCFRCGEVG 259
GG GGG G G A GG G A G GGVGGG G F G G
Sbjct: 273 GGAGGGFGGGAG-GGAGGGAGGGFG------GGAGGGHGGGVGGGFGGGSGGGFGGGAGG 325
Query: 260 HLARDCDGGVAVSGGGGNAG 279
GG GGGG AG
Sbjct: 326 GAGGGAGGGF---GGGGGAG 342
Score = 77.8 bits (190), Expect = 7e-15
Identities = 65/192 (33%), Positives = 70/192 (35%), Gaps = 22/192 (11%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHL 147
G GG GG GGG GGG GG G G L G GGG G G
Sbjct: 148 GAGGGAGGGGGLGGGHGGGIGGGAGGGAGGGLGGGHGGGIGGGAGGGSGGGLGGGIGGGA 207
Query: 148 ARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDC 207
GGGG G + G G +GGG GGG T GGFG
Sbjct: 208 GGGAGGGGGAGGGGGLGGGHGGGFG---------GGAGGGLGGGAGGGTGGGFG------ 252
Query: 208 MRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDG 267
GG GGG G G+ F G G G +GG GGG GH G
Sbjct: 253 --GGAGGGAGGGAGGGFGGGAGGGAGGGFGGGAGGGAGGGAGGGFGGGAGGGH-----GG 305
Query: 268 GVAVSGGGGNAG 279
GV GGG+ G
Sbjct: 306 GVGGGFGGGSGG 317
Score = 77.4 bits (189), Expect = 9e-15
Identities = 69/211 (32%), Positives = 77/211 (35%), Gaps = 10/211 (4%)
Query: 71 GPNGAALQPTRKDSAPRGFGGWRGGERRNGGGGGGGG--GGCYNCGDTGHLARDCHRSNN 128
G +G + A G GG GG G GGG GG GG G G
Sbjct: 103 GGHGGGIGGGAGGGAGGGLGGGHGGGIGGGAGGGSGGGLGGGIGGGAGGGAGGGGGLGGG 162
Query: 129 NGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGG 188
+GGG GG A G G L GGG+G G G A +GGGG
Sbjct: 163 HGGGIGGGA--GGGAGGGLGGGHGGGIGGGAGGGSGGGLGGGIGGGA-----GGGAGGGG 215
Query: 189 GGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVG 248
G GG GG G GG GGG G G+ F G G G +GG G
Sbjct: 216 GAGGGGGLGGGHGGGFGGGAGGGLGGGAGGGTGGGFGGGAGGGAGGGAGGGFGGGAGG-G 274
Query: 249 GGGCFRCGEVGHLARDCDGGVAVSGGGGNAG 279
GG F G G GG GGG+ G
Sbjct: 275 AGGGFGGGAGGGAGGGAGGGFGGGAGGGHGG 305
Score = 77.0 bits (188), Expect = 1e-14
Identities = 69/200 (34%), Positives = 71/200 (35%), Gaps = 10/200 (5%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHL 147
G GG GG G GGGGG GG + G G G GGG G G L
Sbjct: 140 GLGGGIGGGAGGGAGGGGGLGGGHGGGIGGGAGGGAGGGLGGGHGGGIGGGAGGGSGGGL 199
Query: 148 ARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDC 207
GGG G G G +GGG GGG T GGFG A
Sbjct: 200 GGGIGGGAGGGAGGGGGAGGGGGLGG-GHGGGFGGGAGGGLGGGAGGGTGGGFGGGAGGG 258
Query: 208 MRGGNGGGGGPGSASCFRCGGIGHMARDCA--TAKGPSSGGVGG------GGCFRCGEVG 259
GG GGG G A GG G A A A G GG GG GG F G G
Sbjct: 259 A-GGGAGGGFGGGAGGGAGGGFGGGAGGGAGGGAGGGFGGGAGGGHGGGVGGGFGGGSGG 317
Query: 260 HLARDCDGGVAVSGGGGNAG 279
GG GGG G
Sbjct: 318 GFGGGAGGGAGGGAGGGFGG 337
Score = 75.9 bits (185), Expect = 3e-14
Identities = 68/198 (34%), Positives = 79/198 (39%), Gaps = 25/198 (12%)
Query: 87 RGFGGW--RGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDA 144
+GFGG GG G GGG GGG G G + G GGG G
Sbjct: 47 KGFGGGIGAGGGFGGGAGGGAGGGLGGGAGGGGGIGGGAGGGAGGGLGGGAGGGLGGGHG 106
Query: 145 GHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSG-GGGGGGGACYTCGGFGHL 203
G + +GGG G G G + + G GGG GGGA GG G L
Sbjct: 107 GGI-------GGGAGGGAGGGLGGGHGGGIGGGAGGGSGGGLGGGIGGGAGGGAGGGGGL 159
Query: 204 ARDCMRGGNGG--GGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHL 261
GG+GG GGG G + GG GH A G S GG+GGG +G
Sbjct: 160 G-----GGHGGGIGGGAGGGAGGGLGG-GHGGGIGGGAGGGSGGGLGGG-------IGGG 206
Query: 262 ARDCDGGVAVSGGGGNAG 279
A GG +GGGG G
Sbjct: 207 AGGGAGGGGGAGGGGGLG 224
Score = 73.9 bits (180), Expect = 1e-13
Identities = 66/199 (33%), Positives = 73/199 (36%), Gaps = 14/199 (7%)
Query: 88 GFGGWRGGERRNGGGGG----GGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGD 143
GFGG GG G GGG GGGG G G G GGG G
Sbjct: 58 GFGGGAGG----GAGGGLGGGAGGGGGIGGGAGGGAGGGLGGGAGGGLGGGHGGGIGGGA 113
Query: 144 AGHLARDCNRSNNNSGGGGAGCYNCGDT-GHLARDCNRSNNSGGGGGGGGACYTCGGFGH 202
G + GGGAG + G G + GGG GGG GG G
Sbjct: 114 GGGAGGGLGGGHGGGIGGGAGGGSGGGLGGGIGGGAGGGAGGGGGLGGGHGGGIGGGAGG 173
Query: 203 LARDCMRGGNGG--GGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGH 260
A + GG+GG GGG G S GG+G A GG GGGG G G
Sbjct: 174 GAGGGLGGGHGGGIGGGAGGGS---GGGLGGGIGGGAGGGAGGGGGAGGGGGLGGGHGGG 230
Query: 261 LARDCDGGVAVSGGGGNAG 279
GG+ GGG G
Sbjct: 231 FGGGAGGGLGGGAGGGTGG 249
Score = 72.0 bits (175), Expect = 4e-13
Identities = 61/182 (33%), Positives = 68/182 (36%), Gaps = 30/182 (16%)
Query: 100 GGGGGGGGGGCYNCGD--TGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNN 157
GGGGGGG GG + G G + GGG GG G G +
Sbjct: 32 GGGGGGGLGGGFGGGKGFGGGIGAGGGFGGGAGGGAGGGLGGGAGGGGGI-------GGG 84
Query: 158 SGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGG 217
+GGG G G G L G GGG GG G GH GG GGG G
Sbjct: 85 AGGGAGGGLGGGAGGGLGGGHGGGIGGGAGGGAGGGL----GGGH------GGGIGGGAG 134
Query: 218 PGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGGGGN 277
GS GGIG G + GG GGGG G G + GG GGG+
Sbjct: 135 GGSGGGLG-GGIG----------GGAGGGAGGGGGLGGGHGGGIGGGAGGGAGGGLGGGH 183
Query: 278 AG 279
G
Sbjct: 184 GG 185
Score = 58.9 bits (141), Expect = 3e-09
Identities = 57/169 (33%), Positives = 62/169 (35%), Gaps = 56/169 (33%)
Query: 88 GFGGWRGGERRNGGGGG------GGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNC 141
GFGG GG G GGG GG GG G G GG GGGA
Sbjct: 230 GFGGGAGGGLGGGAGGGTGGGFGGGAGGGAGGGAGGGF---------GGGAGGGAGGGFG 280
Query: 142 GDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFG 201
G AG +GGG G + G G GGG GGG + GGFG
Sbjct: 281 GGAG----------GGAGGGAGGGFGGGAGG----------GHGGGVGGGFGGGSGGGFG 320
Query: 202 HLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGG 250
GG GGG G G+ GG G G + GG GGG
Sbjct: 321 --------GGAGGGAGGGAG-----GGFG--------GGGGAGGGFGGG 348
Score = 52.8 bits (125), Expect = 2e-07
Identities = 46/132 (34%), Positives = 52/132 (38%), Gaps = 28/132 (21%)
Query: 150 DCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMR 209
+C R + GGG G G G GGG G GG GGFG A
Sbjct: 22 ECRRFEKETLGGGGG----GGLG---------GGFGGGKGFGGGIGAGGGFGGGAGGGAG 68
Query: 210 GGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGG--GGCFRCGEVGHLARDCDG 267
GG GGG G G GGIG A G + GG+GG GG G G + G
Sbjct: 69 GGLGGGAGGG-------GGIG------GGAGGGAGGGLGGGAGGGLGGGHGGGIGGGAGG 115
Query: 268 GVAVSGGGGNAG 279
G GGG+ G
Sbjct: 116 GAGGGLGGGHGG 127
>At5g49350 unknown protein
Length = 302
Score = 74.3 bits (181), Expect = 7e-14
Identities = 76/236 (32%), Positives = 84/236 (35%), Gaps = 54/236 (22%)
Query: 83 DSAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCG 142
DS G GG GG GG G GGGG G TG + GGG GG G
Sbjct: 35 DSEVGGEGGGIGG----GGTGFGGGGTGVGGGGTGFGGGGLGAGGSGGGGAGGLGSGGTG 90
Query: 143 DAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCG--GF 200
G SGGGGAG G TG GGGG GGG + G GF
Sbjct: 91 VGG----GGGLGAGGSGGGGAGGLGGGGTG-----------VGGGGTGGGTGFNGGGTGF 135
Query: 201 GHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGG--------- 251
G + G GG GG G GG G G GG+GGGG
Sbjct: 136 GPFGGGGLGTGGGGAGGLGGG-----GGNG----------GFGGGGLGGGGLGGEDPPEI 180
Query: 252 ---CFRCGEVGHLARDCDGGVAVSGGGG---NAGRNTCF---NCGKPGHFARECIE 298
C H+ R+CD ++ G GR F C H A CIE
Sbjct: 181 VAKALECLNEKHIYRECDESWRLTLNGDLNIPLGRTEEFCEGPCFSETHLALNCIE 236
Score = 50.4 bits (119), Expect = 1e-06
Identities = 46/142 (32%), Positives = 54/142 (37%), Gaps = 41/142 (28%)
Query: 152 NRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGG---GGGGACYTCGGFGHLARDCM 208
+ +++ GG G G TG GGGG GGGG + GG G
Sbjct: 31 SEEDDSEVGGEGGGIGGGGTGF-----------GGGGTGVGGGGTGFGGGGLG------- 72
Query: 209 RGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGG 268
GG+GGGG GG+G G +GG GGGG G GG
Sbjct: 73 AGGSGGGG---------AGGLGSGGTGVGGGGGLGAGGSGGGGAGGLG---------GGG 114
Query: 269 VAVSGGGGNAGRNTCFNCGKPG 290
V GGG G T FN G G
Sbjct: 115 TGV--GGGGTGGGTGFNGGGTG 134
>At5g19090 unknown protein
Length = 587
Score = 73.6 bits (179), Expect = 1e-13
Identities = 70/206 (33%), Positives = 80/206 (37%), Gaps = 45/206 (21%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHL 147
G GG G ++ G GGGGG G N G G NGG GGG + G +
Sbjct: 313 GGGGGGPGGKKGGPGGGGGNMGNQNQGGGG----------KNGGKGGGGHPLD----GKM 358
Query: 148 ARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDC 207
N N GGGG N G G GGGGGGGG GG R
Sbjct: 359 GGGGGGPNGNKGGGGVQ-MNGGPNG--------GKKGGGGGGGGGGGPMSGGLPPGFRP- 408
Query: 208 MRGGNGGGGGPGSASC--------------FRCGGIGHMARDCATAKG---PSSGGVGGG 250
M GG GGGGGP S S GG G M+ + +G GG GGG
Sbjct: 409 MGGGGGGGGGPQSMSMPMGGAMGGPMGSLPQMGGGPGPMSNNMQAVQGLPAMGPGGGGGG 468
Query: 251 GCFRCGEVGHLARDCDGGVAVSGGGG 276
G G+ G V+ +GGGG
Sbjct: 469 GPSAEAPPGYF----QGQVSGNGGGG 490
Score = 48.9 bits (115), Expect = 3e-06
Identities = 55/182 (30%), Positives = 61/182 (33%), Gaps = 54/182 (29%)
Query: 98 RNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNN 157
+NGGG G GG G + GGGGGG
Sbjct: 289 KNGGGPGPAGGKIEGKGMPFPVQM--------GGGGGGPG------------------GK 322
Query: 158 SGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGG 217
GG G G N G+ N GGGG GG GG GH M GGGGG
Sbjct: 323 KGGPGGGGGNMGN-----------QNQGGGGKNGGK----GGGGHPLDGKM---GGGGGG 364
Query: 218 PGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGGGGN 277
P GG G G +GG GGG G G ++ G GGGG
Sbjct: 365 PNGNK----GGGG------VQMNGGPNGGKKGGGGGGGGGGGPMSGGLPPGFRPMGGGGG 414
Query: 278 AG 279
G
Sbjct: 415 GG 416
Score = 41.6 bits (96), Expect = 5e-04
Identities = 39/112 (34%), Positives = 45/112 (39%), Gaps = 11/112 (9%)
Query: 179 NRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCAT 238
N N GG G GG G + GG GGGGPG GG G+M
Sbjct: 286 NAHKNGGGPGPAGGKIEGKG----MPFPVQMGG--GGGGPGGKKGGPGGGGGNMGNQ-NQ 338
Query: 239 AKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAGRNTCFNCGKPG 290
G +GG GGGG G++G GG + GGG N N GK G
Sbjct: 339 GGGGKNGGKGGGGHPLDGKMG----GGGGGPNGNKGGGGVQMNGGPNGGKKG 386
Score = 40.0 bits (92), Expect = 0.002
Identities = 37/111 (33%), Positives = 42/111 (37%), Gaps = 21/111 (18%)
Query: 185 GGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSS 244
GGGGG GG GG G + +GG G GG G G GH P
Sbjct: 314 GGGGGPGGKKGGPGGGGGNMGNQNQGGGGKNGGKG--------GGGH----------PLD 355
Query: 245 GGVGGGGCFRCGEVGHLARDCDGGV---AVSGGGGNAGRNTCFNCGKPGHF 292
G +GGGG G G +GG GGGG G + G P F
Sbjct: 356 GKMGGGGGGPNGNKGGGGVQMNGGPNGGKKGGGGGGGGGGGPMSGGLPPGF 406
Score = 37.7 bits (86), Expect = 0.008
Identities = 24/71 (33%), Positives = 26/71 (35%), Gaps = 26/71 (36%)
Query: 148 ARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDC 207
A + GGGG G N N+ GGGGGGGG
Sbjct: 96 AMQIDHGGKGGGGGGGGPANN----------NKGQKIGGGGGGGGG-------------- 131
Query: 208 MRGGNGGGGGP 218
GG GGGGGP
Sbjct: 132 --GGGGGGGGP 140
Score = 35.0 bits (79), Expect = 0.050
Identities = 23/60 (38%), Positives = 24/60 (39%), Gaps = 27/60 (45%)
Query: 83 DSAPRGFGGWRGGERRN-------GGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGG 135
D +G GG GG N GGGGGGGGGG GGGGGG
Sbjct: 100 DHGGKGGGGGGGGPANNNKGQKIGGGGGGGGGGG--------------------GGGGGG 139
Score = 31.6 bits (70), Expect = 0.55
Identities = 17/38 (44%), Positives = 21/38 (54%), Gaps = 5/38 (13%)
Query: 182 NNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPG 219
++ G GGGGGG GG + + GG GGGGG G
Sbjct: 100 DHGGKGGGGGG-----GGPANNNKGQKIGGGGGGGGGG 132
Score = 31.6 bits (70), Expect = 0.55
Identities = 22/68 (32%), Positives = 23/68 (33%), Gaps = 29/68 (42%)
Query: 124 HRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNN 183
H GGGGGG A N G + GGGG G
Sbjct: 101 HGGKGGGGGGGGPANNNKGQ---------KIGGGGGGGGGG------------------- 132
Query: 184 SGGGGGGG 191
GGGGGGG
Sbjct: 133 -GGGGGGG 139
Score = 30.8 bits (68), Expect = 0.94
Identities = 22/68 (32%), Positives = 25/68 (36%), Gaps = 32/68 (47%)
Query: 209 RGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGG 268
+GG GGGGGP + + KG GG GGGG GG
Sbjct: 104 KGGGGGGGGPANNN-----------------KGQKIGGGGGGG---------------GG 131
Query: 269 VAVSGGGG 276
GGGG
Sbjct: 132 GGGGGGGG 139
Score = 29.3 bits (64), Expect = 2.7
Identities = 18/46 (39%), Positives = 19/46 (41%), Gaps = 9/46 (19%)
Query: 100 GGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAG 145
G GGGGGGGG N + GGGGGG G G
Sbjct: 103 GKGGGGGGGGPANNNKGQKIG---------GGGGGGGGGGGGGGGG 139
>At5g40490 ribonucleoprotein -like
Length = 423
Score = 70.9 bits (172), Expect = 8e-13
Identities = 84/281 (29%), Positives = 101/281 (35%), Gaps = 60/281 (21%)
Query: 17 KGFGFIKPDDGGEDLFVHQSSIRSDGYRTLLEGDLVEFSIATGDNDKTKAVDVTGPNGAA 76
+GFGF+ + ED+ H L +G+ +E S G + K + PN
Sbjct: 171 RGFGFVTYES--EDMVDH----------LLAKGNRIELS---GTQVEIKKAEPKKPNSVT 215
Query: 77 LQPTRKDSAPRGFGGWRGGERRNGGGGGGGG-------GGCYNCGDTGHLARDCHRSNNN 129
R + FGG G G GGG GG GG Y G +G
Sbjct: 216 TPSKRFGDSRSNFGGGYGDGYGGGHGGGYGGPGGPYKSGGGYGGGRSGGYGGYGGEFGGY 275
Query: 130 GGGG--GGAACYN-------CGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNR 180
GGGG GG Y G G NR + GGGG GD
Sbjct: 276 GGGGYGGGVGPYRGEPALGYSGRYGGGGGGYNRGGYSMGGGGGYGGGPGDM------YGG 329
Query: 181 SNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAK 240
S GGG GG + GG+G GG GG GG G +R GG
Sbjct: 330 SYGEPGGGYGGPSGSYGGGYGSSGIGGYGGGMGGAGGGG----YRGGG------------ 373
Query: 241 GPSSGGVGGGG--CFRCGEVGHLARDCDGGVAVSGGGGNAG 279
G GGVGGGG + G G+ GG GGGG G
Sbjct: 374 GYDMGGVGGGGAGGYGAGGGGN-----GGGSFYGGGGGRGG 409
Score = 54.7 bits (130), Expect = 6e-08
Identities = 49/147 (33%), Positives = 55/147 (37%), Gaps = 36/147 (24%)
Query: 88 GFGGW-RGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGH 146
G GG+ RGG GGGG GGG G G G GG GG + Y G+
Sbjct: 302 GGGGYNRGGYSMGGGGGYGGGPGDMYGGSYGE---------PGGGYGGPSGSYG---GGY 349
Query: 147 LARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARD 206
+ GG G G Y G + GG GGGG Y GG
Sbjct: 350 GSSGIGGYGGGMGGAGGGGYRGGG----------GYDMGGVGGGGAGGYGAGG------- 392
Query: 207 CMRGGNGGG---GGPGSASCFRCGGIG 230
GGNGGG GG G + GG G
Sbjct: 393 ---GGNGGGSFYGGGGGRGGYGGGGSG 416
Score = 43.1 bits (100), Expect = 2e-04
Identities = 28/80 (35%), Positives = 33/80 (41%), Gaps = 25/80 (31%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHL 147
G GG+RGG + GG GGGG G Y G GGG GG + Y G
Sbjct: 365 GGGGYRGGGGYDMGGVGGGGAGGYGAG---------------GGGNGGGSFYGGGG---- 405
Query: 148 ARDCNRSNNNSGGGGAGCYN 167
GGGG+G Y+
Sbjct: 406 ------GRGGYGGGGSGRYH 419
Score = 33.9 bits (76), Expect = 0.11
Identities = 22/59 (37%), Positives = 26/59 (43%), Gaps = 12/59 (20%)
Query: 87 RGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAG 145
RG GG+ G GG GG G GG N G + + GGGGG Y G +G
Sbjct: 370 RGGGGYDMGGVGGGGAGGYGAGGGGNGGGSFY------------GGGGGRGGYGGGGSG 416
>At3g23450 hypothetical protein
Length = 452
Score = 70.9 bits (172), Expect = 8e-13
Identities = 65/197 (32%), Positives = 73/197 (36%), Gaps = 32/197 (16%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHL 147
G GG G GGG G GGG G G + + GGGGG G G +
Sbjct: 279 GIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGGG------FGKGGGI 332
Query: 148 ARDCNRSNNNSGGGGAG-----CYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGH 202
+ GGGG G G G + + GGG GGGG GGFG
Sbjct: 333 GGGIGKGGGIGGGGGFGKGGGIGGGIGKGGGIGGGFGKGGGIGGGIGGGGGFGGGGGFGK 392
Query: 203 LARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLA 262
G G GGG G F GG KG GG+GGGG F G+ G
Sbjct: 393 --------GGGIGGGIGKGGGFGGGG--------GFGKG---GGIGGGGGF--GKGGGFG 431
Query: 263 RDCDGGVAVSGGGGNAG 279
GG GGGG G
Sbjct: 432 GGGFGGGGGGGGGGGGG 448
Score = 69.7 bits (169), Expect = 2e-12
Identities = 65/194 (33%), Positives = 75/194 (38%), Gaps = 35/194 (18%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHL 147
G GG GG GGGGG GGGG + G G + GGG GG G G +
Sbjct: 77 GGGGGGGGGGGGGGGGGFGGGGGFGGGHGGGV------GGGVGGGHGGGVGGGFGKGGGI 130
Query: 148 ARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDC 207
+ GG G G G G + + G GGG GG GG G
Sbjct: 131 GGGIGKGGGVGGGIGKG---GGIGGGIGKGGGVGGGIGKGGGIGGGIGKGGGIG------ 181
Query: 208 MRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDG 267
GG G GGG G GGIG + G GG GGG + G VG G
Sbjct: 182 --GGIGKGGGIG-------GGIG---KGGGIGGGIGKGGGVGGGFGKGGGVG-------G 222
Query: 268 GVAVSGG-GGNAGR 280
G+ GG GG G+
Sbjct: 223 GIGKGGGVGGGFGK 236
Score = 68.9 bits (167), Expect = 3e-12
Identities = 70/233 (30%), Positives = 89/233 (38%), Gaps = 40/233 (17%)
Query: 50 DLVEFSIATGDNDKTKAVDVTGPNGAALQPTRKDSAPRGFGGWRGGERRNGGGGGGGGGG 109
++V +++G ++ K + V G G A G GG GG G GGG GGGG
Sbjct: 22 NVVARDVSSGRDEDEKTL-VGGGKGGGFGGGFGGGAGGGVGGGAGGGFGGGAGGGFGGGG 80
Query: 110 CYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCG 169
G G GGG GG + G G + GGG G + G
Sbjct: 81 GGGGGGGG----------GGGGGFGGGGGFGGGHGGGV-----------GGGVGGGHGGG 119
Query: 170 DTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGI 229
G + G GGG GG GG G GG G GGG G + GGI
Sbjct: 120 VGGGFGKGGGIGGGIGKGGGVGGGIGKGGGIG--------GGIGKGGGVG-GGIGKGGGI 170
Query: 230 -GHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGG-GGNAGR 280
G + + G GG GGG + G +G GG+ GG GG G+
Sbjct: 171 GGGIGKGGGIGGGIGKGGGIGGGIGKGGGIG-------GGIGKGGGVGGGFGK 216
Score = 64.3 bits (155), Expect = 8e-11
Identities = 68/207 (32%), Positives = 78/207 (36%), Gaps = 21/207 (10%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHL 147
G GG G GGG G GGG G G + GGG GG G G +
Sbjct: 219 GVGGGIGKGGGVGGGFGKGGGVGGGIGKGGGIGGGI----GKGGGIGGGIGKGGGIGGGI 274
Query: 148 ARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDC 207
+ GGG G G G + + GGG G GG GGFG
Sbjct: 275 GKGGGIGGGIGKGGGIG-GGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGGGFG--KGGG 331
Query: 208 MRGGNGGGGGPGSASCF-RCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCD 266
+ GG G GGG G F + GGIG KG GG+GGG G+ G +
Sbjct: 332 IGGGIGKGGGIGGGGGFGKGGGIGG-----GIGKG---GGIGGG----FGKGGGIGGGIG 379
Query: 267 GGVAVSGGGG-NAGRNTCFNCGKPGHF 292
GG GGGG G GK G F
Sbjct: 380 GGGGFGGGGGFGKGGGIGGGIGKGGGF 406
Score = 63.9 bits (154), Expect = 1e-10
Identities = 63/195 (32%), Positives = 74/195 (37%), Gaps = 24/195 (12%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGG--GGGGAACYNCGDAG 145
G GG G GGG G GGG G G + + GG G GG G G
Sbjct: 199 GIGGGIGKGGGVGGGFGKGGGVGGGIGKGGGVGGGFGKGGGVGGGIGKGGGIGGGIGKGG 258
Query: 146 HLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLAR 205
+ + GG G G G G + + G GGG GG GG G
Sbjct: 259 GIGGGIGKGGGIGGGIGKG---GGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIG---- 311
Query: 206 DCMRGGNGGGGGPGSASCF-RCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARD 264
GG G GGG G F + GGIG KG GG+GGGG F G+ G +
Sbjct: 312 ----GGIGKGGGIGGGGGFGKGGGIGG-----GIGKG---GGIGGGGGF--GKGGGIGGG 357
Query: 265 CDGGVAVSGGGGNAG 279
G + GG G G
Sbjct: 358 IGKGGGIGGGFGKGG 372
Score = 63.2 bits (152), Expect = 2e-10
Identities = 67/206 (32%), Positives = 75/206 (35%), Gaps = 36/206 (17%)
Query: 90 GGWRGGERRNGGGGGG---GGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGH 146
GG GG + GG GGG GGG G G + + GGGGG G G
Sbjct: 278 GGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGGG------FGKGGG 331
Query: 147 LARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARD 206
+ + GGGG G G G G GGG GG GG G
Sbjct: 332 IGGGIGKGGGIGGGGGFG--KGGGIG---------GGIGKGGGIGGGFGKGGGIG----- 375
Query: 207 CMRGGNGGGGGPGSASCF-RCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDC 265
GG GGGGG G F + GGIG G G GGGG + G +G
Sbjct: 376 ---GGIGGGGGFGGGGGFGKGGGIGG-------GIGKGGGFGGGGGFGKGGGIGGGGGFG 425
Query: 266 DGGVAVSGGGGNAGRNTCFNCGKPGH 291
GG GG G G G GH
Sbjct: 426 KGGGFGGGGFGGGGGGGGGGGGGIGH 451
Score = 60.8 bits (146), Expect = 8e-10
Identities = 50/147 (34%), Positives = 55/147 (37%), Gaps = 3/147 (2%)
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHL 147
G GG GG GGG GGGGG G G + + G G GG G G +
Sbjct: 305 GKGGGIGGGIGKGGGIGGGGGFGKGGGIGGGIGKGGGIGGGGGFGKGGGIGGGIGKGGGI 364
Query: 148 ARDCNRSNNNSGG-GGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARD 206
+ GG GG G + G GGG GGGG GG G
Sbjct: 365 GGGFGKGGGIGGGIGGGGGFGGGGGFGKGGGIGGGIGKGGGFGGGGGFGKGGGIGGGGGF 424
Query: 207 CMRGGNGGGG--GPGSASCFRCGGIGH 231
GG GGGG G G GGIGH
Sbjct: 425 GKGGGFGGGGFGGGGGGGGGGGGGIGH 451
Score = 58.9 bits (141), Expect = 3e-09
Identities = 61/195 (31%), Positives = 72/195 (36%), Gaps = 33/195 (16%)
Query: 90 GGWRGGERRNGGGGGG---GGGGCYNCGDTGHLARDCHRSNNNGGG--GGGAACYNCGDA 144
GG GG + GG GGG GGG G G + + GGG GG G
Sbjct: 158 GGVGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGVGGGFGKG 217
Query: 145 GHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLA 204
G + + GG G G G G + + G GGG GG GG G
Sbjct: 218 GGVGGGIGKGGGVGGGFGKGG---GVGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIG--- 271
Query: 205 RDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARD 264
GG G GGG G GGIG + G GG GGG + G +G
Sbjct: 272 -----GGIGKGGGIG-------GGIG---KGGGIGGGIGKGGGIGGGIGKGGGIG----- 311
Query: 265 CDGGVAVSGGGGNAG 279
GG+ GG G G
Sbjct: 312 --GGIGKGGGIGGGG 324
Score = 50.1 bits (118), Expect = 1e-06
Identities = 49/152 (32%), Positives = 57/152 (37%), Gaps = 28/152 (18%)
Query: 138 CYNCGDAGHLARDCNRSNNNS-----GGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGG 192
C++ +ARD + + GGG G + G G GGG GGG
Sbjct: 15 CFHVFVVNVVARDVSSGRDEDEKTLVGGGKGGGFGGGFGG-----------GAGGGVGGG 63
Query: 193 ACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCG-----GIGHMARDCATAKGPSSGGV 247
A GGFG A GG GGGGG G G G GH G GGV
Sbjct: 64 AG---GGFGGGAGGGFGGGGGGGGGGGGGGGGGFGGGGGFGGGHGGGVGGGVGGGHGGGV 120
Query: 248 GGGGCFRCGEVGHLARDCDGGVAVSGGGGNAG 279
GGG G+ G + G V GG G G
Sbjct: 121 GGG----FGKGGGIGGGIGKGGGVGGGIGKGG 148
Score = 37.0 bits (84), Expect = 0.013
Identities = 38/112 (33%), Positives = 41/112 (35%), Gaps = 46/112 (41%)
Query: 176 RDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARD 235
RD + GGG GGG GGFG GG GGG G G+ GG G
Sbjct: 32 RDEDEKTLVGGGKGGGFG----GGFG--------GGAGGGVGGGAG-----GGFG----- 69
Query: 236 CATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAGRNTCFNCG 287
G + GG GGGG GG GGGG G F G
Sbjct: 70 -----GGAGGGFGGGG---------------GG----GGGGGGGGGGGFGGG 97
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.144 0.482
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,445,799
Number of Sequences: 26719
Number of extensions: 598983
Number of successful extensions: 18202
Number of sequences better than 10.0: 424
Number of HSP's better than 10.0 without gapping: 303
Number of HSP's successfully gapped in prelim test: 126
Number of HSP's that attempted gapping in prelim test: 3478
Number of HSP's gapped (non-prelim): 3442
length of query: 301
length of database: 11,318,596
effective HSP length: 99
effective length of query: 202
effective length of database: 8,673,415
effective search space: 1752029830
effective search space used: 1752029830
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0197.11


