

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0181.1
(325 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
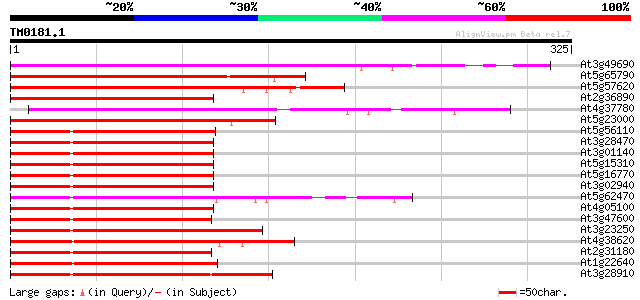
Score E
Sequences producing significant alignments: (bits) Value
At3g49690 AtMYB84 280 8e-76
At5g65790 transcription factor-like protein 260 8e-70
At5g57620 Myb-related transcription factor-like protein 245 2e-65
At2g36890 putative transcription factor (MYB38) 221 3e-58
At4g37780 myb transcription factor like protein 221 4e-58
At5g23000 myb like transcription factor (MYB37) 218 3e-57
At5g56110 Atmyb103 (gb|AAD40692.1) 182 2e-46
At3g28470 putative transcription factor (MYB35) 174 6e-44
At3g01140 putative Myb-related transcription factor 174 8e-44
At5g15310 myb-related protein - like 170 8e-43
At5g16770 putative transcription factor (MYB9) 168 4e-42
At3g02940 MYB family transcription factor like protein 168 4e-42
At5g62470 MYB96 transcription factor-like protein 167 5e-42
At4g05100 MYB - like protein 166 1e-41
At3g47600 putative transcription factor (MYB94) 166 2e-41
At3g23250 myb-related transcription factor like protein 166 2e-41
At4g38620 putative transcription factor (MYB4) 166 2e-41
At2g31180 putative transcription factor (MYB14) 166 2e-41
At1g22640 transcription factor like protein (MYB3) 165 4e-41
At3g28910 MYB family transcription factor (hsr1), putative 164 5e-41
>At3g49690 AtMYB84
Length = 310
Score = 280 bits (716), Expect = 8e-76
Identities = 162/325 (49%), Positives = 197/325 (59%), Gaps = 33/325 (10%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKANVK+GPWSPEEDAKLK+YI++ GTGGNWIALPQKIGLKRCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKANVKKGPWSPEEDAKLKSYIENSGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNIKHGGFSEEE+NIIC+LY++IGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKL+ K
Sbjct: 61 YLRPNIKHGGFSEEEENIICSLYLTIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLINK 120
Query: 121 QRKEQQAQARRVSTMKQEIKRE-TDQNLMVASGVNTTHAAFYWPAEYSIPMPVANASIQD 179
QRKE Q M +KR+ Q + + + F WP + A Q
Sbjct: 121 QRKELQEACMEQQEMMVMMKRQHQQQQIQTSFMMRQDQTMFTWPLHHHNVQVPALFMNQT 180
Query: 180 YDFHNQTSLKSLLINKQGGVRFS-------YDHQQPYSNAITATANSQ---DPCDIYPAS 229
F +Q +K +LI + + HQ +NA + SQ DP + S
Sbjct: 181 NSFCDQEDVKPVLIKNMVKIEDQELEKTNPHHHQDSMTNAFDHLSFSQLLLDPNHNHLGS 240
Query: 230 TMNMINPVAMVSNANIGGTTCQQSNVFQGFENFTSDFSELVCVNPQTIDGFYGMESLEMS 289
+ + +AN SN Q F NF + +T++ F G
Sbjct: 241 GEGF--SMNSILSANTNSPLLNTSNDNQWFGNFQA----------ETVNLFSG------- 281
Query: 290 NVSSTNTPSAEST-SWGDMSSLVYS 313
+ST+T + +ST SW D+SSLVYS
Sbjct: 282 --ASTSTSADQSTISWEDISSLVYS 304
>At5g65790 transcription factor-like protein
Length = 374
Score = 260 bits (664), Expect = 8e-70
Identities = 127/176 (72%), Positives = 141/176 (79%), Gaps = 6/176 (3%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKANVK+GPWSPEEDAKLK YI++ GTGGNWIALPQKIGL+RCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKANVKKGPWSPEEDAKLKDYIENSGTGGNWIALPQKIGLRRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNIKHGGFSEEEDNIICNLYV+IGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLL K
Sbjct: 61 YLRPNIKHGGFSEEEDNIICNLYVTIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLNK 120
Query: 121 QRKEQQAQARRVSTMKQEIKRETDQNLMVASG-----VNTTHAAFYWPAEYSIPMP 171
QRKE Q +AR M +++ Q ++G +N + WP +P P
Sbjct: 121 QRKEFQ-EARMKQEMVMMKRQQQGQGQGQSNGSTDLYLNNMFGSSPWPLLPQLPPP 175
>At5g57620 Myb-related transcription factor-like protein
Length = 333
Score = 245 bits (626), Expect = 2e-65
Identities = 126/213 (59%), Positives = 150/213 (70%), Gaps = 21/213 (9%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKANVK+GPWSPEED KLK YID +GTGGNWIALPQKIGLKRCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKANVKKGPWSPEEDVKLKDYIDKYGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNIKHGGFSEEED II +LY+SIGSRWSIIAAQLPGRTDNDIKNYWNT+LKKKLLG+
Sbjct: 61 YLRPNIKHGGFSEEEDRIILSLYISIGSRWSIIAAQLPGRTDNDIKNYWNTKLKKKLLGR 120
Query: 121 QRKEQQAQARRVST---MKQEIKRETDQNL-----------MVASGVNTTHAAFY----- 161
Q++ + + ST + ++ QNL M + + ++FY
Sbjct: 121 QKQMNRQDSITDSTENNLSNNNNNKSPQNLSNSALERLQLHMQLQNLQSPFSSFYNNPIL 180
Query: 162 WPAEYSIPMPVANASIQDYDFHNQTSLKSLLIN 194
WP + P+ + + Q+ +Q S L +N
Sbjct: 181 WPKLH--PLLQSTTTNQNPKLASQESFHPLGVN 211
>At2g36890 putative transcription factor (MYB38)
Length = 298
Score = 221 bits (564), Expect = 3e-58
Identities = 99/118 (83%), Positives = 108/118 (90%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKANVKRGPWSPEEDAKLK YI+ GTGGNWIALP K GL+RCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKANVKRGPWSPEEDAKLKDYIEKQGTGGNWIALPHKAGLRRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLL 118
YLRPNI+HG F+EEEDNII +L+ SIGSRWS+IAA L GRTDNDIKNYWNT+LKKKL+
Sbjct: 61 YLRPNIRHGDFTEEEDNIIYSLFASIGSRWSVIAAHLQGRTDNDIKNYWNTKLKKKLI 118
>At4g37780 myb transcription factor like protein
Length = 296
Score = 221 bits (563), Expect = 4e-58
Identities = 122/288 (42%), Positives = 168/288 (57%), Gaps = 21/288 (7%)
Query: 12 VKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLNYLRPNIKHGGF 71
VK+GPWS EEDA LK+YI+ HGTG NWI+LPQ+IG+KRCGKSCRLRWLNYLRPN+KHGGF
Sbjct: 3 VKKGPWSTEEDAVLKSYIEKHGTGNNWISLPQRIGIKRCGKSCRLRWLNYLRPNLKHGGF 62
Query: 72 SEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGKQRKEQQAQARR 131
++EED IIC+LY++IGSRWSIIA+QLPGRTDNDIKNYWNTRLKKKLL KQ K Q
Sbjct: 63 TDEEDYIICSLYITIGSRWSIIASQLPGRTDNDIKNYWNTRLKKKLLSKQGKAFHQQLNV 122
Query: 132 VSTMKQEIKRETDQNLMVASGVNTTHAAFYWPAEYSIPMPVANASIQDYDFHNQTSLKSL 191
+ + + NT + ++ V + S++ + Q+ +K+
Sbjct: 123 KFERGTTSSSSSQNQIQIFHDENT-------KSNQTLYNQVVDPSMRAFAMEEQSMIKNQ 175
Query: 192 LIN----KQGGVRFSYDHQ---QPYSNAITATANSQDPCDIYPASTMNMINPVAMVSNAN 244
++ + V F D+ Y + + + NS S++ N +S+
Sbjct: 176 ILEPFSWEPNKVLFDVDYDAAASSYHHHASPSLNSMS-----STSSIGTNNSSLQMSHYT 230
Query: 245 IGGTTCQQSNVF--QGFENFTSDFSELVCVNPQTIDGFYGMESLEMSN 290
+ Q ++F GFENF ++ + + N +GF G E L +N
Sbjct: 231 VNHNDHDQPDMFFMDGFENFQAELFDEIANNNTVENGFDGTEILINNN 278
>At5g23000 myb like transcription factor (MYB37)
Length = 329
Score = 218 bits (556), Expect = 3e-57
Identities = 100/156 (64%), Positives = 120/156 (76%), Gaps = 2/156 (1%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDK VKRGPWSPEED+KL+ YI+ +G GGNWI+ P K GL+RCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKTKVKRGPWSPEEDSKLRDYIEKYGNGGNWISFPLKAGLRRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNIKHG FSEEED II +L+ +IGSRWSIIAA LPGRTDNDIKNYWNT+L+KKLL
Sbjct: 61 YLRPNIKHGDFSEEEDRIIFSLFAAIGSRWSIIAAHLPGRTDNDIKNYWNTKLRKKLLSS 120
Query: 121 QRKEQQA--QARRVSTMKQEIKRETDQNLMVASGVN 154
+ + ++ + Q++KR T + +S N
Sbjct: 121 SSDSSSSAMASPYLNPISQDVKRPTSPTTIPSSSYN 156
>At5g56110 Atmyb103 (gb|AAD40692.1)
Length = 320
Score = 182 bits (463), Expect = 2e-46
Identities = 81/119 (68%), Positives = 97/119 (81%), Gaps = 1/119 (0%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR PCC+K NVKRG W+PEED KL +YI HGT NW +P+ GL+RCGKSCRLRW N
Sbjct: 1 MGRIPCCEKENVKRGQWTPEEDNKLASYIAQHGTR-NWRLIPKNAGLQRCGKSCRLRWTN 59
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLG 119
YLRP++KHG FSE E++II + +G+RWS+IAAQLPGRTDND+KNYWNT+LKKKL G
Sbjct: 60 YLRPDLKHGQFSEAEEHIIVKFHSVLGNRWSLIAAQLPGRTDNDVKNYWNTKLKKKLSG 118
>At3g28470 putative transcription factor (MYB35)
Length = 317
Score = 174 bits (441), Expect = 6e-44
Identities = 76/118 (64%), Positives = 95/118 (80%), Gaps = 1/118 (0%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR PCCDK+NVK+G W+ EEDAK+ AY+ HG G NW +P+K GL RCGKSCRLRW N
Sbjct: 1 MGRPPCCDKSNVKKGLWTEEEDAKILAYVAIHGVG-NWSLIPKKAGLNRCGKSCRLRWTN 59
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLL 118
YLRP++KH FS +E+ +I + +IGSRWS IA +LPGRTDND+KN+WNT+LKKKL+
Sbjct: 60 YLRPDLKHDSFSTQEEELIIECHRAIGSRWSSIARKLPGRTDNDVKNHWNTKLKKKLM 117
>At3g01140 putative Myb-related transcription factor
Length = 345
Score = 174 bits (440), Expect = 8e-44
Identities = 76/118 (64%), Positives = 95/118 (80%), Gaps = 1/118 (0%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR+PCCDKA +K+GPW+PEED KL AYI+ HG G +W +LP+K GL+RCGKSCRLRW N
Sbjct: 1 MGRSPCCDKAGLKKGPWTPEEDQKLLAYIEEHGHG-SWRSLPEKAGLQRCGKSCRLRWTN 59
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLL 118
YLRP+IK G F+ +E+ I L+ +G+RWS IA LP RTDN+IKNYWNT LKK+L+
Sbjct: 60 YLRPDIKRGKFTVQEEQTIIQLHALLGNRWSAIATHLPKRTDNEIKNYWNTHLKKRLI 117
>At5g15310 myb-related protein - like
Length = 326
Score = 170 bits (431), Expect = 8e-43
Identities = 75/118 (63%), Positives = 93/118 (78%), Gaps = 1/118 (0%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR+PCCDK +K+GPW+PEED KL AYI+ HG G +W +LP+K GL RCGKSCRLRW N
Sbjct: 1 MGRSPCCDKLGLKKGPWTPEEDQKLLAYIEEHGHG-SWRSLPEKAGLHRCGKSCRLRWTN 59
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLL 118
YLRP+IK G F+ +E+ I L+ +G+RWS IA LP RTDN+IKNYWNT LKK+L+
Sbjct: 60 YLRPDIKRGKFNLQEEQTIIQLHALLGNRWSAIATHLPKRTDNEIKNYWNTHLKKRLV 117
>At5g16770 putative transcription factor (MYB9)
Length = 336
Score = 168 bits (425), Expect = 4e-42
Identities = 75/118 (63%), Positives = 93/118 (78%), Gaps = 1/118 (0%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR+PCCD+ +K+GPW+ EED KL +I HG G +W ALP++ GL RCGKSCRLRW N
Sbjct: 1 MGRSPCCDENGLKKGPWTQEEDDKLIDHIQKHGHG-SWRALPKQAGLNRCGKSCRLRWTN 59
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLL 118
YLRP+IK G F+EEE+ I NL+ +G++WS IA LPGRTDN+IKNYWNT L+KKLL
Sbjct: 60 YLRPDIKRGNFTEEEEQTIINLHSLLGNKWSSIAGNLPGRTDNEIKNYWNTHLRKKLL 117
>At3g02940 MYB family transcription factor like protein
Length = 321
Score = 168 bits (425), Expect = 4e-42
Identities = 73/118 (61%), Positives = 94/118 (78%), Gaps = 1/118 (0%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR+PCCD++ +K+GPW+PEED KL +I HG G +W ALP++ GL RCGKSCRLRW N
Sbjct: 1 MGRSPCCDESGLKKGPWTPEEDQKLINHIRKHGHG-SWRALPKQAGLNRCGKSCRLRWTN 59
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLL 118
YLRP+IK G F+ EE+ I NL+ +G++WS IA LPGRTDN+IKNYWNT ++KKL+
Sbjct: 60 YLRPDIKRGNFTAEEEQTIINLHSLLGNKWSSIAGHLPGRTDNEIKNYWNTHIRKKLI 117
>At5g62470 MYB96 transcription factor-like protein
Length = 352
Score = 167 bits (424), Expect = 5e-42
Identities = 98/252 (38%), Positives = 133/252 (51%), Gaps = 33/252 (13%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR PCC+K VK+GPW+PEED L +YI HG G NW ++P GL+RC KSCRLRW N
Sbjct: 1 MGRPPCCEKIGVKKGPWTPEEDIILVSYIQEHGPG-NWRSVPTHTGLRRCSKSCRLRWTN 59
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLL-- 118
YLRP IK G F+E E+ I +L +G+RW+ IA+ LP RTDNDIKNYWNT LKKKL
Sbjct: 60 YLRPGIKRGNFTEHEEKTIVHLQALLGNRWAAIASYLPERTDNDIKNYWNTHLKKKLKKI 119
Query: 119 ---------GKQRKEQQAQARRVSTMKQEIKR--ETDQNL---MVASGVNTTHAAFYWPA 164
G +Q ST K + +R +TD N+ + ++ + +
Sbjct: 120 NESGEEDNDGVSSSNTSSQKNHQSTNKGQWERRLQTDINMAKQALCEALSLDKPSSTLSS 179
Query: 165 EYSIPMPVANASIQDYDFHNQTSLKSLLINKQGGVRFSYDHQQPYSNAITATANSQD--- 221
S+P PV N + S L+++ YD S+ T T ++
Sbjct: 180 SSSLPTPVITQ-------QNIRNFSSALLDR------CYDPSSSSSSTTTTTTSNTTNPY 226
Query: 222 PCDIYPASTMNM 233
P +Y +S N+
Sbjct: 227 PSGVYASSAENI 238
>At4g05100 MYB - like protein
Length = 324
Score = 166 bits (421), Expect = 1e-41
Identities = 76/119 (63%), Positives = 95/119 (78%), Gaps = 2/119 (1%)
Query: 1 MGRAPCCDKAN-VKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWL 59
MGR+PCC+K N +K+GPW+PEED KL YI+ HG G NW LP+ GL+RCGKSCRLRW
Sbjct: 1 MGRSPCCEKKNGLKKGPWTPEEDQKLIDYINIHGYG-NWRTLPKNAGLQRCGKSCRLRWT 59
Query: 60 NYLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLL 118
NYLRP+IK G FS EE+ I L+ +G++WS IAA+LPGRTDN+IKNYWNT ++K+LL
Sbjct: 60 NYLRPDIKRGRFSFEEEETIIQLHSIMGNKWSAIAARLPGRTDNEIKNYWNTHIRKRLL 118
>At3g47600 putative transcription factor (MYB94)
Length = 333
Score = 166 bits (420), Expect = 2e-41
Identities = 74/117 (63%), Positives = 88/117 (74%), Gaps = 1/117 (0%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR PCCDK VK+GPW+PEED L +YI HG G NW ++P GL+RC KSCRLRW N
Sbjct: 1 MGRPPCCDKIGVKKGPWTPEEDIILVSYIQEHGPG-NWRSVPTHTGLRRCSKSCRLRWTN 59
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKL 117
YLRP IK G F+E E+ +I +L +G+RW+ IA+ LP RTDNDIKNYWNT LKKKL
Sbjct: 60 YLRPGIKRGNFTEHEEKMILHLQALLGNRWAAIASYLPERTDNDIKNYWNTHLKKKL 116
>At3g23250 myb-related transcription factor like protein
Length = 285
Score = 166 bits (420), Expect = 2e-41
Identities = 77/146 (52%), Positives = 105/146 (71%), Gaps = 1/146 (0%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCC+K +KRGPW+PEED L ++I +HG NW ALP++ GL RCGKSCRLRW+N
Sbjct: 1 MGRAPCCEKMGLKRGPWTPEEDQILVSFILNHGHS-NWRALPKQAGLLRCGKSCRLRWMN 59
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YL+P+IK G F++EE++ I +L+ +G+RWS IAA+LPGRTDN+IKN W+T LKK+L
Sbjct: 60 YLKPDIKRGNFTKEEEDAIISLHQILGNRWSAIAAKLPGRTDNEIKNVWHTHLKKRLEDY 119
Query: 121 QRKEQQAQARRVSTMKQEIKRETDQN 146
Q + + ++ T + T N
Sbjct: 120 QPAKPKTSNKKKGTKPKSESVITSSN 145
>At4g38620 putative transcription factor (MYB4)
Length = 282
Score = 166 bits (419), Expect = 2e-41
Identities = 81/174 (46%), Positives = 116/174 (66%), Gaps = 10/174 (5%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR+PCC+KA+ +G W+ EED +L AYI HG G W +LP+ GL RCGKSCRLRW+N
Sbjct: 1 MGRSPCCEKAHTNKGAWTKEEDERLVAYIKAHGEGC-WRSLPKAAGLLRCGKSCRLRWIN 59
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRP++K G F+EEED +I L+ +G++WS+IA +LPGRTDN+IKNYWNT +++KL+ +
Sbjct: 60 YLRPDLKRGNFTEEEDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIRRKLINR 119
Query: 121 ------QRKEQQAQARRVS--TMKQEIKRET-DQNLMVASGVNTTHAAFYWPAE 165
R Q++ A + S T + + T + + A V T H + +P +
Sbjct: 120 GIDPTSHRPIQESSASQDSKPTQLEPVTSNTINISFTSAPKVETFHESISFPGK 173
>At2g31180 putative transcription factor (MYB14)
Length = 249
Score = 166 bits (419), Expect = 2e-41
Identities = 76/117 (64%), Positives = 92/117 (77%), Gaps = 1/117 (0%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCC+K VKRGPW+PEED L YI +G NW ALP+ GL RCGKSCRLRW+N
Sbjct: 1 MGRAPCCEKMGVKRGPWTPEEDQILINYIHLYGHS-NWRALPKHAGLLRCGKSCRLRWIN 59
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKL 117
YLRP+IK G F+ +E+ I NL+ S+G+RWS IAA+LPGRTDN+IKN W+T LKK+L
Sbjct: 60 YLRPDIKRGNFTPQEEQTIINLHESLGNRWSAIAAKLPGRTDNEIKNVWHTHLKKRL 116
>At1g22640 transcription factor like protein (MYB3)
Length = 257
Score = 165 bits (417), Expect = 4e-41
Identities = 71/120 (59%), Positives = 95/120 (79%), Gaps = 1/120 (0%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR+PCC+KA++ +G W+ EED L YI HG G W +LP+ GL+RCGKSCRLRW+N
Sbjct: 1 MGRSPCCEKAHMNKGAWTKEEDQLLVDYIRKHGEGC-WRSLPRAAGLQRCGKSCRLRWMN 59
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRP++K G F+EEED +I L+ +G++WS+IA +LPGRTDN+IKNYWNT +K+KLL +
Sbjct: 60 YLRPDLKRGNFTEEEDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIKRKLLSR 119
>At3g28910 MYB family transcription factor (hsr1), putative
Length = 323
Score = 164 bits (416), Expect = 5e-41
Identities = 79/152 (51%), Positives = 96/152 (62%), Gaps = 2/152 (1%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
M R PCCDK VK+GPW+PEED L YI HG G NW A+P GL RC KSCRLRW N
Sbjct: 1 MVRPPCCDKGGVKKGPWTPEEDIILVTYIQEHGPG-NWRAVPTNTGLLRCSKSCRLRWTN 59
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRP IK G F+E E+ +I +L +G+RW+ IA+ LP RTDNDIKNYWNT LKKK L K
Sbjct: 60 YLRPGIKRGNFTEHEEKMIVHLQALLGNRWAAIASYLPQRTDNDIKNYWNTHLKKK-LNK 118
Query: 121 QRKEQQAQARRVSTMKQEIKRETDQNLMVASG 152
++ + R S + N ++ G
Sbjct: 119 VNQDSHQELDRSSLSSSPSSSSANSNSNISRG 150
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.130 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,572,450
Number of Sequences: 26719
Number of extensions: 324752
Number of successful extensions: 1219
Number of sequences better than 10.0: 154
Number of HSP's better than 10.0 without gapping: 144
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 871
Number of HSP's gapped (non-prelim): 189
length of query: 325
length of database: 11,318,596
effective HSP length: 100
effective length of query: 225
effective length of database: 8,646,696
effective search space: 1945506600
effective search space used: 1945506600
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0181.1


