

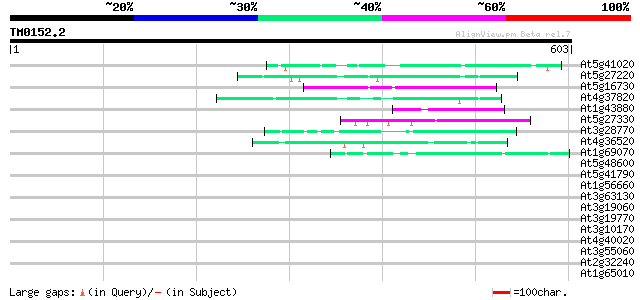
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0152.2
(603 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g41020 putative protein 48 1e-05
At5g27220 putative protein 47 2e-05
At5g16730 putative protein 46 5e-05
At4g37820 unknown protein 45 9e-05
At1g43880 hypothetical protein 43 4e-04
At5g27330 glutamic acid-rich protein 43 6e-04
At3g28770 hypothetical protein 43 6e-04
At4g36520 trichohyalin like protein 42 8e-04
At1g69070 hypothetical protein 42 8e-04
At5g48600 chromosome condensation protein 42 0.001
At5g41790 myosin heavy chain-like protein 42 0.001
At1g56660 hypothetical protein 42 0.001
At3g63130 RAN GTPase like protein 41 0.002
At3g19060 hypothetical protein 41 0.002
At3g19770 unknown protein 40 0.004
At3g10170 hypothetical protein 40 0.004
At4g40020 putative protein 40 0.005
At3g55060 centromere protein - like 40 0.005
At2g32240 putative myosin heavy chain 40 0.005
At1g65010 hypothetical protein 40 0.005
>At5g41020 putative protein
Length = 588
Score = 48.1 bits (113), Expect = 1e-05
Identities = 76/340 (22%), Positives = 135/340 (39%), Gaps = 63/340 (18%)
Query: 277 EQQAAREKRNPAKTLLDAD-------ASHSGTESNRPQKKKRKNETPESAKGKDSSQPSM 329
E++ ++K++ AK +D++ + HS + ++KK K E + G+D
Sbjct: 3 EEKKNKKKKSDAK--VDSEETGEEFISEHSSMKDKEKKRKKNKRENKDGFTGEDMEITGR 60
Query: 330 EKFMVKGNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKIDFNSLVETNLV 389
E + G+ + K S P + SE V ++ K ET
Sbjct: 61 ESEKL-GDEVFIVKKKKKSKKP-----------IRIDSEAVDAV--KKKSKKRSKETKAD 106
Query: 390 FEADREKVKKIGLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKER--EIL 447
EA+ + V+K ++ SK ++S DG+ K+ +KE +++
Sbjct: 107 SEAEDDGVEKKSKEK---------------SKETKVDSEAHDGVKRKKKKSKKESGGDVI 151
Query: 448 KMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKSKAEITHLNS 507
+ + K+ D ++ DL E E + K V+ N K++ V S E +L+S
Sbjct: 152 ENTESSKVSDKKKGKRKRDDTDLGAE-ENIDKEVKRKN----NKKKPSVDSDVEDINLDS 206
Query: 508 SNAELKNENSKLHSEVSELKNSVLDQFEAGFAKAKEQILFLNPQVSINLAGSDPYARIVD 567
+N K K SE SE + + L+ + + K++ +VS SD D
Sbjct: 207 TNDGKKKRKKKKQSEDSETEENGLNSTKDAKKRRKKKKKKKQSEVSEAEEKSDK----SD 262
Query: 568 GKLISPDTG--------------DEEDEGEEDKNEEDENV 593
L +P T ++DEG ED +EE+ V
Sbjct: 263 EDLTTPSTSSKRVKFSDQVEFFPSDDDEGTEDDDEEEVKV 302
>At5g27220 putative protein
Length = 1181
Score = 47.4 bits (111), Expect = 2e-05
Identities = 74/313 (23%), Positives = 117/313 (36%), Gaps = 26/313 (8%)
Query: 246 EFLDAHYENRLGEYVAKMTRLSDDDILRFRREQQAAREKRNPAKTLLDADASHSGT---- 301
E +D E GE M L R R + KR +LD A + T
Sbjct: 308 EQMDIDLERHRGEVNVVMEHLEKSQT-RSRELAEEIERKRKELTAVLDKTAEYGKTIELV 366
Query: 302 -ESNRPQKKK---RKNETPESAKGKDSSQPSMEKFMVKGNPQHMTLKAGSSSAPPPSWKS 357
E Q+K R +E K D +E N T++ S
Sbjct: 367 EEELALQQKLLDIRSSELVSKKKELDGLSLDLELVNSLNNELKETVQRIESKGKE----- 421
Query: 358 LLKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADR-----EKVKKIGLKEACQAIMTK 412
L++ E L E K+ E L + +R E V+K+ L+ + +
Sbjct: 422 -LEDMERLIQERSGHNESIKLLLEEHSE-ELAIKEERHNEIAEAVRKLSLEIVSKEKTIQ 479
Query: 413 GLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLAL 472
L SK L+S + + +L KE E+ +K T + ++ EK+
Sbjct: 480 QLSEKQHSKQTKLDSTEKCLEETTAELVSKENELCSVKDTYRECLQNWEIKEKELKSFQE 539
Query: 473 ENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVLD 532
E ++++ ++D ++KE ELVK K +T + LK + + SE ELK+ LD
Sbjct: 540 EVKKIQDSLKDF----QSKEAELVKLKESLTE-HEKELGLKKKQIHVRSEKIELKDKKLD 594
Query: 533 QFEAGFAKAKEQI 545
E K EQ+
Sbjct: 595 AREERLDKKDEQL 607
Score = 35.4 bits (80), Expect = 0.092
Identities = 65/306 (21%), Positives = 118/306 (38%), Gaps = 22/306 (7%)
Query: 230 EMVDCTFLSSIDLNLT----EFLDAHYENRLGEYVAKMTRLSDDDILRFRREQQAAREKR 285
E+V+ S +DL+L + H E E + L D R E++ R+ +
Sbjct: 134 EIVELLRKSQVDLDLKGEELRQMVTHLERYRVEVKEEKEHLRRTDNGRRELEEEIERKTK 193
Query: 286 NPAKTLLDADASHSGTESNRPQKKKRKNETPESAKGKDSSQPSMEKFMVKGNPQHMTLKA 345
+ + E+ + K + E K D + +EK+ V N + L
Sbjct: 194 DLTLVMNKIVDCDKRIETRSLELIKTQGEVELKEKQLDQMKIDLEKYCVDVNAEKKNLGR 253
Query: 346 GSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIG---L 402
+ + L+E E ++++T + D + L E + E +K G L
Sbjct: 254 TQTH------RRKLEEEIERKTKDLTLVMDKIAECEKLFERRSL-----ELIKTQGEVEL 302
Query: 403 KEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKV 462
K M LE + +E + S + EE ER+ ++ A + K
Sbjct: 303 KGKQLEQMDIDLERHRGEVNVVMEHLEKSQTRSRELAEEIERKRKELTAVLDKTAEYGKT 362
Query: 463 NEKKAADLALENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSE 522
E +LAL+ + L +L +K++EL ++ +NS N ELK ++ S+
Sbjct: 363 IELVEEELALQQKLLDIRSSEL----VSKKKELDGLSLDLELVNSLNNELKETVQRIESK 418
Query: 523 VSELKN 528
EL++
Sbjct: 419 GKELED 424
>At5g16730 putative protein
Length = 853
Score = 46.2 bits (108), Expect = 5e-05
Identities = 48/209 (22%), Positives = 92/209 (43%), Gaps = 5/209 (2%)
Query: 317 ESAKGKDSSQPSMEKFMVKGNPQHMTLKAGSSSAPPPS--WKSLLKEFEELTSEEVTSLW 374
ESA+G ++ E + K N K S+A S W+S KE EE E
Sbjct: 282 ESARGFEAEVKEKEMIVEKLNVDLEAAKMAESNAHSLSNEWQSKAKELEEQLEEANKLER 341
Query: 375 DSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAISKMIDLESADFDGIN 434
+ + S+++ L D+ + + + + I+T LE + DLE ++ +
Sbjct: 342 SASVSLESVMK-QLEGSNDKLHDTETEITDLKERIVT--LETTVAKQKEDLEVSEQRLGS 398
Query: 435 SAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEE 494
+++ + E+E+ K+K+ ++ + KK D +RL + L ++ +EE
Sbjct: 399 VEEEVSKNEKEVEKLKSELETVKEEKNRALKKEQDATSRVQRLSEEKSKLLSDLESSKEE 458
Query: 495 LVKSKAEITHLNSSNAELKNENSKLHSEV 523
KSK + L S+ E+ +E +L ++
Sbjct: 459 EEKSKKAMESLASALHEVSSEGRELKEKL 487
Score = 38.5 bits (88), Expect = 0.011
Identities = 69/347 (19%), Positives = 140/347 (39%), Gaps = 39/347 (11%)
Query: 253 ENRLGEYVAKMTRLSDDDILRFRREQQAAREKRNPAKTLLDADASHSGTESNRPQKKKRK 312
E RLG ++++ ++ ++ + + E + +E++N A L + S + +K K
Sbjct: 393 EQRLGSVEEEVSK-NEKEVEKLKSELETVKEEKNRA--LKKEQDATSRVQRLSEEKSKLL 449
Query: 313 NETPESAKGKDSSQPSMEKFMVKGNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVTS 372
++ S + ++ S+ +ME + H G +E +E +
Sbjct: 450 SDLESSKEEEEKSKKAMESL---ASALHEVSSEG-------------RELKEKLLSQGDH 493
Query: 373 LWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAISKMIDLESADFD- 431
++++ID + LV +A EK + + L EA I + K + D++
Sbjct: 494 EYETQID-----DLKLVIKATNEKYENM-LDEARHEIDVLVSAVEQTKKHFESSKKDWEM 547
Query: 432 -GINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKA 490
N +++ E ++ M M LD+ K E++A + + K ++++
Sbjct: 548 KEANLVNYVKKMEEDVASMGKEMNRLDNLLKRTEEEADAAWKKEAQTKDSLKEVEEEIVY 607
Query: 491 KEEELVKSKAEITHLNSS---------NAELKNENSKLHSEVSELKNSVLDQFEAGFAKA 541
+E L ++KAE L + N +NE+ K +VS K L + A
Sbjct: 608 LQETLGEAKAESMKLKENLLDKETEFQNVIHENEDLKAKEDVSLKKIEELSKLLEEAILA 667
Query: 542 KEQILFLNPQVSINLAGSDPYARIVDGKLISPDTGDEEDEGEEDKNE 588
K+Q N ++S + D ++V+ S + G E + K E
Sbjct: 668 KKQPEEENGELSESEKDYDLLPKVVE---FSSENGHRSVEEKSAKVE 711
Score = 34.3 bits (77), Expect = 0.20
Identities = 96/430 (22%), Positives = 172/430 (39%), Gaps = 96/430 (22%)
Query: 237 LSSIDLNLTEFLDAHYENRLGEYVAKMTRLSDDDILRFRR--EQQAAREKRNPAKTLLDA 294
+SS++ + + LD E + + A+ L DD L+ ++ E+ + EK + ++A
Sbjct: 114 ISSLEKDKAKALD---ELKQAKKEAEQVTLKLDDALKAQKHVEENSEIEKFQAVEAGIEA 170
Query: 295 DASHSGTESNRPQKKKRKNETPESAKGKDSSQ-PSMEKFMVKGNPQHMTL-----KAGSS 348
N ++ K++ ET ++ DS+ ++ + + K N + KA S
Sbjct: 171 -------VQNNEEELKKELETVKNQHASDSAALVAVRQELEKINEELAAAFDAKSKALSQ 223
Query: 349 SAPPPSWKSLLKEFEELTSEEVT---SLWDSKIDFNSLVETNLVFEADREKVKKIGLKEA 405
+ + E ++ S E+T +L DS + ++ + +V + + E V E+
Sbjct: 224 AEDASKTAEIHAEKVDILSSELTRLKALLDSTREKTAISDNEMVAKLEDEIVVLKRDLES 283
Query: 406 CQAIMT--KGLEIAAISKMIDLESADFDGINS----------AKQLEEKEREILKMKATM 453
+ K E+ +DLE+A N+ AK+LEE+ E K++ +
Sbjct: 284 ARGFEAEVKEKEMIVEKLNVDLEAAKMAESNAHSLSNEWQSKAKELEEQLEEANKLERSA 343
Query: 454 ----------------KLLDSANKVNEKKAADLALENERLKKHVEDLNITQK---AKEEE 494
KL D+ ++ + K + LE + K EDL ++++ + EEE
Sbjct: 344 SVSLESVMKQLEGSNDKLHDTETEITDLKERIVTLETT-VAKQKEDLEVSEQRLGSVEEE 402
Query: 495 LVKSKAEITHLNSS--------NAELKNEN-------------SKLHSEVSELKNSVLDQ 533
+ K++ E+ L S N LK E SKL S++ K
Sbjct: 403 VSKNEKEVEKLKSELETVKEEKNRALKKEQDATSRVQRLSEEKSKLLSDLESSKEE---- 458
Query: 534 FEAGFAKAKEQILFLNPQVSINLAGSDPYARIVDGKLISPDTGDEEDEGEED-------- 585
E KA E + +VS R + KL+S GD E E + D
Sbjct: 459 -EEKSKKAMESLASALHEVSSE-------GRELKEKLLS--QGDHEYETQIDDLKLVIKA 508
Query: 586 KNEEDENVND 595
NE+ EN+ D
Sbjct: 509 TNEKYENMLD 518
>At4g37820 unknown protein
Length = 532
Score = 45.4 bits (106), Expect = 9e-05
Identities = 71/314 (22%), Positives = 123/314 (38%), Gaps = 46/314 (14%)
Query: 223 DDSLTPEEMVDCTFLSSIDLNLTE--FLDAHYENRLGEYVAKMTRLSDDDILRFRREQQA 280
D+S E+++ + + + LN TE D + E +K D E
Sbjct: 205 DESGPKNEVLEGSVIKEVSLNTTENGSDDGEQQETKSELDSKTGEKGFSDSNGELPETNL 264
Query: 281 AREKRNPAKTLLDADASHSGTESNRPQKKKRKNETPESAKGKDSSQPSMEKFMVKGNPQH 340
+ N +T + + SG+ ++ KNE E K + S + S K K
Sbjct: 265 STS--NATETTESSGSDESGSSGKSTGYQQTKNEEDEKEKVQSSEEESKVKESGKNEKDA 322
Query: 341 MTLKAGSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKI 400
+ + S P E EE +S + K E + EK +K
Sbjct: 323 SSSQDESKEEKP----------ERKKKEESSSQGEGK-------------EEEPEKREK- 358
Query: 401 GLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSAN 460
+ SK + E+ + + +S ++ E KE EI + + + + N
Sbjct: 359 -----------EDSSSQEESKEEEPENKEKEASSSQEENEIKETEIKEKEESSSQEGNEN 407
Query: 461 KVNEKKAAD-LALENERLKKHVE-----DLNITQKAKEEELVKSKAEITHLNSSNAELKN 514
K EKK+++ EN +K +E D + TQK E++ +SK E + ++SN E ++
Sbjct: 408 KETEKKSSESQRKENTNSEKKIEQVESTDSSNTQKGDEQKTDESKRESGN-DTSNKETED 466
Query: 515 ENSKLHSEVSELKN 528
++SK SE E N
Sbjct: 467 DSSKTESEKKEENN 480
Score = 33.1 bits (74), Expect = 0.46
Identities = 67/324 (20%), Positives = 123/324 (37%), Gaps = 36/324 (11%)
Query: 300 GTESNRPQK-----KKRKNETPESAKGKDSSQPSMEKFMVKGNP-QHMTLKAGSSSAPPP 353
GTE +K + + N T + +D S P E +++G+ + ++L + +
Sbjct: 177 GTEEKSNEKVEVEGESKSNSTENVSVHEDESGPKNE--VLEGSVIKEVSLNTTENGSDDG 234
Query: 354 SWKSLLKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKG 413
+ E + T E+ S DS L ETNL E + G E+ + + G
Sbjct: 235 EQQETKSELDSKTGEKGFS--DSN---GELPETNLSTSNATETTESSGSDESGSSGKSTG 289
Query: 414 LEIAAI--------------SKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSA 459
+ SK+ + + D +S + +E++ E K + + +
Sbjct: 290 YQQTKNEEDEKEKVQSSEEESKVKESGKNEKDASSSQDESKEEKPERKKKEESSSQGEGK 349
Query: 460 NKVNEKKAA-DLALENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSK 518
+ EK+ D + + E ++ E+ + +EE + EI S+++ NEN +
Sbjct: 350 EEEPEKREKEDSSSQEESKEEEPENKEKEASSSQEENEIKETEIKEKEESSSQEGNENKE 409
Query: 519 LHSEVSELKNSVLDQFEAGFAKAKEQILFLNPQVSINLAGSDPYARIVDGKLISPDTGDE 578
+ SE + E K EQ+ S N D + DT ++
Sbjct: 410 TEKKSSESQRKENTNSE----KKIEQV---ESTDSSNTQKGDEQKTDESKRESGNDTSNK 462
Query: 579 EDEGEEDKNE-EDENVNDNEGEGE 601
E E + K E E + N+ GE E
Sbjct: 463 ETEDDSSKTESEKKEENNRNGETE 486
>At1g43880 hypothetical protein
Length = 409
Score = 43.1 bits (100), Expect = 4e-04
Identities = 31/121 (25%), Positives = 54/121 (44%), Gaps = 7/121 (5%)
Query: 412 KGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLA 471
+G IAA +K++ L + SA +LEE K ++ L S+ K + +
Sbjct: 142 EGHAIAATNKLVALYEQRLSQVPSANELEEG-------KTLIRELTSSVKAGQDREVSFQ 194
Query: 472 LENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVL 531
+E ERLK + +K +E++ + EI L + +N+ +L EL V+
Sbjct: 195 VEIERLKMELSTSKDLEKGYDEKIGFMEMEIGGLQADKQTARNQIHRLEQRREELSKKVM 254
Query: 532 D 532
D
Sbjct: 255 D 255
>At5g27330 glutamic acid-rich protein
Length = 628
Score = 42.7 bits (99), Expect = 6e-04
Identities = 54/220 (24%), Positives = 100/220 (44%), Gaps = 17/220 (7%)
Query: 356 KSLLKEFEELTSEEV---TSLWDSKIDFNSL---VETNLVFEADREKVKKIGLKEA---C 406
K+ ++ EL E+ + L ++ NS+ +E +V +D+EK+ + L+E
Sbjct: 322 KARAEQINELVKEKTVKESELEGLMVENNSIKKEIEMAMVQFSDKEKLVEQLLREKNELV 381
Query: 407 QAIMTKGLEIAAISKMIDLESADF-----DGINSAKQLEEKEREILKMKATMKLLDSANK 461
Q ++ + EI +SK+ + D + K E+ + ++K + L++ +
Sbjct: 382 QRVVNQEAEIVELSKLAGEQKHAVAQLRKDYNDQIKNGEKLNCNVSQLKDALALVE-VER 440
Query: 462 VNEKKAADLALENE-RLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLH 520
N KA D N LK+ V L T +A +EL K KAE L EL+N + L
Sbjct: 441 DNAGKALDEEKRNMVALKEKVVALEKTNEATGKELEKIKAERGRLIKEKKELENRSESLR 500
Query: 521 SEVSELKNSVLDQFEA-GFAKAKEQILFLNPQVSINLAGS 559
+E + L+ +++ A G K + + N + S+ + S
Sbjct: 501 NEKAILQKDIVELKRATGVLKTELESAGTNAKQSLTMLKS 540
Score = 35.0 bits (79), Expect = 0.12
Identities = 44/166 (26%), Positives = 73/166 (43%), Gaps = 24/166 (14%)
Query: 384 VETNLVFEADREKV----KKIGLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQL 439
+E +V RE + KK+G +TK E +++ LE + + AK
Sbjct: 266 MEVEMVRRDQREMIVELEKKLGDMNEIVESLTKERE-GLRGQVVGLEKSLDEVTEEAKAR 324
Query: 440 EEKEREILKMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKE---EELV 496
E+ E++K K V E + L +EN +KK +E + KE E+L+
Sbjct: 325 AEQINELVKEKT----------VKESELEGLMVENNSIKKEIEMAMVQFSDKEKLVEQLL 374
Query: 497 KSKAE-ITHLNSSNAELKNENSKLHSE----VSELKNSVLDQFEAG 537
+ K E + + + AE+ E SKL E V++L+ DQ + G
Sbjct: 375 REKNELVQRVVNQEAEIV-ELSKLAGEQKHAVAQLRKDYNDQIKNG 419
>At3g28770 hypothetical protein
Length = 2081
Score = 42.7 bits (99), Expect = 6e-04
Identities = 60/270 (22%), Positives = 102/270 (37%), Gaps = 40/270 (14%)
Query: 275 RREQQAAREKRNPAKTLLDADASHSGTESNRPQKKKRKNETPESAKGKDSSQPSMEKFMV 334
R ++ + +E R+ A + D D ES + +K ++K E + KD+ S ++
Sbjct: 886 RNDKSSTKEVRDFANNM-DIDVQKGSGESVKYKKDEKKEGNKE--ENKDTINTSSKQ--- 939
Query: 335 KGNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADR 394
KG + K +S + K ++ +E + E+ D+K + + L E
Sbjct: 940 KGKDKKKKKKESKNS----NMKKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENKD 995
Query: 395 EKVKKIGLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMK 454
K KK ES D +++K E+KE E K K +
Sbjct: 996 NKEKK--------------------------ESED----SASKNREKKEYEEKKSKTKEE 1025
Query: 455 LLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKN 514
K +KK + E + KK E+ + K+EE K K E + S E K
Sbjct: 1026 AKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKSKKKEDKK 1085
Query: 515 ENSKLHSEVSELKNSVLDQFEAGFAKAKEQ 544
E+ S E + E ++ KE+
Sbjct: 1086 EHEDNKSMKKEEDKKEKKKHEESKSRKKEE 1115
Score = 39.3 bits (90), Expect = 0.006
Identities = 73/337 (21%), Positives = 130/337 (37%), Gaps = 57/337 (16%)
Query: 278 QQAAREKRNPAKTLLDADASHSGTESNRPQKKKRKNETPESAKGKDSSQPSMEKFMVKGN 337
++ +++ AK++ D + NR + K+R E + K + S+E K
Sbjct: 786 EKGEKKESKDAKSVETKDNKKLSSTENRDEAKERSGEDNKEDKEESKDYQSVEA-KEKNE 844
Query: 338 PQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKV 397
+ G+ K K+ ++ S EV + + + + RE+V
Sbjct: 845 NGGVDTNVGN--------KEDSKDLKDDRSVEVKANKEESM------------KKKREEV 884
Query: 398 KKIGLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLD 457
++ TK + A + ID++ + + K E+KE + K T+
Sbjct: 885 QRNDKSS------TKEVRDFANNMDIDVQKGSGESVKYKKD-EKKEGNKEENKDTIN-TS 936
Query: 458 SANKVNEKKAADLALENERLKKHVEDLN--ITQKAKEEELVKSKAEITHLNSSNAELKNE 515
S K +KK +N +KK ED + + K++E +K E T S N++LK E
Sbjct: 937 SKQKGKDKKKKKKESKNSNMKKKEEDKKEYVNNELKKQE--DNKKETT--KSENSKLKEE 992
Query: 516 N----SKLHSEVSELKNSVLDQFEAGFAKAKEQILFLNPQVSINLAGSDPYARIVDGKLI 571
N K SE S KN ++E +K KE+ + D K
Sbjct: 993 NKDNKEKKESEDSASKNREKKEYEEKKSKTKEE-------------AKKEKKKSQDKKRE 1039
Query: 572 SPDTGD-----EEDEGEEDKNEEDENVNDNEGEGENH 603
D+ + E++E + K ++ E + E ENH
Sbjct: 1040 EKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENH 1076
Score = 38.5 bits (88), Expect = 0.011
Identities = 61/339 (17%), Positives = 133/339 (38%), Gaps = 32/339 (9%)
Query: 276 REQQAAREKRNPAKTLLDADASHSGTESNRPQKKKRKNETPESAKGKDSSQPSMEKFMVK 335
+++ +EK+ + + S ++ N KK++K+ + K + + S EK + K
Sbjct: 1148 KKESDKKEKKENEEKSETKEIESSKSQKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKK 1207
Query: 336 GNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADRE 395
+ + K+ +E E+ D K N+ ++ E+
Sbjct: 1208 NEEDRKKQTSVEEN----------KKQKETKKEKNKPKDDKK---NTTKQSGGKKESMES 1254
Query: 396 KVKKIGLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKL 455
+ K+ ++ QA + + ++ +S +S +E + EIL M+A +
Sbjct: 1255 ESKEAENQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESKNEIL-MQADSQA 1313
Query: 456 LDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNE 515
N ++K EN++ K+ E+ N + K+ +S + + S + E +N+
Sbjct: 1314 TTQRNNEEDRKKQTSVAENKKQKETKEEKNKPKDDKKNTTKQSGGKKESMESESKEAENQ 1373
Query: 516 ---NSKLHSEVSELKNSVLDQ----------FEAGFAKAKEQILFLNPQVSINLAGSDPY 562
+ ++ E KN +L Q +A ++K +IL + ++
Sbjct: 1374 QKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESKNEILMQADSQATTQRNNEED 1433
Query: 563 ARIVDGKLISPDTGDEEDEGEEDKNE-EDENVNDNEGEG 600
+ K S ++ E +E+KN+ +D+ N E G
Sbjct: 1434 RK----KQTSVAENKKQKETKEEKNKPKDDKKNTTEQSG 1468
Score = 28.9 bits (63), Expect = 8.6
Identities = 41/173 (23%), Positives = 68/173 (38%), Gaps = 35/173 (20%)
Query: 437 KQLEEKEREILKMKATM-------KLLDSANKVNEKKAADLALENERLKKHVEDLNITQK 489
K LE KE +++A + L A N D E E + +ED +I +
Sbjct: 291 KNLESKEDVKSEVEAAKNDGSSMTENLGEAQGNNGVSTIDNEKEVEGQGESIEDSDIEKN 350
Query: 490 AKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVLDQFEAGFAKAKEQILFLN 549
+ +E VKS+ E S+ K E ++ ++ VS N ++ G ++
Sbjct: 351 LESKEDVKSEVEAAKNAGSSMTGKLEEAQRNNGVS--TNETMNSENKGSGES-------- 400
Query: 550 PQVSINLAGSDPYARIVDGKLISPDTGDEEDEGEEDKNEEDENVNDNEGEGEN 602
+ K+++ T D ED +E+K E EN N +GEN
Sbjct: 401 ----------------TNDKMVNATTND-EDHKKENKEETHEN-NGESVKGEN 435
>At4g36520 trichohyalin like protein
Length = 1432
Score = 42.4 bits (98), Expect = 8e-04
Identities = 64/283 (22%), Positives = 110/283 (38%), Gaps = 23/283 (8%)
Query: 262 KMTRLSDDDILRFRREQQAAREKRNPAKTLLDADASHSGTESNRPQKKKRKNETPESAKG 321
++T + D + RR + RE K HSGT + K+ +N+ E A
Sbjct: 450 ELTWAGNVDWEKQRRRAKGDREDHEARKL-----PKHSGTRKLSSRHKRHENKLAEKAPE 504
Query: 322 KDSSQPSMEKFMVKGNPQHMTLKAGSSSAPPPSWKSL----LKEFEELTSEEVTS-LWDS 376
+ + S M P H + + P K K+ +EL EE T + +
Sbjct: 505 EPKIEKSRHVEMGNDLPDHGGIVKHRNLLKPEENKLFTEKPAKQKKELLCEEKTKRIQNQ 564
Query: 377 KID---FNSLVETNLVFEADREK-VKKIGLKEACQAIMTKGLEIAAISKMIDLESADFDG 432
++D ETN D E+ +K+ ++ + +E+ K +++ S
Sbjct: 565 QLDKKTHQKAAETNQECVYDWEQNARKLREALGNESTLEVSVELNGNGKKMEMRSQSETK 624
Query: 433 INSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKE 492
+N + E+E I + + L N E+ A + A +RLK +E +K KE
Sbjct: 625 LNEPLKRMEEETRIKEAR-----LREENDRRERVAVEKAENEKRLKAALEQEEKERKIKE 679
Query: 493 EELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVLDQFE 535
+ KAE + A K E + E EL+ + + FE
Sbjct: 680 ---AREKAE-NERRAVEAREKAEQERKMKEQQELELQLKEAFE 718
>At1g69070 hypothetical protein
Length = 901
Score = 42.4 bits (98), Expect = 8e-04
Identities = 59/261 (22%), Positives = 105/261 (39%), Gaps = 27/261 (10%)
Query: 346 GSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEA 405
G SS LL + E+L +++ + ++ + N +A E+ ++ KE
Sbjct: 140 GGSSVKDDFDSGLLSD-EDLQDDDLEASASKRLKH---LNRNREVDASGEEERRKSKKEV 195
Query: 406 CQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEK 465
+ I+ K SK+ +E A KQ EEK + + ++ K L ++ +
Sbjct: 196 MEEIIMK-------SKLGRMEKA--------KQKEEKGKLMDELDKNFKSLVNSEAMESL 240
Query: 466 KAADLALENER--LKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAE-LKNENSKLHSE 522
+A EN R + D+++ +A+ E K+ EI E L+ E K E
Sbjct: 241 TKPFVAEENTRDPYLLSLNDMSMEIRARPSERTKTPEEIAQKEREKLEALEEERKKRMQE 300
Query: 523 VSELKNSVLDQFEAGFAKAKEQILFLNPQV--SINLAGSDPYARIVDGKLISPDTGDEED 580
EL + D+ G K + + S ++ P +D L D D
Sbjct: 301 TEELSDG--DEEIGGEESTKRLTVISGDDLGDSFSVEEDKPKRGWIDDVLEREDNVDNS- 357
Query: 581 EGEEDKNEEDENVNDNEGEGE 601
E +ED++ E E D++GE +
Sbjct: 358 ESDEDEDSESEEEEDDDGESD 378
>At5g48600 chromosome condensation protein
Length = 1241
Score = 41.6 bits (96), Expect = 0.001
Identities = 66/288 (22%), Positives = 125/288 (42%), Gaps = 32/288 (11%)
Query: 292 LDADASHSGTESNRPQKKKRKNETPESAKGKDSSQPSMEKFMVKGNPQHMTLKAGSSSAP 351
+ D + TE NR + N+ K + + EK ++G +++ + +
Sbjct: 851 IQTDIDKNNTEINRCNVQIETNQKLIKKLTKGIEEATREKERLEGEKENLHVTFKDITQK 910
Query: 352 PPSWKSLLKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMT 411
+ K+ ++L E L +K D+ +L ++ D K ++ + Q +
Sbjct: 911 AFEIQETYKKTQQLIDEHKDVLTGAKSDYENLKKS-----VDELKASRVDAEFKVQDMKK 965
Query: 412 KGLEIAAISKMIDLESADFDGINSAKQLEEKEREIL---KMKATMKLLDSANKVNE---- 464
K E+ K + D I K +E+ +++++ K++AT L+D N +NE
Sbjct: 966 KYNELEMREKGYKKKLNDLQ-IAFTKHMEQIQKDLVDPDKLQAT--LMD--NNLNEACDL 1020
Query: 465 KKAADL-ALENERLKKHVEDLN-ITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSE 522
K+A ++ AL +LK+ +L+ I + + EL + + LNS + E +
Sbjct: 1021 KRALEMVALLEAQLKELNPNLDSIAEYRSKVELYNGRVD--ELNS----VTQERDDTRKQ 1074
Query: 523 VSELKNSVLDQFEAGF----AKAKE--QILFLNPQVSINLAGS-DPYA 563
EL+ LD+F AGF K KE Q++ L + L S DP++
Sbjct: 1075 YDELRKRRLDEFMAGFNTISLKLKEMYQMITLGGDAELELVDSLDPFS 1122
Score = 34.3 bits (77), Expect = 0.20
Identities = 50/191 (26%), Positives = 81/191 (42%), Gaps = 29/191 (15%)
Query: 347 SSSAPPPSWKSLLKEFEELTSEE---VTSLWDSKIDFNSLVETNLVFEADREKVKKIGLK 403
++S P LKE +++ S+E + +L ++TN+ A EK+K G K
Sbjct: 789 AASQPKTDEIDRLKELKKIISKEEKEIENLEKGSKQLKDKLQTNIE-NAGGEKLK--GQK 845
Query: 404 EACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVN 463
+ I T ID + + + N Q+E ++ I K+ K ++ A +
Sbjct: 846 AKVEKIQTD----------IDKNNTEINRCNV--QIETNQKLIKKLT---KGIEEATREK 890
Query: 464 EKKAADLALENERLKKHVEDLNITQKAKE--EELVKSKAEITHLNSSNAELKNENSKLHS 521
E+ LE E+ HV +ITQKA E E K++ I K++ L
Sbjct: 891 ER------LEGEKENLHVTFKDITQKAFEIQETYKKTQQLIDEHKDVLTGAKSDYENLKK 944
Query: 522 EVSELKNSVLD 532
V ELK S +D
Sbjct: 945 SVDELKASRVD 955
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 41.6 bits (96), Expect = 0.001
Identities = 52/247 (21%), Positives = 104/247 (42%), Gaps = 42/247 (17%)
Query: 385 ETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAI-SKMIDLESADFDGINSAKQLEEKE 443
E+ L +++ ++ +KE + T LE+ ++ +++IDLE+ +QLE +
Sbjct: 730 ESKLFLLTEKDSKSQVQIKELEATVATLELELESVRARIIDLETEIASKTTVVEQLEAQN 789
Query: 444 REIL---------------KMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQ 488
RE++ ++ A + L+ +K + L E + L+ ++ +++ +
Sbjct: 790 REMVARISELEKTMEERGTELSALTQKLEDNDKQSSSSIETLTAEIDGLRAELDSMSVQK 849
Query: 489 KAKEEELVKSKAE------------------ITHLNSSNAELKNENSKLHSEVSELKNSV 530
+ E+++V E + L+S AEL+ + K E+SE + +
Sbjct: 850 EEVEKQMVCKSEEASVKIKRLDDEVNGLRQQVASLDSQRAELEIQLEKKSEEISEYLSQI 909
Query: 531 LDQFEAGFAKAK--EQILFLNPQVSINLAGSDPYARIVDGKLISPDTGDEEDEGEEDKNE 588
+ E K K E IL +S + G + + GK S E DE K E
Sbjct: 910 TNLKEEIINKVKVHESILEEINGLSEKIKGRELELETL-GKQRS-----ELDEELRTKKE 963
Query: 589 EDENVND 595
E+ ++D
Sbjct: 964 ENVQMHD 970
Score = 33.9 bits (76), Expect = 0.27
Identities = 25/101 (24%), Positives = 53/101 (51%), Gaps = 7/101 (6%)
Query: 433 INSAKQLEE----KEREILKMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQ 488
++ + QL+E KERE+ ++ ++ + + +A++L + E K+ V DL+ +
Sbjct: 121 MSESGQLKESHSVKERELFSLRDIHEI---HQRDSSTRASELEAQLESSKQQVSDLSASL 177
Query: 489 KAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNS 529
KA EEE ++ + + +N +L +E+ +LK+S
Sbjct: 178 KAAEEENKAISSKNVETMNKLEQTQNTIQELMAELGKLKDS 218
Score = 33.5 bits (75), Expect = 0.35
Identities = 40/182 (21%), Positives = 81/182 (43%), Gaps = 13/182 (7%)
Query: 359 LKEFEELTSEEVTSLWD-----SKIDFNSLVETNLVFEADREKVKKIGL-----KEACQA 408
LKE + E+ SL D + E E+ +++V + +E +A
Sbjct: 127 LKESHSVKERELFSLRDIHEIHQRDSSTRASELEAQLESSKQQVSDLSASLKAAEEENKA 186
Query: 409 IMTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREI-LKMKATMKLLDSANKVNEKKA 467
I +K +E ++K+ ++ + + +L++ RE ++ + +++ ++ + +
Sbjct: 187 ISSKNVE--TMNKLEQTQNTIQELMAELGKLKDSHREKESELSSLVEVHETHQRDSSIHV 244
Query: 468 ADLALENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELK 527
+L + E KK V +LN T EEE +I L++ E +N +L SE +LK
Sbjct: 245 KELEEQVESSKKLVAELNQTLNNAEEEKKVLSQKIAELSNEIKEAQNTIQELVSESGQLK 304
Query: 528 NS 529
S
Sbjct: 305 ES 306
Score = 33.5 bits (75), Expect = 0.35
Identities = 44/201 (21%), Positives = 87/201 (42%), Gaps = 30/201 (14%)
Query: 403 KEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEE----KEREILKMKATMKLLDS 458
K Q I+ EI K I + ++ ++QL+E KERE+ ++ + ++
Sbjct: 428 KMLSQRILDISNEIQEAQKTIQ------EHMSESEQLKESHGVKERELTGLR---DIHET 478
Query: 459 ANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSK 518
+ + + ++L + + L++ V DL+ + A EEE + I + ELK SK
Sbjct: 479 HQRESSTRLSELETQLKLLEQRVVDLSASLNAAEEEKKSLSSMILEITD---ELKQAQSK 535
Query: 519 LHSEVSELKNSVLDQFEAGFAKAKEQILFLNPQVSINLAGSDPYARIVDGKLISPDTGDE 578
+ V+EL A++K+ + ++S + + + R D +
Sbjct: 536 VQELVTEL------------AESKDTLTQKENELSSFVEVHEAHKR--DSSSQVKELEAR 581
Query: 579 EDEGEEDKNEEDENVNDNEGE 599
+ EE E ++N+N +E E
Sbjct: 582 VESAEEQVKELNQNLNSSEEE 602
Score = 32.0 bits (71), Expect = 1.0
Identities = 48/250 (19%), Positives = 108/250 (43%), Gaps = 19/250 (7%)
Query: 361 EFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIG--LKEACQAIMTKGLEIAA 418
E L T DS I L E E+ ++ V ++ L A + +IA
Sbjct: 225 ELSSLVEVHETHQRDSSIHVKELEEQ---VESSKKLVAELNQTLNNAEEEKKVLSQKIAE 281
Query: 419 ISKMI-DLESADFDGINSAKQLEE----KEREILKMKATMKLLDSANKVNEKKAADLALE 473
+S I + ++ + ++ + QL+E K+R++ ++ + ++ + + + ++L +
Sbjct: 282 LSNEIKEAQNTIQELVSESGQLKESHSVKDRDLFSLR---DIHETHQRESSTRVSELEAQ 338
Query: 474 NERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVLDQ 533
E ++ + DL + K EEE ++ + + +N +L E+ ELK D+
Sbjct: 339 LESSEQRISDLTVDLKDAEEENKAISSKNLEIMDKLEQAQNTIKELMDELGELK----DR 394
Query: 534 FEAGFAKAKEQILFLNPQVSINLAGSDPYARIVDGKLISPDTGDEEDEGEEDKNEEDENV 593
+ ++ + + QV+ D + K++S D +E +E + E++
Sbjct: 395 HKEKESELSSLVKSADQQVADMKQSLDNAEE--EKKMLSQRILDISNEIQEAQKTIQEHM 452
Query: 594 NDNEGEGENH 603
+++E E+H
Sbjct: 453 SESEQLKESH 462
Score = 28.9 bits (63), Expect = 8.6
Identities = 28/120 (23%), Positives = 54/120 (44%), Gaps = 6/120 (5%)
Query: 414 LEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAA----D 469
L+++ IS +I + S +L E + + + ++ L +K +E++++ +
Sbjct: 13 LKVSEISDVIQQGQTTIQELIS--ELGEMKEKYKEKESEHSSLVELHKTHERESSSQVKE 70
Query: 470 LALENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNS 529
L E +K V D + EEE +I L++ E +N +L SE +LK S
Sbjct: 71 LEAHIESSEKLVADFTQSLNNAEEEKKLLSQKIAELSNEIQEAQNTMQELMSESGQLKES 130
>At1g56660 hypothetical protein
Length = 522
Score = 41.6 bits (96), Expect = 0.001
Identities = 68/315 (21%), Positives = 119/315 (37%), Gaps = 49/315 (15%)
Query: 295 DASHSGTESNRPQKKKRKNETPESAKGKDSSQPSMEKFMVKGNPQ-HMTLKAGSSSAPPP 353
D SG +KKK KN E + KD + K + K + + H L+ S
Sbjct: 50 DEESSGKSKKDKEKKKGKNVDSEVKEDKDDDKKKDGKMVSKKHEEGHGDLEVKESDVKVE 109
Query: 354 SWKSLLKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKG 413
+ K+ +E EE+ E ++E KK KE ++ G
Sbjct: 110 EHEKEHKKGKEKKHEEL--------------------EEEKEGKKKKNKKEKDES----G 145
Query: 414 LEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLALE 473
E +K D E D ++LEE++ K N+KK D +
Sbjct: 146 PE--EKNKKADKEKKHEDVSQEKEELEEED----------------GKKNKKKEKDESGT 187
Query: 474 NERLKKHVEDLNITQKAKEEE-----LVKSKAEITHLNSSNAELKNENSKLHSEVSELKN 528
E+ KK ++ +++K E K K E L + E K E+ + E+ E +
Sbjct: 188 EEKKKKPKKEKKQKEESKSNEDKKVKGKKEKGEKGDLEKEDEEKKKEHDETDQEMKEKDS 247
Query: 529 SVLDQFEAGFAKAKEQILFLNPQVSINLAGSDPYARIVDGKLISPDTGDEEDEGEEDKNE 588
+ E + A+E+ + + ++ + + GK + ++EDEG++ K E
Sbjct: 248 KKNKKKEKDESCAEEKKKKPDKEKKEKDESTEKEDKKLKGKKGKGEKPEKEDEGKKTK-E 306
Query: 589 EDENVNDNEGEGENH 603
D + + E +H
Sbjct: 307 HDATEQEMDDEAADH 321
Score = 34.7 bits (78), Expect = 0.16
Identities = 79/337 (23%), Positives = 118/337 (34%), Gaps = 74/337 (21%)
Query: 269 DDILRFRREQQAAREKRNPAKTLLDADASHSGTESNRPQKKKRKNETPESAKGKDSSQPS 328
+D+ + + E + K+N K + SGTE + + KK K + ES +D
Sbjct: 160 EDVSQEKEELEEEDGKKNKKK-----EKDESGTEEKKKKPKKEKKQKEESKSNEDKKVKG 214
Query: 329 MEKFMVKGNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKIDFNSLVETNL 388
++ KG+ L KE EE E + + K
Sbjct: 215 KKEKGEKGD--------------------LEKEDEEKKKEHDETDQEMK----------- 243
Query: 389 VFEADREKVKKIGLKEACQAIMTKGLEIAAISKMIDLESAD--FDGINSAKQLEEKEREI 446
E D +K KK E+C K + K E D G + EKE E
Sbjct: 244 --EKDSKKNKKKEKDESCAEEKKKKPDKEKKEKDESTEKEDKKLKGKKGKGEKPEKEDEG 301
Query: 447 LKMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKSKA---EIT 503
K K D+ + + +AAD E KK +D KAK++E V + E
Sbjct: 302 KKTKEH----DATEQEMDDEAAD---HKEGKKKKNKD-----KAKKKETVIDEVCEKETK 349
Query: 504 HLNSSNAELKNENSKLHSEVSELKNSVLDQFEAGFAKAKEQILFLNPQVSINLAGSDPYA 563
+ E K + +K + S E G KE NP + ++
Sbjct: 350 DKDDDEGETKQKKNKKKEKKS----------EKGEKDVKEDKKKENPLETEVMSR----- 394
Query: 564 RIVDGKLISPDT-GDEEDEGEEDKNEEDENVNDNEGE 599
D KL P+ EED+ EE K + E EG+
Sbjct: 395 ---DIKLEEPEAEKKEEDDTEEKKKSKVEGGESEEGK 428
>At3g63130 RAN GTPase like protein
Length = 535
Score = 41.2 bits (95), Expect = 0.002
Identities = 51/213 (23%), Positives = 83/213 (38%), Gaps = 22/213 (10%)
Query: 410 MTKGLEIAAISKMIDLESADFD---GINSAKQLEEKEREILKMKATMKLLDSANKVNEKK 466
+ + LE + K +DL F GI AK L + + L D + +
Sbjct: 313 LAEALEHCSHLKKLDLRDNMFGVEGGIALAKTLSVLTHLTEIYMSYLNLEDEGTEALSEA 372
Query: 467 AADLALENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKL------- 519
A E L+ D+ + + SK + LN S ELK+E + L
Sbjct: 373 LLKSAPSLEVLELAGNDITVKSTGNLAACIASKQSLAKLNLSENELKDEGTILIAKAVEG 432
Query: 520 HSEVSELKNSVLDQFEAGFAKAKEQILFLNPQVSINLAGS-----------DPYARIVDG 568
H ++ E+ S AG + ++ N +N+ G+ D + +D
Sbjct: 433 HDQLVEVDLSTNMIRRAGARALAQTVVKKNTFKLLNINGNFISEEGIDEVNDMFKDCLD- 491
Query: 569 KLISPDTGDEEDEGEEDKNEEDENVNDNEGEGE 601
KL+ D D E E ED++EE+E + NE E +
Sbjct: 492 KLVPLDDNDPEGEDFEDEDEEEEGEDGNELESK 524
>At3g19060 hypothetical protein
Length = 1647
Score = 41.2 bits (95), Expect = 0.002
Identities = 38/187 (20%), Positives = 89/187 (47%), Gaps = 8/187 (4%)
Query: 414 LEIAAISKMIDLESADF-DGINSAKQLEEKEREILKMKATMKL-LDSANKVNEKKAADLA 471
+ + A+ K + L++ + D ++ A+ LE + +E ++ +++ + A K EK L+
Sbjct: 1261 VHVEALEKTLALKTFELEDAVSHAQMLEVRLQESKEITRNLEVDTEKARKCQEK----LS 1316
Query: 472 LENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVL 531
EN+ ++ EDL + + EEE++++K + L+N +L+ V+ + +
Sbjct: 1317 AENKDIRAEAEDLLAEKCSLEEEMIQTKKVSESMEMELFNLRNALGQLNDTVAFTQRKLN 1376
Query: 532 DQFEAGFAKAKEQILFLNPQV-SINLAGSDPYARIVDGKLISPDTGDEEDEGEEDKNEED 590
D + ++++L L + + + AR ++ + I+ DE EE+ +
Sbjct: 1377 DAIDER-DNLQDEVLNLKEEFGKMKSEAKEMEARYIEAQQIAESRKTYADEREEEVKLLE 1435
Query: 591 ENVNDNE 597
+V + E
Sbjct: 1436 GSVEELE 1442
Score = 30.8 bits (68), Expect = 2.3
Identities = 31/143 (21%), Positives = 59/143 (40%), Gaps = 13/143 (9%)
Query: 360 KEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAI 419
K + + EEV K+ S+ E KV + + Q + + LE+
Sbjct: 1421 KTYADEREEEV------KLLEGSVEELEYTINVLENKVNVVKDEAERQRLQREELEMELH 1474
Query: 420 SKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLALENERLKK 479
+ +ESA + L+EK ++ + K ++ L E+ AD E +L +
Sbjct: 1475 TIRQQMESARNADEEMKRILDEKHMDLAQAKKHIEAL-------ERNTADQKTEITQLSE 1527
Query: 480 HVEDLNITQKAKEEELVKSKAEI 502
H+ +LN+ +A+ E + E+
Sbjct: 1528 HISELNLHAEAQASEYMHKFKEL 1550
>At3g19770 unknown protein
Length = 472
Score = 40.0 bits (92), Expect = 0.004
Identities = 61/269 (22%), Positives = 113/269 (41%), Gaps = 36/269 (13%)
Query: 271 ILRFRREQQAAREKRNPAKTLLDADASHSGTESNRPQKKKRKNE-TPESAKGKDSSQPSM 329
I R+RRE + E +L A++ S ++ + + E ESA+ + S S
Sbjct: 162 IQRYRRESKLVGEAAYFFTNILSAESFISNIDAKSISLDEAEFEKNMESARARISGLDSQ 221
Query: 330 EKFMVKGNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKIDFNSLVETNLV 389
T + G SAPPP +S L++ + L + +L+ SK +SL TN +
Sbjct: 222 ------------TYQTGHGSAPPPRDESTLQKTQSLNPKRENTLFQSK-SSDSLSGTNEL 268
Query: 390 F----EADREKVKKIGLKEACQAIMTKGLEIAAI--------SKMIDLESADFDG-INSA 436
E +K + I E A + K E + + + DL D +G +NS
Sbjct: 269 LNINSETPMKKAESISDLENKGATLLKDTEPSKVFQEYPYIFASAGDLRIGDVEGLLNSY 328
Query: 437 KQLEEKEREILK-MKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKE--E 493
KQL K + K + L S++ + + + E+E ++ D+ +T++ +
Sbjct: 329 KQLVFKYVCLTKGLGDGTSLAPSSSPLQASSGFNTSKESEDHRRSSSDVQMTKETDRSVD 388
Query: 494 ELVKSKAEITHLNSSNAELKNENSKLHSE 522
+L+++ L+ ++ N + H E
Sbjct: 389 DLIRA------LHGEGEDVNNLSDVKHEE 411
>At3g10170 hypothetical protein
Length = 634
Score = 40.0 bits (92), Expect = 0.004
Identities = 34/119 (28%), Positives = 62/119 (51%), Gaps = 17/119 (14%)
Query: 438 QLEEKEREILKMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVK 497
Q+EE +E K + ++K+L ++ +E + +DL E L+K + L+ + AKE+E ++
Sbjct: 164 QIEELNQEAQKHETSLKML---SEHHESERSDLLSHIECLEKDIGSLSSSSLAKEKENLR 220
Query: 498 SKAEITHLNSSNAELKNENSKLHSEVSELKNSVLD--QFEAGFAKAKEQILFLNPQVSI 554
E T +KL S+LKNS+ D + EA A A+ ++ L+ Q ++
Sbjct: 221 KDFEKT------------KTKLKDTESKLKNSMQDKTKLEAEKASAERELKRLHSQKAL 267
Score = 34.7 bits (78), Expect = 0.16
Identities = 44/197 (22%), Positives = 79/197 (39%), Gaps = 23/197 (11%)
Query: 347 SSSAPPPSWKSLLKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEAC 406
SSS+ ++L K+FE+ T L D++ + ++ EA++ ++ E
Sbjct: 208 SSSSLAKEKENLRKDFEKTK----TKLKDTESKLKNSMQDKTKLEAEKASAER----ELK 259
Query: 407 QAIMTKGLEIAAISKMIDLESADFDGI------NSAKQLEEKEREIL--KMKATMKLLDS 458
+ K L ISK D + N + Q E K+ E+L +M+ T+ L+
Sbjct: 260 RLHSQKALLERDISKQESFAGKRRDSLLVERSANQSLQEEFKQLEVLAFEMETTIASLEE 319
Query: 459 ANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSK 518
+ + N+ L + DL E+L S ++ HL + ELK
Sbjct: 320 ELAAERGEKEEALCRNDGLGSEITDLT-------EKLEHSNTKLEHLQNDVTELKTRLEV 372
Query: 519 LHSEVSELKNSVLDQFE 535
S+ +L+ +V E
Sbjct: 373 SSSDQQQLETNVKQLLE 389
Score = 33.1 bits (74), Expect = 0.46
Identities = 40/181 (22%), Positives = 76/181 (41%), Gaps = 43/181 (23%)
Query: 356 KSLLKEFEELTS---------EEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEAC 406
K LL+E EEL EE ++W SK + EA EK++
Sbjct: 385 KQLLEEKEELAMHLANSLLEMEEEKAIWSSK--------EKALTEAVEEKIR-------- 428
Query: 407 QAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKK 466
+ K ++I ++SK + E K+LE E + + ++ + K +++
Sbjct: 429 ---LYKNIQIESLSKEMSEEK---------KELESCRLECVTLADRLRCSEENAKQDKES 476
Query: 467 AADLALENERLKKHVEDLNITQKAKEEELVKS-----KAEITHLNSSNAELKNENSKLHS 521
+ + +LE +RL + + K + +E++KS K+E+ H + + E + S
Sbjct: 477 SLEKSLEIDRLGDELRSADAVSK-QSQEVLKSDIDILKSEVQHACKMSDTFQREMDYVTS 535
Query: 522 E 522
E
Sbjct: 536 E 536
>At4g40020 putative protein
Length = 615
Score = 39.7 bits (91), Expect = 0.005
Identities = 82/391 (20%), Positives = 156/391 (38%), Gaps = 44/391 (11%)
Query: 223 DDSLTPEEMVDCTFLSSIDL-NLTEFLDAHYENR-LGEYVAKMTRLSDDDILRFRREQQA 280
D S EE S +++ +L E +D Y ++ E + + D DI + E ++
Sbjct: 73 DQSRELEETKALVEESKVEIASLKEKIDTSYNSQDSSEEDEDDSSVQDFDIESLKTEMES 132
Query: 281 AREKRNPAKTLLDADASHSGTESNRPQKKKRKNETPESAKGKDSSQPSMEKFMVKGNPQH 340
+E A A AS + K KNE + + +++ +M+
Sbjct: 133 TKESLAQAHEA--AQASSLKVSELLEEMKSVKNELKSATDAEMTNEKAMDDLA------- 183
Query: 341 MTLKAGSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKI-----DFNSLVETN--LVFEAD 393
+ LK ++ K ++ E E + + W K D L T+ L EA+
Sbjct: 184 LALKEVATDCSQTKEKLVIVETELEAARIESQQWKDKYEEVRKDAELLKNTSERLRIEAE 243
Query: 394 REKVKKIGLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATM 453
+ G + + +G + + ++D + + + +A+ L +K +E K
Sbjct: 244 ESLLAWNGKESVFVTCIKRGEDEK--NSLLDENNRLLEALVAAENLSKKAKEE-NHKVRD 300
Query: 454 KLLDSANKVN-EKKAADLA-LENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAE 511
L + N+ N K+AA +A EN LK + D KEEEL + EI + + A
Sbjct: 301 ILKQAINEANVAKEAAGIARAENSNLKDALLD-------KEEELQFALKEIERVKVNEAV 353
Query: 512 LKNENSKLHSEVSELKNSVLDQFEAGFAKAKEQILFLNPQVSINLAGSDPYARIVDGKLI 571
+ KL +SE++ A +E+ LN Q S+ +V+ K+
Sbjct: 354 ANDNIKKLKKMLSEIE----------VAMEEEKQRSLNRQESM----PKEVVEVVEKKIE 399
Query: 572 SPDTGDEEDEGEEDKNEEDENVNDNEGEGEN 602
+ +E+ E +++K E + ++ + E+
Sbjct: 400 EKEKKEEKKENKKEKKESKKEKKEHSEKKED 430
Score = 32.0 bits (71), Expect = 1.0
Identities = 48/202 (23%), Positives = 87/202 (42%), Gaps = 14/202 (6%)
Query: 356 KSLLKEFEELTSE--EVTSLWD-SKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTK 412
K LL F++ + E E +L + SK++ SL E ++ ++ + Q +
Sbjct: 65 KKLLDSFKDQSRELEETKALVEESKVEIASLKEKIDTSYNSQDSSEEDEDDSSVQDFDIE 124
Query: 413 GLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLAL 472
L+ S L A S+ ++ E E+ +K +K A NEK DLAL
Sbjct: 125 SLKTEMESTKESLAQAHEAAQASSLKVSELLEEMKSVKNELKSATDAEMTNEKAMDDLAL 184
Query: 473 ENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVLD 532
LK+ D + T +E+LV + E+ + + K++ ++ + LKN+
Sbjct: 185 ---ALKEVATDCSQT----KEKLVIVETELEAARIESQQWKDKYEEVRKDAELLKNT--- 234
Query: 533 QFEAGFAKAKEQILFLNPQVSI 554
E +A+E +L N + S+
Sbjct: 235 -SERLRIEAEESLLAWNGKESV 255
Score = 29.3 bits (64), Expect = 6.6
Identities = 19/85 (22%), Positives = 43/85 (50%), Gaps = 1/85 (1%)
Query: 461 KVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLH 520
+ + K AD+ E ++K +E+ + ++EL+K I +L ++ +L +
Sbjct: 16 RYSSNKVADIGTELYKMKASLENRENEVVSLKQELLKKDIFIKNLEAAEKKLLDSFKDQS 75
Query: 521 SEVSELKNSVLDQFEAGFAKAKEQI 545
E+ E K +++++ + A KE+I
Sbjct: 76 RELEETK-ALVEESKVEIASLKEKI 99
>At3g55060 centromere protein - like
Length = 896
Score = 39.7 bits (91), Expect = 0.005
Identities = 48/189 (25%), Positives = 84/189 (44%), Gaps = 18/189 (9%)
Query: 368 EEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAISKMIDLES 427
E+ L D D +SLV E +R + E + ++ +E A+ + I
Sbjct: 328 EQQRCLSDCDFDVSSLVGAIRKLEDERLHLAF----ENVNLLRSQIVERASAREEIRWLK 383
Query: 428 ADFDGINSAKQLEEKEREILKMKATM-KLLDSANKVNEKKAADLALENERLKKHVEDLNI 486
+D+D ++ E+E +++A + K LD + K LE ++L++ V +L
Sbjct: 384 SDWD-----LHIQRLEKEKSELQAGLEKELDRRSGEWTSKLEKFQLEEKKLRERVRELAE 438
Query: 487 TQKAKEEEL-------VKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVLDQFEAGFA 539
+ + EL ++K ITHL AEL KLH E + +K + L + + +A
Sbjct: 439 HNVSLQRELSAFHENETENKDMITHLERRVAELTTTADKLHEENNYVKQT-LSKLQESYA 497
Query: 540 KAKEQILFL 548
A E + FL
Sbjct: 498 GATEDLDFL 506
Score = 31.2 bits (69), Expect = 1.7
Identities = 30/132 (22%), Positives = 63/132 (47%), Gaps = 15/132 (11%)
Query: 437 KQLEEKEREILKMKATMKLLDSANKVNEKKAADLALENERLKKH-VEDLNITQKAKEEE- 494
++L KE+EI +++A + N++ + +L+N + H ++DL KEE
Sbjct: 746 EKLYSKEKEIEQLQAELAAAVRGNEILRCEVQS-SLDNLSVTTHELKDLKHQMLKKEESI 804
Query: 495 ------LVKSKAEITHLNSSNAELKNENSKLHSEVSE------LKNSVLDQFEAGFAKAK 542
L ++ E+ LN+ +++ NE ++ SE + L NS + + K +
Sbjct: 805 RRLESNLQEAAKEMARLNALLSKVSNERGQIWSEYKQYGEKNMLLNSENETLKGMVEKLE 864
Query: 543 EQILFLNPQVSI 554
E++L +++I
Sbjct: 865 EKVLEKEGEITI 876
>At2g32240 putative myosin heavy chain
Length = 1333
Score = 39.7 bits (91), Expect = 0.005
Identities = 51/211 (24%), Positives = 95/211 (44%), Gaps = 23/211 (10%)
Query: 357 SLLKEFEELTSEEVTSLWDSK-IDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLE 415
S+ KE EE ++ K + + LVE E E+ KK+ EA T+ +E
Sbjct: 917 SVEKETALKRLEEAIERFNQKETESSDLVEKLKTHENQIEEYKKLA-HEASGVADTRKVE 975
Query: 416 IA-AISKMIDLESADFDGINSAKQLEEKEREI--LKMKATMKLLDSANKVNEKKAADLAL 472
+ A+SK+ +LES + + LE++ ++ + +K ++L + ++ NE + AL
Sbjct: 976 LEDALSKLKNLESTIEELGAKCQGLEKESGDLAEVNLKLNLELANHGSEANELQTKLSAL 1035
Query: 473 ENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVS---ELKNS 529
E E KE+ + +A T + +L +E KL S++S E N
Sbjct: 1036 EAE---------------KEQTANELEASKTTIEDLTKQLTSEGEKLQSQISSHTEENNQ 1080
Query: 530 VLDQFEAGFAKAKEQILFLNPQVSINLAGSD 560
V F++ + + I L Q+++ + +D
Sbjct: 1081 VNAMFQSTKEELQSVIAKLEEQLTVESSKAD 1111
Score = 37.4 bits (85), Expect = 0.024
Identities = 59/254 (23%), Positives = 110/254 (43%), Gaps = 42/254 (16%)
Query: 368 EEVTSLWDSKIDFNSLVETNLVFEADREKVKK-IGLKEACQAIMTKGLEIAAISKMIDLE 426
EE T + S++ V + DR++ K + L +A + K + + I +E
Sbjct: 2 EEATQVTSSEVP---------VVKGDRQRCKYCLDLLQAVNGEVPKEEKEEEDGEFIKVE 52
Query: 427 SADFDGINSAKQLE----EKEREILKMKATMKLLDSANKVNE--KKAADLALENERL--- 477
FD + A++ + E+++E+++ ++ S +++E +KA +L LE ER+
Sbjct: 53 KEAFDAKDDAEKADHVPVEEQKEVIERSSS----GSQRELHESQEKAKELELELERVAGE 108
Query: 478 -------KKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSV 530
H++D ++ K K EE K ++ + E E + HS S+LK S+
Sbjct: 109 LKRYESENTHLKDELLSAKEKLEETEKKHGDLEVVQKKQQEKIVEGEERHS--SQLK-SL 165
Query: 531 LDQFEAGFAKAKE--QILFLNPQVSINLAGSDPYARIVDGKLISPDTGDEEDEGEEDKNE 588
D ++ AK KE ++ + I L S KLI + G + E K E
Sbjct: 166 EDALQSHDAKDKELTEVKEAFDALGIELESSRK-------KLIELEEGLKRSAEEAQKFE 218
Query: 589 EDENVNDNEGEGEN 602
E + + + E+
Sbjct: 219 ELHKQSASHADSES 232
Score = 28.9 bits (63), Expect = 8.6
Identities = 71/363 (19%), Positives = 135/363 (36%), Gaps = 40/363 (11%)
Query: 270 DILRFRREQQAAREKR--NPAKTLLDADASHSGTESNRPQKKKRKNETPESAKGKDS--- 324
D L EQ+ A E R L D DA G ++ +++ ++ E K K+
Sbjct: 316 DELTQELEQKKASESRFKEELSVLQDLDAQTKGLQAKLSEQEGINSKLAEELKEKELLES 375
Query: 325 -SQPSMEKFMVKGNPQHMTLKAGSS--------SAPPPSWKSLLKEFEEL--TSEEVTSL 373
S+ EK LK + ++ + + E EE TS+E S
Sbjct: 376 LSKDQEEKLRTANEKLAEVLKEKEALEANVAEVTSNVATVTEVCNELEEKLKTSDENFSK 435
Query: 374 WDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAISKMIDLESADFDGI 433
D+ + + + N E + ++++ + A AA K ++LE
Sbjct: 436 TDALL--SQALSNNSELEQKLKSLEELHSEAGSAAA-------AATQKNLELEDVVRSSS 486
Query: 434 NSAKQLEE--KEREILKMKATMKLLDSANKVN--EKKAADLALENERLKKHVEDLNITQK 489
+A++ + KE E A K + ++N + K++D E + L + +L +
Sbjct: 487 QAAEEAKSQIKELETKFTAAEQKNAELEQQLNLLQLKSSDAERELKELSEKSSELQTAIE 546
Query: 490 AKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVLDQFEAGFAKAKEQILFLN 549
EEE ++ ++ +EL+ ++ + SEL+ + + G E
Sbjct: 547 VAEEEKKQATTQMQEYKQKASELELSLTQSSARNSELEEDLRIALQKG--AEHEDRANTT 604
Query: 550 PQVSINLAG---------SDPYARIVDGKLISPDTGDEEDEGEEDKNEEDENVNDNEGEG 600
Q SI L G D R+ D +L+ E EE + ++ + E +
Sbjct: 605 HQRSIELEGLCQSSQSKHEDAEGRLKDLELLLQTEKYRIQELEEQVSSLEKKHGETEADS 664
Query: 601 ENH 603
+ +
Sbjct: 665 KGY 667
>At1g65010 hypothetical protein
Length = 1318
Score = 39.7 bits (91), Expect = 0.005
Identities = 54/215 (25%), Positives = 96/215 (44%), Gaps = 38/215 (17%)
Query: 359 LKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAA 418
+K+ EEL++ SL D S+V+ E++ K K+ G + + + +A
Sbjct: 654 MKQIEELSTANA-SLVDEATKLQSIVQ-----ESEDLKEKEAGYLKKIEELSVANESLA- 706
Query: 419 ISKMIDLESADFDGINSAKQLEEKEREILKM-----KATMKLLDSANKVNE--------- 464
+ DL+S + +K L+E+E LK A L+D K+
Sbjct: 707 -DNVTDLQSI----VQESKDLKEREVAYLKKIEELSVANESLVDKETKLQHIDQEAEELR 761
Query: 465 -------KKAADLALENERLKKHVEDL-NITQKAKE--EELVKSKAEITHLNSSNAELKN 514
KK +L+ ENE L +V ++ NI +++K+ E V +I L+++N L +
Sbjct: 762 GREASHLKKIEELSKENENLVDNVANMQNIAEESKDLREREVAYLKKIDELSTANGTLAD 821
Query: 515 ENSKLHSEVSELKNSVLDQFEAGFAKAKEQILFLN 549
+ L + +SE +N L + E K E++ LN
Sbjct: 822 NVTNLQN-ISE-ENKELRERETTLLKKAEELSELN 854
Score = 39.7 bits (91), Expect = 0.005
Identities = 63/286 (22%), Positives = 108/286 (37%), Gaps = 35/286 (12%)
Query: 297 SHSGTESNRPQKKKRKNETPESAKGKDSSQPSMEKFMVK-GNPQHMTLKAGSSSAPPPSW 355
++S E + + + + E ES + K S+ SME M + H+ + S +A
Sbjct: 278 TNSSVEEWKNKVHELEKEVEESNRSKSSASESMESVMKQLAELNHVLHETKSDNAAQKEK 337
Query: 356 KSLLKEFEE--------------LTSEEVTSLWDSKIDFNSLVETNLVFEA---DREKVK 398
LL++ E + EE + L + S +E + + D EK
Sbjct: 338 IELLEKTIEAQRTDLEEYGRQVCIAKEEASKLENLVESIKSELEISQEEKTRALDNEKAA 397
Query: 399 KIGLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDS 458
++ +E+ + D + + A L+E E + KAT+ +
Sbjct: 398 TSNIQNLLDQRTELSIELERCKVEEEKSKKDMESLTLA--LQEASTESSEAKATLLVCQE 455
Query: 459 ANKVNEKKAADLALE----NERLKKHVED-------LNITQKAKEEELVKSKA----EIT 503
K E + L L NE+ +K +ED L T + + E SKA +
Sbjct: 456 ELKNCESQVDSLKLASKETNEKYEKMLEDARNEIDSLKSTVDSIQNEFENSKAGWEQKEL 515
Query: 504 HLNSSNAELKNENSKLHSEVSELKNSVLDQFEAGFAKAKEQILFLN 549
HL + + ENS EVS L N + + E A+ +E+ N
Sbjct: 516 HLMGCVKKSEEENSSSQEEVSRLVNLLKESEEDACARKEEEASLKN 561
Score = 36.2 bits (82), Expect = 0.054
Identities = 58/257 (22%), Positives = 106/257 (40%), Gaps = 33/257 (12%)
Query: 276 REQQAAREKRNPAKTLLDADASHSGTE---SNRPQKKKRKNETPESAKGKDSSQPSMEKF 332
RE++ A K+ + L S T+ SN +++ ++ ET K ++ S+ E
Sbjct: 875 RERETAYLKKIEELSKLHEILSDQETKLQISNHEKEELKERETAYLKKIEELSKVQ-EDL 933
Query: 333 MVKGNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEA 392
+ K N H ++ E E+L S++ SL KI+ S +L+ +
Sbjct: 934 LNKENELH----------------GMVVEIEDLRSKD--SLAQKKIEELSNFNASLLIKE 975
Query: 393 DREKVKKIGLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEK--EREILKMK 450
+ + C+ K +++ + + +L I+ K+L+ E E LK +
Sbjct: 976 NELQAV------VCENEELKSKQVSTLKTIDELSDLKQSLIHKEKELQAAIVENEKLKAE 1029
Query: 451 ATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNA 510
A + L N K+ ++ + + V N KAKE +K E+ HL S
Sbjct: 1030 AALSLQRIEELTNLKQTL---IDKQNELQGVFHENEELKAKEASSLKKIDELLHLEQSWL 1086
Query: 511 ELKNENSKLHSEVSELK 527
E ++E ++ E ELK
Sbjct: 1087 EKESEFQRVTQENLELK 1103
Score = 32.7 bits (73), Expect = 0.60
Identities = 50/197 (25%), Positives = 86/197 (43%), Gaps = 30/197 (15%)
Query: 360 KEFEELTSEEVTSLWDSKIDFNSLVETNLVFE-ADREKVKKIG--LKEACQAIMTKGLEI 416
+E EEL E + L KI+ S NLV A+ + + + L+E A + K E+
Sbjct: 755 QEAEELRGREASHL--KKIEELSKENENLVDNVANMQNIAEESKDLREREVAYLKKIDEL 812
Query: 417 AAISKMIDLESADFDGINSA-KQLEEKEREILKMKATM-----KLLDSANKV------NE 464
+ + + + I+ K+L E+E +LK + L+D A+K+ NE
Sbjct: 813 STANGTLADNVTNLQNISEENKELRERETTLLKKAEELSELNESLVDKASKLQTVVQENE 872
Query: 465 ----------KKAADLALENERLKKHVEDLNITQKAKEEELVKSKA---EITHLNSSNAE 511
KK +L+ +E L L I+ KEE + A +I L+ +
Sbjct: 873 ELRERETAYLKKIEELSKLHEILSDQETKLQISNHEKEELKERETAYLKKIEELSKVQED 932
Query: 512 LKNENSKLHSEVSELKN 528
L N+ ++LH V E+++
Sbjct: 933 LLNKENELHGMVVEIED 949
Score = 31.2 bits (69), Expect = 1.7
Identities = 33/146 (22%), Positives = 67/146 (45%), Gaps = 17/146 (11%)
Query: 415 EIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLALEN 474
EI ++ +D +F+ NS E+KE ++ + +S+++ + +L E+
Sbjct: 488 EIDSLKSTVDSIQNEFE--NSKAGWEQKELHLMGCVKKSEEENSSSQEEVSRLVNLLKES 545
Query: 475 ER-----------LKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEV 523
E LK +++ K +E L ++KAE L S + + + + +E+
Sbjct: 546 EEDACARKEEEASLKNNLKVAEGEVKYLQETLGEAKAESMKLKESLLDKEEDLKNVTAEI 605
Query: 524 SELK---NSVLDQFEAGFAKAKEQIL 546
S L+ SVL++ E +K KE ++
Sbjct: 606 SSLREWEGSVLEKIEE-LSKVKESLV 630
Score = 30.0 bits (66), Expect = 3.9
Identities = 39/202 (19%), Positives = 80/202 (39%), Gaps = 26/202 (12%)
Query: 354 SWKSLLKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEAC------Q 407
SW EF+ +T E + + + E + + E+ EK ++ +EA +
Sbjct: 1084 SWLEKESEFQRVTQENLELKTQDALAAKKIEELSKLKESLLEKETELKCREAAALEKMEE 1143
Query: 408 AIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKER-EILKMKATMKLLDSANKVNEKK 466
E+ +I K DL F +N A +EK + + + ++ ++ + +
Sbjct: 1144 PSKHGNSELNSIGKDYDL--VQFSEVNGASNGDEKTKTDHYQQRSREHMIQE----SPME 1197
Query: 467 AADLALENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSEL 526
A D L ER H + + + E V+ ++E +S E + + +E+ +
Sbjct: 1198 AIDKHLMGERAAIH----KVAHRVEGERNVEKESEFKMWDSYKIEKSEVSPERETELDSV 1253
Query: 527 KNSV---------LDQFEAGFA 539
+ V +DQ+ GF+
Sbjct: 1254 EEEVDSKAESSENMDQYSNGFS 1275
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.130 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,147,513
Number of Sequences: 26719
Number of extensions: 656458
Number of successful extensions: 5452
Number of sequences better than 10.0: 375
Number of HSP's better than 10.0 without gapping: 98
Number of HSP's successfully gapped in prelim test: 283
Number of HSP's that attempted gapping in prelim test: 4096
Number of HSP's gapped (non-prelim): 1143
length of query: 603
length of database: 11,318,596
effective HSP length: 105
effective length of query: 498
effective length of database: 8,513,101
effective search space: 4239524298
effective search space used: 4239524298
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0152.2


