

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0061.10
(276 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
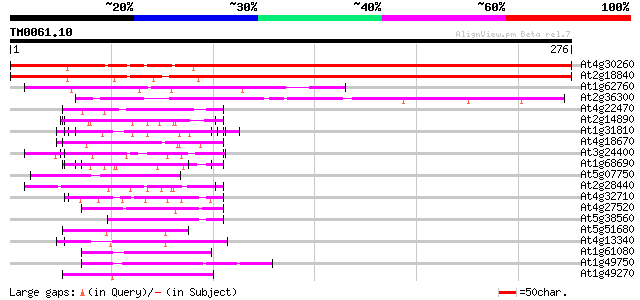
Score E
Sequences producing significant alignments: (bits) Value
At4g30260 unknown protein 441 e-124
At2g18840 unknown protein 435 e-122
At1g62760 hypothetical protein 53 2e-07
At2g36300 unknown protein 51 8e-07
At4g22470 extensin - like protein 48 5e-06
At2g14890 arabinogalactan-protein AGP9 48 5e-06
At1g31810 hypothetical protein 47 9e-06
At4g18670 extensin-like protein 47 1e-05
At3g24400 protein kinase, putative 47 1e-05
At1g68690 protein kinase, putative 46 2e-05
At5g07750 putative protein 46 2e-05
At2g28440 En/Spm-like transposon protein 46 2e-05
At4g32710 putative protein kinase 45 3e-05
At4g27520 early nodulin-like 2 predicted GPI-anchored protein 45 4e-05
At5g38560 putative protein 44 1e-04
At5g51680 unknown protein 43 2e-04
At4g13340 extensin-like protein 43 2e-04
At1g61080 hypothetical protein 43 2e-04
At1g49750 unknown protein 43 2e-04
At1g49270 hypothetical protein 43 2e-04
>At4g30260 unknown protein
Length = 280
Score = 441 bits (1134), Expect = e-124
Identities = 236/284 (83%), Positives = 246/284 (86%), Gaps = 12/284 (4%)
Query: 1 MSHGDTIPLHPSSQSDIDEIENLIYAS----PATVLPARPPSPPRASIPVTTSSPFINSN 56
MSH DTIPL+ SSQSDIDEIEN++ S P TVLPARPPSP R SIPV+ SSPF+ SN
Sbjct: 1 MSHNDTIPLYQSSQSDIDEIENMMNDSFQSGPGTVLPARPPSPIRPSIPVS-SSPFVQSN 59
Query: 57 LPAPLPPKSSISQPQLPKPISLPPAPPPSTRPE----ISSSGFGPAPNTLTEPVWDTVKR 112
LP PLPP SS S Q P+ PP P PS E I SGFG PNTLTEPVWDTVKR
Sbjct: 60 LP-PLPPSSS-SSTQKVMPVPAPP-PLPSAGNEGNKSIGGSGFGSPPNTLTEPVWDTVKR 116
Query: 113 DLSRIVSNLKLVVFPNPYREDPGKALRDWDLWGPFFFIVFLGLILSWSASVKKSEVFAVA 172
DLSRIVSNLKLVVFPNPYREDPGKALRDWDLWGPFFFIVFLGL LSWSASVKKSEVFAVA
Sbjct: 117 DLSRIVSNLKLVVFPNPYREDPGKALRDWDLWGPFFFIVFLGLTLSWSASVKKSEVFAVA 176
Query: 173 FALLAAGAVILTLNVLLLGGHIIFFQSLSLLGYCLFPLDVGALICMLKDNVILKVVVVCV 232
FALLAAGAVILTLNVLLLGGHIIFFQSLSLLGYCLFPLDVGA+ICMLKDNVILK+VVV V
Sbjct: 177 FALLAAGAVILTLNVLLLGGHIIFFQSLSLLGYCLFPLDVGAVICMLKDNVILKMVVVSV 236
Query: 233 TLAWSSWAAYPFMSSAVNPRRKALALYPVFLMYVSVGFLIIAID 276
TLAWSSWAAYPFMS+AVNPRRKALALYPVFLMYVSVGFLIIAI+
Sbjct: 237 TLAWSSWAAYPFMSAAVNPRRKALALYPVFLMYVSVGFLIIAIN 280
>At2g18840 unknown protein
Length = 281
Score = 435 bits (1118), Expect = e-122
Identities = 233/285 (81%), Positives = 243/285 (84%), Gaps = 13/285 (4%)
Query: 1 MSHGDTIPLHPSSQSDIDEIENLIYAS----PATVLPARPPSPPRASIPVTTSS---PFI 53
MS GDT+PLHPSSQSDIDEIENLI S P TVL ARPPSP R SIPV++SS PF+
Sbjct: 1 MSQGDTVPLHPSSQSDIDEIENLINESVQSGPGTVLAARPPSPTRPSIPVSSSSSSSPFM 60
Query: 54 NSNLPAPLPPKSSISQ-PQLPKPISLPPAPPPSTRPEIS-SSGFGPAPNTLTEPVWDTVK 111
SNLP PL P SS + +P P PP P S +S FG PNTLTEPVWDTVK
Sbjct: 61 QSNLP-PLHPSSSAQKVTHVPVP---PPLPAVSNSSNFQGASAFGSPPNTLTEPVWDTVK 116
Query: 112 RDLSRIVSNLKLVVFPNPYREDPGKALRDWDLWGPFFFIVFLGLILSWSASVKKSEVFAV 171
RDLSRIVSNLKLVVFPNPYREDPGKALRDWDLWGPFFFIVFLGL LSWSASVKKSEVFAV
Sbjct: 117 RDLSRIVSNLKLVVFPNPYREDPGKALRDWDLWGPFFFIVFLGLTLSWSASVKKSEVFAV 176
Query: 172 AFALLAAGAVILTLNVLLLGGHIIFFQSLSLLGYCLFPLDVGALICMLKDNVILKVVVVC 231
AFALLAAGAVILTLNVLLLGGHIIFFQSLSLLGYCLFPLDVGA+ICMLKDNVILK+VVV
Sbjct: 177 AFALLAAGAVILTLNVLLLGGHIIFFQSLSLLGYCLFPLDVGAVICMLKDNVILKMVVVS 236
Query: 232 VTLAWSSWAAYPFMSSAVNPRRKALALYPVFLMYVSVGFLIIAID 276
VTLAWSSWAAYPFMSSAVNPRRKALALYPVFLMYVSVGFLIIAI+
Sbjct: 237 VTLAWSSWAAYPFMSSAVNPRRKALALYPVFLMYVSVGFLIIAIN 281
>At1g62760 hypothetical protein
Length = 312
Score = 52.8 bits (125), Expect = 2e-07
Identities = 49/171 (28%), Positives = 76/171 (43%), Gaps = 25/171 (14%)
Query: 8 PLHPSSQSDIDEIENLIYASPA--TVLPARPPSPPRASIPVTTSSPFINSNLPAPLP--- 62
P PSS +L +SP ++ P+ PP PP +S P+++ SP ++ + P+ P
Sbjct: 38 PSPPSSSPSSAPPSSLSPSSPPPLSLSPSSPPPPPPSSSPLSSLSPSLSPSPPSSSPSSA 97
Query: 63 PKSSISQPQLPKPISL----PPAPPPSTRPEISSSGFGPAPNTLTEPVWDTVKRD----L 114
P SS+S P P P+SL PP PPPS+ P S S + + D +K L
Sbjct: 98 PPSSLS-PSSPPPLSLSPSSPPPPPPSSSPLSSLSPSSSSSTYSNQTNLDYIKTSCNITL 156
Query: 115 SRIVSNLKLVVFPNPYREDPGKALRDWDLWGPFFFIVFLGLILSWSASVKK 165
+ + L + + R +P K ++ L L LS + S K
Sbjct: 157 YKTICYNSLSPYASTIRSNPQK-----------LAVIALNLTLSSAKSASK 196
Score = 40.4 bits (93), Expect = 0.001
Identities = 28/86 (32%), Positives = 41/86 (47%), Gaps = 9/86 (10%)
Query: 27 SPATVLPARPPSPPRAS---IPVTTSSPF------INSNLPAPLPPKSSISQPQLPKPIS 77
SP++ P+ PSPP +S P ++ SP ++ + P P PP SS P
Sbjct: 28 SPSSSSPSLSPSPPSSSPSSAPPSSLSPSSPPPLSLSPSSPPPPPPSSSPLSSLSPSLSP 87
Query: 78 LPPAPPPSTRPEISSSGFGPAPNTLT 103
PP+ PS+ P S S P P +L+
Sbjct: 88 SPPSSSPSSAPPSSLSPSSPPPLSLS 113
Score = 29.6 bits (65), Expect = 1.8
Identities = 23/70 (32%), Positives = 30/70 (42%), Gaps = 16/70 (22%)
Query: 46 VTTSSPFINSNLPAP-LPPKSSISQPQLPKPISL--------------PPAPPPSTRPEI 90
+TTSS ++ + +P L P S P P SL PP PPPS+ P
Sbjct: 20 ITTSSSSLSPSSSSPSLSPSPPSSSPSSAPPSSLSPSSPPPLSLSPSSPPPPPPSSSPLS 79
Query: 91 S-SSGFGPAP 99
S S P+P
Sbjct: 80 SLSPSLSPSP 89
>At2g36300 unknown protein
Length = 255
Score = 50.8 bits (120), Expect = 8e-07
Identities = 57/248 (22%), Positives = 103/248 (40%), Gaps = 29/248 (11%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTRPEISS 92
PA P PPR P +++ PF++ ++ + ++ P P T SS
Sbjct: 29 PATPFQPPR---PSSSAIPFMSFDIGSAAASSAT------------PAGPFGGTIASSSS 73
Query: 93 SGFGPAPNTLTEPVWDTVKRDLSRIVSNLKLVVFPNPYREDPGKALRDWDLWGPFFFIVF 152
G G A EP+ D + +I + ++ NP+R + +D DL GP F +
Sbjct: 74 FGGGSASFEDEEPLLDELGIHPDQIWKKTRSIL--NPFRINQA-VHKDSDLSGPIFLYLA 130
Query: 153 LGLILSWSASVKKSEVFAVAFALLAAGAVILTLNVLLLGG---HIIFFQSLSLLGYCLFP 209
L L + ++ F V + ++ L + +L G ++ SL+GYCL P
Sbjct: 131 LCLFQLLAGKIQ----FGVILGWIVVSSIFLYVVFNMLAGRNGNLNLHTCTSLVGYCLLP 186
Query: 210 LDVGALICMLKDNVI--LKVVVVCVTLAWSSWAAYPFMSSAVN--PRRKALALYPVFLMY 265
+ V + + + ++ V+ + + WS+ A + S + + L Y FL+Y
Sbjct: 187 VVVLSAVSLFVPQGAGPVRFVLAALFVLWSTRACSTLVVSLADGGEEHRGLIAYACFLIY 246
Query: 266 VSVGFLII 273
L+I
Sbjct: 247 TLFSLLVI 254
>At4g22470 extensin - like protein
Length = 375
Score = 48.1 bits (113), Expect = 5e-06
Identities = 31/86 (36%), Positives = 43/86 (49%), Gaps = 15/86 (17%)
Query: 27 SPATVLPA-----RPPSPPRASIP--VTTSSPFINSNLPAPLPPKSSISQPQLPKPISLP 79
+P T PA +PP PP+++ P V T+ P + P PLPP S Q P P ++
Sbjct: 55 TPPTFQPAPPANDQPPPPPQSTSPPPVATTPPALP---PKPLPPPLSPPQTTPPPPPAIT 111
Query: 80 PAPPPSTRPEISSSGFGPAPNTLTEP 105
P PPP+ P +S P P +T P
Sbjct: 112 PPPPPAITPPLS-----PPPPAITPP 132
Score = 38.1 bits (87), Expect = 0.005
Identities = 25/78 (32%), Positives = 34/78 (43%), Gaps = 13/78 (16%)
Query: 35 RPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAP------PPSTRP 88
+PP+PP T P +N P PP+S+ P P +LPP P PP T P
Sbjct: 52 QPPTPP-------TFQPAPPANDQPPPPPQSTSPPPVATTPPALPPKPLPPPLSPPQTTP 104
Query: 89 EISSSGFGPAPNTLTEPV 106
+ P P +T P+
Sbjct: 105 PPPPAITPPPPPAITPPL 122
Score = 37.7 bits (86), Expect = 0.007
Identities = 27/82 (32%), Positives = 35/82 (41%), Gaps = 11/82 (13%)
Query: 28 PATVLPARPPS--PPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLP---PAP 82
P + P PP+ PP + P + P + P LPPK LP P+S P P P
Sbjct: 107 PPAITPPPPPAITPPLSPPPPAITPPPPLATTPPALPPKP------LPPPLSPPQTTPPP 160
Query: 83 PPSTRPEISSSGFGPAPNTLTE 104
PP+ P +S G TE
Sbjct: 161 PPAITPPLSPPLVGICSKNDTE 182
Score = 37.7 bits (86), Expect = 0.007
Identities = 25/64 (39%), Positives = 31/64 (48%), Gaps = 10/64 (15%)
Query: 29 ATVLPARPPSP--PRASIPVTTSSPFINSNLPAPL--PPKSSISQPQLPKPISLPPAPPP 84
AT PA PP P P S P TT P P + PP +I+ P P P ++ P PP
Sbjct: 82 ATTPPALPPKPLPPPLSPPQTTPPP------PPAITPPPPPAITPPLSPPPPAITPPPPL 135
Query: 85 STRP 88
+T P
Sbjct: 136 ATTP 139
Score = 33.1 bits (74), Expect = 0.17
Identities = 17/48 (35%), Positives = 21/48 (43%), Gaps = 3/48 (6%)
Query: 58 PAPLPPKSSISQPQLPKPISLPPAPPPSTRPEISSSGFGPAPNTLTEP 105
P P PP+ PQ P P + PAPP + +P P P T P
Sbjct: 42 PGPPPPQPD---PQPPTPPTFQPAPPANDQPPPPPQSTSPPPVATTPP 86
Score = 33.1 bits (74), Expect = 0.17
Identities = 24/72 (33%), Positives = 31/72 (42%), Gaps = 16/72 (22%)
Query: 36 PPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISL-PPAPPPSTRPEISSSG 94
PP PP + P++ P I PP + + P LP P L PP PP T P
Sbjct: 112 PPPPPAITPPLSPPPPAITP------PPPLATTPPALP-PKPLPPPLSPPQTTP------ 158
Query: 95 FGPAPNTLTEPV 106
P P +T P+
Sbjct: 159 --PPPPAITPPL 168
Score = 29.6 bits (65), Expect = 1.8
Identities = 20/71 (28%), Positives = 28/71 (39%), Gaps = 4/71 (5%)
Query: 36 PPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTRPEISSSGF 95
PP PP I +P P P PP QP P PP P ++ P ++++
Sbjct: 31 PPPPPCICI----CNPGPPPPQPDPQPPTPPTFQPAPPANDQPPPPPQSTSPPPVATTPP 86
Query: 96 GPAPNTLTEPV 106
P L P+
Sbjct: 87 ALPPKPLPPPL 97
Score = 29.6 bits (65), Expect = 1.8
Identities = 25/83 (30%), Positives = 30/83 (36%), Gaps = 8/83 (9%)
Query: 28 PATVLPARPPSPPRASIPVTTSSPFINSNLPA---PLPPKSSISQPQLPKPISLPPAPPP 84
P + P PP P + P PA P PP S S P + + PPA PP
Sbjct: 34 PPCICICNPGPPPPQPDPQPPTPPTFQPAPPANDQPPPPPQSTSPPPVA---TTPPALPP 90
Query: 85 STRPEISS--SGFGPAPNTLTEP 105
P S P P +T P
Sbjct: 91 KPLPPPLSPPQTTPPPPPAITPP 113
Score = 28.5 bits (62), Expect = 4.1
Identities = 17/47 (36%), Positives = 19/47 (40%), Gaps = 5/47 (10%)
Query: 58 PAPLPPKSSISQPQLPKPISLPPAPPPSTRPEISSSGFGPAPNTLTE 104
P PLPP Q P P P PPP+ P +S G TE
Sbjct: 262 PPPLPP-----QTLKPPPPQTTPPPPPAITPPLSPPLVGICSKNDTE 303
>At2g14890 arabinogalactan-protein AGP9
Length = 191
Score = 48.1 bits (113), Expect = 5e-06
Identities = 33/89 (37%), Positives = 43/89 (48%), Gaps = 14/89 (15%)
Query: 27 SPATVLPARPPSPPRASIPVTTSSPFINSNLPA--PLPPKSS----ISQPQLPKPISLPP 80
+P T PA P P A PVTTS P + + P P PP SS P P P++ PP
Sbjct: 35 TPTTPPPAATPPPVSAPPPVTTSPPPVTTAPPPANPPPPVSSPPPASPPPATPPPVASPP 94
Query: 81 ----APPPSTRPEISSSGFGPAPNTLTEP 105
+PPP+T P +++ P P L P
Sbjct: 95 PPVASPPPATPPPVAT----PPPAPLASP 119
Score = 45.4 bits (106), Expect = 3e-05
Identities = 30/76 (39%), Positives = 36/76 (46%), Gaps = 6/76 (7%)
Query: 28 PATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLP--PAPPPS 85
PAT P P PP AS P T P P P P S +Q P P + P P+P PS
Sbjct: 84 PATPPPVASPPPPVASPPPATPPPVAT---PPPAPLASPPAQVPAPAPTTKPDSPSPSPS 140
Query: 86 TRPEISSSGFGPAPNT 101
+ P + SS P P+T
Sbjct: 141 SSPPLPSSD-APGPST 155
Score = 41.2 bits (95), Expect = 6e-04
Identities = 30/84 (35%), Positives = 38/84 (44%), Gaps = 4/84 (4%)
Query: 26 ASPATVLPARPP---SPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQL-PKPISLPPA 81
++P V + PP +PP A+ P SSP S PA PP +S P P P + PP
Sbjct: 49 SAPPPVTTSPPPVTTAPPPANPPPPVSSPPPASPPPATPPPVASPPPPVASPPPATPPPV 108
Query: 82 PPPSTRPEISSSGFGPAPNTLTEP 105
P P S PAP T+P
Sbjct: 109 ATPPPAPLASPPAQVPAPAPTTKP 132
Score = 40.8 bits (94), Expect = 8e-04
Identities = 25/82 (30%), Positives = 40/82 (48%), Gaps = 8/82 (9%)
Query: 26 ASPATVLPARPPS--PPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPP 83
A+P + + PP+ PP PV + P + S PA PP ++ L P + PAP
Sbjct: 68 ANPPPPVSSPPPASPPPATPPPVASPPPPVASPPPATPPPVATPPPAPLASPPAQVPAPA 127
Query: 84 PSTRPEISSSGFGPAPNTLTEP 105
P+T+P+ P+P+ + P
Sbjct: 128 PTTKPD------SPSPSPSSSP 143
Score = 40.4 bits (93), Expect = 0.001
Identities = 25/76 (32%), Positives = 36/76 (46%), Gaps = 8/76 (10%)
Query: 33 PARPP----SPPRASIPVTTSSPFINSNLPAPLPPKSS---ISQPQLPKPISLPPAPPPS 85
PA PP SPP AS P T P + P PP ++ ++ P P P++ PPA P+
Sbjct: 67 PANPPPPVSSPPPASPPPATPPPVASPPPPVASPPPATPPPVATPP-PAPLASPPAQVPA 125
Query: 86 TRPEISSSGFGPAPNT 101
P P+P++
Sbjct: 126 PAPTTKPDSPSPSPSS 141
Score = 37.4 bits (85), Expect = 0.009
Identities = 31/98 (31%), Positives = 46/98 (46%), Gaps = 10/98 (10%)
Query: 26 ASPATVLPARPPS-PPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPI-SLPPAPP 83
ASP + + PP+ PP PV T P ++ PA +P + ++P P P S P P
Sbjct: 91 ASPPPPVASPPPATPP----PVATPPPAPLASPPAQVPAPAPTTKPDSPSPSPSSSPPLP 146
Query: 84 PSTRPEISSSGFGPAPNTLTEPVWDTVKRDLSRIVSNL 121
S P S+ PAP+ P + S++VS+L
Sbjct: 147 SSDAPGPSTDSISPAPS----PTDVNDQNGASKMVSSL 180
Score = 35.4 bits (80), Expect = 0.034
Identities = 27/78 (34%), Positives = 33/78 (41%), Gaps = 6/78 (7%)
Query: 29 ATVLPARPPSPPRAS-IPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTR 87
A V P SPP A+ P T ++P PA PP S P P + APPP+
Sbjct: 16 AGVTGQAPTSPPTATPAPPTPTTP-----PPAATPPPVSAPPPVTTSPPPVTTAPPPANP 70
Query: 88 PEISSSGFGPAPNTLTEP 105
P SS +P T P
Sbjct: 71 PPPVSSPPPASPPPATPP 88
Score = 33.9 bits (76), Expect = 0.098
Identities = 22/71 (30%), Positives = 28/71 (38%), Gaps = 10/71 (14%)
Query: 29 ATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTRP 88
AT P P +PP A+ P S+P PP ++ P P P PP S+ P
Sbjct: 29 ATPAPPTPTTPPPAATPPPVSAP----------PPVTTSPPPVTTAPPPANPPPPVSSPP 78
Query: 89 EISSSGFGPAP 99
S P P
Sbjct: 79 PASPPPATPPP 89
>At1g31810 hypothetical protein
Length = 1201
Score = 47.4 bits (111), Expect = 9e-06
Identities = 28/88 (31%), Positives = 40/88 (44%), Gaps = 6/88 (6%)
Query: 24 IYASPATVLPARPPSPPRASI-----PVTTSSPFINSNLPAPLPPKSSISQPQLPKPISL 78
++ S + P++PP PP P+TT IN P P PP + +P P++
Sbjct: 534 LFTSTTSFSPSQPPPPPPLPSFSNRDPLTTLHQPINKTPPPPPPPPPPLPSRSIPPPLAQ 593
Query: 79 PPAP-PPSTRPEISSSGFGPAPNTLTEP 105
PP P PP P SS P+P+ P
Sbjct: 594 PPPPRPPPPPPPPPSSRSIPSPSAPPPP 621
Score = 46.6 bits (109), Expect = 1e-05
Identities = 29/79 (36%), Positives = 36/79 (44%), Gaps = 11/79 (13%)
Query: 24 IYASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSS---ISQPQLPKPISLPP 80
I P + P PP PP+A+I P P PLPP S+ P P P+S P
Sbjct: 672 IRVGPPSTPPPPPPPPPKANISNAPKPP-----APPPLPPSSTRLGAPPPPPPPPLSKTP 726
Query: 81 APPPSTRPEISSSGFGPAP 99
APPP P +S + P P
Sbjct: 727 APPP---PPLSKTPVPPPP 742
Score = 44.7 bits (104), Expect = 6e-05
Identities = 25/70 (35%), Positives = 32/70 (45%), Gaps = 5/70 (7%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTRPEISS 92
P PP PP IP +P P P PP S ++ P + PP PPP + IS+
Sbjct: 640 PPPPPPPPPTRIPAAKCAP-----PPPPPPPTSHSGSIRVGPPSTPPPPPPPPPKANISN 694
Query: 93 SGFGPAPNTL 102
+ PAP L
Sbjct: 695 APKPPAPPPL 704
Score = 43.5 bits (101), Expect = 1e-04
Identities = 33/94 (35%), Positives = 42/94 (44%), Gaps = 17/94 (18%)
Query: 28 PATVLPARPPSPPRASIPVTTSSPFINSNLPA------PLPPKSSISQPQLPKPISLPPA 81
P T +PA +PP P T+ S I P+ P PPK++IS PKP + PP
Sbjct: 647 PPTRIPAAKCAPPPPPPPPTSHSGSIRVGPPSTPPPPPPPPPKANISNA--PKPPAPPPL 704
Query: 82 PPPSTR---------PEISSSGFGPAPNTLTEPV 106
PP STR P +S + P P PV
Sbjct: 705 PPSSTRLGAPPPPPPPPLSKTPAPPPPPLSKTPV 738
Score = 40.8 bits (94), Expect = 8e-04
Identities = 26/84 (30%), Positives = 36/84 (41%), Gaps = 3/84 (3%)
Query: 30 TVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTRPE 89
T+LP PP PP P+ TS+ + + P P PP + S PP PPP
Sbjct: 479 TLLPPPPPPPPP---PLFTSTTSFSPSQPPPPPPPPPLFMSTTSFSPSQPPPPPPPPPLF 535
Query: 90 ISSSGFGPAPNTLTEPVWDTVKRD 113
S++ F P+ P+ RD
Sbjct: 536 TSTTSFSPSQPPPPPPLPSFSNRD 559
Score = 40.4 bits (93), Expect = 0.001
Identities = 29/83 (34%), Positives = 38/83 (44%), Gaps = 6/83 (7%)
Query: 12 SSQSDIDEIENLIYASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQ 71
S++ + + I +P P PP P R SIP + P P P PP SS S
Sbjct: 556 SNRDPLTTLHQPINKTPPPPPPPPPPLPSR-SIPPPLAQPPPPRPPPPPPPPPSSRS--- 611
Query: 72 LPKPISLPPAPPPSTRPEISSSG 94
+P P + PP PPP P S+G
Sbjct: 612 IPSPSAPPPPPPPP--PSFGSTG 632
Score = 37.0 bits (84), Expect = 0.012
Identities = 24/69 (34%), Positives = 31/69 (44%), Gaps = 3/69 (4%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLPAPLPPKS--SISQPQLPKPISLPPAPPPSTRPEI 90
P RPP PP P + S P ++ P P PP S S + +P PP PPP+ P
Sbjct: 596 PPRPPPPPPPP-PSSRSIPSPSAPPPPPPPPPSFGSTGNKRQAQPPPPPPPPPPTRIPAA 654
Query: 91 SSSGFGPAP 99
+ P P
Sbjct: 655 KCAPPPPPP 663
Score = 35.4 bits (80), Expect = 0.034
Identities = 22/74 (29%), Positives = 28/74 (37%), Gaps = 5/74 (6%)
Query: 26 ASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPS 85
A P P PP PP +S + + S P P PP S + PP PPP
Sbjct: 592 AQPPPPRPPPPPPPPPSSRSIPSPSA-----PPPPPPPPPSFGSTGNKRQAQPPPPPPPP 646
Query: 86 TRPEISSSGFGPAP 99
I ++ P P
Sbjct: 647 PPTRIPAAKCAPPP 660
Score = 35.4 bits (80), Expect = 0.034
Identities = 24/83 (28%), Positives = 35/83 (41%), Gaps = 6/83 (7%)
Query: 24 IYASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPP 83
++ S + P++PP PP +++ F S P P PP P S P+ P
Sbjct: 492 LFTSTTSFSPSQPPPPPPPPPLFMSTTSFSPSQPPPPPPP-----PPLFTSTTSFSPSQP 546
Query: 84 PSTRPEISSSGFGPAPNTLTEPV 106
P P S S P TL +P+
Sbjct: 547 PPPPPLPSFSNRDPL-TTLHQPI 568
Score = 34.7 bits (78), Expect = 0.057
Identities = 21/63 (33%), Positives = 26/63 (40%), Gaps = 4/63 (6%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLPAPLP----PKSSISQPQLPKPISLPPAPPPSTRP 88
P PP PP + P P + +P P P SS P K + PP PPP+ R
Sbjct: 714 PPPPPPPPLSKTPAPPPPPLSKTPVPPPPPGLGRGTSSGPPPLGAKGSNAPPPPPPAGRG 773
Query: 89 EIS 91
S
Sbjct: 774 RAS 776
Score = 33.1 bits (74), Expect = 0.17
Identities = 22/69 (31%), Positives = 33/69 (46%), Gaps = 7/69 (10%)
Query: 24 IYASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQ----LPKPISLP 79
++ S + P++PP PP P+ TS+ + + P P PP S S L +PI+
Sbjct: 513 LFMSTTSFSPSQPPPPPPPP-PLFTSTTSFSPSQPPPPPPLPSFSNRDPLTTLHQPIN-- 569
Query: 80 PAPPPSTRP 88
PPP P
Sbjct: 570 KTPPPPPPP 578
Score = 28.5 bits (62), Expect = 4.1
Identities = 23/75 (30%), Positives = 28/75 (36%), Gaps = 3/75 (4%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTRPEISS 92
P PS P +S T P P PL ++ P P P PP PPP S
Sbjct: 464 PLNLPSDPPSSGDHVTLLPPPPPPPPPPLFTSTTSFSPSQPPP---PPPPPPLFMSTTSF 520
Query: 93 SGFGPAPNTLTEPVW 107
S P P P++
Sbjct: 521 SPSQPPPPPPPPPLF 535
>At4g18670 extensin-like protein
Length = 839
Score = 46.6 bits (109), Expect = 1e-05
Identities = 35/83 (42%), Positives = 39/83 (46%), Gaps = 5/83 (6%)
Query: 27 SPATVLPARP-PSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPA--PP 83
SP++ P+ P PSPP S P T SP NS P PP + S P P P LPP P
Sbjct: 548 SPSSPTPSSPIPSPPTPSTPPTPISPGQNSPPIIPSPPFTGPSPPSSPSP-PLPPVIPSP 606
Query: 84 PSTRPEISS-SGFGPAPNTLTEP 105
P P SS P P TL P
Sbjct: 607 PIVGPTPSSPPPSTPTPGTLLHP 629
Score = 41.2 bits (95), Expect = 6e-04
Identities = 29/83 (34%), Positives = 38/83 (44%), Gaps = 2/83 (2%)
Query: 24 IYASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAP- 82
I SP +P+ P +P P + SSP +S +P+P P S+ P P S P P
Sbjct: 525 ISPSPPITVPSPPSTPTSPGSPPSPSSPTPSSPIPSP-PTPSTPPTPISPGQNSPPIIPS 583
Query: 83 PPSTRPEISSSGFGPAPNTLTEP 105
PP T P SS P P + P
Sbjct: 584 PPFTGPSPPSSPSPPLPPVIPSP 606
Score = 37.0 bits (84), Expect = 0.012
Identities = 27/79 (34%), Positives = 33/79 (41%), Gaps = 13/79 (16%)
Query: 28 PATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAP----- 82
P V+P+ P +P P SP I+ P PP + S P P P PP+P
Sbjct: 406 PPVVVPSPPTTPSPGGSP---PSPSIS-----PSPPITVPSPPTTPSPGGSPPSPSIVPS 457
Query: 83 PPSTRPEISSSGFGPAPNT 101
PPST P S P T
Sbjct: 458 PPSTTPSPGSPPTSPTTPT 476
Score = 35.4 bits (80), Expect = 0.034
Identities = 31/84 (36%), Positives = 37/84 (43%), Gaps = 10/84 (11%)
Query: 26 ASPATVLPA-RPPSPPRASIPV-TTSSPFINSNLP--APLPPKSSIS-----QPQLPKPI 76
+SP T P PPS P P + SP I+ + P P PP + S P P P
Sbjct: 496 SSPTTPTPGGSPPSSPTTPSPGGSPPSPSISPSPPITVPSPPSTPTSPGSPPSPSSPTPS 555
Query: 77 SLPPAPP-PSTRPEISSSGFGPAP 99
S P+PP PST P S G P
Sbjct: 556 SPIPSPPTPSTPPTPISPGQNSPP 579
Score = 35.4 bits (80), Expect = 0.034
Identities = 33/102 (32%), Positives = 39/102 (37%), Gaps = 20/102 (19%)
Query: 24 IYASPATVLPARPP------SPPRASI----PVTTSSPFINSNLPA-PLPPKSSISQPQL 72
I SP +P+ P SPP SI P TT SP P P P S S P
Sbjct: 428 ISPSPPITVPSPPTTPSPGGSPPSPSIVPSPPSTTPSPGSPPTSPTTPTPGGSPPSSPTT 487
Query: 73 PKPISLPPA---------PPPSTRPEISSSGFGPAPNTLTEP 105
P P PP+ PPS+ S G P+P+ P
Sbjct: 488 PTPGGSPPSSPTTPTPGGSPPSSPTTPSPGGSPPSPSISPSP 529
Score = 35.4 bits (80), Expect = 0.034
Identities = 31/88 (35%), Positives = 37/88 (41%), Gaps = 10/88 (11%)
Query: 26 ASPATVLPA-RPPSPPRASIPVTTSSPFINSNLPAP--LPPKSSISQPQ---LPKPISLP 79
+SP T P PPS P + P SP + P+P PP SIS +P P S P
Sbjct: 483 SSPTTPTPGGSPPSSP--TTPTPGGSPPSSPTTPSPGGSPPSPSISPSPPITVPSPPSTP 540
Query: 80 PAP--PPSTRPEISSSGFGPAPNTLTEP 105
+P PPS SS P T P
Sbjct: 541 TSPGSPPSPSSPTPSSPIPSPPTPSTPP 568
Score = 35.4 bits (80), Expect = 0.034
Identities = 30/87 (34%), Positives = 37/87 (42%), Gaps = 14/87 (16%)
Query: 27 SPATVLPA-RPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPS 85
SP T P PPS P + P SP S+ P P S S P P P PP+P S
Sbjct: 471 SPTTPTPGGSPPSSP--TTPTPGGSP--PSSPTTPTPGGSPPSSPTTPSPGGSPPSPSIS 526
Query: 86 TRPEI---------SSSGFGPAPNTLT 103
P I +S G P+P++ T
Sbjct: 527 PSPPITVPSPPSTPTSPGSPPSPSSPT 553
Score = 35.4 bits (80), Expect = 0.034
Identities = 26/80 (32%), Positives = 34/80 (42%), Gaps = 11/80 (13%)
Query: 25 YASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPP 84
Y+SP +PP PP S+P T + S P P P S Q P I P PPP
Sbjct: 710 YSSP------QPPPPPHYSLPPPTPTYHYISPPPPPTPIHSPPPQSH-PPCIEYSPPPPP 762
Query: 85 STR----PEISSSGFGPAPN 100
+ P S + + P P+
Sbjct: 763 TVHYNPPPPPSPAHYSPPPS 782
Score = 30.8 bits (68), Expect = 0.83
Identities = 27/74 (36%), Positives = 30/74 (40%), Gaps = 11/74 (14%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLPAPL-------PPKSSISQPQLPKPISLPPAPPPS 85
P PPSP S P + + NS P P PP SQP P PI P PP
Sbjct: 768 PPPPPSPAHYSPPPSPPVYYYNSPPPPPAVHYSPPPPPVIHHSQPP-PPPIYEGPLPP-- 824
Query: 86 TRPEISSSGFGPAP 99
P IS + P P
Sbjct: 825 -IPGISYASPPPPP 837
Score = 28.9 bits (63), Expect = 3.1
Identities = 18/50 (36%), Positives = 20/50 (40%), Gaps = 3/50 (6%)
Query: 36 PPSPPRASI-PVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPP 84
PP PP P SP S P P PP + P P + P PPP
Sbjct: 758 PPPPPTVHYNPPPPPSPAHYS--PPPSPPVYYYNSPPPPPAVHYSPPPPP 805
Score = 28.9 bits (63), Expect = 3.1
Identities = 24/87 (27%), Positives = 32/87 (36%), Gaps = 13/87 (14%)
Query: 32 LPARPPSPPRASIPVTTSSPFINSNL----PAPLPPKSSISQPQLPKPISLPPAPPPSTR 87
LPARP +S P ++ + PP S P P P PP+P S
Sbjct: 372 LPARPAQRSPGQCAAFSSLPPVDCGSFGCGRSTRPPVVVPSPPTTPSPGGSPPSPSISPS 431
Query: 88 PEI---------SSSGFGPAPNTLTEP 105
P I S G P+P+ + P
Sbjct: 432 PPITVPSPPTTPSPGGSPPSPSIVPSP 458
Score = 27.7 bits (60), Expect = 7.0
Identities = 29/81 (35%), Positives = 32/81 (38%), Gaps = 10/81 (12%)
Query: 8 PLH-PSSQSDIDEIENLIYASPATVLPARPPSPPR-ASIPVTTSSPFINSNLPAPLPPKS 65
P+H P QS IE P + PP PP A S P N P P PP
Sbjct: 741 PIHSPPPQSHPPCIE--YSPPPPPTVHYNPPPPPSPAHYSPPPSPPVYYYNSPPP-PPAV 797
Query: 66 SISQPQLPKPI---SLPPAPP 83
S P P P+ S PP PP
Sbjct: 798 HYSPP--PPPVIHHSQPPPPP 816
Score = 27.7 bits (60), Expect = 7.0
Identities = 16/48 (33%), Positives = 20/48 (41%)
Query: 36 PPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPP 83
PP P S P + P I + P P + P P S PP+PP
Sbjct: 737 PPPTPIHSPPPQSHPPCIEYSPPPPPTVHYNPPPPPSPAHYSPPPSPP 784
>At3g24400 protein kinase, putative
Length = 694
Score = 46.6 bits (109), Expect = 1e-05
Identities = 29/81 (35%), Positives = 37/81 (44%), Gaps = 11/81 (13%)
Query: 28 PATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPP--PS 85
P LP PP PP A P P P LPP + P P P ++PP PP PS
Sbjct: 23 PPQPLPVTPPPPPTALPPALPPPP-----PPTALPP----ALPPPPPPTTVPPIPPSTPS 73
Query: 86 TRPEISSSGFGPAPNTLTEPV 106
P ++ S P+P T + P+
Sbjct: 74 PPPPLTPSPLPPSPTTPSPPL 94
Score = 45.8 bits (107), Expect = 2e-05
Identities = 30/86 (34%), Positives = 35/86 (39%), Gaps = 11/86 (12%)
Query: 26 ASPATVLPARPPSP-----PRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPP 80
A P P+ PP P P +PVT P P LPP P P +LPP
Sbjct: 4 APPPGGTPSPPPQPLPIPPPPQPLPVTPPPP------PTALPPALPPPPPPTALPPALPP 57
Query: 81 APPPSTRPEISSSGFGPAPNTLTEPV 106
PPP+T P I S P P P+
Sbjct: 58 PPPPTTVPPIPPSTPSPPPPLTPSPL 83
Score = 45.4 bits (106), Expect = 3e-05
Identities = 37/107 (34%), Positives = 50/107 (46%), Gaps = 10/107 (9%)
Query: 8 PLHPSSQSDIDEIE-NLIYASPATVLPARPPSPPRASIPVTTSSPFINSN----LPAPLP 62
P+ PS+ S + + + SP T P PSP S P+T S P + P+PLP
Sbjct: 66 PIPPSTPSPPPPLTPSPLPPSPTTPSPPLTPSPTTPSPPLTPSPPPAITPSPPLTPSPLP 125
Query: 63 PKSSISQPQLPKPI----SLPPAPPPSTRPEISSSGFGPAPNTLTEP 105
P + P P P L P+PPPS+ P SS P+P T + P
Sbjct: 126 PSPTTPSPPPPSPSIPSPPLTPSPPPSS-PLRPSSPPPPSPATPSTP 171
Score = 45.1 bits (105), Expect = 4e-05
Identities = 27/79 (34%), Positives = 38/79 (47%), Gaps = 8/79 (10%)
Query: 28 PATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTR 87
P + PA PP PP ++P P + +P P+PP S P P P++ P PP T
Sbjct: 35 PTALPPALPPPPPPTALPPALPPPPPPTTVP-PIPP----STPSPPPPLTPSPLPPSPTT 89
Query: 88 PEISSSGFGPAPNTLTEPV 106
P S P+P T + P+
Sbjct: 90 P---SPPLTPSPTTPSPPL 105
Score = 40.4 bits (93), Expect = 0.001
Identities = 32/106 (30%), Positives = 47/106 (44%), Gaps = 6/106 (5%)
Query: 6 TIPLHPSSQSDIDEIENLIYASPATVLPARPPSP-PRASIPVTTSSPFINSNLPAP---- 60
T P P + S L + P + P+ P +P P P T S P + ++P+P
Sbjct: 88 TTPSPPLTPSPTTPSPPLTPSPPPAITPSPPLTPSPLPPSPTTPSPPPPSPSIPSPPLTP 147
Query: 61 LPPKSSISQPQLPKPIS-LPPAPPPSTRPEISSSGFGPAPNTLTEP 105
PP SS +P P P S P+ PP + P S+ P +L+ P
Sbjct: 148 SPPPSSPLRPSSPPPPSPATPSTPPRSPPPPSTPTPPPRVGSLSPP 193
Score = 38.9 bits (89), Expect = 0.003
Identities = 29/82 (35%), Positives = 37/82 (44%), Gaps = 9/82 (10%)
Query: 28 PATVLPARPPSPPRASIPVTTSSPFINSNL----PAPLPPKSSISQPQLPKPISLPPAPP 83
P+ P+ PP P P T SP +S L P P P + + P+ P P S P PP
Sbjct: 126 PSPTTPSPPPPSPSIPSPPLTPSPPPSSPLRPSSPPPPSPATPSTPPRSPPPPSTPTPPP 185
Query: 84 P----STRPEISSSGFGPAPNT 101
S P S SG G +P+T
Sbjct: 186 RVGSLSPPPPASPSG-GRSPST 206
Score = 38.5 bits (88), Expect = 0.004
Identities = 28/90 (31%), Positives = 36/90 (39%), Gaps = 18/90 (20%)
Query: 28 PATVLPARPPSPPRASIPVT---------TSSPFINSNLPAPLPPKSSISQPQLP----- 73
P T +P PPS P P+T T SP + + P PP + P +
Sbjct: 60 PPTTVPPIPPSTPSPPPPLTPSPLPPSPTTPSPPLTPSPTTPSPPLTPSPPPAITPSPPL 119
Query: 74 KPISLPPAP----PPSTRPEISSSGFGPAP 99
P LPP+P PP P I S P+P
Sbjct: 120 TPSPLPPSPTTPSPPPPSPSIPSPPLTPSP 149
Score = 37.7 bits (86), Expect = 0.007
Identities = 28/85 (32%), Positives = 38/85 (43%), Gaps = 18/85 (21%)
Query: 28 PATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQL-PKPIS----LPPAP 82
P TV PP PP P + P+PLPP + P L P P + L P+P
Sbjct: 61 PTTV----PPIPPSTPSPPPPLT-------PSPLPPSPTTPSPPLTPSPTTPSPPLTPSP 109
Query: 83 PPSTRPE--ISSSGFGPAPNTLTEP 105
PP+ P ++ S P+P T + P
Sbjct: 110 PPAITPSPPLTPSPLPPSPTTPSPP 134
Score = 35.8 bits (81), Expect = 0.026
Identities = 21/69 (30%), Positives = 31/69 (44%)
Query: 26 ASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPS 85
+SP PA P +PPR+ P +T +P +P PP S P + P + PP+
Sbjct: 158 SSPPPPSPATPSTPPRSPPPPSTPTPPPRVGSLSPPPPASPSGGRSPSTPSTTPGSSPPA 217
Query: 86 TRPEISSSG 94
+ S G
Sbjct: 218 QSSKELSKG 226
Score = 30.0 bits (66), Expect = 1.4
Identities = 21/64 (32%), Positives = 24/64 (36%), Gaps = 2/64 (3%)
Query: 27 SPATVLPARP--PSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPP 84
SP P RP P PP + P T + P P P S+S P P P
Sbjct: 148 SPPPSSPLRPSSPPPPSPATPSTPPRSPPPPSTPTPPPRVGSLSPPPPASPSGGRSPSTP 207
Query: 85 STRP 88
ST P
Sbjct: 208 STTP 211
>At1g68690 protein kinase, putative
Length = 708
Score = 46.2 bits (108), Expect = 2e-05
Identities = 33/84 (39%), Positives = 41/84 (48%), Gaps = 11/84 (13%)
Query: 28 PATVLPARPPS--PPRASIPVTTSSP----FINSNLPAPLPPKSSISQPQLPKPISLPPA 81
P V+P+ PPS PP A +P SSP + P+P PP S P +PI PP
Sbjct: 98 PQPVIPSPPPSASPPPALVPPLPSSPPPPASVPPPRPSPSPPILVRSPPPSVRPIQSPP- 156
Query: 82 PPPSTRPEISSSGFGPAPNTLTEP 105
PPPS RP S P P+ + P
Sbjct: 157 PPPSDRPTQSP----PPPSPPSPP 176
Score = 46.2 bits (108), Expect = 2e-05
Identities = 32/88 (36%), Positives = 38/88 (42%), Gaps = 11/88 (12%)
Query: 27 SPATVLPARPPSPPRASIP---------VTTSSPFINSNLPAPLPPKSSISQPQLPKPIS 77
SPAT P P PP A P VTTS P + + P P PK S P+P+
Sbjct: 28 SPATPPPVTSPLPPSAPPPNRAPPPPPPVTTSPPPVANGAPPPPLPKPPESSSPPPQPVI 87
Query: 78 LPPAPPPSTRPEISSSGFGPAPNTLTEP 105
P+PPPST P P P+ P
Sbjct: 88 --PSPPPSTSPPPQPVIPSPPPSASPPP 113
Score = 43.9 bits (102), Expect = 9e-05
Identities = 26/73 (35%), Positives = 30/73 (40%), Gaps = 2/73 (2%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTRPEISS 92
P PPSPP S T S P S P PP S P +P PP PP T+P+
Sbjct: 169 PPSPPSPP--SERPTQSPPSPPSERPTQSPPPPSPPSPPSDRPSQSPPPPPEDTKPQPPR 226
Query: 93 SGFGPAPNTLTEP 105
P T + P
Sbjct: 227 RSPNSPPPTFSSP 239
Score = 43.9 bits (102), Expect = 9e-05
Identities = 26/67 (38%), Positives = 33/67 (48%), Gaps = 3/67 (4%)
Query: 36 PPSPPRASIPVTTSSP--FINSNLPAPLPPKSSISQPQLPKPISLPPAPPP-STRPEISS 92
P PP ++ P TS P N+ PA PP +S P P P PP PPP +T P +
Sbjct: 5 PVQPPVSNSPPVTSPPPPLNNATSPATPPPVTSPLPPSAPPPNRAPPPPPPVTTSPPPVA 64
Query: 93 SGFGPAP 99
+G P P
Sbjct: 65 NGAPPPP 71
Score = 42.0 bits (97), Expect = 4e-04
Identities = 27/69 (39%), Positives = 36/69 (52%), Gaps = 8/69 (11%)
Query: 28 PATVLPARPPS---PPRASIPVTTSS----PFINSNLPAPLPPKSSISQPQ-LPKPISLP 79
P V+P+ PPS PP+ IP S P + LP+ PP +S+ P+ P P L
Sbjct: 83 PQPVIPSPPPSTSPPPQPVIPSPPPSASPPPALVPPLPSSPPPPASVPPPRPSPSPPILV 142
Query: 80 PAPPPSTRP 88
+PPPS RP
Sbjct: 143 RSPPPSVRP 151
Score = 39.3 bits (90), Expect = 0.002
Identities = 29/78 (37%), Positives = 38/78 (48%), Gaps = 9/78 (11%)
Query: 24 IYASPATVLPARPPSP-PRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPP-- 80
+ SP V PP P P+ P +SSP +P+P P S QP +P P PP
Sbjct: 56 VTTSPPPVANGAPPPPLPK---PPESSSPPPQPVIPSPPPSTSPPPQPVIPSP---PPSA 109
Query: 81 APPPSTRPEISSSGFGPA 98
+PPP+ P + SS PA
Sbjct: 110 SPPPALVPPLPSSPPPPA 127
Score = 36.6 bits (83), Expect = 0.015
Identities = 22/66 (33%), Positives = 27/66 (40%), Gaps = 5/66 (7%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPA-----PPPSTR 87
P P PP + + P + S LP PP + P P S PP PPP +
Sbjct: 15 PVTSPPPPLNNATSPATPPPVTSPLPPSAPPPNRAPPPPPPVTTSPPPVANGAPPPPLPK 74
Query: 88 PEISSS 93
P SSS
Sbjct: 75 PPESSS 80
Score = 34.7 bits (78), Expect = 0.057
Identities = 39/123 (31%), Positives = 47/123 (37%), Gaps = 20/123 (16%)
Query: 28 PATVLPARP-PSPPRASIPVTTSSPFINSNLPAPLPPKSS--ISQPQLPKPISLP----- 79
PA+V P RP PSPP I V + P + + +P PP S P P P S P
Sbjct: 126 PASVPPPRPSPSPP---ILVRSPPPSVRP-IQSPPPPPSDRPTQSPPPPSPPSPPSERPT 181
Query: 80 --PAPPPSTRPEISSSGFGP------APNTLTEPVWDTVKRDLSRIVSNLKLVVFPNPYR 131
P PPS RP S P P+ P + K R N F +P R
Sbjct: 182 QSPPSPPSERPTQSPPPPSPPSPPSDRPSQSPPPPPEDTKPQPPRRSPNSPPPTFSSPPR 241
Query: 132 EDP 134
P
Sbjct: 242 SPP 244
>At5g07750 putative protein
Length = 1289
Score = 45.8 bits (107), Expect = 2e-05
Identities = 26/74 (35%), Positives = 34/74 (45%), Gaps = 4/74 (5%)
Query: 11 PSSQSDIDEIENLIYASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQP 70
P S E + ++++S A P PP PP P+ T S + S LP P PP S
Sbjct: 625 PPLPSLTSEAKTVLHSSQAVASPPPPPPPP----PLPTYSHYQTSQLPPPPPPPPPFSSE 680
Query: 71 QLPKPISLPPAPPP 84
+ LPP PPP
Sbjct: 681 RPNSGTVLPPPPPP 694
Score = 37.7 bits (86), Expect = 0.007
Identities = 24/69 (34%), Positives = 31/69 (44%), Gaps = 4/69 (5%)
Query: 30 TVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTRPE 89
TVLP PP PP P ++ P + LP P PP S + LPP P P +
Sbjct: 686 TVLPPPPPPPP----PFSSERPNSGTVLPPPPPPPLPFSSERPNSGTVLPPPPSPPWKSV 741
Query: 90 ISSSGFGPA 98
+S+ PA
Sbjct: 742 YASALAIPA 750
Score = 36.6 bits (83), Expect = 0.015
Identities = 29/95 (30%), Positives = 36/95 (37%), Gaps = 14/95 (14%)
Query: 7 IPLHPSSQSDIDEIENLIYASPATVLPARPPSPP--RASIPVTTSSPFINSNLPAPLPPK 64
+PLH S + + P P PP PP A ++ P S P P PP
Sbjct: 901 LPLHGISSAP---------SPPVKTAPPPPPPPPFSNAHSVLSPPPPSYGSPPPPPPPPP 951
Query: 65 SSISQPQLPKPISLPPAPPPSTRPEISSSGFGPAP 99
S S P P P +PPP P G+G P
Sbjct: 952 SYGSPPPPPPPPPSYGSPPPPPPP---PPGYGSPP 983
Score = 36.6 bits (83), Expect = 0.015
Identities = 19/57 (33%), Positives = 21/57 (36%), Gaps = 2/57 (3%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPI--SLPPAPPPSTR 87
P PP P P P P P PP + P P P+ PP PPP R
Sbjct: 1050 PPPPPPPMHGGAPPPPPPPMFGGAQPPPPPPMRGGAPPPPPPPMRGGAPPPPPPPMR 1106
Score = 36.2 bits (82), Expect = 0.020
Identities = 25/73 (34%), Positives = 28/73 (38%), Gaps = 9/73 (12%)
Query: 34 ARPPSPP--RASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPI--SLPPAPPPSTR-- 87
A+PP PP R P P P P PP + P P P+ PP PPP R
Sbjct: 1073 AQPPPPPPMRGGAPPPPPPPMRGGAPPPPPPPMRGGAPPPPPPPMHGGAPPPPPPPMRGG 1132
Query: 88 ---PEISSSGFGP 97
P G GP
Sbjct: 1133 APPPPPPPGGRGP 1145
Score = 36.2 bits (82), Expect = 0.020
Identities = 26/67 (38%), Positives = 28/67 (40%), Gaps = 11/67 (16%)
Query: 25 YASPATVLPARPPSPPR--ASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKP-----IS 77
Y SP P PP PP + P P S P P PP S S P P P S
Sbjct: 953 YGSP----PPPPPPPPSYGSPPPPPPPPPGYGSPPPPPPPPPSYGSPPPPPPPPFSHVSS 1008
Query: 78 LPPAPPP 84
+PP PPP
Sbjct: 1009 IPPPPPP 1015
Score = 35.8 bits (81), Expect = 0.026
Identities = 25/79 (31%), Positives = 28/79 (34%), Gaps = 8/79 (10%)
Query: 25 YASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPP---- 80
Y SP P PP PP P P + P P PP +P P PP
Sbjct: 966 YGSP----PPPPPPPPGYGSPPPPPPPPPSYGSPPPPPPPPFSHVSSIPPPPPPPPMHGG 1021
Query: 81 APPPSTRPEISSSGFGPAP 99
APPP P + P P
Sbjct: 1022 APPPPPPPPMHGGAPPPPP 1040
Score = 35.8 bits (81), Expect = 0.026
Identities = 23/68 (33%), Positives = 25/68 (35%), Gaps = 1/68 (1%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISL-PPAPPPSTRPEIS 91
P PP P R P P P P PP + P P P P APPP P
Sbjct: 1098 PPPPPPPMRGGAPPPPPPPMHGGAPPPPPPPMRGGAPPPPPPPGGRGPGAPPPPPPPGGR 1157
Query: 92 SSGFGPAP 99
+ G P P
Sbjct: 1158 APGPPPPP 1165
Score = 35.0 bits (79), Expect = 0.044
Identities = 22/67 (32%), Positives = 23/67 (33%), Gaps = 1/67 (1%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTRPEISS 92
P PP P R P P + P P PP P P P APPP P
Sbjct: 1086 PPPPPPPMRGGAPPPPPPP-MRGGAPPPPPPPMHGGAPPPPPPPMRGGAPPPPPPPGGRG 1144
Query: 93 SGFGPAP 99
G P P
Sbjct: 1145 PGAPPPP 1151
Score = 35.0 bits (79), Expect = 0.044
Identities = 28/81 (34%), Positives = 31/81 (37%), Gaps = 12/81 (14%)
Query: 25 YASPATVLPARPPSPPRASIPVTTSSPF--INSNLPAPLPPKSSISQPQLPKPISLPP-- 80
Y SP P PP P S P PF ++S P P PP P P P PP
Sbjct: 979 YGSPP---PPPPPPPSYGSPPPPPPPPFSHVSSIPPPPPPPPMHGGAPPPPPP---PPMH 1032
Query: 81 --APPPSTRPEISSSGFGPAP 99
APPP P + P P
Sbjct: 1033 GGAPPPPPPPPMHGGAPPPPP 1053
Score = 34.7 bits (78), Expect = 0.057
Identities = 20/58 (34%), Positives = 24/58 (40%), Gaps = 3/58 (5%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLPAPLPPKS-SISQPQLPKPI--SLPPAPPPSTR 87
P PP P P P ++ P P PP +QP P P+ PP PPP R
Sbjct: 1037 PPPPPPPMHGGAPPPPPPPPMHGGAPPPPPPPMFGGAQPPPPPPMRGGAPPPPPPPMR 1094
Score = 34.7 bits (78), Expect = 0.057
Identities = 26/77 (33%), Positives = 28/77 (35%), Gaps = 6/77 (7%)
Query: 25 YASPATVLPARPPSPPR--ASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAP 82
Y SP P PP PP + P P S P P PP S P P P +P
Sbjct: 940 YGSP----PPPPPPPPSYGSPPPPPPPPPSYGSPPPPPPPPPGYGSPPPPPPPPPSYGSP 995
Query: 83 PPSTRPEISSSGFGPAP 99
PP P S P P
Sbjct: 996 PPPPPPPFSHVSSIPPP 1012
Score = 34.7 bits (78), Expect = 0.057
Identities = 21/69 (30%), Positives = 23/69 (32%), Gaps = 2/69 (2%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLPAPLPP--KSSISQPQLPKPISLPPAPPPSTRPEI 90
P PP P P P P P PP + + P P P P PPP P
Sbjct: 1110 PPPPPPPMHGGAPPPPPPPMRGGAPPPPPPPGGRGPGAPPPPPPPGGRAPGPPPPPGPRP 1169
Query: 91 SSSGFGPAP 99
G P P
Sbjct: 1170 PGGGPPPPP 1178
Score = 33.9 bits (76), Expect = 0.098
Identities = 27/78 (34%), Positives = 32/78 (40%), Gaps = 10/78 (12%)
Query: 30 TVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPK-----SSI---SQPQLPKPISLPPA 81
T P PP PP SS S LP+P PP +S+ S+ LP P PP
Sbjct: 759 TSSPTPPPPPPAYYSVGQKSSDLQTSQLPSPPPPPPPPPFASVRRNSETLLPPP--PPPP 816
Query: 82 PPPSTRPEISSSGFGPAP 99
PPP +S P P
Sbjct: 817 PPPFASVRRNSETLLPPP 834
Score = 33.1 bits (74), Expect = 0.17
Identities = 25/90 (27%), Positives = 37/90 (40%), Gaps = 4/90 (4%)
Query: 6 TIPLHPSSQSDIDEIENLIYASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKS 65
T P P + + + + + S P PP PP AS+ S + P P PP +
Sbjct: 763 TPPPPPPAYYSVGQKSSDLQTSQLPSPPPPPPPPPFASVR-RNSETLLPPPPPPPPPPFA 821
Query: 66 SISQPQLPKPISLPPAPPPSTRPEISSSGF 95
S+ + LPP PPP + +S F
Sbjct: 822 SVRRNS---ETLLPPPPPPPPWKSLYASTF 848
Score = 33.1 bits (74), Expect = 0.17
Identities = 27/89 (30%), Positives = 32/89 (35%), Gaps = 9/89 (10%)
Query: 11 PSSQSDIDEIENLIYASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQP 70
PS I + + A V A PP PP PF N++ PP S S P
Sbjct: 894 PSPSVKILPLHGISSAPSPPVKTAPPPPPP---------PPFSNAHSVLSPPPPSYGSPP 944
Query: 71 QLPKPISLPPAPPPSTRPEISSSGFGPAP 99
P P +PPP P S P P
Sbjct: 945 PPPPPPPSYGSPPPPPPPPPSYGSPPPPP 973
Score = 33.1 bits (74), Expect = 0.17
Identities = 34/125 (27%), Positives = 48/125 (38%), Gaps = 25/125 (20%)
Query: 30 TVLPARPPSPPRASIPV----------TTSSPFINSNLPAPLPPKSSISQPQLPKPISLP 79
TVLP PPSPP S+ T+ +P + P P P S+ Q S
Sbjct: 728 TVLPP-PPSPPWKSVYASALAIPAICSTSQAPTSSPTPPPPPPAYYSVGQKSSDLQTSQL 786
Query: 80 PAPPPSTRP------EISSSGFGPAPNTLTEPVWDTVKRDLSRIVSNLKLVVFPNPYRED 133
P+PPP P +S P P P + +V+R+ ++ P P
Sbjct: 787 PSPPPPPPPPPFASVRRNSETLLPPPPPPPPPPFASVRRNSETLL--------PPPPPPP 838
Query: 134 PGKAL 138
P K+L
Sbjct: 839 PWKSL 843
Score = 32.7 bits (73), Expect = 0.22
Identities = 24/77 (31%), Positives = 30/77 (38%), Gaps = 5/77 (6%)
Query: 28 PATVLPARPPSPPRA---SIPVTTSSPFINSNLPAPLPPKSSI--SQPQLPKPISLPPAP 82
P+ P PP PP + SIP P ++ P P PP + P P P AP
Sbjct: 990 PSYGSPPPPPPPPFSHVSSIPPPPPPPPMHGGAPPPPPPPPMHGGAPPPPPPPPMHGGAP 1049
Query: 83 PPSTRPEISSSGFGPAP 99
PP P + P P
Sbjct: 1050 PPPPPPPMHGGAPPPPP 1066
Score = 32.7 bits (73), Expect = 0.22
Identities = 19/53 (35%), Positives = 20/53 (36%), Gaps = 4/53 (7%)
Query: 36 PPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKP----ISLPPAPPP 84
PP P S P P + P P PP S P P P PP PPP
Sbjct: 935 PPPPSYGSPPPPPPPPPSYGSPPPPPPPPPSYGSPPPPPPPPPGYGSPPPPPP 987
Score = 32.3 bits (72), Expect = 0.28
Identities = 23/68 (33%), Positives = 29/68 (41%), Gaps = 16/68 (23%)
Query: 30 TVLPARPPSPPRASIPVTTSSPFINSNLPAP-LPPKSSI-----------SQPQLPKPIS 77
TVLP PP P +P ++ P + LP P PP S+ S Q P
Sbjct: 707 TVLPPPPPPP----LPFSSERPNSGTVLPPPPSPPWKSVYASALAIPAICSTSQAPTSSP 762
Query: 78 LPPAPPPS 85
PP PPP+
Sbjct: 763 TPPPPPPA 770
Score = 32.3 bits (72), Expect = 0.28
Identities = 18/65 (27%), Positives = 26/65 (39%), Gaps = 5/65 (7%)
Query: 25 YASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPP---KSSISQPQLPKPI--SLP 79
++ +++ P PP P P P ++ P P PP P P P+ P
Sbjct: 1003 FSHVSSIPPPPPPPPMHGGAPPPPPPPPMHGGAPPPPPPPPMHGGAPPPPPPPPMHGGAP 1062
Query: 80 PAPPP 84
P PPP
Sbjct: 1063 PPPPP 1067
Score = 31.2 bits (69), Expect = 0.63
Identities = 27/86 (31%), Positives = 37/86 (42%), Gaps = 16/86 (18%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISL--------PP---A 81
P PP PP + + T ++ +I LP P P +SI+ K + L PP A
Sbjct: 861 PPPPPPPPFSPLNTTKANDYI---LPPPPLPYTSIAPSPSVKILPLHGISSAPSPPVKTA 917
Query: 82 PPPSTRPEISS--SGFGPAPNTLTEP 105
PPP P S+ S P P + P
Sbjct: 918 PPPPPPPPFSNAHSVLSPPPPSYGSP 943
Score = 30.4 bits (67), Expect = 1.1
Identities = 22/62 (35%), Positives = 26/62 (41%), Gaps = 5/62 (8%)
Query: 30 TVLPARPPSPPRASIPVTTSSPF---INSNLPAPLPPKSSISQPQLPK--PISLPPAPPP 84
T+LP PP PP S+ +T S+ P P PP S K LPP P P
Sbjct: 829 TLLPPPPPPPPWKSLYASTFETHEACSTSSSPPPPPPPPPFSPLNTTKANDYILPPPPLP 888
Query: 85 ST 86
T
Sbjct: 889 YT 890
Score = 30.4 bits (67), Expect = 1.1
Identities = 18/57 (31%), Positives = 23/57 (39%), Gaps = 5/57 (8%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLP--APLPPKSSISQPQLPKPI---SLPPAPPP 84
P PP P P P ++ P P PP + P P P+ + PP PPP
Sbjct: 1024 PPPPPPPMHGGAPPPPPPPPMHGGAPPPPPPPPMHGGAPPPPPPPMFGGAQPPPPPP 1080
Score = 30.4 bits (67), Expect = 1.1
Identities = 19/64 (29%), Positives = 21/64 (32%)
Query: 36 PPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTRPEISSSGF 95
PP PP P + P P PP P P P APPP P +
Sbjct: 1064 PPPPPMFGGAQPPPPPPMRGGAPPPPPPPMRGGAPPPPPPPMRGGAPPPPPPPMHGGAPP 1123
Query: 96 GPAP 99
P P
Sbjct: 1124 PPPP 1127
Score = 29.6 bits (65), Expect = 1.8
Identities = 17/54 (31%), Positives = 19/54 (34%), Gaps = 2/54 (3%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPI--SLPPAPPP 84
P PP P P P P PP + P P P+ PP PPP
Sbjct: 1062 PPPPPPPMFGGAQPPPPPPMRGGAPPPPPPPMRGGAPPPPPPPMRGGAPPPPPP 1115
Score = 28.5 bits (62), Expect = 4.1
Identities = 21/67 (31%), Positives = 22/67 (32%), Gaps = 4/67 (5%)
Query: 37 PSPPRASIPVTTSSPFINSNLPAPLPPK----SSISQPQLPKPISLPPAPPPSTRPEISS 92
P PP P P + P P PP S P P P P PPP P S
Sbjct: 935 PPPPSYGSPPPPPPPPPSYGSPPPPPPPPPSYGSPPPPPPPPPGYGSPPPPPPPPPSYGS 994
Query: 93 SGFGPAP 99
P P
Sbjct: 995 PPPPPPP 1001
Score = 28.1 bits (61), Expect = 5.4
Identities = 19/68 (27%), Positives = 22/68 (31%), Gaps = 1/68 (1%)
Query: 33 PARPPSPPRASIPVTTSSPFINS-NLPAPLPPKSSISQPQLPKPISLPPAPPPSTRPEIS 91
P PP P R P P P P PP + P P PP P P +
Sbjct: 1122 PPPPPPPMRGGAPPPPPPPGGRGPGAPPPPPPPGGRAPGPPPPPGPRPPGGGPPPPPMLG 1181
Query: 92 SSGFGPAP 99
+ G P
Sbjct: 1182 ARGAAVDP 1189
>At2g28440 En/Spm-like transposon protein
Length = 268
Score = 45.8 bits (107), Expect = 2e-05
Identities = 42/116 (36%), Positives = 52/116 (44%), Gaps = 19/116 (16%)
Query: 8 PLHPSSQSDIDEIENLIYASPATVLPARPPSPPRASIPVT-TSSPFINSNLP-------- 58
PL PSS + D +SP P P S P P+ +SSP ++S P
Sbjct: 56 PLSPSSSPEEDS-PLPPSSSPEEDSPLAPSSSPEVDSPLAPSSSPEVDSPQPPSSSPEAD 114
Query: 59 APLPPKSS--ISQPQLP----KPISL---PPAPPPSTRPEISSSGFGPAPNTLTEP 105
+PLPP SS + PQ P KP SL P PPP +PE SS P P + P
Sbjct: 115 SPLPPSSSPEANSPQSPASSPKPESLADSPSPPPPPPQPESPSSPSYPEPAPVPAP 170
Score = 40.8 bits (94), Expect = 8e-04
Identities = 35/96 (36%), Positives = 44/96 (45%), Gaps = 6/96 (6%)
Query: 8 PLHPSSQSDIDEIENLIYASPATVLPARPPSPPRASIPVT-TSSPFINSNLPAPLPPKSS 66
PL PSS ++D + +SP P P S P A+ P + SSP S +P PP
Sbjct: 92 PLAPSSSPEVDSPQPPS-SSPEADSPLPPSSSPEANSPQSPASSPKPESLADSPSPPPPP 150
Query: 67 ISQPQLPKPISLP-PAPPPSTRPEISSSGFGPAPNT 101
QP+ P S P PAP P+ P S P P T
Sbjct: 151 -PQPESPSSPSYPEPAPVPA--PSDDDSDDDPEPET 183
Score = 35.8 bits (81), Expect = 0.026
Identities = 29/87 (33%), Positives = 39/87 (44%), Gaps = 21/87 (24%)
Query: 27 SPATVLPARPPSPPRASIPVTT---SSPFINSNLP---APLPPKSS---------ISQPQ 71
SPA V P R PS P ++ SP S+ P +PLPP SS S P+
Sbjct: 29 SPAAVSPGREPSTDSPLSPSSSPEEDSPLSPSSSPEEDSPLPPSSSPEEDSPLAPSSSPE 88
Query: 72 LPKPISLPPAP------PPSTRPEISS 92
+ P++ +P PPS+ PE S
Sbjct: 89 VDSPLAPSSSPEVDSPQPPSSSPEADS 115
Score = 32.7 bits (73), Expect = 0.22
Identities = 27/87 (31%), Positives = 39/87 (44%), Gaps = 11/87 (12%)
Query: 8 PLHPSSQSDIDEIENLIYASPATVL---PARPPSPPRASIPVTTSSPFINSNLPAPLPPK 64
PL PSS + + ++ + L P+ PP PP+ P + S P PAP+P
Sbjct: 116 PLPPSSSPEANSPQSPASSPKPESLADSPSPPPPPPQPESPSSPSYP-----EPAPVPAP 170
Query: 65 SSISQPQLPKP-ISLPPAPPPSTRPEI 90
S P+P P+P PS PE+
Sbjct: 171 SDDDSDDDPEPETEYFPSPAPS--PEL 195
>At4g32710 putative protein kinase
Length = 731
Score = 45.4 bits (106), Expect = 3e-05
Identities = 29/78 (37%), Positives = 39/78 (49%), Gaps = 3/78 (3%)
Query: 30 TVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKS-SISQPQLPKPI-SLPPAPPPSTR 87
TV P SPP S+P ++ P ++ P P PP+ S +P P PI S PP P P+T
Sbjct: 129 TVPPGNTISPPPRSLPSESTPPVNTASPPPPSPPRRRSGPKPSFPPPINSSPPNPSPNT- 187
Query: 88 PEISSSGFGPAPNTLTEP 105
P + + P P T P
Sbjct: 188 PSLPETSPPPKPPLSTTP 205
Score = 43.1 bits (100), Expect = 2e-04
Identities = 30/83 (36%), Positives = 39/83 (46%), Gaps = 6/83 (7%)
Query: 28 PATVLPARPPSPPRASIPVTTSSPFIN-----SNLPAPLPPKSSISQPQLPKPISLPPAP 82
P LPA+P SPP +S P T P +LP+ P + + P P P P
Sbjct: 110 PLPFLPAKP-SPPPSSPPSETVPPGNTISPPPRSLPSESTPPVNTASPPPPSPPRRRSGP 168
Query: 83 PPSTRPEISSSGFGPAPNTLTEP 105
PS P I+SS P+PNT + P
Sbjct: 169 KPSFPPPINSSPPNPSPNTPSLP 191
Score = 42.4 bits (98), Expect = 3e-04
Identities = 34/81 (41%), Positives = 40/81 (48%), Gaps = 9/81 (11%)
Query: 28 PATVLPARPPSPPRA-SIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPST 86
P PPSPPR S P + P INS+ P P P + P LP+ S PP PP ST
Sbjct: 150 PVNTASPPPPSPPRRRSGPKPSFPPPINSSPPNPSP-----NTPSLPE-TSPPPKPPLST 203
Query: 87 RPEISSSGFGP--APNTLTEP 105
P SSS P +P +T P
Sbjct: 204 TPFPSSSTPPPKKSPAAVTLP 224
Score = 41.2 bits (95), Expect = 6e-04
Identities = 30/81 (37%), Positives = 41/81 (50%), Gaps = 10/81 (12%)
Query: 28 PATVLP--ARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPP-P 84
PA +P + PP+PP + +P SSP LP+P PP S+ + P P+ PP+PP
Sbjct: 22 PADSVPDTSSPPAPPLSPLPPPLSSP---PPLPSP-PPLSAPTASPPPLPVESPPSPPIE 77
Query: 85 STRPEISSSGFGPAPNTLTEP 105
S P + S P P L P
Sbjct: 78 SPPPPLLES---PPPPPLESP 95
Score = 37.7 bits (86), Expect = 0.007
Identities = 30/84 (35%), Positives = 35/84 (40%), Gaps = 9/84 (10%)
Query: 26 ASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAP--- 82
ASP + PPSPP S P P + S P PL S S P + P PP P
Sbjct: 61 ASPPPLPVESPPSPPIESPP----PPLLESPPPPPLESPSPPS-PHVSAPSGSPPLPFLP 115
Query: 83 -PPSTRPEISSSGFGPAPNTLTEP 105
PS P S P NT++ P
Sbjct: 116 AKPSPPPSSPPSETVPPGNTISPP 139
Score = 37.0 bits (84), Expect = 0.012
Identities = 28/84 (33%), Positives = 31/84 (36%), Gaps = 13/84 (15%)
Query: 28 PATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTR 87
P P PSPP S P + P LP PP I P P P+ P PPP
Sbjct: 42 PPLSSPPPLPSPPPLSAPTASPPP-----LPVESPPSPPIESP--PPPLLESPPPPPLES 94
Query: 88 PEISS------SGFGPAPNTLTEP 105
P S SG P P +P
Sbjct: 95 PSPPSPHVSAPSGSPPLPFLPAKP 118
Score = 37.0 bits (84), Expect = 0.012
Identities = 24/62 (38%), Positives = 29/62 (46%), Gaps = 11/62 (17%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLP---PAPPPSTRPE 89
PA SPP S+P S P +S PL P LP P+S P P+PPP + P
Sbjct: 9 PAPATSPPAMSLPPADSVPDTSSPPAPPLSP--------LPPPLSSPPPLPSPPPLSAPT 60
Query: 90 IS 91
S
Sbjct: 61 AS 62
Score = 34.3 bits (77), Expect = 0.075
Identities = 23/77 (29%), Positives = 31/77 (39%), Gaps = 14/77 (18%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPP----APPPSTRP 88
P PSPP + + SP P P P P P ++PP +PPP + P
Sbjct: 91 PLESPSPPSPHVSAPSGSP------PLPFLPAKPSPPPSSPPSETVPPGNTISPPPRSLP 144
Query: 89 EISSSGFGPAPNTLTEP 105
S+ P NT + P
Sbjct: 145 SEST----PPVNTASPP 157
Score = 30.0 bits (66), Expect = 1.4
Identities = 20/53 (37%), Positives = 25/53 (46%), Gaps = 1/53 (1%)
Query: 54 NSNLPAPLPPKSSISQPQLPKPISLPPAPPPS-TRPEISSSGFGPAPNTLTEP 105
+S PA PP S+ S PPAPP S P +SS P+P L+ P
Sbjct: 7 SSPAPATSPPAMSLPPADSVPDTSSPPAPPLSPLPPPLSSPPPLPSPPPLSAP 59
>At4g27520 early nodulin-like 2 predicted GPI-anchored protein
Length = 349
Score = 45.1 bits (105), Expect = 4e-05
Identities = 28/77 (36%), Positives = 42/77 (54%), Gaps = 9/77 (11%)
Query: 36 PPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPP-------APPPSTRP 88
PP+PP+++ PV+ SS + S PAP+ PKSS + P P++ PP + P S P
Sbjct: 217 PPAPPKSTSPVSPSSAPMTSP-PAPMAPKSSSTIPPSSAPMTSPPGSMAPKSSSPVSNSP 275
Query: 89 EISSSGFGPAPNTLTEP 105
+S S P +T + P
Sbjct: 276 TVSPS-LAPGGSTSSSP 291
Score = 35.0 bits (79), Expect = 0.044
Identities = 24/77 (31%), Positives = 33/77 (42%), Gaps = 10/77 (12%)
Query: 27 SPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAP---- 82
SP + P + P S+ + SP + P P PPKS+ P++ PPAP
Sbjct: 186 SPKSSSAVSPATSPPGSMAPKSGSPVSPTTSP-PAPPKSTSPVSPSSAPMTSPPAPMAPK 244
Query: 83 -----PPSTRPEISSSG 94
PPS+ P S G
Sbjct: 245 SSSTIPPSSAPMTSPPG 261
Score = 34.3 bits (77), Expect = 0.075
Identities = 23/67 (34%), Positives = 30/67 (44%), Gaps = 4/67 (5%)
Query: 27 SPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPST 86
SP T P P A P ++S+ ++ P + PKS P + PPAPP ST
Sbjct: 169 SPTTSPPGSTTPPGGAHSPKSSSAVSPATSPPGSMAPKSG----SPVSPTTSPPAPPKST 224
Query: 87 RPEISSS 93
P SS
Sbjct: 225 SPVSPSS 231
Score = 32.0 bits (71), Expect = 0.37
Identities = 27/95 (28%), Positives = 39/95 (40%), Gaps = 11/95 (11%)
Query: 8 PLHPSSQSDIDEIENLIYASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSS- 66
P P S S + + + PA + P + P +S P+T+ P + PKSS
Sbjct: 218 PAPPKSTSPVSPSSAPMTSPPAPMAPKSSSTIPPSSAPMTSP--------PGSMAPKSSS 269
Query: 67 -ISQPQLPKPISLPPAPPPSTRPEISSSGFGPAPN 100
+S P SL P S+ P S SG P+
Sbjct: 270 PVSNSPTVSP-SLAPGGSTSSSPSDSPSGSAMGPS 303
Score = 31.6 bits (70), Expect = 0.49
Identities = 22/66 (33%), Positives = 27/66 (40%), Gaps = 9/66 (13%)
Query: 42 ASIPVTTSSPFI---NSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTRP------EISS 92
A IP T SP S+ P + P P+ P+S +PP ST P SS
Sbjct: 131 ARIPSTAQSPHAAAPGSSTPGSMTPPGGAHSPKSSSPVSPTTSPPGSTTPPGGAHSPKSS 190
Query: 93 SGFGPA 98
S PA
Sbjct: 191 SAVSPA 196
>At5g38560 putative protein
Length = 681
Score = 43.5 bits (101), Expect = 1e-04
Identities = 22/57 (38%), Positives = 32/57 (55%), Gaps = 3/57 (5%)
Query: 49 SSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTRPEISSSGFGPAPNTLTEP 105
S P NS+ AP P ++ + P P P++ PP+PP S P +SSS P P ++ P
Sbjct: 11 SPPSSNSSTTAPPPLQTQPTTPSAPPPVTPPPSPPQSPPPVVSSS---PPPPVVSSP 64
Score = 38.9 bits (89), Expect = 0.003
Identities = 27/85 (31%), Positives = 41/85 (47%), Gaps = 7/85 (8%)
Query: 26 ASPATVLPARPPSPPRASIPVTTSSP---FINSNLPAPLPPKSSISQPQLPKPISLPPAP 82
++P V P PPSPP++ PV +SSP ++S P+ PP S P ++ P P
Sbjct: 33 SAPPPVTP--PPSPPQSPPPVVSSSPPPPVVSSPPPSSSPPPSPPVITSPPPTVASSPPP 90
Query: 83 PP--STRPEISSSGFGPAPNTLTEP 105
P ++ P + + PAP P
Sbjct: 91 PVVIASPPPSTPATTPPAPPQTVSP 115
Score = 38.9 bits (89), Expect = 0.003
Identities = 27/85 (31%), Positives = 37/85 (42%), Gaps = 14/85 (16%)
Query: 28 PATVLPARPPS--PPRASIPVTTSSPFINSNLPAPL-----PPKSSISQPQLPKPISLPP 80
P V+ + PPS PP + +T+ P + S+ P P+ PP + + P P PP
Sbjct: 57 PPPVVSSPPPSSSPPPSPPVITSPPPTVASSPPPPVVIASPPPSTPATTPPAPPQTVSPP 116
Query: 81 APPPSTRPEISSSGFGPAPNTLTEP 105
PPP P PAP T P
Sbjct: 117 -PPPDASPS------PPAPTTTNPP 134
Score = 38.5 bits (88), Expect = 0.004
Identities = 30/88 (34%), Positives = 37/88 (41%), Gaps = 13/88 (14%)
Query: 23 LIYASPATVLPAR-PPSPPRASIPVTTSSPFINSNLPAPL----PPKSSISQPQLPKPIS 77
++ ASP PA PP+PP+ P P + + PAP PPK S P P
Sbjct: 92 VVIASPPPSTPATTPPAPPQTVSP--PPPPDASPSPPAPTTTNPPPKPS------PSPPG 143
Query: 78 LPPAPPPSTRPEISSSGFGPAPNTLTEP 105
P+PP T S P P T T P
Sbjct: 144 ETPSPPGETPSPPKPSPSTPTPTTTTSP 171
Score = 38.5 bits (88), Expect = 0.004
Identities = 36/101 (35%), Positives = 40/101 (38%), Gaps = 25/101 (24%)
Query: 26 ASPATVLPA-----RPPSPPRAS----IPVTTSSPFINSNLP----------APLPPKSS 66
++PAT PA PP PP AS P TT+ P S P P PPK S
Sbjct: 100 STPATTPPAPPQTVSPPPPPDASPSPPAPTTTNPPPKPSPSPPGETPSPPGETPSPPKPS 159
Query: 67 ISQPQLPKPISLPPAP-----PPSTRPEISSSGFGPAPNTL 102
S P S PP P PPS+ P S P P L
Sbjct: 160 PSTPTPTTTTSPPPPPATSASPPSSNP-TDPSTLAPPPTPL 199
Score = 36.2 bits (82), Expect = 0.020
Identities = 25/79 (31%), Positives = 32/79 (39%), Gaps = 9/79 (11%)
Query: 27 SPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPST 86
SP + + PP + P T S+P P PP S PQ P P+ PPP
Sbjct: 11 SPPSSNSSTTAPPPLQTQPTTPSAP------PPVTPPPSP---PQSPPPVVSSSPPPPVV 61
Query: 87 RPEISSSGFGPAPNTLTEP 105
SS P+P +T P
Sbjct: 62 SSPPPSSSPPPSPPVITSP 80
Score = 36.2 bits (82), Expect = 0.020
Identities = 25/80 (31%), Positives = 35/80 (43%), Gaps = 10/80 (12%)
Query: 23 LIYASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAP 82
+I + P TV + PP PV +SP ++ P P ++S P P PPAP
Sbjct: 76 VITSPPPTVASSPPP-------PVVIASPPPSTPATTPPAPPQTVSPPPPPDASPSPPAP 128
Query: 83 P---PSTRPEISSSGFGPAP 99
P +P S G P+P
Sbjct: 129 TTTNPPPKPSPSPPGETPSP 148
Score = 35.4 bits (80), Expect = 0.034
Identities = 27/77 (35%), Positives = 34/77 (44%), Gaps = 17/77 (22%)
Query: 33 PARPPSPPRAS----IPVTTSSPFINSNLPAPLPPKSSISQPQL----PKPISLPPAP-P 83
P PSPP+ S P TT+SP P PP +S S P P ++ PP P P
Sbjct: 149 PGETPSPPKPSPSTPTPTTTTSP--------PPPPATSASPPSSNPTDPSTLAPPPTPLP 200
Query: 84 PSTRPEISSSGFGPAPN 100
R + + GPA N
Sbjct: 201 VVPREKPIAKPTGPASN 217
>At5g51680 unknown protein
Length = 343
Score = 42.7 bits (99), Expect = 2e-04
Identities = 28/68 (41%), Positives = 32/68 (46%), Gaps = 6/68 (8%)
Query: 27 SPATVLPARPPSPPRASIPV--TTSSPFINSNLPAPLPPKSSISQPQLPKP----ISLPP 80
S AT +P+ PP PP +PV TS PF P P SS PQLP P + P
Sbjct: 40 SLATFVPSPPPLPPPIPVPVKLVTSVPFRWEETPGKPLPASSNDPPQLPHPPLETATPTP 99
Query: 81 APPPSTRP 88
PPP P
Sbjct: 100 LPPPVPVP 107
>At4g13340 extensin-like protein
Length = 760
Score = 42.7 bits (99), Expect = 2e-04
Identities = 30/85 (35%), Positives = 35/85 (40%), Gaps = 5/85 (5%)
Query: 28 PATVLPARPPSPPRASIPVTT-SSPFINSNLPAPLPPKSSISQPQLPKP----ISLPPAP 82
P V P PPS P P S+P ++ P P PP S P P P S PP P
Sbjct: 396 PPVVTPLPPPSLPSPPPPAPIFSTPPTLTSPPPPSPPPPVYSPPPPPPPPPPVYSPPPPP 455
Query: 83 PPSTRPEISSSGFGPAPNTLTEPVW 107
PP P + S P P PV+
Sbjct: 456 PPPPPPPVYSPPPPPPPPPPPPPVY 480
Score = 41.6 bits (96), Expect = 5e-04
Identities = 30/90 (33%), Positives = 37/90 (40%), Gaps = 16/90 (17%)
Query: 24 IYASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKP------IS 77
I+++P T+ PPSPP P + S P P PP S P P P S
Sbjct: 416 IFSTPPTLTSPPPPSPP----------PPVYSPPPPPPPPPPVYSPPPPPPPPPPPPVYS 465
Query: 78 LPPAPPPSTRPEISSSGFGPAPNTLTEPVW 107
PP PPP P S P+P PV+
Sbjct: 466 PPPPPPPPPPPPPVYSPPPPSPPPPPPPVY 495
Score = 39.3 bits (90), Expect = 0.002
Identities = 26/79 (32%), Positives = 32/79 (39%), Gaps = 7/79 (8%)
Query: 28 PATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPP-PST 86
P V PPSPP PV + P P P PP + P P S PP PP P+
Sbjct: 476 PPPVYSPPPPSPPPPPPPVYSPPP------PPPPPPPPPVYSPPPPPVYSSPPPPPSPAP 529
Query: 87 RPEISSSGFGPAPNTLTEP 105
P + P P++ P
Sbjct: 530 TPVYCTRPPPPPPHSPPPP 548
Score = 37.4 bits (85), Expect = 0.009
Identities = 24/68 (35%), Positives = 28/68 (40%), Gaps = 5/68 (7%)
Query: 21 ENLIYASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPP 80
E Y+SP P PP + P + P I L P PP S P P P+ PP
Sbjct: 556 EPYYYSSPP---PPHSSPPPHSPPPPHSPPPPIYPYLSPPPPPTPVSSPP--PTPVYSPP 610
Query: 81 APPPSTRP 88
PPP P
Sbjct: 611 PPPPCIEP 618
Score = 37.0 bits (84), Expect = 0.012
Identities = 27/95 (28%), Positives = 36/95 (37%), Gaps = 16/95 (16%)
Query: 24 IYASPATVLPARPP---SPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKP----- 75
+Y+ P P PP SPP P P + P+P PP + P P P
Sbjct: 448 VYSPPPPPPPPPPPPVYSPPPPPPPPPPPPPVYSPPPPSPPPPPPPVYSPPPPPPPPPPP 507
Query: 76 -ISLPPAPPPSTRPEISSSGFGPAPNTLTEPVWDT 109
+ PP PP + P P P+ PV+ T
Sbjct: 508 PVYSPPPPPVYSSPP-------PPPSPAPTPVYCT 535
Score = 36.6 bits (83), Expect = 0.015
Identities = 20/60 (33%), Positives = 25/60 (41%), Gaps = 3/60 (5%)
Query: 25 YASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPP 84
++SP P P SPP P + P P PP + + P P P PP PPP
Sbjct: 567 HSSPPPHSPPPPHSPPPPIYPYLSPPP---PPTPVSSPPPTPVYSPPPPPPCIEPPPPPP 623
Score = 35.4 bits (80), Expect = 0.034
Identities = 27/89 (30%), Positives = 35/89 (38%), Gaps = 9/89 (10%)
Query: 24 IYASPATVLPARPPSPPRASIPVTTSSPFINSNLPAP--LPPKSSISQPQLPKP-----I 76
+Y + P P PP+ S P P+ S+ P P PP S P P P +
Sbjct: 532 VYCTRPPPPPPHSPPPPQFSPP--PPEPYYYSSPPPPHSSPPPHSPPPPHSPPPPIYPYL 589
Query: 77 SLPPAPPPSTRPEISSSGFGPAPNTLTEP 105
S PP P P + P + P P EP
Sbjct: 590 SPPPPPTPVSSPPPTPVYSPPPPPPCIEP 618
Score = 34.3 bits (77), Expect = 0.075
Identities = 26/83 (31%), Positives = 30/83 (35%), Gaps = 4/83 (4%)
Query: 27 SPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISL----PPAP 82
SP P PP P + P P P P P SS P P P + PP P
Sbjct: 481 SPPPPSPPPPPPPVYSPPPPPPPPPPPPVYSPPPPPVYSSPPPPPSPAPTPVYCTRPPPP 540
Query: 83 PPSTRPEISSSGFGPAPNTLTEP 105
PP + P S P P + P
Sbjct: 541 PPHSPPPPQFSPPPPEPYYYSSP 563
Score = 33.9 bits (76), Expect = 0.098
Identities = 30/92 (32%), Positives = 36/92 (38%), Gaps = 13/92 (14%)
Query: 27 SPATVLPARPPSPPRASIPVTTSSP-----FINSNLPAP--LPPKSSISQPQLPKP---- 75
+P V RPP PP S P SP + S+ P P PP S P P P
Sbjct: 528 APTPVYCTRPPPPPPHSPPPPQFSPPPPEPYYYSSPPPPHSSPPPHSPPPPHSPPPPIYP 587
Query: 76 -ISLPPAPPP-STRPEISSSGFGPAPNTLTEP 105
+S PP P P S+ P P P + P
Sbjct: 588 YLSPPPPPTPVSSPPPTPVYSPPPPPPCIEPP 619
Score = 32.7 bits (73), Expect = 0.22
Identities = 24/78 (30%), Positives = 33/78 (41%), Gaps = 8/78 (10%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSI--SQPQLPKPISLPP---APPPSTR 87
P PP PP + P +S P P P + + ++P P P S PP +PPP
Sbjct: 498 PPPPPPPPPPPVYSPPPPPVYSSPPPPPSPAPTPVYCTRPPPPPPHSPPPPQFSPPP--- 554
Query: 88 PEISSSGFGPAPNTLTEP 105
PE P P++ P
Sbjct: 555 PEPYYYSSPPPPHSSPPP 572
Score = 31.2 bits (69), Expect = 0.63
Identities = 26/86 (30%), Positives = 33/86 (38%), Gaps = 8/86 (9%)
Query: 27 SPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPI--SLPPAPPP 84
SP P P PP I + P + +P PP S P P P+ S PP PPP
Sbjct: 608 SPPPPPPCIEPPPPPPCIEYSPPPPPPVVHYSSPPPPPVYYSSPP-PPPVYYSSPPPPPP 666
Query: 85 -----STRPEISSSGFGPAPNTLTEP 105
PE+ P+P + P
Sbjct: 667 VHYSSPPPPEVHYHSPPPSPVHYSSP 692
Score = 30.4 bits (67), Expect = 1.1
Identities = 20/60 (33%), Positives = 25/60 (41%), Gaps = 2/60 (3%)
Query: 24 IYASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPP 83
+ + P T + + PP PP P P I + P P P S P P S PP PP
Sbjct: 598 VSSPPPTPVYSPPPPPPCIEPP--PPPPCIEYSPPPPPPVVHYSSPPPPPVYYSSPPPPP 655
Score = 28.9 bits (63), Expect = 3.1
Identities = 21/80 (26%), Positives = 28/80 (34%), Gaps = 5/80 (6%)
Query: 25 YASPATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPP 84
Y+SP P SPP + + P P PP + + P P+ PPP
Sbjct: 658 YSSPPPPPPVHYSSPPPPEVHYHSPPPSPVHYSSPPPPPSAPCEESPPPAPVVHHSPPPP 717
Query: 85 STR-----PEISSSGFGPAP 99
P I S P+P
Sbjct: 718 MVHHSPPPPVIHQSPPPPSP 737
Score = 27.3 bits (59), Expect = 9.1
Identities = 23/65 (35%), Positives = 27/65 (41%), Gaps = 17/65 (26%)
Query: 28 PATVLPARPPSPPRASIPVTTSSPFINSNLPAP-----LPPKSSISQPQLPKPISLP--P 80
P V + PP PP PV SSP P P PP S + P P S P
Sbjct: 653 PPPVYYSSPPPPP----PVHYSSP------PPPEVHYHSPPPSPVHYSSPPPPPSAPCEE 702
Query: 81 APPPS 85
+PPP+
Sbjct: 703 SPPPA 707
>At1g61080 hypothetical protein
Length = 907
Score = 42.7 bits (99), Expect = 2e-04
Identities = 21/64 (32%), Positives = 31/64 (47%), Gaps = 5/64 (7%)
Query: 36 PPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTRPEISSSGF 95
PP PP +P T ++P P P PP+++++ P P P APPP P + +
Sbjct: 508 PPPPPPPPLPTTIAAP-----PPPPPPPRAAVAPPPPPPPPGTAAAPPPPPPPPGTQAAP 562
Query: 96 GPAP 99
P P
Sbjct: 563 PPPP 566
Score = 40.4 bits (93), Expect = 0.001
Identities = 24/78 (30%), Positives = 33/78 (41%), Gaps = 9/78 (11%)
Query: 28 PATVLPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTR 87
P V+P + +PP + P + + P P PP P LP I+ PP PPP R
Sbjct: 481 PPAVMPLKHFAPPPPTPPAFKP---LKGSAPPPPPP------PPLPTTIAAPPPPPPPPR 531
Query: 88 PEISSSGFGPAPNTLTEP 105
++ P P T P
Sbjct: 532 AAVAPPPPPPPPGTAAAP 549
Score = 39.7 bits (91), Expect = 0.002
Identities = 23/73 (31%), Positives = 31/73 (41%), Gaps = 11/73 (15%)
Query: 28 PATVL-PARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPST 86
P T+ P PP PPRA++ P P PP ++ + P P P APPP
Sbjct: 517 PTTIAAPPPPPPPPRAAVAPP----------PPPPPPGTAAAPPPPPPPPGTQAAPPPPP 566
Query: 87 RPEISSSGFGPAP 99
P + + P P
Sbjct: 567 PPPMQNRAPSPPP 579
Score = 38.9 bits (89), Expect = 0.003
Identities = 23/70 (32%), Positives = 31/70 (43%), Gaps = 10/70 (14%)
Query: 32 LPARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTRPEIS 91
LP+ PP+PP A I ++ P P P PP ++ +P PP PPP +
Sbjct: 437 LPSPPPTPPIADIAISMPPP------PPPPPPPPAV----MPLKHFAPPPPPPLPPAVMP 486
Query: 92 SSGFGPAPNT 101
F P P T
Sbjct: 487 LKHFAPPPPT 496
Score = 34.7 bits (78), Expect = 0.057
Identities = 29/92 (31%), Positives = 42/92 (45%), Gaps = 10/92 (10%)
Query: 9 LHPSSQSDIDEIENLIYASPATVLPARPPS-PPRASIPVTTSSPFINSNLPAPLPPKSSI 67
L +S+S ++ E Y + + + P PP PP + T+S + S P P PP + I
Sbjct: 394 LESTSESKLNHSEK--YENSSQLFPPPPPPPPPPPLSFIKTASLPLPS--PPPTPPIADI 449
Query: 68 SQPQLPKPISLPPAPPPSTRPEISSSGFGPAP 99
+ P P PP PPP+ P F P P
Sbjct: 450 AISMPPPP--PPPPPPPAVMP---LKHFAPPP 476
Score = 33.1 bits (74), Expect = 0.17
Identities = 25/86 (29%), Positives = 35/86 (40%), Gaps = 16/86 (18%)
Query: 28 PATVLPARPPSPP--RASIPVTTSSPFINSNL------PAPLPPKSSISQPQLPKPISL- 78
P T PP PP + P P NS P P+P + + P P P+++
Sbjct: 556 PGTQAAPPPPPPPPMQNRAPSPPPMPMGNSGSGGPPPPPPPMPLANGATPPPPPPPMAMA 615
Query: 79 -----PPAPPPSTRPEISSSGFGPAP 99
PP PPP R +++ GP P
Sbjct: 616 NGAAGPPPPPP--RMGMANGAAGPPP 639
>At1g49750 unknown protein
Length = 494
Score = 42.7 bits (99), Expect = 2e-04
Identities = 30/94 (31%), Positives = 42/94 (43%), Gaps = 6/94 (6%)
Query: 36 PPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTRPEISSSGF 95
PP PP P S P P P P S PQLP P LPP PP +P +
Sbjct: 62 PPPPPPPPCPPPPSPP----PCPPPPSPPPSPPPPQLPPPPQLPPPAPPKPQPSPPTPDL 117
Query: 96 GPAPNTLTEPVWDTVKRDLSRIVSNLKLVVFPNP 129
P ++L + V+ ++R +V++ KL + P
Sbjct: 118 -PFASSLLKKVYPVLQR-FKDLVADDKLKSWEGP 149
Score = 35.4 bits (80), Expect = 0.034
Identities = 21/53 (39%), Positives = 22/53 (40%), Gaps = 4/53 (7%)
Query: 36 PPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTRP 88
PPSP P P P P PP S P P P S PP+PPP P
Sbjct: 47 PPSPSPEPEPEPADCPPPPPPPPCPPPP----SPPPCPPPPSPPPSPPPPQLP 95
Score = 33.1 bits (74), Expect = 0.17
Identities = 22/68 (32%), Positives = 27/68 (39%), Gaps = 9/68 (13%)
Query: 38 SPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTRPEISSSGFGP 97
+PP + P P ++ P P PP P P P S PP PPP + P P
Sbjct: 45 NPPPSPSPEPEPEP---ADCPPPPPP------PPCPPPPSPPPCPPPPSPPPSPPPPQLP 95
Query: 98 APNTLTEP 105
P L P
Sbjct: 96 PPPQLPPP 103
Score = 28.5 bits (62), Expect = 4.1
Identities = 14/36 (38%), Positives = 16/36 (43%), Gaps = 1/36 (2%)
Query: 54 NSNLPAPLPPKSSISQP-QLPKPISLPPAPPPSTRP 88
N N P P P +P P P PP PPP + P
Sbjct: 42 NDNNPPPSPSPEPEPEPADCPPPPPPPPCPPPPSPP 77
>At1g49270 hypothetical protein
Length = 699
Score = 42.7 bits (99), Expect = 2e-04
Identities = 27/75 (36%), Positives = 33/75 (44%), Gaps = 1/75 (1%)
Query: 27 SPATVLPARPPSPPRASIPVTTS-SPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPS 85
SP P PPSPP ++ TTS P N +P PP S P + PP P P
Sbjct: 10 SPPAPPPPSPPSPPSSNDQQTTSPPPSDNQETTSPPPPSSPDIAPPPQQQQESPPPPLPE 69
Query: 86 TRPEISSSGFGPAPN 100
+ SSS P P+
Sbjct: 70 NSSDGSSSSSPPPPS 84
Score = 30.4 bits (67), Expect = 1.1
Identities = 22/68 (32%), Positives = 27/68 (39%), Gaps = 11/68 (16%)
Query: 33 PARPPSPPRASIPVTTSSPFINSNLPAPLPPKSSISQPQLPKPISLPPAPPPSTRPEISS 92
P + P +P +S S+ +P PP S SQ Q P PPPST P S
Sbjct: 56 PQQQQESPPPPLPENSSD---GSSSSSPPPPSDSSSQSQSP--------PPPSTSPPQQS 104
Query: 93 SGFGPAPN 100
G N
Sbjct: 105 DNNGNKGN 112
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.139 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,822,694
Number of Sequences: 26719
Number of extensions: 339645
Number of successful extensions: 5966
Number of sequences better than 10.0: 390
Number of HSP's better than 10.0 without gapping: 174
Number of HSP's successfully gapped in prelim test: 226
Number of HSP's that attempted gapping in prelim test: 2038
Number of HSP's gapped (non-prelim): 1803
length of query: 276
length of database: 11,318,596
effective HSP length: 98
effective length of query: 178
effective length of database: 8,700,134
effective search space: 1548623852
effective search space used: 1548623852
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0061.10


