

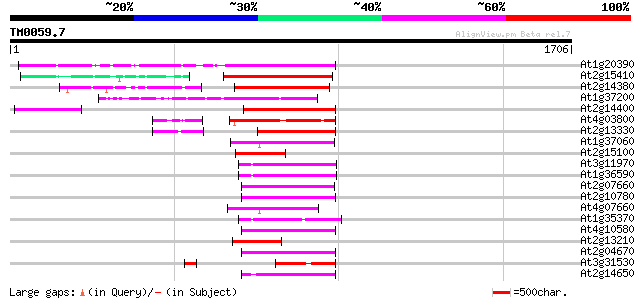
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0059.7
(1706 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g20390 hypothetical protein 512 e-145
At2g15410 putative retroelement pol polyprotein 364 e-100
At2g14380 putative retroelement pol polyprotein 342 1e-93
At1g37200 hypothetical protein 332 1e-90
At2g14400 putative retroelement pol polyprotein 321 3e-87
At4g03800 280 5e-75
At2g13330 F14O4.9 268 2e-71
At1g37060 Athila retroelment ORF 1, putative 201 4e-51
At2g15100 putative retroelement pol polyprotein 190 7e-48
At3g11970 hypothetical protein 189 9e-48
At1g36590 hypothetical protein 189 9e-48
At2g07660 putative retroelement pol polyprotein 189 2e-47
At2g10780 pseudogene 188 2e-47
At4g07660 putative athila transposon protein 183 7e-46
At1g35370 hypothetical protein 178 3e-44
At4g10580 putative reverse-transcriptase -like protein 177 5e-44
At2g13210 pseudogene 173 9e-43
At2g04670 putative retroelement pol polyprotein 172 2e-42
At3g31530 hypothetical protein 171 4e-42
At2g14650 putative retroelement pol polyprotein 165 2e-40
>At1g20390 hypothetical protein
Length = 1791
Score = 512 bits (1318), Expect = e-145
Identities = 336/983 (34%), Positives = 507/983 (51%), Gaps = 105/983 (10%)
Query: 27 QGWARPKVKLYEGDSDPQEHVNFFVGAMQYAGAS----DPLYCRCFPMSLGKGPMNWFQN 82
+G + K++ Y G +DP+E + F A+ A + D C+ F L NWF
Sbjct: 167 RGAQKIKLESYNGRNDPKEFLTSFNVAINRAELTIDNFDAGRCQIFIEHLTGPAHNWFSR 226
Query: 83 LPSNSLHDWNGVVTCFLAQYSSVRSIPKTAQTLALVKQKEKESLKAFLNRFNKEAGDITG 142
L NS+ ++ + + FL Y+ + + L + Q KESL++F++RF +IT
Sbjct: 227 LKPNSIDSFHQLTSSFLKHYAPLIENQTSNADLWSISQGAKESLRSFVDRFKLVVTNIT- 285
Query: 143 LLPDTRLVLA--TAALAPGPFLTSLDGKPANTLEEFLARAEKFINMEDAVTLRA----AS 196
+PD ++A A F + +TLE+ L RA +FI +E+ + A ++
Sbjct: 286 -VPDEAAIVALRNAVWYDSRFRDDITLHAPSTLEDALHRASRFIELEEEKLILARKHNST 344
Query: 197 QTLAIKGPQKMKEHKDQPSTRESR--RKGQDERKPKRKKYDSYTPLNSSLSRILREKAST 254
+T A K +K D + R RKP + TP ++ +R+
Sbjct: 345 KTPACKDAVVIKVGPDDSNEPRQHLDRNPSAGRKPTSFLVSTETPDAKPWNKYIRDA--- 401
Query: 255 DLRDRPPPLLTRGDKLDSKRFCEFHDSPGHNTDECLNLKDKVEELIRAGRLSRYVAVSTG 314
D P +CE+H S H+T+ C L+ + ++G + T
Sbjct: 402 ---DSPAA---------GPMYCEYHKSRAHSTENCRFLQGLLMAKYKSGGI-------TI 442
Query: 315 ALPRP--RSPPPRRTPTPPRH--RDQTPPKRRSAERRDKTPPRRRSPERRRRSRERRRSP 370
RP + RR T R DQT P AE+ T + +R+R + P
Sbjct: 443 ECDRPPINNKNQRRNETTARQYLNDQTKPPT-PAEQGIITSADDPAAKRQRNGKAIAAEP 501
Query: 371 DRHGRDEVRRQHGSNIVDVGSIAGGWAAGGPTNNSQKR-STRVIMSAAGRPRPGSLHPPR 429
VR+ H I GG + S K+ + M A + P
Sbjct: 502 V-----VVRQVH--------VIMGGLQNCSDSVRSIKQYRKKAEMVVAWPSSTSTTRNPN 548
Query: 430 QKVAITFTEDDY-GEDTGEEDDPIVIEALIGNGKIRRTLIDTGSSADIMFYDAYKSLGLS 488
Q I+FT+ D G DT +DP+V+E +I + ++ R LIDTGSS D++F D ++ ++
Sbjct: 549 QSAPISFTDVDLEGLDT-PHNDPLVVELIISDSRVTRVLIDTGSSVDLIFKDVLTAMNIT 607
Query: 489 VKDLLPYDHDLVGFTGDRVLPLGYFDTCLSLGDHRICRTIKARFLVVECPTAYNAILGRP 548
+ + P L GF GD V+ +G + +G +F+V+ P YN ILG P
Sbjct: 608 DRQIKPVSKPLAGFDGDFVMTIGTIKLPIFVGG----LIAWVKFVVIGKPAVYNVILGTP 663
Query: 549 SLNTFRAIISTHHLMLKYPWAGRAVPVRGNLEMARSCYNSSCRLAREERKRKKSKAHDQN 608
++ +AI ST+H +K+P +N L + + S++++++
Sbjct: 664 WIHQMQAIPSTYHQCVKFP-----------------THNGIFTLRAPKEAKTPSRSYEES 706
Query: 609 GNYHVQHANLLTDLDPRVDPSRDDHRLKPDGESHPI*VGPLPENTTNLARGLPRDLSKRM 668
+ N+ DP+R + + + +
Sbjct: 707 ELCRTEMVNI-----DESDPTR----------------------CVGVGAEISPSIRLEL 739
Query: 669 EKLLLSNGKLFAWSSEDMPGIDPAFCSHRLSVDRKYKPVAQKKRQMSAEKQQVIQQQTTE 728
LL N K FAWS EDM GIDPA +H L+VD +KPV QK+R++ E+ + + ++ +
Sbjct: 740 IALLKRNSKTFAWSIEDMKGIDPAITAHELNVDPTFKPVKQKRRKLGPERARAVNEEVEK 799
Query: 729 LLRAGIIREVKYTTWLSNVVLVKKANGKWRMCVDYTDLNKACPKDPFPLPSIDALVDNSS 788
LL+AG I EVKY WL+N V+VKK NGKWR+CVDYTDLNKACPKD +PLP ID LV+ +S
Sbjct: 800 LLKAGQIIEVKYPEWLANPVVVKKKNGKWRVCVDYTDLNKACPKDSYPLPHIDRLVEATS 859
Query: 789 GYEYLSLMDAYSGYNQIPMHRDDEEKTAFITDRGTYCYTMLPFGLKNAVATYQRMMTRIF 848
G LS MDA+SGYNQI MH+DD+EKT+F+TDRGTYCY ++ FGLKNA ATYQR + ++
Sbjct: 860 GNGLLSFMDAFSGYNQILMHKDDQEKTSFVTDRGTYCYKVMSFGLKNAGATYQRFVNKML 919
Query: 849 GDLMGKSVEVYIDDIIVKTPKGGDHAADLAVVFEQLKKHNMRLNPDKCTFGVRSGKFLGY 908
D +G++VEVYIDD++VK+ K DH L+ F+ L + M+LNP KCTFGV SG+FLGY
Sbjct: 920 ADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKCFDVLNTYGMKLNPTKCTFGVTSGEFLGY 979
Query: 909 MLTNRGIELNPDKCQAIMNMKSPRTVKEVQQLAGRMAAIGRFLPKAALRALPLYTLLKNG 968
++T RGIE NP + +AI+ + SPR +EVQ+L GR+AA+ RF+ ++ + LP Y LLK
Sbjct: 980 VVTKRGIEANPKQIRAILELPSPRNAREVQRLTGRIAALNRFISRSTDKCLPFYNLLKRR 1039
Query: 969 ATFEWSAEADAAFTQLKEILSSP 991
A F+W +++ AF +LK+ LS+P
Sbjct: 1040 AQFDWDKDSEEAFEKLKDYLSTP 1062
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 364 bits (934), Expect = e-100
Identities = 170/333 (51%), Positives = 232/333 (69%)
Query: 650 PENTTNLARGLPRDLSKRMEKLLLSNGKLFAWSSEDMPGIDPAFCSHRLSVDRKYKPVAQ 709
P + LP +L + L N FAWS EDMPGID A H L+VD YKP+ Q
Sbjct: 726 PSRCVGIRIDLPSELQNELVNFLRQNAATFAWSVEDMPGIDSAVTCHELNVDPTYKPLKQ 785
Query: 710 KKRQMSAEKQQVIQQQTTELLRAGIIREVKYTTWLSNVVLVKKANGKWRMCVDYTDLNKA 769
K+R++ ++ + + ++ +LL AG I EV+Y WL N V+VKK NGKWR+C+D+TDLNKA
Sbjct: 786 KRRKLGPDRTKDVNEEVKKLLDAGSIVEVRYPDWLRNPVVVKKKNGKWRVCIDFTDLNKA 845
Query: 770 CPKDPFPLPSIDALVDNSSGYEYLSLMDAYSGYNQIPMHRDDEEKTAFITDRGTYCYTML 829
CPKD FPLP ID LV+ ++G E LS MDA+SGYNQI MH++D EKT FITD+GTYCY ++
Sbjct: 846 CPKDSFPLPHIDRLVEATAGNELLSFMDAFSGYNQILMHQNDREKTVFITDQGTYCYKVM 905
Query: 830 PFGLKNAVATYQRMMTRIFGDLMGKSVEVYIDDIIVKTPKGGDHAADLAVVFEQLKKHNM 889
PFGLKNA ATY R++ ++F D + S+EVYIDD++VK+ + +H L F+ L ++NM
Sbjct: 906 PFGLKNAGATYPRLVNQMFTDQLDHSMEVYIDDMLVKSLRAEEHITHLRQCFQVLNRYNM 965
Query: 890 RLNPDKCTFGVRSGKFLGYMLTNRGIELNPDKCQAIMNMKSPRTVKEVQQLAGRMAAIGR 949
+LNP KCTFGV SG+FLGY++T RGIE NP + AI+++ SPR +EVQ+L GR+AA+ R
Sbjct: 966 KLNPSKCTFGVTSGEFLGYLVTRRGIEANPKQISAIIDLPSPRNTREVQRLIGRIAALNR 1025
Query: 950 FLPKAALRALPLYTLLKNGATFEWSAEADAAFT 982
F+ ++ + LP Y LL+ FEW + + T
Sbjct: 1026 FISRSTDKCLPFYQLLRANKRFEWDEKCEEGET 1058
Score = 83.2 bits (204), Expect = 1e-15
Identities = 126/536 (23%), Positives = 189/536 (34%), Gaps = 109/536 (20%)
Query: 33 KVKL--YEGDSDPQEH---VNFFVGAMQYAGAS-DPLYCRCFPMSLGKGPMNWFQNLPSN 86
K+KL YEG DP+ V+ +G ++ D C+ F L + WF L +N
Sbjct: 252 KIKLANYEGLVDPRPFLTSVSIAIGRAHFSDEDRDAGSCQLFVKHLSGAALTWFSRLEAN 311
Query: 87 SLHDWNGVVTCFLAQYSSVRSIPKTAQTLALVKQKEKESLKAFLNRFNKEAGDITGLLPD 146
S+ + + T FL Y + L + Q KESL++F+ RF + + PD
Sbjct: 312 SIDSVHALTTSFLKNYGVFMEKGASNVDLWTMAQTAKESLRSFIGRFKEIVTSVA--TPD 369
Query: 147 TRLVLATAALAPGPFLTSLDGKPANTLEEFLARAEKFINMEDAVTLRAASQTLAIKGPQK 206
+ A + +A+ +S+ + P
Sbjct: 370 DAAIAA---------------------------------LRNALWHGDSSRCTTLCKPFH 396
Query: 207 MKEHKDQPSTRESRRKGQDE-RKPKRKKYDSYTPLNSSLSRILREKASTDLRDRPPPLLT 265
+ P +E ++G E R+ Y L S + + A D+P
Sbjct: 397 RQGSAKAPIAKEKPKEGHHESRQHYDADYAKEEKLKKGTSYYVGDTAPRS--DKPWNKWN 454
Query: 266 RG-DKLDSKRFCEFHDSPGHNTDECLNLKDKVEELIRAGRLSRYVAVSTGALPRPRSPPP 324
R D +++ EFH PGH+TDEC L+ + + G L P
Sbjct: 455 RSADAKGDQKYYEFHKIPGHSTDECRQLQTLLLTKFKKGDLDI-------------EPDR 501
Query: 325 RRTPTPPRH----------RDQTPPKRRSAERRDKTPPRRRSPERRRRSRERRRSPDRHG 374
RRT T R RD KRR E D+ R +RR +R R
Sbjct: 502 RRTGTNDRDNTHRRVENTDRDNQDDKRRQDET-DRRAERIHDDDRRAEPHKRNHDAPRQF 560
Query: 375 RDEVRRQHGSNIVDVGSIAGGWAAGGPTNNSQKRSTRVIMSAAGRPRPGSLHPPRQKVA- 433
DE + N+ I GG A + S K R G + + V
Sbjct: 561 EDEPASKRRRNM-----IMGGLTACRDSVRSIKSYRR-----QGEKQRAWIEKNTSAVEC 610
Query: 434 ---ITFTEDDYGEDTGEEDDPIVIEALIGNGKIRRTLIDTGSSADIMFYDAYKSLGLSVK 490
ITF+E D G +DP+V+E IG + + ID S D +
Sbjct: 611 NEPITFSEADIALP-GSHNDPLVVELKIGENVLLQLEIDVRSFQDKV------------- 656
Query: 491 DLLPYDHDLVGFTGDRVLPLGYFDTCLSLGDHRICRTIK-ARFLVVECPTAYNAIL 545
L GF GD ++ +G + +G T+ F VV+ P YN IL
Sbjct: 657 ------QPLTGFDGDTIMTVGTITLPIYVGG-----TVSWFEFAVVDKPIIYNVIL 701
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 342 bits (877), Expect = 1e-93
Identities = 159/291 (54%), Positives = 217/291 (73%), Gaps = 1/291 (0%)
Query: 683 SEDMPGIDPAFCSHRLSVDRKYKPVAQKKRQMSAEKQQVIQQQTTELLRAGIIREVKYTT 742
+ DM GIDP H L+VD +K V QK+R++ E+ + + + +LL AG I EVKY
Sbjct: 474 AHDMVGIDPEVACHELNVDPTFKLVKQKRRKLGPERSKAVNDEVDKLLDAGSIVEVKYPE 533
Query: 743 WLSNVVLVKKANGKWRMCVDYTDLNKACPKDPFPLPSIDALVDNSSGYEYLSLMDAYSGY 802
WL+N V+VKK N KWR+C+D+TDLNKACPKD FPLP ID +V+ ++G E LS MDA+SGY
Sbjct: 534 WLANPVVVKKKNDKWRVCIDFTDLNKACPKDSFPLPHIDRMVEATTGNELLSFMDAFSGY 593
Query: 803 NQIPMHRDDEEKTAFITDRGTYCYTMLPFGLKNAVATYQRMMTRIFGDLMGKSVEVYIDD 862
NQIPMH+DD+EKT+FI DRGTYCY ++PFGLKN A YQR++ ++F +GK++EVYIDD
Sbjct: 594 NQIPMHKDDQEKTSFIIDRGTYCYKVMPFGLKNVGARYQRLVNQMFAPQLGKTMEVYIDD 653
Query: 863 IIVKTPKGGDHAADLAVVFEQLKKHNMRLNPDKCTFGVRSGKFLGYMLTNRGIELNPDKC 922
++VK+ + DH L FE L K+NM+LNP KC FGV SG+FLGY++T RGIE NP +
Sbjct: 654 MLVKSTRSADHIDHLKACFETLNKYNMKLNPAKCLFGVTSGEFLGYIVTKRGIEANPKQI 713
Query: 923 QAIMNMKSPRTVKEVQQLAGRMAAIGRFLPKAALRALPLYTLLKN-GATFE 972
+AI++++SPR KEVQ+L GR+A + RF+ ++ ++LP Y LL++ TFE
Sbjct: 714 RAILDLQSPRNKKEVQRLTGRIAGLNRFIARSTDKSLPFYQLLRSANKTFE 764
Score = 102 bits (255), Expect = 1e-21
Identities = 110/453 (24%), Positives = 185/453 (40%), Gaps = 59/453 (13%)
Query: 153 TAALAPGPFLTSLDGKPANT-----------LEEFLARAEKFINMEDAVTLRAASQTLAI 201
T L PG + ++G+PA T + LA+ ++ + A++ + T +
Sbjct: 58 TVGLEPG--VQGMEGRPALTPPARPDDPGALMRNILAQIQELRSTIQAMSQKVHQATSST 115
Query: 202 KGPQKM--KEHKDQPSTRESRRKGQDERKPKRKKYDSYTPLNSSLSRILREKASTDLRDR 259
+++ K K + S + + K K K Y+ T LS +T
Sbjct: 116 PELEQILEKNQKTPFAVHISNVSVRHDNKIKLKSYEGNTDPQQFLSSF---SVATQRDHF 172
Query: 260 PPPLLTRGDKLDSKRFCEFHDSPGHNTDECLNL---------KDKVEELIRAGRLSRYVA 310
P ++LD ++C+FH GH+T C +L K +E R +
Sbjct: 173 HPSEAEVENRLD--QYCDFHKRSGHSTAACRHLQSILLNKYKKGDIEVQHRQYKSHNNTY 230
Query: 311 VSTGALPRPRSPPPRRTPTPPRHRDQTPPKRRSAERRDKTPPRRRSPERRRRSRERRRSP 370
+ G R + R + + PP D PP++ R +
Sbjct: 231 AARGG--RDGNNVFHRLGPHTGRQQEAPPANEEERHPDMEPPKKN-----------REND 277
Query: 371 DRHGRDEVRRQHGSNIVDVGSIAGGWAAGGPTNNSQKRSTRVIMSAAGRPRPGSLHPPRQ 430
+H V R+ V I GG A + S K + +G S ++
Sbjct: 278 QQHNDAPVPRRR------VNMIMGGLTACRDSFRSIKEYIK-----SGAATLWSSPATKE 326
Query: 431 KVAITFTEDD-YGEDTGEEDDPIVIEALIGNGKIRRTLIDTGSSADIMFYDAYKSLGLSV 489
+TFT +D +G D +DP+VIE IG ++ R LIDTGSS +++F D + + +
Sbjct: 327 MTPLTFTSEDLFGVDL-PHNDPLVIELHIGESEVTRILIDTGSSVNVVFKDVLQKMKVHD 385
Query: 490 KDLLPYDHDLVGFTGDRVLPLGYFDTCLSLGDHRICRTIKARFLVVECPTAYNAILGRPS 549
+ + P L GF G+ ++ G + LG +F+VV+ PT YN ILG P
Sbjct: 386 RHIKPSVRPLTGFDGNTMMTNGTIKLPIYLGGAATWH----KFVVVDKPTIYNIILGTPW 441
Query: 550 LNTFRAIISTHHLMLKYPWAGRAVPVRGNLEMA 582
++ +AI S++H +K P + +RGN +A
Sbjct: 442 IHDMQAIPSSYHQCIKIPTSIGIETIRGNQNLA 474
>At1g37200 hypothetical protein
Length = 1564
Score = 332 bits (850), Expect = 1e-90
Identities = 223/674 (33%), Positives = 344/674 (50%), Gaps = 85/674 (12%)
Query: 269 KLDSKRFCEFHDSPGHNTDECLNLKDKVEELIRAGRLSRYVAVSTGALPRPRSPPPRRTP 328
+LD ++C++H+ GH+T+EC +K +LI AG ++ G+ P+ +PPP
Sbjct: 317 ELDLGKYCKYHNKRGHSTEECRAVK----KLIAAGGKTK-----KGSNPKVETPPPDE-- 365
Query: 329 TPPRHRDQTPPKRRSAERRDKTPPRRRSPERRRRSRERRRSPDRHGRDEVRRQHGSNIVD 388
+ +QTP +++ R R+ E SP R+ + +D
Sbjct: 366 ---QEEEQTPKQKK-----------------RERTLEGGDSPPPARRERIDLVFTE--LD 403
Query: 389 VGSIAGGWAAGGPTNNSQKRSTRVIMSAAGRPRPGSLHPPRQKVAITFTEDDYGEDTGEE 448
+G G + P + K++ R + + R + H +++ D+
Sbjct: 404 LGGKTGR-SVTTPPSAPHKKNMRFPLRLSKFCRSATEHLSPKRIDFIMGGSQLCNDS--- 459
Query: 449 DDPIVIEALIGNGKIRRTLIDTGSSADIMFYDAYKSLGLSVKDLLPYDHDLVGFTGDRV- 507
I + K + D+ Y KSL ++ DH + + +
Sbjct: 460 ---------INSIKTHQRKADS--------YTKGKSL------MMGPDHKITLWESETTD 496
Query: 508 LPLGYFDTC---LSLGDHRICRT-IKARFLVVECPTAYNAILGRPSLNTFRAIISTHHLM 563
L + D + +G++++ R I V + P +NAILGRP L+ +A+ ST+H
Sbjct: 497 LDKPHDDAIVIRIDVGNYKLSRIMIDTGSSVDKAP--FNAILGRPWLHAMKAVPSTYHQC 554
Query: 564 LKYPWAGRAVPVRGNLEMARSCYNSSCRLAREERKRKKSKAHDQNGNYHVQHANLLTDLD 623
+K+P + G+ +R CY S L KK++ + + D
Sbjct: 555 IKFPSEKGIAVIYGSQRSSRRCYMGSYELI------KKAELVVLMIEDKLAEMKTVRSSD 608
Query: 624 P-RVDPSRDDHRLKPDGESHPI*VGPLPENTTNLARGLPRDLSKRMEKLLLSNGKLFAWS 682
P + P + ES P + + + L + + L N FAWS
Sbjct: 609 PSQCGPQKSSITQVYIDESDP-------KQCVGVGQDLDPAIREDFITFLKENKDSFAWS 661
Query: 683 SEDMPGIDPAFCSHRLSVDRKYKPVAQKKRQMSAEKQQVIQQQTTELLRAGIIREVKYTT 742
S ++ GI SH L+VD Y+P+ QK+R++ E+ + +Q + LL+ G IREVKY
Sbjct: 662 SANLQGISLEVTSHELNVDPTYRPIKQKRRKLGPERAKAVQDEVDRLLKIGSIREVKYPD 721
Query: 743 WLSNVVLVKKANGKWRMCVDYTDLNKACPKDPFPLPSIDALVDNSSGYEYLSLMDAYSGY 802
WL+N V+VK NGKWR+C+D+ DLNKACPKD FPLP ID LV ++G+E LS MDA+SGY
Sbjct: 722 WLANPVVVKNKNGKWRVCIDFMDLNKACPKDSFPLPHIDRLVKATAGHELLSFMDAFSGY 781
Query: 803 NQIPMHRDDEEKTAFITDRGTYCYTMLPFGLKNAVATYQRMMTRIFGDLMGKSVEVYIDD 862
NQI M DD+EKTAFITD CY ++PFGLKN ATYQR++ R+F D +GK++E+YIDD
Sbjct: 782 NQILMRPDDQEKTAFITD----CYKVMPFGLKNTGATYQRLVNRMFADQLGKTMELYIDD 837
Query: 863 IIVKTPKGGDHAADLAVVFEQLKKHNMRLNPDKCTFGVRSGKFLGYMLTNRGIELNPDKC 922
++VK+ DH L F+ L K M+LNP+KC+FGV SG+FLGY++T RGIE NP +
Sbjct: 838 MLVKSAHEKDHLPQLRECFKILNKFEMKLNPEKCSFGVPSGEFLGYLVTQRGIEANPKQI 897
Query: 923 QAIMNMKSPRTVKE 936
A ++M SP+ ++
Sbjct: 898 AAFIDMPSPKMARD 911
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 321 bits (822), Expect = 3e-87
Identities = 148/281 (52%), Positives = 208/281 (73%)
Query: 711 KRQMSAEKQQVIQQQTTELLRAGIIREVKYTTWLSNVVLVKKANGKWRMCVDYTDLNKAC 770
+R++ E+ + + + +LL+ G IREV+Y WL+N V+VKK NGK R+C+D+TDLNKAC
Sbjct: 469 RRKLGVERAKAVNDEVDKLLKIGSIREVQYPDWLANTVVVKKKNGKDRVCIDFTDLNKAC 528
Query: 771 PKDPFPLPSIDALVDNSSGYEYLSLMDAYSGYNQIPMHRDDEEKTAFITDRGTYCYTMLP 830
PKD FPLP ID LV++++G E LS MDA+SGYNQI M+ +D+EKT FITDRG YCY ++P
Sbjct: 529 PKDSFPLPHIDRLVESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRGIYCYKVMP 588
Query: 831 FGLKNAVATYQRMMTRIFGDLMGKSVEVYIDDIIVKTPKGGDHAADLAVVFEQLKKHNMR 890
FGL+NA ATY R++ ++F + +GK++EVYIDD+++K+ K DH L F L ++ M+
Sbjct: 589 FGLRNAGATYPRLVNKMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECFAILNQYQMK 648
Query: 891 LNPDKCTFGVRSGKFLGYMLTNRGIELNPDKCQAIMNMKSPRTVKEVQQLAGRMAAIGRF 950
LNP KCTFGV SG+FLGY++T RGIE NP++ A +NM SP+ KEVQ+L GR+AA+ RF
Sbjct: 649 LNPAKCTFGVPSGEFLGYIVTKRGIEANPNQINAFLNMPSPKNFKEVQRLTGRIAALNRF 708
Query: 951 LPKAALRALPLYTLLKNGATFEWSAEADAAFTQLKEILSSP 991
+ ++ ++LP Y +LK F W + + AF QLK L+SP
Sbjct: 709 ISRSTDKSLPFYQILKGNKEFLWDEKCEEAFGQLKAYLTSP 749
Score = 48.9 bits (115), Expect = 3e-05
Identities = 47/212 (22%), Positives = 88/212 (41%), Gaps = 9/212 (4%)
Query: 15 PLSPEIMAFPFPQGWARPKVKLYEGDSDPQEHVNFFVGAMQYAGAS----DPLYCRCFPM 70
P +P+I + + ++ Y G DP+ ++ F+ A + D YC+ F
Sbjct: 144 PFTPQITSLRIRDS-RKLNLESYNGLEDPKGYLAAFLIAAGRVDLNEADEDVRYCKLFSE 202
Query: 71 SLGKGPMNWFQNLPSNSLHDWNGVVTCFLAQYSSVRSIPKTAQTLALVKQKEKESLKAFL 130
+L + WF L S+ ++N + FL QYS + + L + Q E+L+AF+
Sbjct: 203 NLCGQALMWFTQLEPGSISNFNELSVVFLKQYSILMDKSISDTDLWNLSQGPNETLRAFI 262
Query: 131 NRFNKEAGDITGLLPDTRL-VLATAALAPGPFLTSLDGKPANTLEEFLARAEKFINMED- 188
+F ++ + + L L F L +T+++ L RA ++ +ED
Sbjct: 263 TKFKYVLSKLSRISQQSALSALRKGLWYDSRFKEDLILHKLDTIQDALFRANNWMEVEDE 322
Query: 189 --AVTLRAASQTLAIKGPQKMKEHKDQPSTRE 218
+ R A+ P K E ++ R+
Sbjct: 323 KESFAKRDKQAKPAVTFPPKKFEPRENQGPRK 354
Score = 42.7 bits (99), Expect = 0.002
Identities = 21/63 (33%), Positives = 34/63 (53%)
Query: 430 QKVAITFTEDDYGEDTGEEDDPIVIEALIGNGKIRRTLIDTGSSADIMFYDAYKSLGLSV 489
Q +I F E + DD +V+ + N ++ R LIDTGSS D++F + +G+S
Sbjct: 405 QSTSILFDEKETQHLERSHDDALVVTLDVANFEVSRILIDTGSSVDLIFLSTLERMGISR 464
Query: 490 KDL 492
D+
Sbjct: 465 ADV 467
>At4g03800
Length = 637
Score = 280 bits (716), Expect = 5e-75
Identities = 147/329 (44%), Positives = 210/329 (63%), Gaps = 28/329 (8%)
Query: 670 KLLLSNGKLFAW-------SSEDMPGIDPAFCSHRLSVDRKYKPVAQKKRQMSAEKQQVI 722
K SNG W +S MP IDP+ H L+VD ++KP+ QK+R++ E+ + +
Sbjct: 298 KFPTSNGVETIWGDLEGSRTSAYMPDIDPSIICHELNVDPRFKPLKQKRRKLGVERAKAV 357
Query: 723 QQQTTELLRAGIIREVKYTTWLSNVVLVKKANGKWRMCVDYTDLNKACPKDPFPLPSIDA 782
+ +LL+ G IREV+Y W++ V+VKK NGK R+C+D+TDLNKACPKD FPLP ID
Sbjct: 358 NGKIDKLLKIGSIREVQYPDWVAITVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHIDR 417
Query: 783 LVDNSSGYEYLSLMDAYSGYNQIPMHRDDEEKTAFITDRGTYCYTMLPFGLKNAVATYQR 842
LV++++G E L+ MDA+ GYNQI M+ +D+EKT+FITDRG ATYQ
Sbjct: 418 LVESTAGNELLTFMDAFLGYNQIMMNPEDQEKTSFITDRG---------------ATYQW 462
Query: 843 MMTRIFGDLMGKSVEVYIDDIIVKTPKGGDHAADLAVVFEQLKKHNMRLNPDKCTFGVRS 902
++ ++F + + K++EV IDD +VK+ K DH L FE L ++ M+LN KCTFGV S
Sbjct: 463 LVNKMFNEHLRKTMEVSIDDTLVKSLKKEDHVKHLGECFEILNQYQMKLNLAKCTFGVPS 522
Query: 903 GKFLGYMLTNRGIELNPDKCQAIMNMKSPRTVKEVQQLAGRMAAIGRFLPKAALRALPLY 962
G+FLGY++T RGIE NP++ A + S R KEVQ+L GR+A+ ++ ++LP Y
Sbjct: 523 GEFLGYIVTKRGIEANPNQINAFLKTPSLRNFKEVQRLTGRIAS------RSTDKSLPFY 576
Query: 963 TLLKNGATFEWSAEADAAFTQLKEILSSP 991
+LK F W + + AF QLK L++P
Sbjct: 577 QILKGNNGFLWDEKCEEAFRQLKAYLTTP 605
Score = 67.4 bits (163), Expect = 7e-11
Identities = 45/152 (29%), Positives = 75/152 (48%), Gaps = 18/152 (11%)
Query: 433 AITFTEDDYGEDTGEEDDPIVIEALIGNGKIRRTLIDTGSSADIMFYDAYKSLGLSVKDL 492
+++F E++ DD ++I + N KI R L+DTGSS D++F
Sbjct: 184 SVSFDEEETRHIERPHDDALIITLDVANFKISRILVDTGSSVDLIF-------------- 229
Query: 493 LPYDHDLVGFTGDRVLPLGYFDTCLSLGDHRICRTIKARFLVVECPTAYNAILGRPSLNT 552
L LV FT + + LG + G ++ + + F+V + P YN ILG P +
Sbjct: 230 LGPPSPLVAFTSESAMSLGTIKLPVLAG--KMSKIVD--FVVFDKPATYNIILGTPWIFQ 285
Query: 553 FRAIISTHHLMLKYPWAGRAVPVRGNLEMARS 584
+A+ ST+H LK+P + + G+LE +R+
Sbjct: 286 MKAVPSTYHQWLKFPTSNGVETIWGDLEGSRT 317
>At2g13330 F14O4.9
Length = 889
Score = 268 bits (686), Expect = 2e-71
Identities = 126/238 (52%), Positives = 171/238 (70%)
Query: 754 NGKWRMCVDYTDLNKACPKDPFPLPSIDALVDNSSGYEYLSLMDAYSGYNQIPMHRDDEE 813
NGKWR+C+D+ DLNKACPKD FPLP ID LV+ + +E LS MDA+ GYNQI M RD +E
Sbjct: 291 NGKWRVCIDFRDLNKACPKDSFPLPHIDRLVEATVEHEKLSFMDAFYGYNQILMRRDGQE 350
Query: 814 KTAFITDRGTYCYTMLPFGLKNAVATYQRMMTRIFGDLMGKSVEVYIDDIIVKTPKGGDH 873
KTAFITDRGTY Y ++PFGLKNA TYQR++ R+F D +GK++EVYIDD++VK+ DH
Sbjct: 351 KTAFITDRGTYYYKVMPFGLKNAGTTYQRLVNRMFVDQLGKTIEVYIDDMLVKSAHEKDH 410
Query: 874 AADLAVVFEQLKKHNMRLNPDKCTFGVRSGKFLGYMLTNRGIELNPDKCQAIMNMKSPRT 933
L F+ L K M+LNP+KC+F V+S +FL Y++T RGIE NP + A + M SP+
Sbjct: 411 VPQLRECFKILIKFEMKLNPEKCSFEVQSREFLEYLVTERGIEANPKQIAAFIEMPSPKM 470
Query: 934 VKEVQQLAGRMAAIGRFLPKAALRALPLYTLLKNGATFEWSAEADAAFTQLKEILSSP 991
+EVQ+L GR+ A+ F+ ++A + +P Y L+ G F+W+ + + AF QLK L+ P
Sbjct: 471 AREVQRLTGRIPALNGFISRSANKCVPFYQPLRKGKEFDWNKDCEQAFKQLKAYLTEP 528
Score = 89.7 bits (221), Expect = 1e-17
Identities = 55/158 (34%), Positives = 81/158 (50%), Gaps = 8/158 (5%)
Query: 434 ITFTEDDYGEDTGEEDDPIVIEALIGNGKIRRTLIDTGSSADIMFYDAYKSLGLSVKDLL 493
ITF E + + DD +VI +GN ++ ++DTGSS D++FYDA+K G L
Sbjct: 47 ITFWESEITDLDKPHDDALVIRIDVGNYELSCIMVDTGSSVDVLFYDAFKRTGHLDSKLQ 106
Query: 494 PYDHDLVGFTGDRVLPLGYFDTCLSLGDHRICRTIK--ARFLVVECPTAYNAILGRPSLN 551
L GF GD +G ++ I R ++ FLVV+ +NAILGRP L+
Sbjct: 107 GRKTPLTGFAGDTTFSIG------TIQLPTIARGVRQLTNFLVVDKKAPFNAILGRPWLH 160
Query: 552 TFRAIISTHHLMLKYPWAGRAVPVRGNLEMARSCYNSS 589
+A+ ST+H +K+P V G+ +R CY S
Sbjct: 161 VMKAVPSTYHQCIKFPSYKGIAVVYGSQRSSRKCYMGS 198
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 201 bits (510), Expect = 4e-51
Identities = 112/335 (33%), Positives = 180/335 (53%), Gaps = 18/335 (5%)
Query: 672 LLSNGKLFAWSSEDMPGIDPAFCSHRLSVDRKYKPVAQKKRQMSAEKQQVIQQQTTELLR 731
L+ K +S +D+ GI P C+HR+ ++ + + +R+++ ++V++++ +LL
Sbjct: 865 LMKYRKAIGYSLDDIKGISPTLCTHRIHLENESYSSIEPQRRLNPNLKEVVKKEILKLLD 924
Query: 732 AGIIREVKYTTWLSNVVLVKKANGKW------------------RMCVDYTDLNKACPKD 773
AG+I + +TW+S V V K G RMC++Y LN A K+
Sbjct: 925 AGVIYPISDSTWVSPVHCVPKKGGMTVVKNSKDELIPTRTITGHRMCIEYRKLNVASRKE 984
Query: 774 PFPLPSIDALVDNSSGYEYLSLMDAYSGYNQIPMHRDDEEKTAFITDRGTYCYTMLPFGL 833
FPLP ID +++ + + Y +D+YSG+ QIP+H +D+ KT F GT+ Y +PFGL
Sbjct: 985 HFPLPFIDHMLERLANHPYYCFLDSYSGFFQIPIHPNDQGKTTFTCPYGTFAYKRMPFGL 1044
Query: 834 KNAVATYQRMMTRIFGDLMGKSVEVYIDDIIVKTPKGGDHAADLAVVFEQLKKHNMRLNP 893
NA AT+QR MT IF DL+ + VEV++DD V +L V ++ ++ N+ LN
Sbjct: 1045 CNAPATFQRCMTSIFSDLIEEMVEVFMDDFSVYGSSFSSCLLNLCRVLKRCEETNLVLNW 1104
Query: 894 DKCTFGVRSGKFLGYMLTNRGIELNPDKCQAIMNMKSPRTVKEVQQLAGRMAAIGRFLPK 953
+KC F VR G LG ++ GIE++ K +M ++ P+TVK+++ G F+
Sbjct: 1105 EKCHFMVREGIVLGRKISEEGIEVDKAKIDVMMQLQPPKTVKDIRSFLGHAGFYRIFIKD 1164
Query: 954 AALRALPLYTLLKNGATFEWSAEADAAFTQLKEIL 988
+ A PL LL F + E AF +KE L
Sbjct: 1165 FSKLARPLTRLLCKETEFAFDDECLTAFKLIKEAL 1199
Score = 38.5 bits (88), Expect = 0.034
Identities = 25/96 (26%), Positives = 38/96 (39%), Gaps = 4/96 (4%)
Query: 42 DPQEHVNFFVGAMQYAG----ASDPLYCRCFPMSLGKGPMNWFQNLPSNSLHDWNGVVTC 97
DP +H++ F + D R FP SLG W ++LP S+ WN
Sbjct: 61 DPLDHLDEFDRLCSLTKINRVSEDGFKLRLFPFSLGDKAHQWEKSLPQGSITSWNDCKKA 120
Query: 98 FLAQYSSVRSIPKTAQTLALVKQKEKESLKAFLNRF 133
FLA++ S + ++ Q E+ RF
Sbjct: 121 FLAKFFSNSRTARLRNDISGFTQTNNETFYEAWERF 156
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 190 bits (482), Expect = 7e-48
Identities = 89/152 (58%), Positives = 115/152 (75%)
Query: 686 MPGIDPAFCSHRLSVDRKYKPVAQKKRQMSAEKQQVIQQQTTELLRAGIIREVKYTTWLS 745
M GI SH L+VD +KPV QK+R++ ++ Q + + LL G IREVKY WL+
Sbjct: 412 MTGISTEVISHELNVDPTFKPVKQKRRKLGPDRAQAVNIEVVRLLEVGRIREVKYPEWLA 471
Query: 746 NVVLVKKANGKWRMCVDYTDLNKACPKDPFPLPSIDALVDNSSGYEYLSLMDAYSGYNQI 805
N V+VKK NGKWR+CVD+TDLNKAC KD FPLP ID LV++++G+E LS MDA+SGYNQI
Sbjct: 472 NPVVVKKKNGKWRVCVDFTDLNKACSKDFFPLPHIDRLVESTTGHEMLSFMDAFSGYNQI 531
Query: 806 PMHRDDEEKTAFITDRGTYCYTMLPFGLKNAV 837
M+ +D+EKT+FIT+ GTY Y ++PFGLKNAV
Sbjct: 532 LMNPEDQEKTSFITECGTYYYKVMPFGLKNAV 563
Score = 36.6 bits (83), Expect = 0.13
Identities = 16/36 (44%), Positives = 22/36 (60%)
Query: 532 FLVVECPTAYNAILGRPSLNTFRAIISTHHLMLKYP 567
F V + P AYN ILG P L + + ST+H +K+P
Sbjct: 371 FTVFDRPAAYNVILGTPWLYEMKVVPSTYHQCVKFP 406
Score = 35.4 bits (80), Expect = 0.29
Identities = 27/108 (25%), Positives = 43/108 (39%), Gaps = 12/108 (11%)
Query: 33 KVKLYEGDSDPQEHVNFFVGAMQYAGASDPL--------YCRCFPMSLGKGPMNWFQNLP 84
K+ +Y G D +EH+ F Q PL C+ F +L + WF L
Sbjct: 174 KLPVYNGKGDLKEHLTSF----QVIAGRVPLEPHEEDAGLCKLFSENLFGLALTWFTQLE 229
Query: 85 SNSLHDWNGVVTCFLAQYSSVRSIPKTAQTLALVKQKEKESLKAFLNR 132
S+ ++ + T F+ QY + T L Q E L+ ++ R
Sbjct: 230 EGSIDNFKQLSTAFIKQYEYFINSDITEAHLWNFSQSADEPLRTYIYR 277
>At3g11970 hypothetical protein
Length = 1499
Score = 189 bits (481), Expect = 9e-48
Identities = 111/305 (36%), Positives = 168/305 (54%), Gaps = 8/305 (2%)
Query: 695 SHRLSVDRKYKPVAQKKRQMSAEKQQVIQQQTTELLRAGIIREVKYTTWLSNVVLVKKAN 754
+H++ + PV Q+ + S ++ I + +LL G + + + + S VVLVKK +
Sbjct: 590 NHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTV-QASSSPYASPVVLVKKKD 648
Query: 755 GKWRMCVDYTDLNKACPKDPFPLPSIDALVDNSSGYEYLSLMDAYSGYNQIPMHRDDEEK 814
G WR+CVDY +LN KD FP+P I+ L+D G S +D +GY+Q+ M DD +K
Sbjct: 649 GTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQK 708
Query: 815 TAFITDRGTYCYTMLPFGLKNAVATYQRMMTRIFGDLMGKSVEVYIDDIIVKTPKGGDHA 874
TAF T G + Y ++PFGL NA AT+Q +M IF + K V V+ DDI+V + +H
Sbjct: 709 TAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHR 768
Query: 875 ADLAVVFEQLKKHNMRLNPDKCTFGVRSGKFLGYMLTNRGIELNPDKCQAIMNMKSPRTV 934
L VFE ++ + + KC F V ++LG+ ++ +GIE +P K +A+ P T+
Sbjct: 769 QHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTL 828
Query: 935 KEVQQLAGRMAAIGRFLPKAALRALPLYTLLKNGATFEWSAEADAAFTQLK------EIL 988
K+++ G RF+ + A PL+ L K A FEW+A A AF LK +L
Sbjct: 829 KQLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTDA-FEWTAVAQQAFEDLKAALCQAPVL 887
Query: 989 SSPLF 993
S PLF
Sbjct: 888 SLPLF 892
>At1g36590 hypothetical protein
Length = 1499
Score = 189 bits (481), Expect = 9e-48
Identities = 111/305 (36%), Positives = 168/305 (54%), Gaps = 8/305 (2%)
Query: 695 SHRLSVDRKYKPVAQKKRQMSAEKQQVIQQQTTELLRAGIIREVKYTTWLSNVVLVKKAN 754
+H++ + PV Q+ + S ++ I + +LL G + + + + S VVLVKK +
Sbjct: 590 NHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTV-QASSSPYASPVVLVKKKD 648
Query: 755 GKWRMCVDYTDLNKACPKDPFPLPSIDALVDNSSGYEYLSLMDAYSGYNQIPMHRDDEEK 814
G WR+CVDY +LN KD FP+P I+ L+D G S +D +GY+Q+ M DD +K
Sbjct: 649 GTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQK 708
Query: 815 TAFITDRGTYCYTMLPFGLKNAVATYQRMMTRIFGDLMGKSVEVYIDDIIVKTPKGGDHA 874
TAF T G + Y ++PFGL NA AT+Q +M IF + K V V+ DDI+V + +H
Sbjct: 709 TAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHR 768
Query: 875 ADLAVVFEQLKKHNMRLNPDKCTFGVRSGKFLGYMLTNRGIELNPDKCQAIMNMKSPRTV 934
L VFE ++ + + KC F V ++LG+ ++ +GIE +P K +A+ P T+
Sbjct: 769 QHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTL 828
Query: 935 KEVQQLAGRMAAIGRFLPKAALRALPLYTLLKNGATFEWSAEADAAFTQLK------EIL 988
K+++ G RF+ + A PL+ L K A FEW+A A AF LK +L
Sbjct: 829 KQLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTDA-FEWTAVAQQAFEDLKAALCQAPVL 887
Query: 989 SSPLF 993
S PLF
Sbjct: 888 SLPLF 892
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 189 bits (479), Expect = 2e-47
Identities = 99/283 (34%), Positives = 160/283 (55%), Gaps = 1/283 (0%)
Query: 706 PVAQKKRQMSAEKQQVIQQQTTELLRAGIIREVKYTTWLSNVVLVKKANGKWRMCVDYTD 765
P+++ +M+ + +++Q ELL G IR + W + V+ VKK +G +R+C+DY
Sbjct: 154 PISKAPYRMAPAEMAELKKQLEELLAKGFIRPSS-SPWGAPVLFVKKKDGSFRLCIDYRG 212
Query: 766 LNKACPKDPFPLPSIDALVDNSSGYEYLSLMDAYSGYNQIPMHRDDEEKTAFITDRGTYC 825
LNK K+ +PLP ID L+D G ++ S +D SGY+QIP+ D KTAF T G +
Sbjct: 213 LNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYGHFE 272
Query: 826 YTMLPFGLKNAVATYQRMMTRIFGDLMGKSVEVYIDDIIVKTPKGGDHAADLAVVFEQLK 885
+ ++PFGL NA A + +MM +F D + + V ++IDDI+V + H L V E+L+
Sbjct: 273 FVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVHSKSWEAHQEHLRAVLERLR 332
Query: 886 KHNMRLNPDKCTFGVRSGKFLGYMLTNRGIELNPDKCQAIMNMKSPRTVKEVQQLAGRMA 945
+H + K +F RS FLG++++++G+ ++P+K ++I PR E++ G
Sbjct: 333 EHELFAKLSKFSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEIRSFLGLAG 392
Query: 946 AIGRFLPKAALRALPLYTLLKNGATFEWSAEADAAFTQLKEIL 988
RF+ A A PL L F WS E + +F +LK +L
Sbjct: 393 YYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAML 435
>At2g10780 pseudogene
Length = 1611
Score = 188 bits (478), Expect = 2e-47
Identities = 98/285 (34%), Positives = 162/285 (56%), Gaps = 1/285 (0%)
Query: 706 PVAQKKRQMSAEKQQVIQQQTTELLRAGIIREVKYTTWLSNVVLVKKANGKWRMCVDYTD 765
P+++ +M+ + +++Q ELL G IR + W + V+ VKK +G +R+C+DY
Sbjct: 659 PISKAPYRMAPAEMAKLKKQLEELLDKGFIRPSS-SPWGAPVLFVKKKDGSFRLCIDYRG 717
Query: 766 LNKACPKDPFPLPSIDALVDNSSGYEYLSLMDAYSGYNQIPMHRDDEEKTAFITDRGTYC 825
LNK K+ +PLP ID L+D G ++ S +D SGY+QIP+ D KTAF T +
Sbjct: 718 LNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYDHFE 777
Query: 826 YTMLPFGLKNAVATYQRMMTRIFGDLMGKSVEVYIDDIIVKTPKGGDHAADLAVVFEQLK 885
+ ++PFGL NA A + +MM +F D + + V ++I+DI+V + H L V E+L+
Sbjct: 778 FVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFINDILVYSKSWEAHQEHLRAVLERLR 837
Query: 886 KHNMRLNPDKCTFGVRSGKFLGYMLTNRGIELNPDKCQAIMNMKSPRTVKEVQQLAGRMA 945
+H + KC+F RS FLG++++++G+ ++P+K ++I PR E++ G
Sbjct: 838 EHELFAKLSKCSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEIRSFLGLAG 897
Query: 946 AIGRFLPKAALRALPLYTLLKNGATFEWSAEADAAFTQLKEILSS 990
RF+ A A PL L F WS E + +F +LK +L++
Sbjct: 898 YYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLTN 942
>At4g07660 putative athila transposon protein
Length = 724
Score = 183 bits (465), Expect = 7e-46
Identities = 99/297 (33%), Positives = 167/297 (55%), Gaps = 22/297 (7%)
Query: 663 DLSKRMEKLLLSNGKLF----AWSSEDMPGIDPAFCSHRLSVDRKYKPVAQKKRQMSAEK 718
+L+ LLLS + + +S D+ I P+ C+HR+ ++ + + +R+++
Sbjct: 302 ELNDNEVNLLLSELRKYKRAIGYSLSDIKRISPSLCNHRIHLENESYSSIEPQRRLNLNL 361
Query: 719 QQVIQQQTTELLRAGIIREVKYTTWLSNVVLVKKANGKW------------------RMC 760
++V++++ +LL AG+I + +TW+ V V K G R+C
Sbjct: 362 KEVVKKEILKLLDAGVIYPISDSTWVFPVHCVPKKGGMTVVKNEKDELIPTRTITGHRVC 421
Query: 761 VDYTDLNKACPKDPFPLPSIDALVDNSSGYEYLSLMDAYSGYNQIPMHRDDEEKTAFITD 820
+DY LN A KD FPLP + +++ + + Y +D YSG+ QIP+H +D+EKT F
Sbjct: 422 IDYRKLNAASRKDHFPLPFTNQMLEGLANHLYNCFLDGYSGFFQIPIHPNDQEKTTFTCP 481
Query: 821 RGTYCYTMLPFGLKNAVATYQRMMTRIFGDLMGKSVEVYIDDIIVKTPKGGDHAADLAVV 880
GT+ Y +PFGL NA T+QR MT IF DL+ K VEV++DD V P +L V
Sbjct: 482 YGTFAYKRMPFGLCNAPTTFQRCMTSIFSDLIEKMVEVFMDDFSVYGPSFSSCLLNLGRV 541
Query: 881 FEQLKKHNMRLNPDKCTFGVRSGKFLGYMLTNRGIELNPDKCQAIMNMKSPRTVKEV 937
+ ++ N+ LN +KC F V+ G LG+ ++ +GIE++ +K + +M ++ P+TVK++
Sbjct: 542 LTKWEETNLVLNWEKCYFMVKEGIVLGHKISEKGIEVDKEKIKVMMQLQPPKTVKDI 598
>At1g35370 hypothetical protein
Length = 1447
Score = 178 bits (451), Expect = 3e-44
Identities = 109/315 (34%), Positives = 171/315 (53%), Gaps = 11/315 (3%)
Query: 696 HRLSVDRKYKPVAQKKRQMSAEKQQVIQQQTTELLRAGIIREVKYTTWLSNVVLVKKANG 755
H++ + PV Q+ + ++ I + +++++G I +V + + S VVLVKK +G
Sbjct: 570 HKIKLLEGANPVNQRPYRYVVHQKDEIDKIVQDMIKSGTI-QVSSSPFASPVVLVKKKDG 628
Query: 756 KWRMCVDYTDLNKACPKDPFPLPSIDALVDNSSGYEYLSLMDAYSGYNQIPMHRDDEEKT 815
WR+CVDYT+LN KD F +P I+ L+D G S +D +GY+Q+ M DD +KT
Sbjct: 629 TWRLCVDYTELNGMTVKDRFLIPLIEDLMDELGGSVVFSKIDLRAGYHQVRMDPDDIQKT 688
Query: 816 AFITDRGTYCYTMLPFGLKNAVATYQRMMTRIFGDLMGKSVEVYIDDIIVKTPKGGDHAA 875
AF T G + Y ++ FGL NA AT+Q +M +F D + K V V+ DDI++ + +H
Sbjct: 689 AFKTHNGHFEYLVMLFGLTNAPATFQSLMNSVFRDFLRKFVLVFFDDILIYSSSIEEHKE 748
Query: 876 DLAVVFEQLKKHNMRLNPDKCTFGVRSGKFLGYMLTNRGIELNPDKCQAIMNMKSPRTVK 935
L +VFE ++ H + F S + LG+ ++ R IE +P K QA+ +P TVK
Sbjct: 749 HLRLVFEVMRLHKL--------FAKGSKEHLGHFISAREIETDPAKIQAVKEWPTPTTVK 800
Query: 936 EVQQLAGRMAAIGRFLPKAALRALPLYTLLKNGATFEWSAEADAAFTQLKEIL-SSPLFW 994
+V+ G RF+ + A PL+ L K F WS EA +AF LK +L ++P+
Sbjct: 801 QVRGFLGFAGYYRRFVRNFGVIAGPLHALTKTDG-FCWSLEAQSAFDTLKAVLCNAPVLA 859
Query: 995 LALSQGRLCTYT*PC 1009
L + + T C
Sbjct: 860 LPVFDKQFMVETDAC 874
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 177 bits (449), Expect = 5e-44
Identities = 95/285 (33%), Positives = 157/285 (54%), Gaps = 1/285 (0%)
Query: 706 PVAQKKRQMSAEKQQVIQQQTTELLRAGIIREVKYTTWLSNVVLVKKANGKWRMCVDYTD 765
P+++ +M+ + +++Q +LL G IR + W + V+ VKK +G +R+C+DY +
Sbjct: 491 PLSKAPYRMAPAEMAELKKQLKDLLGKGFIRP-STSPWGAPVLFVKKKDGSFRLCIDYRE 549
Query: 766 LNKACPKDPFPLPSIDALVDNSSGYEYLSLMDAYSGYNQIPMHRDDEEKTAFITDRGTYC 825
LN+ K+ +PLP ID L+D G S +D SGY+QIP+ D KTAF T G +
Sbjct: 550 LNRVTVKNRYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFE 609
Query: 826 YTMLPFGLKNAVATYQRMMTRIFGDLMGKSVEVYIDDIIVKTPKGGDHAADLAVVFEQLK 885
+ ++PFGL NA A + R+M +F + + + V ++IDDI+V + + L V E+L+
Sbjct: 610 FVVMPFGLTNAPAVFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEQEVHLRRVMEKLR 669
Query: 886 KHNMRLNPDKCTFGVRSGKFLGYMLTNRGIELNPDKCQAIMNMKSPRTVKEVQQLAGRMA 945
+ + KC+F R FLG++++ G+ ++P+K +AI + P E++ G
Sbjct: 670 EQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSFLGWAG 729
Query: 946 AIGRFLPKAALRALPLYTLLKNGATFEWSAEADAAFTQLKEILSS 990
RF+ A A P+ L F WS E + F LKE+L+S
Sbjct: 730 YYRRFVKGFASMAQPMTKLTGKDVPFVWSQECEEGFVSLKEMLTS 774
>At2g13210 pseudogene
Length = 152
Score = 173 bits (438), Expect = 9e-43
Identities = 75/147 (51%), Positives = 107/147 (72%)
Query: 679 FAWSSEDMPGIDPAFCSHRLSVDRKYKPVAQKKRQMSAEKQQVIQQQTTELLRAGIIREV 738
F WS ED+PG+ +H L+VD +KPV QK+R++ E+ + ++ + +LLR G I +
Sbjct: 6 FVWSPEDLPGVSVEIVAHELNVDPTFKPVKQKRRKLGRERAEAVKAEVEKLLRIGSITKA 65
Query: 739 KYTTWLSNVVLVKKANGKWRMCVDYTDLNKACPKDPFPLPSIDALVDNSSGYEYLSLMDA 798
KY WL+N V+VKK NGKWR+C+D+TDLNKACPKD FP+ I LV+++SG + LS MDA
Sbjct: 66 KYPEWLANPVVVKKKNGKWRVCIDFTDLNKACPKDSFPMSHIGRLVESTSGDKLLSFMDA 125
Query: 799 YSGYNQIPMHRDDEEKTAFITDRGTYC 825
++GYNQI M+ +D+EKT F T++G C
Sbjct: 126 FAGYNQIMMNSEDQEKTTFYTEQGIVC 152
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 172 bits (436), Expect = 2e-42
Identities = 94/285 (32%), Positives = 154/285 (53%), Gaps = 1/285 (0%)
Query: 706 PVAQKKRQMSAEKQQVIQQQTTELLRAGIIREVKYTTWLSNVVLVKKANGKWRMCVDYTD 765
P+++ +M+ + +++Q +LL G IR + W + V+ VKK +G +R+C+DY
Sbjct: 517 PLSKAPYRMAPAEMTELKKQLEDLLGKGFIRP-STSPWGAPVLFVKKKDGSFRLCIDYRG 575
Query: 766 LNKACPKDPFPLPSIDALVDNSSGYEYLSLMDAYSGYNQIPMHRDDEEKTAFITDRGTYC 825
LN K+ +PLP ID L+D G S +D SGY+QIP+ D KTAF T G +
Sbjct: 576 LNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFE 635
Query: 826 YTMLPFGLKNAVATYQRMMTRIFGDLMGKSVEVYIDDIIVKTPKGGDHAADLAVVFEQLK 885
+ ++PF L NA A + R+M +F + + + V ++IDDI+V + +H L V E+L+
Sbjct: 636 FVVMPFALTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEHEVHLRRVMEKLR 695
Query: 886 KHNMRLNPDKCTFGVRSGKFLGYMLTNRGIELNPDKCQAIMNMKSPRTVKEVQQLAGRMA 945
+ + KC+F R FLG++++ G+ ++P+K +AI + P E++
Sbjct: 696 EQKLFAKLSKCSFWQREIGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSFLRLTG 755
Query: 946 AIGRFLPKAALRALPLYTLLKNGATFEWSAEADAAFTQLKEILSS 990
RF+ A A P+ L F WS E + F LKE+L+S
Sbjct: 756 YYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTS 800
>At3g31530 hypothetical protein
Length = 831
Score = 171 bits (432), Expect = 4e-42
Identities = 82/185 (44%), Positives = 124/185 (66%), Gaps = 11/185 (5%)
Query: 807 MHRDDEEKTAFITDRGTYCYTMLPFGLKNAVATYQRMMTRIFGDLMGKSVEVYIDDIIVK 866
M+ +D+EKTAF T++G +CY ++PFGLKNA ATY+R + +IF +GK++EVYIDD++VK
Sbjct: 96 MNPEDQEKTAFYTEQGIFCYRVMPFGLKNAGATYERFVNKIFALQIGKTMEVYIDDMLVK 155
Query: 867 TPKGGDHAADLAVVFEQLKKHNMRLNPDKCTFGVRSGKFLGYMLTNRGIELNPDKCQAIM 926
+ DH + L F+QL +N++LNP KC FGVRS GIE NP + +A++
Sbjct: 156 SMTEKDHISHLRECFKQLNLYNVKLNPAKCRFGVRS-----------GIEANPKQIEALL 204
Query: 927 NMKSPRTVKEVQQLAGRMAAIGRFLPKAALRALPLYTLLKNGATFEWSAEADAAFTQLKE 986
M SP+ +EVQ L GR+AA+ RF+ ++ + L Y +L+ FEW+ + AF +LK+
Sbjct: 205 GMASPQNKREVQCLTGRVAALNRFISRSTEKCLAFYDVLRRNKKFEWTTRCEEAFQELKK 264
Query: 987 ILSSP 991
L++P
Sbjct: 265 YLATP 269
Score = 49.7 bits (117), Expect = 1e-05
Identities = 21/36 (58%), Positives = 27/36 (74%)
Query: 532 FLVVECPTAYNAILGRPSLNTFRAIISTHHLMLKYP 567
F + + PT YNAI+G P LN FRA+ ST+HL LK+P
Sbjct: 21 FSITDQPTVYNAIIGTPWLNQFRAVASTYHLCLKFP 56
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 165 bits (418), Expect = 2e-40
Identities = 91/285 (31%), Positives = 152/285 (52%), Gaps = 13/285 (4%)
Query: 706 PVAQKKRQMSAEKQQVIQQQTTELLRAGIIREVKYTTWLSNVVLVKKANGKWRMCVDYTD 765
P+++ +M+ + +++Q +LL A ++ VKK +G +R+C+DY
Sbjct: 478 PLSKAPYRMAPAEMAELKKQLEDLLGAPVL-------------FVKKKDGSFRLCIDYRG 524
Query: 766 LNKACPKDPFPLPSIDALVDNSSGYEYLSLMDAYSGYNQIPMHRDDEEKTAFITDRGTYC 825
LN K+ +PLP ID L+D G S +D SGY+ IP+ D KTAF T G +
Sbjct: 525 LNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHLIPIAEADVRKTAFRTRYGHFE 584
Query: 826 YTMLPFGLKNAVATYQRMMTRIFGDLMGKSVEVYIDDIIVKTPKGGDHAADLAVVFEQLK 885
+ ++PFGL NA A + R+M +F +++ + V ++IDDI+V + +H L V E+L+
Sbjct: 585 FVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDILVYSKSLEEHEVHLRRVMEKLR 644
Query: 886 KHNMRLNPDKCTFGVRSGKFLGYMLTNRGIELNPDKCQAIMNMKSPRTVKEVQQLAGRMA 945
+ + KC+F R FLG++++ G+ ++P+K +AI + +P E++ G
Sbjct: 645 EQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWHTPTNATEIRSFLGLAG 704
Query: 946 AIGRFLPKAALRALPLYTLLKNGATFEWSAEADAAFTQLKEILSS 990
RF+ A A P+ L F WS E + F LKE+L+S
Sbjct: 705 YYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTS 749
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.138 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,103,546
Number of Sequences: 26719
Number of extensions: 1834381
Number of successful extensions: 9091
Number of sequences better than 10.0: 175
Number of HSP's better than 10.0 without gapping: 100
Number of HSP's successfully gapped in prelim test: 77
Number of HSP's that attempted gapping in prelim test: 7922
Number of HSP's gapped (non-prelim): 749
length of query: 1706
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1593
effective length of database: 8,299,349
effective search space: 13220862957
effective search space used: 13220862957
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0059.7


