

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0058a.9
(407 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
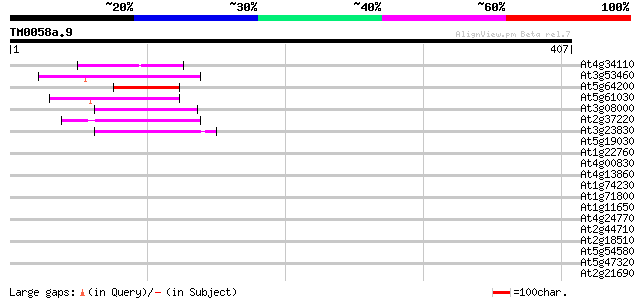
Score E
Sequences producing significant alignments: (bits) Value
At4g34110 poly(A)-binding protein 46 3e-05
At3g53460 RNA-binding protein cp29 protein 45 9e-05
At5g64200 unknown protein 44 1e-04
At5g61030 RNA-binding protein - like 43 4e-04
At3g08000 RNA-binding protein like 42 5e-04
At2g37220 putative RNA-binding protein 42 5e-04
At3g23830 glycine-rich RNA binding protein, putative 42 8e-04
At5g19030 unknown protein 41 0.001
At1g22760 putative polyA-binding protein, PAB3 40 0.003
At4g00830 unknown protein 39 0.004
At4g13860 RNA-binding protein like 39 0.005
At1g74230 putative RNA-binding protein 39 0.005
At1g71800 cleavage stimulation factor like protein 39 0.005
At1g11650 putative DNA binding protein 39 0.005
At4g24770 31 kDa RNA binding protein (rbp31) 39 0.007
At2g44710 putative heterogeneous nuclear ribonucleoprotein 39 0.007
At2g18510 putative spliceosome associated protein 39 0.007
At5g54580 unknown protein 38 0.011
At5g47320 40S ribosomal protein S19 38 0.011
At2g21690 putative glycine-rich RNA binding protein 38 0.011
>At4g34110 poly(A)-binding protein
Length = 629
Score = 46.2 bits (108), Expect = 3e-05
Identities = 28/77 (36%), Positives = 40/77 (51%), Gaps = 1/77 (1%)
Query: 50 RSCSAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYG 109
R S R ++FI ++ES D+ L D S FG +V+ V SG+ +GF +Y
Sbjct: 114 RDPSVRRSGAGNIFIKNLDESIDHKALHDTFSSFGNIVSCKVAVD-SSGQSKGYGFVQYA 172
Query: 110 LRDQALKAIQGLNGFRL 126
+ A KAI+ LNG L
Sbjct: 173 NEESAQKAIEKLNGMLL 189
Score = 35.8 bits (81), Expect = 0.044
Identities = 19/76 (25%), Positives = 39/76 (51%), Gaps = 1/76 (1%)
Query: 50 RSCSAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYG 109
R +A + +V++ + ES L++ ++GK+ + V + + G+ FGF +
Sbjct: 205 RDSTANKTKFTNVYVKNLAESTTDDDLKNAFGEYGKITSAVVMKDGE-GKSKGFGFVNFE 263
Query: 110 LRDQALKAIQGLNGFR 125
D A +A++ LNG +
Sbjct: 264 NADDAARAVESLNGHK 279
Score = 32.0 bits (71), Expect = 0.63
Identities = 20/76 (26%), Positives = 39/76 (51%), Gaps = 1/76 (1%)
Query: 61 SVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQG 120
++++ ++ S L++I S FG V + V R +G GF + ++A +A+
Sbjct: 319 NLYVKNLDPSISDEKLKEIFSPFGTVTSSKVMRD-PNGTSKGSGFVAFATPEEATEAMSQ 377
Query: 121 LNGFRLGSAPLSASLA 136
L+G + S PL ++A
Sbjct: 378 LSGKMIESKPLYVAIA 393
>At3g53460 RNA-binding protein cp29 protein
Length = 342
Score = 44.7 bits (104), Expect = 9e-05
Identities = 37/125 (29%), Positives = 51/125 (40%), Gaps = 8/125 (6%)
Query: 22 GSTTARGVGDVAGEEEGGELDGEADGGSRSCS--------AGRRSCFSVFIDGIEESFDY 73
GS G G G G + G GGS+ S +G S +++ + D
Sbjct: 211 GSERGGGYGSERGGGYGSQRSGGGYGGSQRSSYGSGSGSGSGSGSGNRLYVGNLSWGVDD 270
Query: 74 LHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNGFRLGSAPLSA 133
+ L ++ ++ GKVV V R SGR FGF + KAI LNG L +
Sbjct: 271 MALENLFNEQGKVVEARVIYDRDSGRSKGFGFVTLSSSQEVQKAINSLNGADLDGRQIRV 330
Query: 134 SLAWA 138
S A A
Sbjct: 331 SEAEA 335
Score = 28.5 bits (62), Expect = 6.9
Identities = 28/112 (25%), Positives = 43/112 (38%), Gaps = 6/112 (5%)
Query: 32 VAGEEEGGELDGEADGGSRSCSAGRRSCFS----VFIDGIEESFDYLHLRDICSKFGKVV 87
V+ + E E D ADG + R+ FS +F+ + + D L + G V
Sbjct: 69 VSSDFEVEEDDMFADGDDSA--PVERNSFSPDLKLFVGNLSFNVDSAQLAQLFESAGNVE 126
Query: 88 NLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNGFRLGSAPLSASLAWAP 139
+ V + +GR FGF + A Q NG+ PL + P
Sbjct: 127 MVEVIYDKVTGRSRGFGFVTMSTAAEVEAAAQQFNGYEFEGRPLRVNAGPPP 178
>At5g64200 unknown protein
Length = 303
Score = 44.3 bits (103), Expect = 1e-04
Identities = 20/48 (41%), Positives = 33/48 (68%)
Query: 76 LRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNG 123
L + +K+GKVV++F+ R R++G F F RY +D+A KA++ L+G
Sbjct: 32 LYPLFAKYGKVVDVFIPRDRRTGDSRGFAFVRYKYKDEAHKAVERLDG 79
>At5g61030 RNA-binding protein - like
Length = 309
Score = 42.7 bits (99), Expect = 4e-04
Identities = 30/96 (31%), Positives = 48/96 (49%), Gaps = 2/96 (2%)
Query: 30 GDVAGEEEGGELDGEADGGSRSCSAGRR--SCFSVFIDGIEESFDYLHLRDICSKFGKVV 87
G++ + +L+ + S S R S +FI G+ S D LR+ +K+G+VV
Sbjct: 8 GNILKQTTNKQLNAQVSLSSPSLFQAIRCMSSSKLFIGGMAYSMDEDSLREAFTKYGEVV 67
Query: 88 NLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNG 123
+ V R++GR FGF + + A AIQ L+G
Sbjct: 68 DTRVILDRETGRSRGFGFVTFTSSEAASSAIQALDG 103
>At3g08000 RNA-binding protein like
Length = 143
Score = 42.4 bits (98), Expect = 5e-04
Identities = 26/75 (34%), Positives = 36/75 (47%)
Query: 62 VFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGL 121
+FI G+ S D L+D S FG+V + + + SGR FGF + AL A +
Sbjct: 43 LFIGGLSWSVDEQSLKDAFSSFGEVAEVRIAYDKGSGRSRGFGFVDFAEEGDALSAKDAM 102
Query: 122 NGFRLGSAPLSASLA 136
+G L PL S A
Sbjct: 103 DGKGLLGRPLRISFA 117
>At2g37220 putative RNA-binding protein
Length = 289
Score = 42.4 bits (98), Expect = 5e-04
Identities = 31/101 (30%), Positives = 42/101 (40%), Gaps = 4/101 (3%)
Query: 38 GGELDGEADGGSRSCSAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKS 97
G G GG +G R V++ + D + L + S+ GKVV V R S
Sbjct: 186 GSSGSGYGGGGGSGAGSGNR----VYVGNLSWGVDDMALESLFSEQGKVVEARVIYDRDS 241
Query: 98 GRFFRFGFFRYGLRDQALKAIQGLNGFRLGSAPLSASLAWA 138
GR FGF Y + AI+ L+G L + S A A
Sbjct: 242 GRSKGFGFVTYDSSQEVQNAIKSLDGADLDGRQIRVSEAEA 282
Score = 32.3 bits (72), Expect = 0.48
Identities = 43/176 (24%), Positives = 65/176 (36%), Gaps = 23/176 (13%)
Query: 7 ASRSFAPISAAARQIGSTTARGVGDVAGEEEGGELDGEADGGSRSCSAGRRSCFSVFIDG 66
A++ +P S AR + T+ V EE G D A +S SA + +F+
Sbjct: 49 AAKWNSPASRFARNVAITSEFEV------EEDGFAD-VAPPKEQSFSADLK----LFVGN 97
Query: 67 IEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNGFRL 126
+ + D L + G V + V + +GR FGF + A Q NG+ L
Sbjct: 98 LPFNVDSAQLAQLFESAGNVEMVEVIYDKITGRSRGFGFVTMSSVSEVEAAAQQFNGYEL 157
Query: 127 GSAPLSASLAWAP------------WVCGSPRRGFGLCSQASSAQGQRLAARVLSW 170
PL + P GS G+G + + G R+ LSW
Sbjct: 158 DGRPLRVNAGPPPPKREDGFSRGPRSSFGSSGSGYGGGGGSGAGSGNRVYVGNLSW 213
>At3g23830 glycine-rich RNA binding protein, putative
Length = 136
Score = 41.6 bits (96), Expect = 8e-04
Identities = 26/89 (29%), Positives = 41/89 (45%), Gaps = 2/89 (2%)
Query: 62 VFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGL 121
+F+ G+ D L+ + FG+V V R++GR FGF + D A AI+ +
Sbjct: 37 LFVGGLSWGTDDSSLKQAFTSFGEVTEATVIADRETGRSRGFGFVSFSCEDSANNAIKEM 96
Query: 122 NGFRLGSAPLSASLAWAPWVCGSPRRGFG 150
+G L + +LA +PR FG
Sbjct: 97 DGKELNGRQIRVNLATER--SSAPRSSFG 123
>At5g19030 unknown protein
Length = 172
Score = 40.8 bits (94), Expect = 0.001
Identities = 23/97 (23%), Positives = 48/97 (48%), Gaps = 11/97 (11%)
Query: 27 RGVGDVAGEEEGGELDGEADGGSRSCSAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKV 86
R + VAG++E +R S+ S S+F+ G +S L+ + S+FG+V
Sbjct: 55 RTISCVAGDDE-----------TREASSLPSSISSLFVKGFSDSVSEGRLKKVFSEFGQV 103
Query: 87 VNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNG 123
N+ + ++ + +G+ + ++ A A++ +NG
Sbjct: 104 TNVKIIANERTRQSLGYGYVWFNSKEDAQSAVEAMNG 140
>At1g22760 putative polyA-binding protein, PAB3
Length = 660
Score = 39.7 bits (91), Expect = 0.003
Identities = 22/76 (28%), Positives = 43/76 (55%), Gaps = 1/76 (1%)
Query: 61 SVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQG 120
++++ +++S D L+++ S++G V + V + G FGF Y ++AL+A+
Sbjct: 333 NLYLKNLDDSVDDEKLKEMFSEYGNVTSSKVMLNPQ-GMSRGFGFVAYSNPEEALRALSE 391
Query: 121 LNGFRLGSAPLSASLA 136
+NG +G PL +LA
Sbjct: 392 MNGKMIGRKPLYIALA 407
Score = 33.5 bits (75), Expect = 0.22
Identities = 19/67 (28%), Positives = 32/67 (47%), Gaps = 1/67 (1%)
Query: 61 SVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQG 120
+V++ + + LR KFG + + V R + SG FGF + + A A++
Sbjct: 230 NVYVKNLPKEIGEDELRKTFGKFGVISSAVVMRDQ-SGNSRCFGFVNFECTEAAASAVEK 288
Query: 121 LNGFRLG 127
+NG LG
Sbjct: 289 MNGISLG 295
Score = 30.8 bits (68), Expect = 1.4
Identities = 19/63 (30%), Positives = 32/63 (50%), Gaps = 1/63 (1%)
Query: 61 SVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQG 120
++FI ++ S D L + S FG +++ V +GR +GF ++ + A AI
Sbjct: 137 NIFIKNLDASIDNKALFETFSSFGTILSCKVAMD-VTGRSKGYGFVQFEKEESAQAAIDK 195
Query: 121 LNG 123
LNG
Sbjct: 196 LNG 198
>At4g00830 unknown protein
Length = 495
Score = 39.3 bits (90), Expect = 0.004
Identities = 21/75 (28%), Positives = 36/75 (48%)
Query: 62 VFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGL 121
VFI G+ LRD+C + G++ + + + R SG + F + +D A KAI+ L
Sbjct: 118 VFIGGLPRDVGEEDLRDLCEEIGEIFEVRLMKDRDSGDSKGYAFVAFKTKDVAQKAIEEL 177
Query: 122 NGFRLGSAPLSASLA 136
+ + SL+
Sbjct: 178 HSKEFKGKTIRCSLS 192
Score = 34.7 bits (78), Expect = 0.097
Identities = 21/84 (25%), Positives = 37/84 (44%), Gaps = 2/84 (2%)
Query: 53 SAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRD 112
SA ++++ I E+ L+++ + G+V + + R FGF Y R
Sbjct: 285 SAAAAQVKALYVKNIPENTSTEQLKELFQRHGEVTKIVTPPGKGGKR--DFGFVHYAERS 342
Query: 113 QALKAIQGLNGFRLGSAPLSASLA 136
ALKA++ + + PL LA
Sbjct: 343 SALKAVKDTERYEVNGQPLEVVLA 366
>At4g13860 RNA-binding protein like
Length = 126
Score = 38.9 bits (89), Expect = 0.005
Identities = 21/60 (35%), Positives = 30/60 (50%)
Query: 76 LRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNGFRLGSAPLSASL 135
LR+ S +G VV+ V R R + R FGF Y +A A+ G++G L +S L
Sbjct: 58 LREAFSGYGNVVDAIVMRDRYTDRSRGFGFVTYSSHSEAEAAVSGMDGKELNGRRVSVKL 117
>At1g74230 putative RNA-binding protein
Length = 289
Score = 38.9 bits (89), Expect = 0.005
Identities = 30/93 (32%), Positives = 46/93 (49%), Gaps = 2/93 (2%)
Query: 58 SCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKA 117
S +F+ GI S D LR+ SK+G+VV+ + R++GR F F + ++A A
Sbjct: 32 SSSKIFVGGISYSTDEFGLREAFSKYGEVVDAKIIVDRETGRSRGFAFVTFTSTEEASNA 91
Query: 118 IQGLNGFRLGSAPLSASLAWAPWVCGSPRRGFG 150
+Q L+G L + + A G RGFG
Sbjct: 92 MQ-LDGQDLHGRRIRVNYA-TERGSGFGGRGFG 122
>At1g71800 cleavage stimulation factor like protein
Length = 461
Score = 38.9 bits (89), Expect = 0.005
Identities = 24/84 (28%), Positives = 39/84 (45%), Gaps = 2/84 (2%)
Query: 53 SAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRD 112
S+ +R C VF+ I LR+IC + G VV+ + R++G+ +GF Y +
Sbjct: 4 SSSQRRC--VFVGNIPYDATEEQLREICGEVGPVVSFRLVTDRETGKPKGYGFCEYKDEE 61
Query: 113 QALKAIQGLNGFRLGSAPLSASLA 136
AL A + L + + L A
Sbjct: 62 TALSARRNLQSYEINGRQLRVDFA 85
>At1g11650 putative DNA binding protein
Length = 405
Score = 38.9 bits (89), Expect = 0.005
Identities = 21/79 (26%), Positives = 45/79 (56%), Gaps = 6/79 (7%)
Query: 61 SVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQG 120
+VF+ G++ S HL+++ S++G++V++ + + R GF ++ + A +A++
Sbjct: 262 TVFVGGLDASVTDDHLKNVFSQYGEIVHVKIPAGK------RCGFVQFSEKSCAEEALRM 315
Query: 121 LNGFRLGSAPLSASLAWAP 139
LNG +LG + S +P
Sbjct: 316 LNGVQLGGTTVRLSWGRSP 334
Score = 29.6 bits (65), Expect = 3.1
Identities = 17/73 (23%), Positives = 35/73 (47%), Gaps = 1/73 (1%)
Query: 60 FSVFI-DGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAI 118
+++F+ D + DY+ L + + V V R +GR +GF R+ + ++A+
Sbjct: 155 YTIFVGDLAADVTDYILLETFRASYPSVKGAKVVIDRVTGRTKGYGFVRFSDESEQIRAM 214
Query: 119 QGLNGFRLGSAPL 131
+NG + P+
Sbjct: 215 TEMNGVPCSTRPM 227
>At4g24770 31 kDa RNA binding protein (rbp31)
Length = 329
Score = 38.5 bits (88), Expect = 0.007
Identities = 33/107 (30%), Positives = 47/107 (43%), Gaps = 6/107 (5%)
Query: 45 ADGGSRSCSAGR--RSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFR 102
A GSR A R F V++ + D L + S+ GKVV V R++GR
Sbjct: 227 APRGSRPERAPRVYEPAFRVYVGNLPWDVDNGRLEQLFSEHGKVVEARVVYDRETGRSRG 286
Query: 103 FGFFRYGLRDQALKAIQGLNGFRLGSAPLSASLAWAPWVCGSPRRGF 149
FGF D+ +AI L+G L + ++A PRRG+
Sbjct: 287 FGFVTMSDVDELNEAISALDGQNLEGRAIRVNVAEE----RPPRRGY 329
>At2g44710 putative heterogeneous nuclear ribonucleoprotein
Length = 300
Score = 38.5 bits (88), Expect = 0.007
Identities = 19/67 (28%), Positives = 35/67 (51%)
Query: 61 SVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQG 120
++FIDG+ S++ +RD+ +GK+ + + R S R FGF + + A+ +
Sbjct: 225 TIFIDGLLPSWNEERVRDLLKPYGKLEKVELARNMPSARRKDFGFVTFDTHEAAVSCAKF 284
Query: 121 LNGFRLG 127
+N LG
Sbjct: 285 INNSELG 291
>At2g18510 putative spliceosome associated protein
Length = 363
Score = 38.5 bits (88), Expect = 0.007
Identities = 23/84 (27%), Positives = 41/84 (48%)
Query: 53 SAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRD 112
SA R +V++ G++ L ++ + G VVN++V + R + +GF Y +
Sbjct: 18 SAERNQDATVYVGGLDAQLSEELLWELFVQAGPVVNVYVPKDRVTNLHQNYGFIEYRSEE 77
Query: 113 QALKAIQGLNGFRLGSAPLSASLA 136
A AI+ LN +L P+ + A
Sbjct: 78 DADYAIKVLNMIKLHGKPIRVNKA 101
>At5g54580 unknown protein
Length = 156
Score = 37.7 bits (86), Expect = 0.011
Identities = 20/63 (31%), Positives = 33/63 (51%)
Query: 61 SVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQG 120
++F+ G+ + LR ++FG+V + V R SG FGF RY + + K I G
Sbjct: 57 NLFVSGLSKRTTSEGLRTAFAQFGEVADAKVVTDRVSGYSKGFGFVRYATLEDSAKGIAG 116
Query: 121 LNG 123
++G
Sbjct: 117 MDG 119
>At5g47320 40S ribosomal protein S19
Length = 212
Score = 37.7 bits (86), Expect = 0.011
Identities = 22/75 (29%), Positives = 34/75 (45%)
Query: 62 VFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGL 121
++I G+ D L+D S F V V + +GR +GF + D A AI +
Sbjct: 33 LYIGGLSPGTDEHSLKDAFSSFNGVTEARVMTNKVTGRSRGYGFVNFISEDSANSAISAM 92
Query: 122 NGFRLGSAPLSASLA 136
NG L +S ++A
Sbjct: 93 NGQELNGFNISVNVA 107
>At2g21690 putative glycine-rich RNA binding protein
Length = 117
Score = 37.7 bits (86), Expect = 0.011
Identities = 17/49 (34%), Positives = 29/49 (58%)
Query: 60 FSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRY 108
++ F+ G+++ D L DI SKFG V++ + R +G+ RFGF +
Sbjct: 7 YTCFVRGLDQDTDEKDLTDIFSKFGNVIDSKIIYDRDTGKSRRFGFVTF 55
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.142 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,436,205
Number of Sequences: 26719
Number of extensions: 425799
Number of successful extensions: 1530
Number of sequences better than 10.0: 95
Number of HSP's better than 10.0 without gapping: 77
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 1372
Number of HSP's gapped (non-prelim): 162
length of query: 407
length of database: 11,318,596
effective HSP length: 102
effective length of query: 305
effective length of database: 8,593,258
effective search space: 2620943690
effective search space used: 2620943690
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0058a.9


