

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0058a.3
(579 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
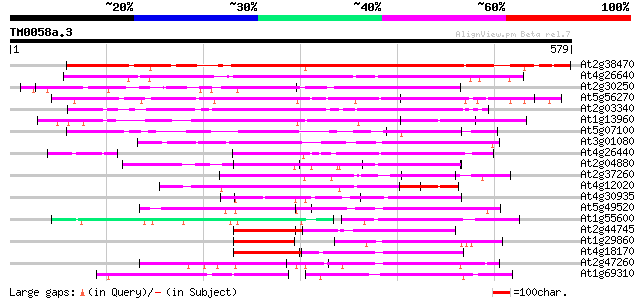
Score E
Sequences producing significant alignments: (bits) Value
At2g38470 putative WRKY-type DNA binding protein 434 e-122
At4g26640 WRKY transcription factor 20 (WRKY20) 340 1e-93
At2g30250 putative WRKY-type DNA binding protein 335 4e-92
At5g56270 transcription factor NtWRKY4-like 320 2e-87
At2g03340 putative WRKY DNA-binding protein 318 4e-87
At1g13960 WRKY DNA-binding protein 4 (WRKY4) 310 2e-84
At5g07100 SPF1-like protein 302 3e-82
At3g01080 WRKY transcription factor 58 (WRKY58) 265 7e-71
At4g26440 putative protein 249 4e-66
At2g04880 WRKY transcription factor 1 splice variant 1 (WRKY1) 211 7e-55
At2g37260 putative WRKY-type DNA binding protein 211 1e-54
At4g12020 putative disease resistance protein 197 1e-50
At4g30935 putative WRKY family transcription factor 180 2e-45
At5g49520 unknown protein 127 2e-29
At1g55600 putative protein 126 4e-29
At2g44745 WRKY transcription factor 12 (WRKY12) 119 4e-27
At1g29860 putative protein 119 6e-27
At4g18170 DNA binding like protein 116 4e-26
At2g47260 putative WRKY-type DNA binding protein 115 9e-26
At1g69310 putative WRKY transcription factor 112 4e-25
>At2g38470 putative WRKY-type DNA binding protein
Length = 512
Score = 434 bits (1116), Expect = e-122
Identities = 254/538 (47%), Positives = 327/538 (60%), Gaps = 77/538 (14%)
Query: 59 VPKFKSTPPPSLPLSPPTIFIPPGLSPAELLDSPVLLNSS-NILPSPTTGAFAAQSFNWK 117
+PKF+S P S+ +SP + SP+ LDSP ++SS N+L SPTTGA N K
Sbjct: 33 IPKFRSFAPSSISISPSLVSPSTCFSPSLFLDSPAFVSSSANVLASPTTGALITNVTNQK 92
Query: 118 SSSGGNQQVVKEEDKSFS--NFSFQTQQ----APPPATTSTATYQSSNVASQTQQQPWSY 171
+ G+ K + +F+ +FSF TQ AP TT+T T ++N + Q+
Sbjct: 93 GINEGD----KSNNNNFNLFDFSFHTQSSGVSAPTTTTTTTTTTTTTNSSIFQSQE---- 144
Query: 172 QEATKQDSMMMKTENSSSMQSFSPEIASVQTNHNNGFQSDYNNYQPQQQQLQPQTLNRRS 231
Q+ Q +TE + Q+ S Y ++Q R+
Sbjct: 145 QQKKNQSEQWSQTETRPNNQAVS--------------------YNGREQ--------RKG 176
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDGQITEIVYKGTHNHPKPQ 291
+DGYNWRKYGQKQVKGSENPRSYYKCT+PNCPTKKKVERSL+GQITEIVYKG+HNHPKPQ
Sbjct: 177 EDGYNWRKYGQKQVKGSENPRSYYKCTFPNCPTKKKVERSLEGQITEIVYKGSHNHPKPQ 236
Query: 292 ATRRNSSSSSSL-------PIPHSNHISNDIQDQSYATHGSGQMDSVATPENSSISIGDD 344
+TRR+SSSSS+ + H+ S+D + + + H S +S S+GDD
Sbjct: 237 STRRSSSSSSTFHSAVYNASLDHNRQASSDQPNSNNSFHQSDSFGMQQEDNTTSDSVGDD 296
Query: 345 DFEQSSQKCRSGGDELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILD 404
+FEQ S S +E EP++KRWK + E G + GS+TVREPR+VVQTTSDIDILD
Sbjct: 297 EFEQGSSIV-SRDEEDCGSEPEAKRWKGDNETNGGNGGGSKTVREPRIVVQTTSDIDILD 355
Query: 405 DGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPA 464
DGYRWRKYGQKVVKGNPNPRSYYKCT GCPVRKHVERASHD+RAVITTYEGKHNHDVPA
Sbjct: 356 DGYRWRKYGQKVVKGNPNPRSYYKCTTIGCPVRKHVERASHDMRAVITTYEGKHNHDVPA 415
Query: 465 ARGSGNHSVTRPLPNNTANAIRPSSVTHQHHHNTNNNSLQSLRPQQAPDQVQSSPFTLEM 524
ARGSG ++ R ++++ IRP+++ ++ T++ +P+TL+M
Sbjct: 416 ARGSG-YATNRAPQDSSSVPIRPAAIAGHSNYTTSS----------------QAPYTLQM 458
Query: 525 LQQNQ---GGFGFSSGFGNSMGSYNMNNQQQQQLPDNVFSSRAKEEPRDDT-FFDSLL 578
L N G FG++ N+ N N Q QQ F SRAKEEP ++T FFDS +
Sbjct: 459 LHNNNTNTGPFGYAMNNNNN----NSNLQTQQNFVGGGF-SRAKEEPNEETSFFDSFM 511
>At4g26640 WRKY transcription factor 20 (WRKY20)
Length = 557
Score = 340 bits (872), Expect = 1e-93
Identities = 217/507 (42%), Positives = 278/507 (54%), Gaps = 52/507 (10%)
Query: 56 GSGVPKFKSTPPPSLPLSPPT-IFIPPGLSPAELLDSPVLLNSSNILPSPTTGA-FAAQS 113
G G ++K P LP+S T I IPPGLSP L+SPV +++ PSPTTG+ F +
Sbjct: 32 GGGGARYKLMSPAKLPISRSTDITIPPGLSPTSFLESPVFISNIKPEPSPTTGSLFKPRP 91
Query: 114 FNWKSSSG-----GNQQVVKEEDKSFSNFSFQTQ------------QAPPPATTSTATYQ 156
+ +SS G Q E KS S F F+ ++ PP +
Sbjct: 92 VHISASSSSYTGRGFHQNTFTEQKS-SEFEFRPPASNMVYAELGKIRSEPPVHFQGQGHG 150
Query: 157 SSNVASQTQQQPWSYQEATKQDSMMMKTENSSSMQSFSPEIASVQTNHNNGFQSDYNNYQ 216
SS+ S S E ++ T SS + + S + S+QT+ N+ S
Sbjct: 151 SSHSPSSISDAAGSSSELSRPTPPCQMTPTSSDIPAGSDQEESIQTSQNDSRGST----- 205
Query: 217 PQQQQLQPQTLNRRSDDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDGQI 276
P L +DDGYNWRKYGQK VKGSE PRSYYKCT+PNC KK ERS DGQI
Sbjct: 206 -------PSIL---ADDGYNWRKYGQKHVKGSEFPRSYYKCTHPNCEVKKLFERSHDGQI 255
Query: 277 TEIVYKGTHNHPKPQATRRNSSSSSSLPIPHSNHISNDIQDQ--SYATHGSGQMDSVATP 334
T+I+YKGTH+HPKPQ RRNS ++ + S+ +D+ S + S + P
Sbjct: 256 TDIIYKGTHDHPKPQPGRRNSGGMAAQEERLDKYPSSTGRDEKGSGVYNLSNPNEQTGNP 315
Query: 335 ENSSISIGDDDFEQSSQKCRSGGDELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVV 394
E IS DD E ++ DE D+D+P SKR ++EG E P + +REPRVVV
Sbjct: 316 EVPPISASDDGGEAAASNRNK--DEPDDDDPFSKRRRMEGAME--ITPLVKPIREPRVVV 371
Query: 395 QTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTY 454
QT S++DILDDGYRWRKYGQKVV+GNPNPRSYYKCT GCPVRKHVERASHD +AVITTY
Sbjct: 372 QTLSEVDILDDGYRWRKYGQKVVRGNPNPRSYYKCTAHGCPVRKHVERASHDPKAVITTY 431
Query: 455 EGKHNHDVPAARGSGNHSVT---RPLPNNTAN---AIRPSSVTHQHHHNTNNNSLQSLRP 508
EGKH+HDVP ++ S NH + RP +T + + SS H N + + Q L
Sbjct: 432 EGKHDHDVPTSKSSSNHEIQPRFRPDETDTISLNLGVGISSDGPNHASNEHQHQNQQLVN 491
Query: 509 QQAPDQ-----VQSSPFTLEMLQQNQG 530
Q P+ V +SP + N G
Sbjct: 492 QTHPNGVNFRFVHASPMSSYYASLNSG 518
>At2g30250 putative WRKY-type DNA binding protein
Length = 393
Score = 335 bits (859), Expect = 4e-92
Identities = 204/447 (45%), Positives = 262/447 (57%), Gaps = 68/447 (15%)
Query: 27 TTSFSDLLASP------ISEDKPRGGLS--DRIAERTGSGVPKFKSTPPPSLPLSPPTIF 78
+TSF+DLL S ED G S ERTGSG+PKFK+ PP LP+S +
Sbjct: 3 STSFTDLLGSSGVDCYEDDEDLRVSGSSFGGYYPERTGSGLPKFKTAQPPPLPISQSS-- 60
Query: 79 IPPGLSPAELLDSPVLLNSSNILPSPTTGAFAAQSFNWKSSSGGNQQVVKEEDKSFSNFS 138
+ ++ LDSP+LL+SS+ L SPTTG F Q FN +++ S+F
Sbjct: 61 --HNFTFSDYLDSPLLLSSSHSLISPTTGTFPLQGFNGTTNN-------------HSDFP 105
Query: 139 FQTQQAPPPATTSTATYQSSNVASQTQQQPWSYQEATKQDSMMMKTENSSSMQSFSPEIA 198
+Q Q P SN AS Q+ + Q+ K+ M+ EIA
Sbjct: 106 WQLQSQP------------SN-ASSALQETYGVQDHEKKQEMIPN------------EIA 140
Query: 199 SVQTNHNNGFQSDYNNYQPQQQQLQPQTLNRRSDDGYNWRKYGQKQVKGSENPRSYYKCT 258
+ N + G + +Q ++ ++R S+DGY WRKYGQKQVK SENPRSY+KCT
Sbjct: 141 TQNNNQSFGTE--------RQIKIPAYMVSRNSNDGYGWRKYGQKQVKKSENPRSYFKCT 192
Query: 259 YPNCPTKKKVERSLDGQITEIVYKGTHNHPKPQATRRNSSSSSSLPIPHSNHISNDIQDQ 318
YP+C +KK VE + DGQITEI+YKG HNHPKP+ T+R S SS +P S +
Sbjct: 193 YPDCVSKKIVETASDGQITEIIYKGGHNHPKPEFTKRPSQSS----LPSSVNGRRLFNPA 248
Query: 319 SYATHGSGQMDSVATPENSSISIGDDDFEQSSQKCRSGGDELDEDEPDSKRWKVEGENEG 378
S + Q ENSSIS D EQ S K G + +E++P+ KR K EGE+EG
Sbjct: 249 SVVSEPHDQS------ENSSISFDYSDLEQKSFKSEYGEIDEEEEQPEMKRMKREGEDEG 302
Query: 379 ISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRK 438
+S S+ V+EPRVVVQT SDID+L DG+RWRKYGQKVVKGN NPRSYYKCT GC V+K
Sbjct: 303 MSIEVSKGVKEPRVVVQTISDIDVLIDGFRWRKYGQKVVKGNTNPRSYYKCTFQGCGVKK 362
Query: 439 HVERASHDLRAVITTYEGKHNHDVPAA 465
VER++ D RAV+TTYEG+HNHD+P A
Sbjct: 363 QVERSAADERAVLTTYEGRHNHDIPTA 389
Score = 83.2 bits (204), Expect = 4e-16
Identities = 82/344 (23%), Positives = 142/344 (40%), Gaps = 66/344 (19%)
Query: 12 ASSFNNFTFSTH---PFMTTSFSDLLASPISEDKPRGGLSDRIAERTGSGVP-KFKSTPP 67
+ S +NFTFS + P + +S S L SP + P G + S P + +S P
Sbjct: 57 SQSSHNFTFSDYLDSPLLLSS-SHSLISPTTGTFPLQGFNG--TTNNHSDFPWQLQSQPS 113
Query: 68 PSLPLSPPTIFIPPGLSPAELLDSPVLLNSSNI---------LPSPTTGAFAAQSFNWKS 118
+ T + E++ + + ++N +P+ + + W+
Sbjct: 114 NASSALQETYGVQDHEKKQEMIPNEIATQNNNQSFGTERQIKIPAYMVSRNSNDGYGWRK 173
Query: 119 SSGGNQQVVKEED-KSFSNFSFQTQQAPPPATTSTATYQSSNVASQTQQQPWSYQEATKQ 177
G +QV K E+ +S+ ++ + T++ + + P E TK+
Sbjct: 174 Y--GQKQVKKSENPRSYFKCTYPDCVSKKIVETASDGQITEIIYKGGHNHP--KPEFTKR 229
Query: 178 DSMMMKTENSSSMQSFSP-----------EIASVQTNHNN----GFQSDYNNYQPQQQQL 222
S + + + F+P E +S+ ++++ F+S+Y +++Q
Sbjct: 230 PSQSSLPSSVNGRRLFNPASVVSEPHDQSENSSISFDYSDLEQKSFKSEYGEIDEEEEQP 289
Query: 223 QPQTLNRRSDD-----------------------------GYNWRKYGQKQVKGSENPRS 253
+ + + R +D G+ WRKYGQK VKG+ NPRS
Sbjct: 290 EMKRMKREGEDEGMSIEVSKGVKEPRVVVQTISDIDVLIDGFRWRKYGQKVVKGNTNPRS 349
Query: 254 YYKCTYPNCPTKKKVERSL-DGQITEIVYKGTHNHPKPQATRRN 296
YYKCT+ C KK+VERS D + Y+G HNH P A RR+
Sbjct: 350 YYKCTFQGCGVKKQVERSAADERAVLTTYEGRHNHDIPTALRRS 393
>At5g56270 transcription factor NtWRKY4-like
Length = 687
Score = 320 bits (819), Expect = 2e-87
Identities = 217/570 (38%), Positives = 290/570 (50%), Gaps = 91/570 (15%)
Query: 44 RGGLSDRIAERTGSGVPKFKST--------PPPSLPLSPPTIFIPPGLSPAELLDSPVLL 95
RGGLS+RIA R G P+ + S SP PGLSPA LL+SPV L
Sbjct: 72 RGGLSERIAARAGFNAPRLNTENIRTNTDFSIDSNLRSPCLTISSPGLSPATLLESPVFL 131
Query: 96 NSSNILPSPTTGAFAAQSFNWKSSSGGNQQVVKEEDKSF----SNFSFQTQQAPPPATTS 151
++ PSPTTG F +G K +D+ F ++FSF P + +S
Sbjct: 132 SNPLAQPSPTTGKFPFLP----GVNGNALSSEKAKDEFFDDIGASFSFH-----PVSRSS 182
Query: 152 TATYQSSNVASQTQQQPWSYQEATKQDSMMMKTENSSSMQSFSPEIASVQTNHNNGFQS- 210
++ +Q + ++ + ++ Q + +K + + S + ++T++ NG
Sbjct: 183 SSFFQGTTEMMSVDYGNYNNRSSSHQSAEEVKPGSENIESS---NLYGIETDNQNGQNKT 239
Query: 211 ------------DYNNYQPQQQQLQPQTLNRRSDDGYNWRKYGQKQVKGSENPRSYYKCT 258
D+ + +Q++ ++DGYNWRKYGQK VKGSE PRSYYKCT
Sbjct: 240 SDVTTNTSLETVDHQEEEEEQRRGDSMAGGAPAEDGYNWRKYGQKLVKGSEYPRSYYKCT 299
Query: 259 YPNCPTKKKVERSLDGQITEIVYKGTHNHPKPQATRR----------------------- 295
PNC KKKVERS +G ITEI+YKG HNH KP RR
Sbjct: 300 NPNCQVKKKVERSREGHITEIIYKGAHNHLKPPPNRRSGMQVDGTEQVEQQQQQRDSAAT 359
Query: 296 ---------------NSSSSSSLPIPHSNHISNDIQDQSYATHGSGQMDSVATPENSSIS 340
N+ S + N S IQ Q+ + SG V +S+ S
Sbjct: 360 WVSCNNTQQQGGSNENNVEEGSTRFEYGNQ-SGSIQAQTGGQYESGD-PVVVVDASSTFS 417
Query: 341 IGDDDFEQSSQKCRS-------GGDELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVV 393
+D+ ++ + S GG + DE +SKR K+E +S +R +REPRVV
Sbjct: 418 NDEDEDDRGTHGSVSLGYDGGGGGGGGEGDESESKRRKLEAFAAEMSG-STRAIREPRVV 476
Query: 394 VQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITT 453
VQTTSD+DILDDGYRWRKYGQKVVKGNPNPRSYYKCT PGC VRKHVERASHDL++VITT
Sbjct: 477 VQTTSDVDILDDGYRWRKYGQKVVKGNPNPRSYYKCTAPGCTVRKHVERASHDLKSVITT 536
Query: 454 YEGKHNHDVPAARGSGNHSVTRPLPNNTANAIRPSSVTHQHHHNTNNNSLQSLRPQQAPD 513
YEGKHNHDVPAAR S + + N+ ++V+H H+HN +++ R +
Sbjct: 537 YEGKHNHDVPAARNSSHGGGG---DSGNGNSGGSAAVSH-HYHNGHHSEPPRGRFDRQVT 592
Query: 514 QVQSSPFTLEMLQQNQGG--FGFSSGFGNS 541
SPF+ Q G GFS G G +
Sbjct: 593 TNNQSPFSRPFSFQPHLGPPSGFSFGLGQT 622
Score = 78.2 bits (191), Expect = 1e-14
Identities = 62/203 (30%), Positives = 94/203 (45%), Gaps = 38/203 (18%)
Query: 404 DDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNH-DV 462
+DGY WRKYGQK+VKG+ PRSYYKCT+P C V+K VER S + Y+G HNH
Sbjct: 273 EDGYNWRKYGQKLVKGSEYPRSYYKCTNPNCQVKKKVER-SREGHITEIIYKGAHNHLKP 331
Query: 463 PAARGSG----------------NHSVTRPLPNNT-------ANAIRPSSVTHQHHHNTN 499
P R SG + + T NNT N + S ++ + +
Sbjct: 332 PPNRRSGMQVDGTEQVEQQQQQRDSAATWVSCNNTQQQGGSNENNVEEGSTRFEYGNQSG 391
Query: 500 NNSLQSLRPQQAPDQV----QSSPFTLEMLQQNQG-----GFGFSSGFGNSMGSYNMNNQ 550
+ Q+ ++ D V SS F+ + + ++G G+ G G G + +
Sbjct: 392 SIQAQTGGQYESGDPVVVVDASSTFSNDEDEDDRGTHGSVSLGYDGGGGGGGGEGDESES 451
Query: 551 QQQQL----PDNVFSSRAKEEPR 569
++++L + S+RA EPR
Sbjct: 452 KRRKLEAFAAEMSGSTRAIREPR 474
>At2g03340 putative WRKY DNA-binding protein
Length = 513
Score = 318 bits (816), Expect = 4e-87
Identities = 200/437 (45%), Positives = 244/437 (55%), Gaps = 48/437 (10%)
Query: 60 PKFKSTPPPSLPLS-PPTIF-IPPGLSPAELLDSPVLLNSSNILPSPTTGAFAAQSFNWK 117
P+FK + P L ++ PP +F +PPGLSPA LLDSP L SP G F
Sbjct: 108 PRFKQSRPTGLMITQPPGMFTVPPGLSPATLLDSPSFFG----LFSPLQGTFGMT----- 158
Query: 118 SSSGGNQQVVKEEDKSFSNFSFQTQQAPPPATTSTATYQSSNVASQTQQQPWSYQEATKQ 177
+QQ + + T QA Q S S TQQQ Q+A+
Sbjct: 159 -----HQQALAQV----------TAQAVQGNNVHMQQSQQSEYPSSTQQQQQQQQQASLT 203
Query: 178 DSMMMKTENSSSMQSFSPEIASVQTNHNNGFQSDYNNYQPQQQQLQPQTLNRRSDDGYNW 237
+ + S ++ ASVQ Q + + + + QPQ ++ +DDGYNW
Sbjct: 204 EIPSFSSAPRSQIR------ASVQETSQG--QRETSEISVFEHRSQPQNADKPADDGYNW 255
Query: 238 RKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDGQITEIVYKGTHNHPKPQATRRNS 297
RKYGQKQVKGS+ PRSYYKCT+P CP KKKVERSLDGQ+TEI+YKG HNH PQ N+
Sbjct: 256 RKYGQKQVKGSDFPRSYYKCTHPACPVKKKVERSLDGQVTEIIYKGQHNHELPQKRGNNN 315
Query: 298 SSSSSLPIPHSNHISNDIQDQSYATHGSGQMDSVATPENSSISIGDDDFEQSSQKCRSGG 357
S S I + SN ++S + Q V T E S + D E+ S G
Sbjct: 316 GSCKSSDIANQFQTSNSSLNKSKRDQETSQ---VTTTEQMSEA---SDSEEVGNAETSVG 369
Query: 358 DELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVV 417
E EDEPD KR E A RTV EPR++VQTTS++D+LDDGYRWRKYGQKVV
Sbjct: 370 -ERHEDEPDPKRRNTEVRVSEPVASSHRTVTEPRIIVQTTSEVDLLDDGYRWRKYGQKVV 428
Query: 418 KGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSGNHSVTRPL 477
KGNP PRSYYKCT P C VRKHVERA+ D +AV+TTYEGKHNHDVPAAR S +H +
Sbjct: 429 KGNPYPRSYYKCTTPDCGVRKHVERAATDPKAVVTTYEGKHNHDVPAARTS-SHQLR--- 484
Query: 478 PNNTANAIRPSSVTHQH 494
PNN N S+V H
Sbjct: 485 PNNQHNT---STVNFNH 498
>At1g13960 WRKY DNA-binding protein 4 (WRKY4)
Length = 514
Score = 310 bits (793), Expect = 2e-84
Identities = 198/479 (41%), Positives = 259/479 (53%), Gaps = 79/479 (16%)
Query: 29 SFSDLLASPISEDKPRGGLS--------DRIAERTGSGV----PKFKSTPPPSLPLSP-- 74
SFS LLA +S + R+ E T S P+FK P L +S
Sbjct: 60 SFSQLLAGAMSSPATAAAAAAAATASDYQRLGEGTNSSSGDVDPRFKQNRPTGLMISQSQ 119
Query: 75 -PTIF-IPPGLSPAELLDSPVLLNSSNILPSPTTGAFAAQSFNWKSSSGGNQQVVKEEDK 132
P++F +PPGLSPA LLDSP L L SP G++
Sbjct: 120 SPSMFTVPPGLSPAMLLDSPSFLG----LFSPVQGSYG---------------------- 153
Query: 133 SFSNFSFQTQQAPPPATTSTATYQSSNVASQTQQQPWSYQEATKQDSMMMKTENSSSMQS 192
T Q T+ A ++N+ QT+ P S +QS
Sbjct: 154 -------MTHQQALAQVTAQAVQANANMQPQTEYPP------------------PSQVQS 188
Query: 193 FSPEIASVQTNHNNGFQSDYNNYQP-QQQQLQPQTLNRRSDDGYNWRKYGQKQVKGSENP 251
FS A + T+ Q + ++ + + QP +++ +DDGYNWRKYGQKQVKGSE P
Sbjct: 189 FSSGQAQIPTSAPLPAQRETSDVTIIEHRSQQPLNVDKPADDGYNWRKYGQKQVKGSEFP 248
Query: 252 RSYYKCTYPNCPTKKKVERSLDGQITEIVYKGTHNHPKPQATRR----NSSSSSSLPIPH 307
RSYYKCT P CP KKKVERSLDGQ+TEI+YKG HNH PQ T+R N+++ + I +
Sbjct: 249 RSYYKCTNPGCPVKKKVERSLDGQVTEIIYKGQHNHEPPQNTKRGNKDNTANINGSSI-N 307
Query: 308 SNHISNDIQDQSYATHGSGQMDSVATPENSSISIGDDDFEQSSQKCRSGGD----ELDED 363
+N S+++ + T+ S + S + + E S + G+ E DE+
Sbjct: 308 NNRGSSELGASQFQTNSSNKTKREQHEAVSQATTTEHLSEASDGEEVGNGETDVREKDEN 367
Query: 364 EPDSKRWKVEGE-NEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPN 422
EPD KR E +E A RTV EPR++VQTTS++D+LDDGYRWRKYGQKVVKGNP
Sbjct: 368 EPDPKRRSTEVRISEPAPAASHRTVTEPRIIVQTTSEVDLLDDGYRWRKYGQKVVKGNPY 427
Query: 423 PRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSGNHSVTRPL-PNN 480
PRSYYKCT PGC VRKHVERA+ D +AV+TTYEGKHNHD+PAA+ S + + L P+N
Sbjct: 428 PRSYYKCTTPGCGVRKHVERAATDPKAVVTTYEGKHNHDLPAAKSSSHAAAAAQLRPDN 486
Score = 97.8 bits (242), Expect = 1e-20
Identities = 53/130 (40%), Positives = 70/130 (53%), Gaps = 1/130 (0%)
Query: 404 DDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVP 463
DDGY WRKYGQK VKG+ PRSYYKCT+PGCPV+K VER S D + Y+G+HNH+ P
Sbjct: 229 DDGYNWRKYGQKQVKGSEFPRSYYKCTNPGCPVKKKVER-SLDGQVTEIIYKGQHNHEPP 287
Query: 464 AARGSGNHSVTRPLPNNTANAIRPSSVTHQHHHNTNNNSLQSLRPQQAPDQVQSSPFTLE 523
GN T + ++ N R SS TN+++ +A Q ++ E
Sbjct: 288 QNTKRGNKDNTANINGSSINNNRGSSELGASQFQTNSSNKTKREQHEAVSQATTTEHLSE 347
Query: 524 MLQQNQGGFG 533
+ G G
Sbjct: 348 ASDGEEVGNG 357
>At5g07100 SPF1-like protein
Length = 309
Score = 302 bits (774), Expect = 3e-82
Identities = 182/410 (44%), Positives = 218/410 (52%), Gaps = 125/410 (30%)
Query: 59 VPKFKSTPPPSLPLSPPTIF-IPPGLSPAELLDSPVLLNSSNILPSPTTGAFAAQSFNWK 117
VPKFK+ P LPLSP F +PPGL+PA+ LDSP+L SSNILPSPTTG F AQS N+
Sbjct: 10 VPKFKTATPSPLPLSPSPYFTMPPGLTPADFLDSPLLFTSSNILPSPTTGTFPAQSLNYN 69
Query: 118 SSSGGNQQVVKEEDKSFSNFSFQTQQAPPPATTSTATYQSSNVASQTQQQPWSYQEATKQ 177
+ N ++ + + + + PP S
Sbjct: 70 N----NGLLIDKNEIKYED------TTPPLFLPS-------------------------- 93
Query: 178 DSMMMKTENSSSMQSFSPEIASVQTNHNNGFQSDYNNYQPQQQQLQPQTLNRRSDDGYNW 237
M T+ + F EI S ++G+ NW
Sbjct: 94 ----MVTQPLPQLDLFKSEIMSSNKTSDDGY---------------------------NW 122
Query: 238 RKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSL-DGQITEIVYKGTHNHPKPQATRRN 296
RKYGQKQVKGSENPRSY+KCTYPNC TKKKVE SL GQ+ EIVYKG+HNHPKPQ+T+R+
Sbjct: 123 RKYGQKQVKGSENPRSYFKCTYPNCLTKKKVETSLVKGQMIEIVYKGSHNHPKPQSTKRS 182
Query: 297 SSSSSSLPIPHSNHISNDIQDQSYATHGSGQMDSVATPENSSISIGDDDFEQSSQKCRSG 356
SS+ ++A +NSS
Sbjct: 183 SST------------------------------AIAAHQNSS---------------NGD 197
Query: 357 GDELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKV 416
G ++ EDE ++KRWK E V+EPRVVVQTTSDIDILDDGYRWRKYGQKV
Sbjct: 198 GKDIGEDETEAKRWKRE-----------ENVKEPRVVVQTTSDIDILDDGYRWRKYGQKV 246
Query: 417 VKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAAR 466
VKGNPNPRSYYKCT GC VRKHVERA D ++VITTYEGKH H +P R
Sbjct: 247 VKGNPNPRSYYKCTFTGCFVRKHVERAFQDPKSVITTYEGKHKHQIPTPR 296
Score = 79.7 bits (195), Expect = 4e-15
Identities = 42/125 (33%), Positives = 59/125 (46%), Gaps = 26/125 (20%)
Query: 390 PRVVVQTTSDIDIL-----------DDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRK 438
P +V Q +D+ DDGY WRKYGQK VKG+ NPRSY+KCT+P C +K
Sbjct: 92 PSMVTQPLPQLDLFKSEIMSSNKTSDDGYNWRKYGQKQVKGSENPRSYFKCTYPNCLTKK 151
Query: 439 HVERASHDLRAVITTYEGKHNHDVPAARGSGNHSVTRPLPNNTANAIRPSSVTHQHHHNT 498
VE + + + Y+G HNH P P +T + + HQ+ N
Sbjct: 152 KVETSLVKGQMIEIVYKGSHNH---------------PKPQSTKRSSSTAIAAHQNSSNG 196
Query: 499 NNNSL 503
+ +
Sbjct: 197 DGKDI 201
>At3g01080 WRKY transcription factor 58 (WRKY58)
Length = 423
Score = 265 bits (676), Expect = 7e-71
Identities = 156/376 (41%), Positives = 204/376 (53%), Gaps = 44/376 (11%)
Query: 133 SFSNFSFQTQQAPPPATTSTATYQSSNVASQTQQQPWSYQEATKQDSMMMKTENSSSMQS 192
SF++ PP ++ T Q + Q QP + T M T + +
Sbjct: 76 SFTHLLTSPMFFPPQSSAHTGFIQP-----RQQSQPQPQRPDTFPHHMPPSTSVAVHGRQ 130
Query: 193 FSPEIASVQTNHNNGFQSDYNNYQPQQQQLQPQTLNRRSDDGYNWRKYGQKQVKGSENPR 252
S +++ V N + + NN + + +++ +DDGYNWRKYGQK +KG E PR
Sbjct: 131 -SLDVSQVDQRARNHYNNPGNNNNNRSYNVV--NVDKPADDGYNWRKYGQKPIKGCEYPR 187
Query: 253 SYYKCTYPNCPTKKKVERSLDGQITEIVYKGTHNHPKPQATRRNSSSSSSLPIPHSNHIS 312
SYYKCT+ NCP KKKVERS DGQIT+I+YKG H+H +PQ R S+
Sbjct: 188 SYYKCTHVNCPVKKKVERSSDGQITQIIYKGQHDHERPQNRRGGGGRDST---------- 237
Query: 313 NDIQDQSYATHGSGQMDSVATPENSSISIGDDDFEQSSQKCRSGGDELDEDEPDSKRWKV 372
G+GQM + DDD + DE DED P SK ++
Sbjct: 238 --------EVGGAGQMMESSDDSGYRKDHDDDDDD----------DEDDEDLPASKIRRI 279
Query: 373 EGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHP 432
+G + RTV EP+++VQT S++D+LDDGYRWRKYGQKVVKGNP+PRSYYKCT P
Sbjct: 280 DGVSTT-----HRTVTEPKIIVQTKSEVDLLDDGYRWRKYGQKVVKGNPHPRSYYKCTTP 334
Query: 433 GCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSGNHSVTRPLPNNTANAIRPSSVTH 492
C VRKHVERAS D +AVITTYEGKHNHDVPAAR + + + + +R S +
Sbjct: 335 NCTVRKHVERASTDAKAVITTYEGKHNHDVPAARNGTAAATAAAVGPSDHHRMRSMSGNN 394
Query: 493 QHHH---NTNNNSLQS 505
H NNN+ QS
Sbjct: 395 MQQHMSFGNNNNTGQS 410
>At4g26440 putative protein
Length = 568
Score = 249 bits (635), Expect = 4e-66
Identities = 142/298 (47%), Positives = 178/298 (59%), Gaps = 42/298 (14%)
Query: 231 SDDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDGQITEIVYKGTHNHPKP 290
+DDGYNWRKYGQK VKGSE PRSYYKCT+PNC KKKVERS +G I EI+Y G H H KP
Sbjct: 177 ADDGYNWRKYGQKLVKGSEYPRSYYKCTHPNCEAKKKVERSREGHIIEIIYTGDHIHSKP 236
Query: 291 QATRRNSSSSS-----------------------------SLPIPHSNHISNDIQDQSYA 321
RR+ SS S + + +H S +Q Q+
Sbjct: 237 PPNRRSGIGSSGTGQDMQIDATEYEGFAGTNENIEWTSPVSAELEYGSH-SGSMQVQN-G 294
Query: 322 THGSGQMDSVATPENSSISIGDDDFEQSSQKCRSGGDELDEDEPDSKRWKVEGENEGISA 381
TH G D+ A ++ +++ +++S S + + +E +SKR K+E S
Sbjct: 295 THQFGYGDAAA----DALYRDENEDDRTSHMSVSLTYDGEVEESESKRRKLEAYATETSG 350
Query: 382 PGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVE 441
+R REPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCT GC V KHVE
Sbjct: 351 -STRASREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTANGCTVTKHVE 409
Query: 442 RASHDLRAVITTYEGKHNHDVPAARGSGNHSVTRPLPNNTANAIRPSSVTHQHHHNTN 499
RAS D ++V+TTY GKH H VPAAR S + + ++ ++ S T H+HN +
Sbjct: 410 RASDDFKSVLTTYIGKHTHVVPAARNSSH------VGAGSSGTLQGSLATQTHNHNVH 461
Score = 46.2 bits (108), Expect = 5e-05
Identities = 30/72 (41%), Positives = 39/72 (53%), Gaps = 5/72 (6%)
Query: 40 EDKPRGGLSDRIAERTGSGVPKFKSTPPPSLPLSPPTIFIPPGLSPAELLDSPVLLNSSN 99
++ RG L +RIA R+G P + + S PGLSPA LL+SPV L SN
Sbjct: 50 QESSRGALRERIAARSGFNAPWLNTE---DILQSKSLTISSPGLSPATLLESPVFL--SN 104
Query: 100 ILPSPTTGAFAA 111
L SPTTG ++
Sbjct: 105 PLLSPTTGKLSS 116
>At2g04880 WRKY transcription factor 1 splice variant 1 (WRKY1)
Length = 487
Score = 211 bits (538), Expect = 7e-55
Identities = 117/261 (44%), Positives = 149/261 (56%), Gaps = 28/261 (10%)
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDGQITEIVYKGTHNHPKPQ 291
+DGYNWRKYGQK VKG+E RSYY+CT+PNC KK++ERS GQ+ + VY G H+HPKP
Sbjct: 111 EDGYNWRKYGQKLVKGNEFVRSYYRCTHPNCKAKKQLERSAGGQVVDTVYFGEHDHPKPL 170
Query: 292 A----TRRNSSSSSSLPIPHSNH-------------ISNDIQDQSYATHGSGQMDSVATP 334
A ++ S + +S DI + S Q P
Sbjct: 171 AGAVPINQDKRSDVFTAVSKGEQRIDIVSLIYKLCIVSYDIMFVEKTSGSSVQTLRQTEP 230
Query: 335 EN-------SSISIGDDDFEQSSQKCRSGGDELDED--EPDSKRWKVEGENEGISAPGSR 385
S I DD SQ R GD +D P +KR K G E +P R
Sbjct: 231 PKIHGGLHVSVIPPADDVKTDISQSSRITGDNTHKDYNSPTAKRRKKGGNIE--LSPVER 288
Query: 386 TVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASH 445
+ + R+VV T + DI++DGYRWRKYGQK VKG+P PRSYY+C+ PGCPV+KHVER+SH
Sbjct: 289 STNDSRIVVHTQTLFDIVNDGYRWRKYGQKSVKGSPYPRSYYRCSSPGCPVKKHVERSSH 348
Query: 446 DLRAVITTYEGKHNHDVPAAR 466
D + +ITTYEGKH+HD+P R
Sbjct: 349 DTKLLITTYEGKHDHDMPPGR 369
Score = 87.0 bits (214), Expect = 3e-17
Identities = 65/252 (25%), Positives = 113/252 (44%), Gaps = 45/252 (17%)
Query: 117 KSSSGGNQQVVKEEDKSFSNFSFQTQQAPPPATTSTATYQSSNVASQTQQQPWSYQEATK 176
+ +SG + Q +++ + + PP T QSS + + ++ A +
Sbjct: 215 EKTSGSSVQTLRQTEPPKIHGGLHVSVIPPADDVKTDISQSSRITGDNTHKDYNSPTAKR 274
Query: 177 QDSMMMKTENSSSMQSFSPEIASVQTNHNNGFQSDYNNYQPQQQQLQPQTLNRRSDDGYN 236
+ + + E++ V+ + N+ + + QTL +DGY
Sbjct: 275 RK------------KGGNIELSPVERSTNDS-----------RIVVHTQTLFDIVNDGYR 311
Query: 237 WRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSL-DGQITEIVYKGTHNHPKPQATRR 295
WRKYGQK VKGS PRSYY+C+ P CP KK VERS D ++ Y+G H+H P
Sbjct: 312 WRKYGQKSVKGSPYPRSYYRCSSPGCPVKKHVERSSHDTKLLITTYEGKHDHDMPPGR-- 369
Query: 296 NSSSSSSLPIPHSNHISNDIQDQSYATHGSGQMDSVATPENSSI-SIGDDDF--EQSSQK 352
+ H+N + +++ D+ + D+ TP++S++ SI D + +K
Sbjct: 370 --------VVTHNNMLDSEVDDK--------EGDANKTPQSSTLQSITKDQHVEDHLRKK 413
Query: 353 CRSGGDELDEDE 364
++ G E D+
Sbjct: 414 TKTNGFEKSLDQ 425
Score = 80.5 bits (197), Expect = 2e-15
Identities = 52/171 (30%), Positives = 82/171 (47%), Gaps = 13/171 (7%)
Query: 300 SSSLPIPHSNHISNDIQDQSYATHGSGQMDSVATPENSSI-----SIGDDDFEQSSQKCR 354
+S + + HSN D AT ++ VA ++ S + E+
Sbjct: 9 ASDMELDHSNETK--AVDDVVATTDKAEVIPVAVTRTETVVESLESTDCKELEKLVPHTV 66
Query: 355 SGGDELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQ 414
+ E+D P S++ E+ G + S + + + +++DGY WRKYGQ
Sbjct: 67 ASQSEVDVASPVSEKAPKVSESSGALSLQSGSEGNSPFIREK-----VMEDGYNWRKYGQ 121
Query: 415 KVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAA 465
K+VKGN RSYY+CTHP C +K +ER++ + V T Y G+H+H P A
Sbjct: 122 KLVKGNEFVRSYYRCTHPNCKAKKQLERSAGG-QVVDTVYFGEHDHPKPLA 171
>At2g37260 putative WRKY-type DNA binding protein
Length = 349
Score = 211 bits (536), Expect = 1e-54
Identities = 128/271 (47%), Positives = 152/271 (55%), Gaps = 42/271 (15%)
Query: 217 PQQQQLQPQTLNRRSDDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDGQI 276
P + + T +R S DGYNWRKYGQKQVKGSE PRSYYKCT+P CP KKKVERS++GQ+
Sbjct: 70 PSFTESETSTGDRSSVDGYNWRKYGQKQVKGSECPRSYYKCTHPKCPVKKKVERSVEGQV 129
Query: 277 TEIVYKGTHNHPKPQA--TRRNSSSSSS----------------------LPIPHSNHIS 312
+EIVY+G HNH KP RR SSS SS L P N+ S
Sbjct: 130 SEIVYQGEHNHSKPSCPLPRRASSSISSGFQKPPKSIASEGSMGQDPNNNLYSPLWNNQS 189
Query: 313 NDIQDQSYATHGSGQMDS---VATPENSSISIGDDDFEQSSQKCRSGGDELDEDEPDSKR 369
ND G + + A P +++ + G D S +C G ELD D SKR
Sbjct: 190 NDSTQNRTEKMSEGCVITPFEFAVPRSTNSNPGTSDSGCKSSQCDEG--ELD-DPSRSKR 246
Query: 370 WKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKC 429
K E ++ V Q + + D L+DG+RWRKYGQKVV GN PRSYY+C
Sbjct: 247 RKNEKQSSEAG------------VSQGSVESDSLEDGFRWRKYGQKVVGGNAYPRSYYRC 294
Query: 430 THPGCPVRKHVERASHDLRAVITTYEGKHNH 460
T C RKHVERAS D RA ITTYEGKHNH
Sbjct: 295 TSANCRARKHVERASDDPRAFITTYEGKHNH 325
Score = 88.2 bits (217), Expect = 1e-17
Identities = 50/121 (41%), Positives = 65/121 (53%), Gaps = 19/121 (15%)
Query: 405 DGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPA 464
DGY WRKYGQK VKG+ PRSYYKCTHP CPV+K VER+ + I Y+G+HNH P+
Sbjct: 86 DGYNWRKYGQKQVKGSECPRSYYKCTHPKCPVKKKVERSVEGQVSEIV-YQGEHNHSKPS 144
Query: 465 ARGSGNHSVTRPLPNNTANAIR------PSSVTHQHH--HNTNNNSLQSLRPQQAPDQVQ 516
PLP +++I P S+ + + NNN L Q+ D Q
Sbjct: 145 C----------PLPRRASSSISSGFQKPPKSIASEGSMGQDPNNNLYSPLWNNQSNDSTQ 194
Query: 517 S 517
+
Sbjct: 195 N 195
>At4g12020 putative disease resistance protein
Length = 1895
Score = 197 bits (501), Expect = 1e-50
Identities = 115/277 (41%), Positives = 162/277 (57%), Gaps = 16/277 (5%)
Query: 155 YQSSNVASQTQQQPWSYQEATKQDSMMMKTENSSSMQSFSPEIASVQTNHNNGFQSDYNN 214
Y+ + +QT Q + Q T+ S SS++QSFS + T+ + +N
Sbjct: 394 YEMTYQQAQTVQANANMQPQTEYPS-------SSAVQSFSSGQPQIPTSAPDSSLLAKSN 446
Query: 215 YQP----QQQQLQPQTLNRRSDDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVER 270
+ QP ++++ +DGYNW+KYGQK+VKGS+ P SYYKCTY CP+K+KVER
Sbjct: 447 TSGITIIEHMSQQPLNVDKQVNDGYNWQKYGQKKVKGSKFPLSYYKCTYLGCPSKRKVER 506
Query: 271 SLDGQITEIVYKGTHNHPKPQATRRNSSSSSSLPIPHSNHISNDIQDQSYATHGSGQMDS 330
SLDGQ+ EIVYK HNH P + S++ S H N +S+++ ++++ +
Sbjct: 507 SLDGQVAEIVYKDRHNHEPPNQGKDGSTTYLSGSSTHINCMSSELTASQFSSNKTKIEQQ 566
Query: 331 VATPENSSI---SIGDDDFEQSSQKCRSGGDELDEDEPDSKRWKVEGENEGISAPGSRTV 387
A ++I S D+ E S+ + G E DEDEP+ KR E + ++ RTV
Sbjct: 567 EAASLATTIEYMSEASDNEEDSNGETSEG--EKDEDEPEPKRRITEVQVSELADASDRTV 624
Query: 388 REPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPR 424
REPRV+ QTTS++D LDDGYRWRKYGQKVVKGNP PR
Sbjct: 625 REPRVIFQTTSEVDNLDDGYRWRKYGQKVVKGNPYPR 661
Score = 72.0 bits (175), Expect = 8e-13
Identities = 32/61 (52%), Positives = 43/61 (70%), Gaps = 1/61 (1%)
Query: 403 LDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDV 462
++DGY W+KYGQK VKG+ P SYYKCT+ GCP ++ VER S D + Y+ +HNH+
Sbjct: 467 VNDGYNWQKYGQKKVKGSKFPLSYYKCTYLGCPSKRKVER-SLDGQVAEIVYKDRHNHEP 525
Query: 463 P 463
P
Sbjct: 526 P 526
>At4g30935 putative WRKY family transcription factor
Length = 466
Score = 180 bits (456), Expect = 2e-45
Identities = 104/264 (39%), Positives = 145/264 (54%), Gaps = 30/264 (11%)
Query: 218 QQQQLQPQTLNRRS---------DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKV 268
Q+Q+ +NR S DGYNWRKYGQKQVK + RSYY+CTY C KK+
Sbjct: 145 QEQRSDSPVVNRLSVTPVPRTPARDGYNWRKYGQKQVKSPKGSRSYYRCTYTEC-CAKKI 203
Query: 269 ERSLD-GQITEIVYKGTHNHPKPQAT----RRNSSSSSSLPIPHSNHISNDIQDQSYATH 323
E S D G + EIV KG H H P+ T R +++ P+ + + +++ S
Sbjct: 204 ECSNDSGNVVEIVNKGLHTHEPPRKTSFSPREIRVTTAIRPVSEDDTV---VEELSIVPS 260
Query: 324 GSGQMDSVATPENSSISIGDDDFEQSSQKCRSGGDELDEDEPDSKRWKVEGEN-EGISAP 382
GS S S ++ D +K + ++E EP + K ++ + +S P
Sbjct: 261 GSDPSASTKEYICESQTLVD-------RKRHCENEAVEEPEPKRRLKKDNSQSSDSVSKP 313
Query: 383 GSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVER 442
G + + VV D+ I DGYRWRKYGQK+VKGNP+PR+YY+CT GCPVRKH+E
Sbjct: 314 GKKN----KFVVHAAGDVGICGDGYRWRKYGQKMVKGNPHPRNYYRCTSAGCPVRKHIET 369
Query: 443 ASHDLRAVITTYEGKHNHDVPAAR 466
A + +AVI TY+G HNHD+P +
Sbjct: 370 AVENTKAVIITYKGVHNHDMPVPK 393
Score = 89.4 bits (220), Expect = 5e-18
Identities = 49/134 (36%), Positives = 69/134 (50%), Gaps = 18/134 (13%)
Query: 233 DGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDGQITEIV-YKGTHNHPKPQ 291
DGY WRKYGQK VKG+ +PR+YY+CT CP +K +E +++ I+ YKG HNH P
Sbjct: 332 DGYRWRKYGQKMVKGNPHPRNYYRCTSAGCPVRKHIETAVENTKAVIITYKGVHNHDMPV 391
Query: 292 ATRRNSSSSSSL---PIPHSNHISNDIQDQSYATHGSGQMDSVATPENSSISIGDDDFEQ 348
+R+ SS L P S D D V P +S S+G + +Q
Sbjct: 392 PKKRHGPPSSMLVAAAAPTSMRTRTD--------------DQVNIPTSSQCSVGRESEKQ 437
Query: 349 SSQKCRSGGDELDE 362
S + GG+++ E
Sbjct: 438 SKEALDVGGEKVME 451
>At5g49520 unknown protein
Length = 399
Score = 127 bits (319), Expect = 2e-29
Identities = 80/224 (35%), Positives = 110/224 (48%), Gaps = 28/224 (12%)
Query: 296 NSSSSSSLPIPH--SNHISNDIQDQSYATHGSGQMDSVATPENSSISIGDDDFEQSSQKC 353
N+S + LP+P S+ + N +T S + A NS + D E+
Sbjct: 126 NTSFFTDLPLPQAESSEVVNTTPTSPNSTSVSSSSNEAANDNNSGKEVTVKDQEE----- 180
Query: 354 RSGGDELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYG 413
GD+ E + + K + +N+ + RE R T SDID LDDGYRWRKYG
Sbjct: 181 ---GDQQQEQKGTKPQLKAKKKNQ-------KKAREARFAFLTKSDIDNLDDGYRWRKYG 230
Query: 414 QKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSGNHSV 473
QK VK +P PRSYY+CT GC V+K VER+S D V+TTYEG+H H P +
Sbjct: 231 QKAVKNSPYPRSYYRCTTVGCGVKKRVERSSDDPSIVMTTYEGQHTHPFPMTPRGHIGML 290
Query: 474 TRPLPNNTANAIRPSSVT-----------HQHHHNTNNNSLQSL 506
T P+ ++ A SS + HQ ++ NNNSL +
Sbjct: 291 TSPILDHGATTASSSSFSIPQPRYLLTQHHQPYNMYNNNSLSMI 334
Score = 83.2 bits (204), Expect = 4e-16
Identities = 65/215 (30%), Positives = 89/215 (41%), Gaps = 48/215 (22%)
Query: 135 SNFSFQTQQAPPPATTSTATYQSSNVASQTQQQPWSYQEATKQDSMMMKTENSSSMQSFS 194
S+FSF P ++N S P E+++ + + NS+S+ S S
Sbjct: 111 SSFSFDAFPLP----------NNNNNTSFFTDLPLPQAESSEVVNTTPTSPNSTSVSSSS 160
Query: 195 PEIASVQTNHNNGFQSDYNNYQPQQQQ------LQPQTLNRRS----------------- 231
E A+ + D QQ+Q L+ + N++
Sbjct: 161 NEAANDNNSGKEVTVKDQEEGDQQQEQKGTKPQLKAKKKNQKKAREARFAFLTKSDIDNL 220
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDG-QITEIVYKGTHNHPKP 290
DDGY WRKYGQK VK S PRSYY+CT C KK+VERS D I Y+G H HP P
Sbjct: 221 DDGYRWRKYGQKAVKNSPYPRSYYRCTTVGCGVKKRVERSSDDPSIVMTTYEGQHTHPFP 280
Query: 291 QATR--------------RNSSSSSSLPIPHSNHI 311
R ++SSSS IP ++
Sbjct: 281 MTPRGHIGMLTSPILDHGATTASSSSFSIPQPRYL 315
>At1g55600 putative protein
Length = 485
Score = 126 bits (316), Expect = 4e-29
Identities = 72/192 (37%), Positives = 101/192 (52%), Gaps = 22/192 (11%)
Query: 343 DDDFEQSSQKCRSGGDELDEDE--------PDSKRWKVEGENEGISAPGSRTVREPRVVV 394
+D ++Q +E DED P KR + E N +RT + R+++
Sbjct: 241 EDQYDQDQDVDEDEEEEKDEDNVALDDPQPPPPKRRRYEVSN---MIGATRTSKTQRIIL 297
Query: 395 QTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTY 454
Q SD D +DGYRWRKYGQKVVKGNPNPRSY+KCT+ C V+KHVER + +++ V+TTY
Sbjct: 298 QMESDEDNPNDGYRWRKYGQKVVKGNPNPRSYFKCTNIECRVKKHVERGADNIKLVVTTY 357
Query: 455 EGKHNHDVPAARGSGNHSVTRPLPNNTANAIRPSSVTHQHHHNTNNNSLQSLRPQQAPDQ 514
+G HNH P AR S N+++ R + T + N + L P P
Sbjct: 358 DGIHNHPSPPARRS-----------NSSSRNRSAGATIPQNQNDRTSRLGRAPPTPTPPT 406
Query: 515 VQSSPFTLEMLQ 526
S +T E ++
Sbjct: 407 PPPSSYTPEEMR 418
Score = 92.4 bits (228), Expect = 6e-19
Identities = 92/355 (25%), Positives = 134/355 (36%), Gaps = 74/355 (20%)
Query: 44 RGGLSDRIAERTGSGVPKFKSTPPPSLPL----------SPPTIFIPPGLSPAELLDSP- 92
R GL +R+A R G +P + S PL + P + I PG SP+ LL +P
Sbjct: 60 RSGLRERLAARVGFNLPTLNTEENMS-PLDAFFRSSNVPNSPVVAISPGFSPSALLHTPN 118
Query: 93 VLLNSSNILPSPTTGAFAAQSFNWKSSSGGNQQVVKEEDKSFSNF--------------- 137
++ +SS I+P + + S ++ D + +
Sbjct: 119 MVSDSSQIIPPSSATNYGPLEMVETSGEDNAAMMMFNNDLPYQPYNVDLPSLEVFDDIAT 178
Query: 138 --SFQTQQAPP---PATTSTATYQSSNVASQTQQQPWSYQEATKQD----------SMMM 182
SF P P T T S + S +++ +D
Sbjct: 179 EESFYIPSYEPHVDPIGTPLVTSFESELVDDAHTDIISIEDSESEDGNKDDDDEDFQYED 238
Query: 183 KTENSSSMQSFSPEIASVQTNHNNGFQSDYNNYQPQQQQLQPQT--------------LN 228
+ E+ E + + +N D P++++ + L
Sbjct: 239 EDEDQYDQDQDVDEDEEEEKDEDNVALDDPQPPPPKRRRYEVSNMIGATRTSKTQRIILQ 298
Query: 229 RRSDD-----GYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDG-QITEIVYK 282
SD+ GY WRKYGQK VKG+ NPRSY+KCT C KK VER D ++ Y
Sbjct: 299 MESDEDNPNDGYRWRKYGQKVVKGNPNPRSYFKCTNIECRVKKHVERGADNIKLVVTTYD 358
Query: 283 GTHNHPKPQATRRNSSS---SSSLPIPHSNHISNDIQDQSYATHGSGQMDSVATP 334
G HNHP P A R NSSS S+ IP Q+Q+ T G+ TP
Sbjct: 359 GIHNHPSPPARRSNSSSRNRSAGATIP---------QNQNDRTSRLGRAPPTPTP 404
>At2g44745 WRKY transcription factor 12 (WRKY12)
Length = 218
Score = 119 bits (299), Expect = 4e-27
Identities = 69/167 (41%), Positives = 91/167 (54%), Gaps = 11/167 (6%)
Query: 296 NSSSSSSLPIPHSNHISNDIQDQSYATHGSGQMDSVATPENSSISIGDDDFEQSSQKCRS 355
+SSSSSSL P ++ ++A G P S + + DD +
Sbjct: 44 SSSSSSSLSSPSFPIHNSSSTTTTHAPLGFSNNLQGGGPLGSKV-VNDDQ--------EN 94
Query: 356 GGDELDEDEPDSKRWKVEGENEGISAPGS--RTVREPRVVVQTTSDIDILDDGYRWRKYG 413
G + D + W+ + + R +REPR QT SD+D+LDDGY+WRKYG
Sbjct: 95 FGGGTNNDAHSNSWWRSNSGSGDMKNKVKIRRKLREPRFCFQTKSDVDVLDDGYKWRKYG 154
Query: 414 QKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNH 460
QKVVK + +PRSYY+CTH C V+K VER S D R VITTYEG+HNH
Sbjct: 155 QKVVKNSLHPRSYYRCTHNNCRVKKRVERLSEDCRMVITTYEGRHNH 201
Score = 77.8 bits (190), Expect = 2e-14
Identities = 39/73 (53%), Positives = 48/73 (65%), Gaps = 2/73 (2%)
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVER-SLDGQITEIVYKGTHNH-PK 289
DDGY WRKYGQK VK S +PRSYY+CT+ NC KK+VER S D ++ Y+G HNH P
Sbjct: 145 DDGYKWRKYGQKVVKNSLHPRSYYRCTHNNCRVKKRVERLSEDCRMVITTYEGRHNHIPS 204
Query: 290 PQATRRNSSSSSS 302
+T + SS
Sbjct: 205 DDSTSPDHDCLSS 217
>At1g29860 putative protein
Length = 282
Score = 119 bits (297), Expect = 6e-27
Identities = 71/202 (35%), Positives = 105/202 (51%), Gaps = 34/202 (16%)
Query: 336 NSSISIGDDDFEQSSQKCRSGGDELDEDEPD-------SKRWKVEGENEGISAPGSRTVR 388
NS + + + + D+++++E D SK+ +G+ +G + R
Sbjct: 66 NSPVVSSSSNEGEPKENTNDKSDQMEDNEGDLHGVGESSKQLTKQGKKKG-----EKKER 120
Query: 389 EPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLR 448
E RV T S+ID L+DGYRWRKYGQK VK +P PRSYY+CT C V+K VER+ D
Sbjct: 121 EVRVAFMTKSEIDHLEDGYRWRKYGQKAVKNSPYPRSYYRCTTQKCNVKKRVERSFQDPS 180
Query: 449 AVITTYEGKHNHDVPAA-------------RGSGN---HSVTR------PLPNNTANAIR 486
VITTYEGKHNH +P+ RG G HS R + ++ AN
Sbjct: 181 IVITTYEGKHNHPIPSTLRGTVAAEHLLVHRGGGGSLLHSFPRHHQDFLMMKHSPANYQS 240
Query: 487 PSSVTHQHHHNTNNNSLQSLRP 508
S++++H H T++ + + +P
Sbjct: 241 VGSLSYEHGHGTSSYNFNNNQP 262
Score = 80.1 bits (196), Expect = 3e-15
Identities = 37/64 (57%), Positives = 41/64 (63%), Gaps = 1/64 (1%)
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSL-DGQITEIVYKGTHNHPKP 290
+DGY WRKYGQK VK S PRSYY+CT C KK+VERS D I Y+G HNHP P
Sbjct: 136 EDGYRWRKYGQKAVKNSPYPRSYYRCTTQKCNVKKRVERSFQDPSIVITTYEGKHNHPIP 195
Query: 291 QATR 294
R
Sbjct: 196 STLR 199
>At4g18170 DNA binding like protein
Length = 318
Score = 116 bits (290), Expect = 4e-26
Identities = 68/170 (40%), Positives = 93/170 (54%), Gaps = 12/170 (7%)
Query: 300 SSSLPIPHSNHISNDIQDQSYATHGSGQMDSVATPENSSISIGDDDFEQSSQKCRSGGDE 359
+SS+ + ++ND+ + + ++ +P NSS S D E S + R
Sbjct: 79 NSSIDQEPNRDVTNDVINGG----ACNETETRVSPSNSSSSEADHPGEDSGKSRRKRELV 134
Query: 360 LDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKG 419
+ED+ K K + + REPRV T S++D L+DGYRWRKYGQK VK
Sbjct: 135 GEEDQISKKVGKTKKTEV-------KKQREPRVSFMTKSEVDHLEDGYRWRKYGQKAVKN 187
Query: 420 NPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPA-ARGS 468
+P PRSYY+CT C V+K VER+ D VITTYEG+HNH +P RGS
Sbjct: 188 SPYPRSYYRCTTQKCNVKKRVERSFQDPTVVITTYEGQHNHPIPTNLRGS 237
Score = 80.9 bits (198), Expect = 2e-15
Identities = 37/71 (52%), Positives = 47/71 (66%), Gaps = 1/71 (1%)
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSL-DGQITEIVYKGTHNHPKP 290
+DGY WRKYGQK VK S PRSYY+CT C KK+VERS D + Y+G HNHP P
Sbjct: 172 EDGYRWRKYGQKAVKNSPYPRSYYRCTTQKCNVKKRVERSFQDPTVVITTYEGQHNHPIP 231
Query: 291 QATRRNSSSSS 301
R +S++++
Sbjct: 232 TNLRGSSAAAA 242
>At2g47260 putative WRKY-type DNA binding protein
Length = 337
Score = 115 bits (287), Expect = 9e-26
Identities = 77/241 (31%), Positives = 112/241 (45%), Gaps = 31/241 (12%)
Query: 286 NHPKPQATRRNSSSSSSLPIPHSNHISNDIQDQSYATHGSGQMDSVATPENSSISIG--- 342
+ P+ Q SSS + P S + + + H ++ ATP +SSIS
Sbjct: 60 SQPQTQTQPSAKLSSSIIQAPPSEQLVTSKVESLCSDHLL--INPPATPNSSSISSASSE 117
Query: 343 --DDDFEQSSQKCRSGGDELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDI 400
+++ ++ GG++ E K+ K + N+ + RE RV T S++
Sbjct: 118 ALNEEKPKTEDNEEEGGEDQQEKSHTKKQLKAKKNNQ-------KRQREARVAFMTKSEV 170
Query: 401 DILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNH 460
D L+DGYRWRKYGQK VK +P PRSYY+CT C V+K VER+ D V+TTYEG+H H
Sbjct: 171 DHLEDGYRWRKYGQKAVKNSPFPRSYYRCTTASCNVKKRVERSFRDPSTVVTTYEGQHTH 230
Query: 461 DVP----------------AARGSGNHSVTRPLPNNTANAIRPSSVTHQHHHNTNNNSLQ 504
P AA GN P+ +T + + + Q+HH L
Sbjct: 231 ISPLTSRPISTGGFFGSSGAASSLGNGCFGFPIDGSTLISPQFQQLV-QYHHQQQQQELM 289
Query: 505 S 505
S
Sbjct: 290 S 290
Score = 83.2 bits (204), Expect = 4e-16
Identities = 72/237 (30%), Positives = 102/237 (42%), Gaps = 42/237 (17%)
Query: 135 SNFSFQTQQAPPPATTSTATYQSSNV--ASQTQQQPWSYQEATKQDSMMMK---TENSSS 189
S SF Q + P T + SS++ A ++Q S E+ D +++ T NSSS
Sbjct: 51 SPHSFLLQTSQPQTQTQPSAKLSSSIIQAPPSEQLVTSKVESLCSDHLLINPPATPNSSS 110
Query: 190 MQSFSPEIAS-----VQTNHNNGFQSDYN------------NYQPQQQQLQPQTLNRRS- 231
+ S S E + + N G + N Q +Q++ + + +
Sbjct: 111 ISSASSEALNEEKPKTEDNEEEGGEDQQEKSHTKKQLKAKKNNQKRQREARVAFMTKSEV 170
Query: 232 ---DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSL-DGQITEIVYKGTHNH 287
+DGY WRKYGQK VK S PRSYY+CT +C KK+VERS D Y+G H H
Sbjct: 171 DHLEDGYRWRKYGQKAVKNSPFPRSYYRCTTASCNVKKRVERSFRDPSTVVTTYEGQHTH 230
Query: 288 PKPQATR--------RNSSSSSSL-------PIPHSNHISNDIQDQSYATHGSGQMD 329
P +R +S ++SSL PI S IS Q H Q +
Sbjct: 231 ISPLTSRPISTGGFFGSSGAASSLGNGCFGFPIDGSTLISPQFQQLVQYHHQQQQQE 287
>At1g69310 putative WRKY transcription factor
Length = 287
Score = 112 bits (281), Expect = 4e-25
Identities = 74/234 (31%), Positives = 105/234 (44%), Gaps = 34/234 (14%)
Query: 306 PHSNHISNDIQDQS--------YATHGSGQMDSVATPENSSISIGDDDFEQSSQKCRSGG 357
PHS +D+ + + S + T N++ S E ++ +
Sbjct: 51 PHSLRFDSDLTQTTGVKPTTVTSSCSSSAAVSVAVTSTNNNPSATSSSSEDPAENSTASA 110
Query: 358 DELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVV 417
++ E K K + +R+PR T SD+D L+DGYRWRKYGQK V
Sbjct: 111 EKTPPPETPVKEKK----------KAQKRIRQPRFAFMTKSDVDNLEDGYRWRKYGQKAV 160
Query: 418 KGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHD-VPAARG--------- 467
K +P PRSYY+CT+ C V+K VER+S D VITTYEG+H H + RG
Sbjct: 161 KNSPFPRSYYRCTNSRCTVKKRVERSSDDPSIVITTYEGQHCHQTIGFPRGGILTAHDPH 220
Query: 468 --SGNHSVTRPLPNNTANAIRPSSVTHQHHHNTNNNSLQSLRPQQAPDQVQSSP 519
+ +H + PLP N + HQ H + N S + RP S+P
Sbjct: 221 SFTSHHHLPPPLP----NPYYYQELLHQLHRDNNAPSPRLPRPTTEDTPAVSTP 270
Score = 73.6 bits (179), Expect = 3e-13
Identities = 61/204 (29%), Positives = 85/204 (40%), Gaps = 11/204 (5%)
Query: 90 DSPVLLNSSNI-----LPSPTTGAFAAQSFNWKSSSGGNQQVVKEEDKSFSNFSFQTQQA 144
D+P L N + L + + F + N S G N + S S TQ
Sbjct: 5 DNPDLSNDDSAWRELTLTAQDSDFFDRDTSNILSDFGWNLHHSSDHPHSLRFDSDLTQTT 64
Query: 145 PPPATTSTATYQSSNVASQTQQQPWSYQEATKQDSMMMKTENSSSMQSFSPEIASVQTNH 204
TT T++ SS S + AT S +++S + P V+
Sbjct: 65 GVKPTTVTSSCSSSAAVSVAVTSTNNNPSATSSSSEDPAENSTASAEKTPPPETPVKEKK 124
Query: 205 NNGFQSDYNNYQPQQQQLQPQTLNRRSDDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPT 264
++ QP+ + ++ +DGY WRKYGQK VK S PRSYY+CT C
Sbjct: 125 ----KAQKRIRQPRFAFMTKSDVDNL-EDGYRWRKYGQKAVKNSPFPRSYYRCTNSRCTV 179
Query: 265 KKKVERSLDG-QITEIVYKGTHNH 287
KK+VERS D I Y+G H H
Sbjct: 180 KKRVERSSDDPSIVITTYEGQHCH 203
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.309 0.125 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,777,496
Number of Sequences: 26719
Number of extensions: 720538
Number of successful extensions: 3893
Number of sequences better than 10.0: 194
Number of HSP's better than 10.0 without gapping: 90
Number of HSP's successfully gapped in prelim test: 106
Number of HSP's that attempted gapping in prelim test: 3296
Number of HSP's gapped (non-prelim): 488
length of query: 579
length of database: 11,318,596
effective HSP length: 105
effective length of query: 474
effective length of database: 8,513,101
effective search space: 4035209874
effective search space used: 4035209874
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0058a.3


