

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0058a.1
(1183 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
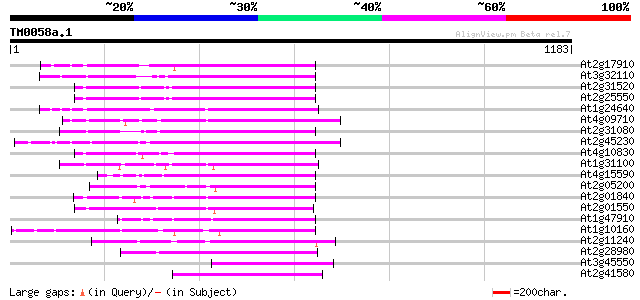
At2g17910 putative non-LTR retroelement reverse transcriptase 241 1e-63
At3g32110 non-LTR reverse transcriptase, putative 240 3e-63
At2g31520 putative non-LTR retroelement reverse transcriptase 231 2e-60
At2g25550 putative non-LTR retroelement reverse transcriptase 230 3e-60
At1g24640 hypothetical protein 229 7e-60
At4g09710 RNA-directed DNA polymerase -like protein 224 2e-58
At2g31080 putative non-LTR retroelement reverse transcriptase 220 4e-57
At2g45230 putative non-LTR retroelement reverse transcriptase 219 6e-57
At4g10830 putative protein 214 2e-55
At1g31100 hypothetical protein 214 2e-55
At4g15590 reverse transcriptase like protein 213 7e-55
At2g05200 putative non-LTR retroelement reverse transcriptase 209 8e-54
At2g01840 putative non-LTR retroelement reverse transcriptase 206 5e-53
At2g01550 putative non-LTR retroelement reverse transcriptase 201 2e-51
At1g47910 reverse transcriptase, putative 200 4e-51
At1g10160 putative reverse transcriptase 198 2e-50
At2g11240 pseudogene 187 4e-47
At2g28980 putative non-LTR retroelement reverse transcriptase 184 3e-46
At3g45550 putative protein 183 6e-46
At2g41580 putative non-LTR retroelement reverse transcriptase 183 6e-46
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 241 bits (616), Expect = 1e-63
Identities = 179/604 (29%), Positives = 277/604 (45%), Gaps = 55/604 (9%)
Query: 66 RGGGLLCIWNSKLFVLEESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRA-MWDA 124
R GGL W S L + D LM +++ + I+ + R +W+
Sbjct: 42 RSGGLAIFWKSHLEIEFLYADK----NLMDLQVSSRNKVWFISCVYGLPVTHMRPKLWEH 97
Query: 125 LCAWRGTCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRREMREFNVF---IANMDLVDI 181
L + G WCL GDFN +R +E+ G RR F F + N + ++
Sbjct: 98 LNSI-GLKRAEAWCLIGDFNDIRSNDEKLGGP------RRSPSSFQCFEHMLLNCSMHEL 150
Query: 182 PLTGRRFTWR-RANDQ-AQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQVVA 239
TG FTW NDQ Q ++DR + +W ++PN L + SDH P+L++
Sbjct: 151 GSTGNSFTWGGNRNDQWVQCKLDRCFGNPAWFSIFPNAHQWFLEKFGSDHRPVLVKFTND 210
Query: 240 -DWGPKPFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKEVF 298
+ FR DDP + +SW QG + F
Sbjct: 211 NELFRGQFRYDKRLDDDPYCIEVIHRSWNSAMSQGT-------------------HSSFF 251
Query: 299 GDLKTRREKVVQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRY-----------QE 347
++ RR V K + + + ++L + + I W + Y +E
Sbjct: 252 SLIECRRAISVWKHSSDTNAQSRIKRLRKDLDAEKSIQIPCWPRIEYIKDQLSLAYGDEE 311
Query: 348 SFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSIVGLMVDGMWEDEPVRVKAAI-HDFF 406
F QK+ KWL GD N+GFFH+T++ +R + + L+ + E K I FF
Sbjct: 312 LFWRQKSRQKWLAGGDKNTGFFHATVHSERLKNELSFLLDENDQEFTRNSDKGKIASSFF 371
Query: 407 QKKFKSVAWLRPK--LDGVQFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFN 464
+ F S L L+G+Q + +T N L+ E+E+ AV+ +PGPDGF
Sbjct: 372 ENLFTSTYILTHNNHLEGLQ-AKVTSEMNHNLIQEVTELEVYNAVFSINKESAPGPDGFT 430
Query: 465 FRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGC 524
F Q+ WD++ ++ + F+ G P+ N + I LIPK+ PQ ++D RPISL
Sbjct: 431 ALFFQQHWDLVKHQILTEIFGFFETGVLPQDWNHTHICLIPKITSPQRMSDLRPISLCSV 490
Query: 525 IYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKKRPS---IF 581
+YK++ K+ RL+ + ++ Q FV R + D+++V +E++H + R S +
Sbjct: 491 LYKIISKILTQRLKKHLPAIVSTTQSAFVPQRLISDNILVAHEMIHSLRTNDRISKEHMA 550
Query: 582 FKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKG 641
FK D KAY+ VEW FLE MM + F KWI WIM C+ S + SVL+NG P G +G
Sbjct: 551 FKTDMSKAYDRVEWPFLETMMTALGFNNKWISWIMNCVTSVSYSVLINGQPYGHIIPTRG 610
Query: 642 LRQG 645
+RQG
Sbjct: 611 IRQG 614
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 240 bits (613), Expect = 3e-63
Identities = 176/594 (29%), Positives = 279/594 (46%), Gaps = 55/594 (9%)
Query: 63 AVNRGGGLLCIWNSKLFVLEESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRAMW 122
AV GGL +W + + + F+ G V++ Y+ + +W
Sbjct: 626 AVGHSGGLWLLWRTGIGEVSVVDSTDQFIHAKDVNGKDNVNLVVV--YAAPTASRRSGLW 683
Query: 123 DALCAWRGTCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRREMREFNVFIANMDLVDIP 182
D L + D G + GDFN + +ER G +G LSS + F +I + L+D+
Sbjct: 684 DRLGDVIRSMD-GPVVIGGDFNTIVRLDERSGGNGRLSS---DSLAFGEWINDHSLIDLG 739
Query: 183 LTGRRFTWRRANDQ---AQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMR---Q 236
G +FTW+R ++ R+DR L W S L L SDH PL ++ +
Sbjct: 740 FKGNKFTWKRGREERFFVAKRLDRVLCCAHARLKWQEASVLHLPFLASDHAPLYVQLTPE 799
Query: 237 VVADWGPKPFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKE 296
V + G +PFR WL P + SW NK
Sbjct: 800 VSGNRGRRPFRFEAAWLSHPGFKELLLTSW---------------------------NK- 831
Query: 297 VFGDLKTRREKVVQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQKACM 356
D+ T VQ+ LLD+ + + L+ EE +LL +F VL +E QK+
Sbjct: 832 ---DISTPEALKVQE--LLDLHQSDDLLKKEE-----ELLKDFDVVLEQEEVVWMQKSRE 881
Query: 357 KWLVEGDLNSGFFHSTINWKRRADSIVGLMV-DGMWEDEPVRVKAAIHDFFQKKFK--SV 413
KW V GD N+ FFH++ +RR + I L DG W ++ D++++ + +
Sbjct: 882 KWFVHGDRNTKFFHTSTIIRRRRNQIEMLQDNDGRWLSNAQELETHAIDYYKRLYSLDDL 941
Query: 414 AWLRPKLDGVQFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWD 473
+ +L F+ +++ D + L PF +E++ A+ G K+PGPDGF F Q+ W+
Sbjct: 942 DAVVEQLPQEGFTALSEADFSSLTKPFSPLEVEGAIRSMGKYKAPGPDGFQPVFYQQGWE 1001
Query: 474 VLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLF 533
V+ V K V +F+ G+ P+ +N +VLI KV P+ + FRPISL ++K + K+
Sbjct: 1002 VVGESVTKFVMDFFSSGSFPQETNDVLVVLIAKVLKPEKITQFRPISLCNVLFKTITKVM 1061
Query: 534 AIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKK--RPSIFFKVDYEKAYN 591
RL+ VI K+I Q +F+ R D++VV+ EV+H + KK + + K+D EKAY+
Sbjct: 1062 VGRLKGVINKLIGPAQTSFIPGRLSTDNIVVVQEVVHSMRRKKGVKGWMLLKLDLEKAYD 1121
Query: 592 SVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQG 645
+ W LE + W+ WIM C+ ++ +L NG + F +GLRQG
Sbjct: 1122 RIRWDLLEDTLKAAGLPGTWVQWIMKCVEGPSMRLLWNGEKTDAFKPLRGLRQG 1175
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 231 bits (588), Expect = 2e-60
Identities = 155/518 (29%), Positives = 249/518 (47%), Gaps = 18/518 (3%)
Query: 136 EWCLAGDFNAVRYAEERRGSSGVLSSHRREMREFNVFIANMDLVDIPLTGRRFTW--RRA 193
EW L GDFN + E+ G R F +++ D+ D+ G RF+W R
Sbjct: 285 EWLLVGDFNEILSNAEKIGGP---MREEWTFRNFRNMVSHCDIEDMRSKGDRFSWVGERH 341
Query: 194 NDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMR-QVVADWGPKPFRVLNCW 252
+ +DR ++++W +P L+ SDH P+L+ K FR N
Sbjct: 342 THTVKCCLDRVFINSAWTATFPYAETEFLDFTGSDHKPVLVHFNESFPRRSKLFRFDNRL 401
Query: 253 LDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKEVFGDLKTRREKVVQKI 312
+D P V+ SW+ + + + E++ R ++ + + R +K+ +
Sbjct: 402 IDIPTFKRIVQTSWRTNRNSR--STPITERISSCRQAMARLKHASNLNSEQRIKKLQSSL 459
Query: 313 NLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQKACMKWLVEGDLNSGFFHST 372
N V Q + ++++ L + +E + QK+ +W+ EGD N+G+FH+
Sbjct: 460 NRAMESTRRVDRQL--IPQLQESLA---KAFSDEEIYWKQKSRNQWMKEGDQNTGYFHAC 514
Query: 373 INWKRRADSIVGLMVD-GMWEDEPVRVKAAIHDFFQKKFKSVAWLRPKLDGVQF-STITD 430
+ + + +M D G + DFF F + +D F ST+T+
Sbjct: 515 TKTRYSQNRVNTIMDDQGRMFTGDKEIGNHAQDFFTNIFSTNGIKVSPIDFADFKSTVTN 574
Query: 431 RDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRG 490
N +L F + EI A+ + G K+PGPDG RF + WD++ DV+ V++F+
Sbjct: 575 TVNLDLTKEFSDTEIYDAICQIGDDKAPGPDGLTARFYKNCWDIVGYDVILEVKKFFETS 634
Query: 491 A*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQF 550
N + I +IPK+ P L+D+RPI+L +YKV+ K RL+ + ++ + Q
Sbjct: 635 FMKPSINHTNICMIPKITNPTTLSDYRPIALCNVLYKVISKCLVNRLKSHLNSIVSDSQA 694
Query: 551 TFVGNRNMLDSVVVLNEVMHEAKVKKRPS---IFFKVDYEKAYNSVEWGFLEYMMGRMNF 607
F+ R + D+V++ +EVMH KV+KR S + K D KAY+ VEW FLE M F
Sbjct: 695 AFIPGRIINDNVMIAHEVMHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGF 754
Query: 608 CAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQG 645
C KWIGWIM + S SVL+NGSP G +G+RQG
Sbjct: 755 CNKWIGWIMAAVKSVHYSVLINGSPHGYITPTRGIRQG 792
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 230 bits (587), Expect = 3e-60
Identities = 155/518 (29%), Positives = 249/518 (47%), Gaps = 18/518 (3%)
Query: 136 EWCLAGDFNAVRYAEERRGSSGVLSSHRREMREFNVFIANMDLVDIPLTGRRFTW--RRA 193
EW L GDFN + E+ G R F +++ D+ D+ G RF+W R
Sbjct: 511 EWLLVGDFNEILSNAEKIGGP---MREEWTFRNFRNMVSHCDIEDMRSKGDRFSWVGERH 567
Query: 194 NDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMR-QVVADWGPKPFRVLNCW 252
+ +DR ++++W +P L+ SDH P+L+ K FR N
Sbjct: 568 THTVKCCLDRVFINSAWTATFPYAEIEFLDFTGSDHKPVLVHFNESFPRRSKLFRFDNRL 627
Query: 253 LDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKEVFGDLKTRREKVVQKI 312
+D P V+ SW+ + + + E++ R ++ + + R +K+ +
Sbjct: 628 IDIPTFKRIVQTSWRTNRNSR--STPITERISSCRQAMARLKHASNLNSEQRIKKLQSSL 685
Query: 313 NLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQKACMKWLVEGDLNSGFFHST 372
N V Q + ++++ L + +E + QK+ +W+ EGD N+G+FH+
Sbjct: 686 NRAMESTRRVDRQL--IPQLQESLA---KAFSDEEIYWKQKSRNQWMKEGDQNTGYFHAC 740
Query: 373 INWKRRADSIVGLMVD-GMWEDEPVRVKAAIHDFFQKKFKSVAWLRPKLDGVQF-STITD 430
+ + + +M D G + DFF F + +D F ST+T+
Sbjct: 741 TKTRYSQNRVNTIMDDQGRMFTGDKEIGNHAQDFFTNIFSTNGIKVSPIDFADFKSTVTN 800
Query: 431 RDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRG 490
N +L F + EI A+ + G K+PGPDG RF + WD++ DV+ V++F+
Sbjct: 801 TVNLDLTKEFSDTEIYDAICQIGDDKAPGPDGLTARFYKNCWDIVGYDVILEVKKFFETS 860
Query: 491 A*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQF 550
N + I +IPK+ P L+D+RPI+L +YKV+ K RL+ + ++ + Q
Sbjct: 861 FMKPSINHTNICMIPKITNPTTLSDYRPIALCNVLYKVISKCLVNRLKSHLNSIVSDSQA 920
Query: 551 TFVGNRNMLDSVVVLNEVMHEAKVKKRPS---IFFKVDYEKAYNSVEWGFLEYMMGRMNF 607
F+ R + D+V++ +EVMH KV+KR S + K D KAY+ VEW FLE M F
Sbjct: 921 AFIPGRIINDNVMIAHEVMHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGF 980
Query: 608 CAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQG 645
C KWIGWIM + S SVL+NGSP G +G+RQG
Sbjct: 981 CNKWIGWIMAAVKSVHYSVLINGSPHGYITPTRGIRQG 1018
>At1g24640 hypothetical protein
Length = 1270
Score = 229 bits (584), Expect = 7e-60
Identities = 182/603 (30%), Positives = 283/603 (46%), Gaps = 37/603 (6%)
Query: 64 VNRGGGLLCIWNSKLFVLEESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRAMWD 123
V + GGL +W S + V + VD + ++GA CV +Y D + W+
Sbjct: 25 VGKCGGLALLWKSSVQVDLKFVD-KNLMDAQVQFGAVNF-CVSC-VYGDPDRSKRSQAWE 81
Query: 124 ALCAWRGTCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRR---EMREFNVFIANMDLVD 180
+ G +WC+ GDFN + + E+ G RR + + FN I DLV+
Sbjct: 82 RISRI-GVGRRDKWCMFGDFNDILHNGEKNGGP------RRSDLDCKAFNEMIKGCDLVE 134
Query: 181 IPLTGRRFTW--RRANDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQVV 238
+P G FTW RR + Q R+DR + W +P + L+ SDH P+L++ +
Sbjct: 135 MPAHGNGFTWAGRRGDHWIQCRLDRAFGNKEWFCFFPVSNQTFLDFRGSDHRPVLIKLMS 194
Query: 239 A-DWGPKPFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKEV 297
+ D FR +L + + ++W K G V ++L+ R SL K+
Sbjct: 195 SQDSYRGQFRFDKRFLFKEDVKEAIIRTWSRGK-HGTNISVA-DRLRACRKSLSSWKKQ- 251
Query: 298 FGDLKTRREKVVQKINLLDVK-EEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQKACM 356
+ KIN L+ E+E L + L + + R +E++ QK+
Sbjct: 252 ------NNLNSLDKINQLEAALEKEQSLVWPIFQRVSVLKKDLAKAYREEEAYWKQKSRQ 305
Query: 357 KWLVEGDLNSGFFHSTINWKRRADSIVGLM-VDGMWEDEPVRVKAAIHDFFQKKFKSVAW 415
KWL G+ NS +FH+ + R+ I L V+G + +F FKS
Sbjct: 306 KWLRSGNRNSKYFHAAVKQNRQRKRIEKLKDVNGNMQTSEAAKGEVAAAYFGNLFKSS-- 363
Query: 416 LRPKLDGVQFSTITDRD----NTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRF 471
P FS + R N LVG EIK+AV+ +PGPDG + F Q +
Sbjct: 364 -NPSGFTDWFSGLVPRVSEVMNESLVGEVSAQEIKEAVFSIKPASAPGPDGMSALFFQHY 422
Query: 472 WDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPK 531
W + V V++F+ G P N + + LIPK + P + D RPISL +YK++ K
Sbjct: 423 WSTVGNQVTSEVKKFFADGIMPAEWNYTHLCLIPKTQHPTEMVDLRPISLCSVLYKIISK 482
Query: 532 LFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKKRPSIFF---KVDYEK 588
+ A RL+ + +++ + Q FV R + D+++V +E++H KV R S F K D K
Sbjct: 483 IMAKRLQPWLPEIVSDTQSAFVSERLITDNILVAHELVHSLKVHPRISSEFMAVKSDMSK 542
Query: 589 AYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQGILL 648
AY+ VEW +L ++ + F KW+ WIM C++S T SVL+N P G +++GLRQG L
Sbjct: 543 AYDRVEWSYLRSLLLSLGFHLKWVNWIMVCVSSVTYSVLINDCPFGLIILQRGLRQGDPL 602
Query: 649 RRF 651
F
Sbjct: 603 SPF 605
>At4g09710 RNA-directed DNA polymerase -like protein
Length = 1274
Score = 224 bits (571), Expect = 2e-58
Identities = 171/605 (28%), Positives = 284/605 (46%), Gaps = 43/605 (7%)
Query: 111 SPCDIDGKRAMWDALCAWRGTCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRREMREFN 170
+P ID + WD + + G W L GDFN + E++G F
Sbjct: 50 APNFIDNRSVFWDKISSL-GAQRSSAWLLTGDFNDILDNSEKQGGPLRWEGF---FLAFR 105
Query: 171 VFIANMDLVDIPLTGRRFTWR--RANDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSD 228
F++ L DI TG +WR R + +SR+DR L + SW +++P L + SD
Sbjct: 106 SFVSQNGLWDINHTGNSLSWRGTRYSHFIKSRLDRALGNCSWSELFPMSKCEYLRFEGSD 165
Query: 229 HCPLLMRQVVADWGP------KPFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEK 282
H PL V +G KPFR + + V++ W+ + + K
Sbjct: 166 HRPL-----VTYFGAPPLKRSKPFRFDRRLREKEEIRALVKEVWELARQDS-----VLYK 215
Query: 283 LKGLRGSLRI*NKEVFGDLKTRREKVVQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRV 342
+ R S+ KE + +K Q + E + + + + E
Sbjct: 216 ISRCRQSIIKWTKEQNSNSAKAIKKAQQAL------ESALSADIPDPSLIGSITQELEAA 269
Query: 343 LRYQESFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSIVGLMVDGMWED--EPVRVKA 400
R +E F Q + ++WL GD N G+FH+T +R +++ ++ DG ++ E ++ +
Sbjct: 270 YRQEELFWKQWSRVQWLNSGDRNKGYFHATTRTRRMLNNL-SVIEDGSGQEFHEEEQIAS 328
Query: 401 AIHDFFQKKFKSVAWLRPKLDGVQFST---ITDRDNTELVGPFDEVEIKQAVWECGGTKS 457
I +FQ F + L VQ + I+ N EL+ +EIK+A++ K+
Sbjct: 329 TISSYFQNIFTTSN--NSDLQVVQEALSPIISSHCNEELIKISSLLEIKEALFSISADKA 386
Query: 458 PGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFR 517
PGPDGF+ F +WD++ DV + +R F++ N + + LIPK+ P+ ++D+R
Sbjct: 387 PGPDGFSASFFHAYWDIIEADVSRDIRSFFVDSCLSPRLNETHVTLIPKISAPRKVSDYR 446
Query: 518 PISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKV--- 574
PI+L YK+V K+ RL+ + ++I Q FV R + D+V++ +E++H +V
Sbjct: 447 PIALCNVQYKIVAKILTRRLQPWLSELISLHQSAFVPGRAIADNVLITHEILHFLRVSGA 506
Query: 575 KKRPSIFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSG 634
KK S+ K D KAY+ ++W FL+ ++ R+ F KWI W+M C+ + + S L+NGSP G
Sbjct: 507 KKYCSMAIKTDMSKAYDRIKWNFLQEVLMRLGFHDKWIRWVMQCVCTVSYSFLINGSPQG 566
Query: 635 EFHMEKGLRQGILLRRFFS*LLLKA*MGYSRMQLELG---NLKVSRLGQEAILWCLYCSL 691
+GLRQG L + L + G R E G ++V+R G + L+
Sbjct: 567 SVVPSRGLRQGDPLSPYLFILCTEVLSGLCRKAQEKGVMVGIRVAR-GSPQVNHLLFADD 625
Query: 692 LMILC 696
M C
Sbjct: 626 TMFFC 630
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 220 bits (560), Expect = 4e-57
Identities = 160/552 (28%), Positives = 260/552 (46%), Gaps = 67/552 (12%)
Query: 106 IINIYSPCDIDGKRAMWDALCAWRGTCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRRE 165
+I +Y+ + + +W L + G + GDFN + + +ER G +G LS +
Sbjct: 3 LIVVYAAPSVSRRSGLWGELKDVVNGLE-GPLLIGGDFNTILWVDERMGGNGRLSP---D 58
Query: 166 MREFNVFIANMDLVDIPLTGRRFTWRRANDQAQ---SRIDRFLVSTSWCDVWPNCSHLVL 222
F +I + L+D+ G +FTWRR ++ R+DR V W L
Sbjct: 59 SLAFGDWINELSLIDLGFKGNKFTWRRGRQESTVVAKRLDRVFVCAHARLKWQEAVVSHL 118
Query: 223 NRDVSDHCPLLMRQVVADWGPKPFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEK 282
SDH PL YV E
Sbjct: 119 PFMASDHAPL--------------------------------------------YVQLEP 134
Query: 283 LKGLRGSLRI*NKEVFGDLKTRREKVVQKINLLDVKEEE----VGLQPEELVEMRDLLIE 338
L+ + LR N+EVFGD+ R+EK+V D+KE + V L + L + LL E
Sbjct: 135 LQ--QRKLRKWNREVFGDIHVRKEKLVA-----DIKEVQDLLGVVLSDDLLAKEEVLLKE 187
Query: 339 FWRVLRYQESFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSIVGLMVDG-MWEDEPVR 397
VL +E+ QK+ K++ GD N+ FFH++ +RR + I L D W + V
Sbjct: 188 MDLVLEQEETLWFQKSREKYIELGDRNTTFFHTSTIIRRRRNRIESLKGDDDRWVTDKVE 247
Query: 398 VKAAIHDFFQKKF--KSVAWLRPKLDGVQFSTITDRDNTELVGPFDEVEIKQAVWECGGT 455
++A ++++ + + V+ +R L F++I++ + L+ F + E+ AV G
Sbjct: 248 LEAMALTYYKRLYSLEDVSEVRNMLPTGGFASISEAEKAALLQAFTKAEVVSAVKSMGRF 307
Query: 456 KSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLND 515
K+PGPDG+ F Q+ W+ + V + V EF+ G P +N + +VLI KV P+ +
Sbjct: 308 KAPGPDGYQPVFYQQCWETVGPSVTRFVLEFFETGVLPASTNDALLVLIAKVAKPERIQQ 367
Query: 516 FRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVK 575
FRP+SL ++K++ K+ RL+ VI K+I Q +F+ R +D++V++ E +H + K
Sbjct: 368 FRPVSLCNVLFKIITKMMVTRLKNVISKLIGPAQASFIPGRLSIDNIVLVQEAVHSMRRK 427
Query: 576 K--RPSIFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPS 633
K + + K+D EKAY+ V W FL+ + W IM + ++SVL NG +
Sbjct: 428 KGRKGWMLLKLDLEKAYDRVRWDFLQETLEAAGLSEGWTSRIMAGVTDPSMSVLWNGERT 487
Query: 634 GEFHMEKGLRQG 645
F +GLRQG
Sbjct: 488 DSFVPARGLRQG 499
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 219 bits (559), Expect = 6e-57
Identities = 186/703 (26%), Positives = 315/703 (44%), Gaps = 39/703 (5%)
Query: 10 GVGGNAKKRVVRELVCQQHVELLCIQETKCQVVDRRMCGLL*GD-SEFEWKATPAVNRGG 68
GVG R +RE+ E++ + ETK + R + G F+ + + G
Sbjct: 10 GVGNTPTVRHLREIRGLYFPEVIFLCETKKR---RNYLENVVGHLGFFDLHTVEPIGKSG 66
Query: 69 GLLCIWNSKLFVLEESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRAMWDALCAW 128
GL +W + + D + + W + + IY + +W+ L
Sbjct: 67 GLALMWKDSVQIKVLQSD-KRLIDALLIW--QDKEFYLTCIYGEPVQAERGELWERLTRL 123
Query: 129 RGTCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRREMREFNVFIANMDLVDIPLTGRRF 188
G G W L GDFN + E+ G S E R+ + + L ++ +G +F
Sbjct: 124 -GLSRSGPWMLTGDFNELVDPSEKIGGPARKESSCLEFRQM---LNSCGLWEVNHSGYQF 179
Query: 189 TW--RRANDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQVVADWGP-KP 245
+W R ++ Q R+DR + + +W +++P L + SDH PL+ V +W
Sbjct: 180 SWYGNRNDELVQCRLDRTVANQAWMELFPQAKATYLQKICSDHSPLINNLVGDNWRKWAG 239
Query: 246 FRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKEVFGDLKTRR 305
F+ W+ + W + ++ EK+ R +E+ + +
Sbjct: 240 FKYDKRWVQREGFKDLLCNFWSQQSTK--TNALMMEKIASCR-------REISKWKRVSK 290
Query: 306 EKVVQKINLLDVKEEEVGLQ-PEELVEMRDLLIEFWRVLRYQESFLCQKACMKWLVEGDL 364
+I L K + Q P + E+ L E + +E F +K+ + W+ GD
Sbjct: 291 PSSAVRIQELQFKLDAATKQIPFDRRELARLKKELSQEYNNEEQFWQEKSRIMWMRNGDR 350
Query: 365 NSGFFHSTINWKRRADSIVGLMVD---GMWEDEPVRVKAAIHDFFQKKFKS--VAWLRPK 419
N+ +FH+ RRA + + ++D W + + A +F+K F S V + +
Sbjct: 351 NTKYFHAATK-NRRAQNRIQKLIDEEGREWTSDEDLGRVA-EAYFKKLFASEDVGYTVEE 408
Query: 420 LDGVQFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDV 479
L+ + ++D+ N L+ P + E+++A + K PGPDG N Q+FW+ + +
Sbjct: 409 LENLT-PLVSDQMNNNLLAPITKEEVQRATFSINPHKCPGPDGMNGFLYQQFWETMGDQI 467
Query: 480 VKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRL 539
+ V+ F+ G+ G N + I LIPK+ + + DFRPISL IYKV+ KL A RL+
Sbjct: 468 TEMVQAFFRSGSIEEGMNKTNICLIPKILKAEKMTDFRPISLCNVIYKVIGKLMANRLKK 527
Query: 540 VIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKKRPS---IFFKVDYEKAYNSVEWG 596
++ +I E Q FV R + D++++ +E++H + S I K D KAY+ VEW
Sbjct: 528 ILPSLISETQAAFVKGRLISDNILIAHELLHALSSNNKCSEEFIAIKTDISKAYDRVEWP 587
Query: 597 FLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQGILLRRFFS*L- 655
FLE M + F WI IM C+ S VL+NG+P GE +GLRQG L + +
Sbjct: 588 FLEKAMRGLGFADHWIRLIMECVKSVRYQVLINGTPHGEIIPSRGLRQGDPLSPYLFVIC 647
Query: 656 --LLKA*MGYSRMQLELGNLKVSRLGQEAILWCLYCSLLMILC 696
+L + + + ++ LKV+R G I L+ M C
Sbjct: 648 TEMLVKMLQSAEQKNQITGLKVAR-GAPPISHLLFADDSMFYC 689
>At4g10830 putative protein
Length = 1294
Score = 214 bits (546), Expect = 2e-55
Identities = 149/523 (28%), Positives = 245/523 (46%), Gaps = 30/523 (5%)
Query: 137 WCLAGDFNAVRYAEERRGSSGVLSSHRREMREFNVFIANMDLVDIPLTGRRFTW--RRAN 194
W L GDFN + E+ G R F ++ DL DI G RF+W R +
Sbjct: 492 WILIGDFNEILSNNEKIGGP---QRDEWTFRGFRNMVSTCDLKDIRSIGDRFSWVGERHS 548
Query: 195 DQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQVVADWGP-KPFRVLNCWL 253
+ +DR +++ ++P L SDH PL + + +PFR L
Sbjct: 549 HTVKCCLDRAFINSEGAFLFPFAELEFLEFTGSDHKPLFLSLEKTETRKMRPFRFDKRLL 608
Query: 254 DDPRLFPFVEQSWKGMKIQGWGAYV------LKEKLKGLRGSLRI*NKEVFGDLKTRREK 307
+ P +V+ W I G ++ ++ + L+ + ++ L+ +K
Sbjct: 609 EVPHFKTYVKAGWN-KAINGQRKHLPDQVRTCRQAMAKLKHKSNLNSRIRINQLQAALDK 667
Query: 308 VVQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQKACMKWLVEGDLNSG 367
+ +N + + +Q E V RD +E + QK+ +W+ EGD N+
Sbjct: 668 AMSSVNRTE-RRTISHIQRELTVAYRD-----------EERYWQQKSRNQWMKEGDRNTE 715
Query: 368 FFHSTINWKRRADSIVGLM-VDGMWEDEPVRVKAAIHDFFQKKFKSVAWLRPKLDGVQFS 426
FFH+ + + +V + +GM + +FF K ++S +D F
Sbjct: 716 FFHACTKTRFSVNRLVTIKDEEGMIYRGDKEIGVHAQEFFTKVYESNGRPVSIIDFAGFK 775
Query: 427 TI-TDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVRE 485
I T++ N +L ++EI A+ G K+PGPDG RF + W+++ DV+K V+
Sbjct: 776 PIVTEQINDDLTKDLSDLEIYNAICHIGDDKAPGPDGLTARFYKSCWEIVGPDVIKEVKI 835
Query: 486 FWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVI 545
F+ + N + I +IPK+ P+ L+D+RPI+L +YK++ K RL+ + ++
Sbjct: 836 FFRTSYMKQSINHTNICMIPKITNPETLSDYRPIALCNVLYKIISKCLVERLKGHLDAIV 895
Query: 546 DERQFTFVGNRNMLDSVVVLNEVMHEAKVKKRPS---IFFKVDYEKAYNSVEWGFLEYMM 602
+ Q F+ R + D+V++ +E+MH K +KR S + K D KAY+ VEW FLE M
Sbjct: 896 SDSQAAFIPGRLVNDNVMIAHEMMHSLKTRKRVSQSYMAVKTDVSKAYDRVEWNFLETTM 955
Query: 603 GRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQG 645
F WI WIMG + S SVLVNG P G ++G+RQG
Sbjct: 956 RLFGFSETWIKWIMGAVKSVNYSVLVNGIPHGTIQPQRGIRQG 998
>At1g31100 hypothetical protein
Length = 1090
Score = 214 bits (546), Expect = 2e-55
Identities = 158/576 (27%), Positives = 270/576 (46%), Gaps = 46/576 (7%)
Query: 105 VIINIYSPCDIDGKRAMWDALCAWRGTCDVG--EWCLAGDFNAVRYAEERRGSSGVLSSH 162
V+ +Y+ + ++ +W+ L + W + GDFN V E ++ + +
Sbjct: 16 VVSIVYAANEAITRKELWEELLLLSVSLSGNGKPWIMLGDFNQVLCPAEHSQATSL--NV 73
Query: 163 RREMREFNVFIANMDLVDIPLTGRRFTW--RRANDQAQSRIDRFLVSTSWCDVWPNCSHL 220
R M+ F + +L D+ G FTW + A ++DR LV+ SWC +P+ +
Sbjct: 74 NRRMKVFRDCLFEAELCDLVFKGNTFTWWNKSATRPVAKKLDRILVNESWCSRFPSAYAV 133
Query: 221 VLNRDVSDHC-------PLLMRQVVADWGPKPFRVLNCWLDDPRLFPFVEQSWKGMKIQG 273
D SDH PL+ R+ +PFR N L +P V + W + + G
Sbjct: 134 FGEPDFSDHASCGVIINPLMHRE------KRPFRFYNFLLQNPDFISLVGELWYSINVVG 187
Query: 274 WGAYVLKEKLKGLRGSLRI*NKEVFGDLKTRREKVVQKINLLDVKEEEVGLQPE------ 327
+ + +KLK L+ +R + E F +L+ R V + NL+ ++ + P
Sbjct: 188 SSMFKMSKKLKALKNPIRTFSMENFSNLEKR---VKEAHNLVLYRQNKTLSDPTIPNAAL 244
Query: 328 ELVEMRDLLIEFWRVLRYQESFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSIVGLMV 387
E+ R LI +++ +ESF CQ++ + W+ EGD N+ +FH + R+A + + +++
Sbjct: 245 EMEAQRKWLI----LVKAEESFFCQRSRVTWMGEGDSNTSYFHRMAD-SRKAVNTIHIII 299
Query: 388 D--GMWEDEPVRVKAAIHDFFQKKFKSVAWLRPKLDGVQFSTI-----TDRDNTELVGPF 440
D G+ D + +K ++F P L F + + EL F
Sbjct: 300 DDNGVKIDTQLGIKEHCIEYFSNLLGGEVG-PPMLIQEDFDLLLPFRCSHDQKKELAMSF 358
Query: 441 DEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASF 500
+IK A + K+ GPDGF F + W V+ +V AV EF+ + NA+
Sbjct: 359 SRQDIKSAFFSFPSNKTSGPDGFPVEFFKETWSVIGTEVTDAVSEFFTSSVLLKQWNATT 418
Query: 501 IVLIPKVKIPQLLNDFRPISLIG----CIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNR 556
+VLIPK+ +NDFRPIS +YKV+ +L RL+ ++ +VI Q F+ R
Sbjct: 419 LVLIPKITNASKMNDFRPISCNDFGPITLYKVIARLLTNRLQCLLSQVISPFQSAFLPGR 478
Query: 557 NMLDSVVVLNEVMHEAKVKK-RPSIFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWI 615
+ ++V++ E++ + P KVD KA++S+ W F+ + + +++ WI
Sbjct: 479 FLAENVLLATELVQGYNRQNIDPRGMLKVDLRKAFDSIRWDFIISALKAIGIPDRFVYWI 538
Query: 616 MGCLNSATVSVLVNGSPSGEFHMEKGLRQGILLRRF 651
C+++ T SV VNG+ G F +GLRQG L F
Sbjct: 539 TQCISTPTFSVCVNGNTGGFFKSTRGLRQGNPLSPF 574
>At4g15590 reverse transcriptase like protein
Length = 929
Score = 213 bits (541), Expect = 7e-55
Identities = 153/470 (32%), Positives = 230/470 (48%), Gaps = 32/470 (6%)
Query: 185 GRRFTWRRANDQAQ---SRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQVVADW 241
G RFTWRR ++ R+DR L C+H L + CP D
Sbjct: 5 GNRFTWRRGLVESTFVAKRLDRVLF----------CAHARLKWQEALLCPAQN----VDA 50
Query: 242 GPKPFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKEVFGDL 301
+PFR WL + SW G V L LR L+ NKEVFG++
Sbjct: 51 RRRPFRFEAAWLSHEGFKELLTASWD----TGLSTPVA---LNRLRWQLKKWNKEVFGNI 103
Query: 302 KTRREKVVQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRYQESFLC-QKACMKWLV 360
R+EKVV + + E V Q ++L+ D L++ + VL +QE L QK+ K L
Sbjct: 104 HVRKEKVVSDLKAVQDLLEVV--QTDDLLMKEDTLLKEFDVLLHQEETLWFQKSREKLLA 161
Query: 361 EGDLNSGFFHSTINWKRRADSIVGLM-VDGMWEDEPVRVKAAIHDFFQKKF--KSVAWLR 417
GD N+ FFH++ +RR + I L + W E ++ D+++K + + V+ +R
Sbjct: 162 LGDRNTTFFHTSTVIRRRRNRIEMLKDSEDRWVTEKEALEKLAMDYYRKLYSLEDVSVVR 221
Query: 418 PKLDGVQFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSG 477
L F +T + L PF E+ AV G K+PGPDG+ F Q+ W+ +
Sbjct: 222 GTLPTEGFPRLTREEKNNLNRPFTRDEVVVAVRSMGRFKAPGPDGYQPVFYQQCWETVGE 281
Query: 478 DVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRL 537
V K V EF+ G P+ +N +VL+ KV P+ + FRP+SL ++K++ K+ IRL
Sbjct: 282 SVSKFVMEFFESGVLPKSTNDVLLVLLAKVAKPERITQFRPVSLCNVLFKIITKMMVIRL 341
Query: 538 RLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKK--RPSIFFKVDYEKAYNSVEW 595
+ VI K+I Q +F+ R D++VV+ E +H + KK + + K+D EKAY+ + W
Sbjct: 342 KNVISKLIGPAQASFIPGRLSFDNIVVVQEAVHSMRRKKGRKGWMLLKLDLEKAYDRIRW 401
Query: 596 GFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQG 645
FL + WI IM C+ +S+L NG + F E+GLRQG
Sbjct: 402 DFLAETLEAAGLSEGWIKRIMECVAGPEMSLLWNGEKTDSFTPERGLRQG 451
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 209 bits (532), Expect = 8e-54
Identities = 147/495 (29%), Positives = 245/495 (48%), Gaps = 37/495 (7%)
Query: 168 EFNVFIANMDLVDIPLTGRRFTWR--RANDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRD 225
+F FI+ L D+ +G F+WR R + + R+DR + + SW + +P+ L +
Sbjct: 57 DFRSFISKNGLWDLKYSGNPFSWRGMRYDWFVRQRLDRAMSNNSWLESFPSGRSEYLRFE 116
Query: 226 VSDHCPLLMRQVVADWGPK-PFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLK 284
SDH PL++ A + FR N D+ + ++++W G + K+
Sbjct: 117 GSDHRPLVVFVDEARVKRRGQFRFDNRLRDNDVVNALIQETWTNA-----GDASVLTKMN 171
Query: 285 GLRGSLRI*NKEVFGDLKTRREKVVQKINLLD-VKEEEVGLQPEELVEMRDLLIEFWRVL 343
R +E+ + + + I EE + P + L
Sbjct: 172 QCR-------REIINWTRLQNLNSAELIEKTQKALEEALTADPPNPTTIGALTATLEHAY 224
Query: 344 RYQESFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSIVGLMVD--GMWEDEPVRVKAA 401
+ +E F Q++ + WL GD N+G+FH+ + RR + + +M D G+ + E ++
Sbjct: 225 KLEEQFWKQRSRVLWLHSGDRNTGYFHA-VTRNRRTQNRLTVMEDINGVAQHEEHQISQI 283
Query: 402 IHDFFQKKFKSVAWLRPKLDGVQFSTITDR--------DNTELVGPFDEVEIKQAVWECG 453
I +FQ+ F S + DG FS + + DN L ++ E+K AV+
Sbjct: 284 ISGYFQQIFTSES------DG-DFSVVDEAIEPMVSQGDNDFLTRIPNDEEVKDAVFSIN 336
Query: 454 GTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLL 513
+K+PGPDGF F +W ++S DV + +R F+ PR N + I LIPK P+ +
Sbjct: 337 ASKAPGPDGFTAGFYHSYWHIISTDVGREIRLFFTSKNFPRRMNETHIRLIPKDLGPRKV 396
Query: 514 NDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMH--- 570
D+RPI+L YK+V K+ R++L++ K+I E Q FV R + D+V++ +EV+H
Sbjct: 397 ADYRPIALCNIFYKIVAKIMTKRMQLILPKLISENQSAFVPGRVISDNVLITHEVLHFLR 456
Query: 571 EAKVKKRPSIFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNG 630
+ KK S+ K D KAY+ VEW FL+ ++ R F + WI W++ C+ S + S L+NG
Sbjct: 457 TSSAKKHCSMAVKTDMSKAYDRVEWDFLKKVLQRFGFHSIWIDWVLECVTSVSYSFLING 516
Query: 631 SPSGEFHMEKGLRQG 645
+P G+ +GLRQG
Sbjct: 517 TPQGKVVPTRGLRQG 531
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 206 bits (525), Expect = 5e-53
Identities = 146/530 (27%), Positives = 252/530 (47%), Gaps = 41/530 (7%)
Query: 135 GEWCLAGDFNAVRYAEERRGSSGVLSSHRRE---MREFNVFIANMDLVDIPLTGRRFTW- 190
G W + GDFN + E++G RR ++ F I ++ D+ G ++W
Sbjct: 491 GPWMMCGDFNEILNLNEKKGG------RRRSIGSLQNFTNMINCCNMKDLKSKGNPYSWV 544
Query: 191 -RRANDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQVVADWGPKPFRVL 249
+R N+ +S +DR +++ W +P L SDH P V+ D +
Sbjct: 545 GKRQNETIESCLDRVFINSDWQASFPAFETEFLPIAGSDHAP-----VIIDIAEEVCTKR 599
Query: 250 NCWLDDPRLFPF------VEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKEVFGDLKT 303
+ D R F F V++ W + G Y EKL R L + KT
Sbjct: 600 GQFRYDRRHFQFEDFVDSVQRGWNRGRSDSHGGYY--EKLHCCRQELAKWKRRT----KT 653
Query: 304 RREKVVQKINL-LDVKEEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQKACMKWLVEG 362
+ ++ + +D E + L + ++ +R L + +R +E + K+ +W++ G
Sbjct: 654 NTAEKIETLKYRVDAAERDHTLPHQTILRLRQDLNQAYRD---EELYWHLKSRNRWMLLG 710
Query: 363 DLNSGFFHSTINWKRRADSIVGLM-VDGMWEDEPVRVKAAIHDFFQKKFKSVA---WLRP 418
D N+ FF+++ ++ + I + G+ + ++F F + W
Sbjct: 711 DRNTMFFYASTKLRKSRNRIKAITDAQGIENFRDDTIGKVAENYFADLFTTTQTSDW-EE 769
Query: 419 KLDGVQFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGD 478
+ G+ +T++ N EL+ + E++ AV+ G ++PG DGF F WD++ D
Sbjct: 770 IISGIA-PKVTEQMNHELLQSVTDQEVRDAVFAIGADRAPGFDGFTAAFYHHLWDLIGND 828
Query: 479 VVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLR 538
V VR F+ N + I LIPK+ P+ ++D+RPISL YK++ K+ RL+
Sbjct: 829 VCLMVRHFFESDVMDNQINQTQICLIPKIIDPKHMSDYRPISLCTASYKIISKILIKRLK 888
Query: 539 LVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKKRPS---IFFKVDYEKAYNSVEW 595
+G VI + Q FV +N+ D+V+V +E++H K ++ + K D KAY+ VEW
Sbjct: 889 QCLGDVISDSQAAFVPGQNISDNVLVAHELLHSLKSRRECQSGYVAVKTDISKAYDRVEW 948
Query: 596 GFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQG 645
FLE +M ++ F +W+ WIM C+ S + VL+NGSP G+ +G+RQG
Sbjct: 949 NFLEKVMIQLGFAPRWVKWIMTCVTSVSYEVLINGSPYGKIFPSRGIRQG 998
>At2g01550 putative non-LTR retroelement reverse transcriptase
Length = 1449
Score = 201 bits (512), Expect = 2e-51
Identities = 146/528 (27%), Positives = 249/528 (46%), Gaps = 34/528 (6%)
Query: 137 WCLAGDFNAVRYAEE--RRGSSGVLSSHRREMREFNVFIANMDLVDIPLTGRRFTW--RR 192
W + GDFN + +E R ++S MR+F + D+ G FTW +R
Sbjct: 552 WIIFGDFNEILDMDEHSRMEDHPAVTSG---MRDFQSLVNYCSFSDLASHGPLFTWCNKR 608
Query: 193 ANDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCP----LLMRQVVADWGPKPFRV 248
ND ++DR +V+ +W V+P ++ SDH L M G KPF+
Sbjct: 609 DNDPIWKKLDRVMVNEAWKMVYPQSYNVFEAGGCSDHLRCRINLNMNSGAQVRGNKPFKF 668
Query: 249 LNCWLDDPRLFPFVEQSWKGMK---IQGWGAYVLKEKLKGLRGSLRI*NKEVFGDLKTRR 305
+N D P VE W+ + + + +KLK L+ LR KE G+L R
Sbjct: 669 VNAVADMEEFKPLVENFWRETEPIHMSTSSLFRFTKKLKALKPKLRGLAKEKMGNLVKRT 728
Query: 306 EKVVQKINLLDVKEEEVGLQPEELVEMRDLLIEFW-RVLRYQESFLCQKACMKWLVEGDL 364
+ ++L ++ + +E+ W R+ +E +L Q + + WL GD
Sbjct: 729 REAY--LSLCQAQQSNSQNPSQRAMEIESEAYVRWDRIASIEEKYLKQVSKLHWLKVGDK 786
Query: 365 NSGFFHSTINWKRRADSIVGLMV-DGMWEDEPVRVKAAIHDFFQKKFKSVAWLRPKLDGV 423
N+ FH + +SI + DG +K FFQ+ + + +G+
Sbjct: 787 NNKTFHRAATARAAQNSIREIQKEDGSTATTKDDIKNETERFFQEFLQLIP---NDYEGI 843
Query: 424 QFSTITD--------RDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVL 475
+T + L EI+ A++ KSPGPDG+ F +R WD++
Sbjct: 844 TVEKLTSLLPYHCSPAEKDMLTASVSAKEIRGALFSMPNDKSPGPDGYTSEFYKRAWDII 903
Query: 476 SGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAI 535
+ V AV+ F+ +G P+G N + + LIPK + + D+RPIS IYKV+ K+ A
Sbjct: 904 GAEFVLAVKSFFEKGFLPKGVNTTILALIPKKLEAKEMKDYRPISCCNVIYKVISKIIAN 963
Query: 536 RLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVM---HEAKVKKRPSIFFKVDYEKAYNS 592
RL+ V+ I Q FV +R +++++++ E++ H+ + R +I K+D KA++S
Sbjct: 964 RLKHVLPNFIAGNQSAFVKDRLLIENLLLATELVKDYHKDTISGRCAI--KIDISKAFDS 1021
Query: 593 VEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEK 640
V+W FL+ ++ ++F +++ W+M C+ +A+ SV VNG +G F +
Sbjct: 1022 VQWSFLKNVLSALDFPPEFVHWVMLCVTTASFSVQVNGELAGYFQSSR 1069
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 200 bits (509), Expect = 4e-51
Identities = 137/428 (32%), Positives = 205/428 (47%), Gaps = 20/428 (4%)
Query: 227 SDHCPLLMRQVVADW---GPKPFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKL 283
SDH P++ +AD G + FR W+ L + Q W G +V EKL
Sbjct: 4 SDHSPVIA--TIADKIPRGKQNFRFDKRWIGKDGLLEAISQGWNLDSGFREGQFV--EKL 59
Query: 284 KGLRGSLRI*NKEVFGDLKTRREKVVQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVL 343
R ++ K + + R+ + LDV + + EE+ E+ L E +R
Sbjct: 60 TNCRRAISKWRKSL---IPFGRQTIEDLKAELDVAQRDDDRSREEITELTLRLKEAYRD- 115
Query: 344 RYQESFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSIVGLMVD-GMWEDEPVRVKAAI 402
+E + QK+ W+ GD NS FFH+ +R + I GL + G+W E ++
Sbjct: 116 --EEQYWYQKSRSLWMKLGDNNSKFFHALTKQRRARNRITGLHDENGIWSIEDDDIQNIA 173
Query: 403 HDFFQKKFKSV--AWLRPKLDGVQFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGP 460
+FQ F + L VQ ITDR N L E E++ A++ K+PGP
Sbjct: 174 VSYFQNLFTTANPQVFDEALGEVQV-LITDRINDLLTADATECEVRAALFMIHPEKAPGP 232
Query: 461 DGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPIS 520
DG F Q+ W ++ D++ V F G + N + I LIPK + P + + RPIS
Sbjct: 233 DGMTALFFQKSWAIIKSDLLSLVNSFLQEGVFDKRLNTTNICLIPKTERPTRMTELRPIS 292
Query: 521 LIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVK---KR 577
L YKV+ K+ RL+ V+ +I E Q FV R + D++++ E+ H + K
Sbjct: 293 LCNVGYKVISKILCQRLKTVLPNLISETQSAFVDGRLISDNILIAQEMFHGLRTNSSCKD 352
Query: 578 PSIFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFH 637
+ K D KAY+ VEW F+E ++ +M FC KWI WIM C+ + VL+NG P G
Sbjct: 353 KFMAIKTDMSKAYDQVEWNFIEALLRKMGFCEKWISWIMWCITTVQYKVLINGQPKGLII 412
Query: 638 MEKGLRQG 645
E+GLRQG
Sbjct: 413 PERGLRQG 420
>At1g10160 putative reverse transcriptase
Length = 1118
Score = 198 bits (503), Expect = 2e-50
Identities = 174/678 (25%), Positives = 300/678 (43%), Gaps = 68/678 (10%)
Query: 5 NVRGLGVGGNAKKRVVRELVCQQHVELLCIQETKC--QVVDRRMCGLL*GDSEFEWKATP 62
N+RGL ++RVVR + ++ + C ET + + + L G + +
Sbjct: 7 NIRGLN--SRNRQRVVRSWIASNNLLVGCFLETHVAQENANSVLASTLPG---WRMDSNY 61
Query: 63 AVNRGGGLLCIWNSKLFVLE-ESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRAM 121
+ G + +W+ + VL + D F + + + Q + +Y +R++
Sbjct: 62 CCSELGRIWIVWDPSISVLVFKRTDQIMFCSI--KIPSLLQSFAVAFVYGRNSELDRRSL 119
Query: 122 WDALCAWRGTC--DVGEWCLAGDFNAVRYAEERRGSSGVLSSHRREMREFNVFIANMDLV 179
W+ + T V W L GDFN + A E + L + R M + + + L
Sbjct: 120 WEDILVLSRTSPLSVTPWLLLGDFNQIAAASEHYSINQSLLN-LRGMEDLQCCLRDSQLS 178
Query: 180 DIPLTGRRFTW--RRANDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQV 237
D+P G FTW + ++ ++DR L + W V+P+ + SDH P + +
Sbjct: 179 DLPSRGVFFTWSNHQQDNPILRKLDRALANGEWFAVFPSALAVFDPPGDSDHAPCI---I 235
Query: 238 VADWGPKP----FRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI* 293
+ D P P F+ + P + +W+ + G + L++ LK + R
Sbjct: 236 LIDNQPPPSKKSFKYFSFLSSHPSYLAALSTAWEENTLVGSHMFSLRQHLKVAKLCCRTL 295
Query: 294 NKEVFGDLKTRREKVVQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRYQ------- 346
N+ F +++ R + + ++ +Q E L D L V R Q
Sbjct: 296 NRLRFSNIQQRTAQSLTRLE---------DIQVELLTSPSDTLFRREHVARKQWIFFAAA 346
Query: 347 -ESFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSIVGLMVDGMWEDEPV-RVKAAIHD 404
ESF QK+ ++WL EGD N+ FFH + + + I L D + E V ++K +
Sbjct: 347 LESFFRQKSRIRWLHEGDANTRFFHRAVIAHQATNLIKFLRGDDGFRVENVDQIKGMLIA 406
Query: 405 FFQKKFKSVAWLRPKLDGVQFSTITDRDNTELVG--PF--------------DEVEIKQA 448
++ L G+ +T ++ G PF E EI Q
Sbjct: 407 YYSH-----------LLGIPSENVTPFSVEKIKGLLPFRCDSFLASQLTTIPSEEEITQV 455
Query: 449 VWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVK 508
++ K+PGPDGF F W ++ VV A+REF++ G PRG NA+ I LIPKV
Sbjct: 456 LFSMPRNKAPGPDGFPVEFFIEAWAIVKSSVVAAIREFFISGNLPRGFNATAITLIPKVT 515
Query: 509 IPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEV 568
L FRP++ IYKV+ ++ + RL+L I + + Q F+ R + ++V++ +E+
Sbjct: 516 GADRLTQFRPVACCTTIYKVITRIISRRLKLFIDQAVQANQVGFIKGRLLCENVLLASEL 575
Query: 569 MHEAKVKKRPSI-FFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVL 627
+ + + +VD KAY++V W FL ++ ++ +I WI C++SA+ S+
Sbjct: 576 VDNFEADGETTRGCLQVDISKAYDNVNWEFLINILKALDLPLVFIHWIWVCISSASYSIA 635
Query: 628 VNGSPSGEFHMEKGLRQG 645
NG G F +KG+RQG
Sbjct: 636 FNGELIGFFQGKKGIRQG 653
>At2g11240 pseudogene
Length = 1044
Score = 187 bits (474), Expect = 4e-47
Identities = 145/537 (27%), Positives = 253/537 (47%), Gaps = 45/537 (8%)
Query: 173 IANMDLVDIPLTGRRFTWR--RANDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHC 230
+A DL D+ +G +WR R + R+DR L + +W + +P + L + SDH
Sbjct: 1 MAECDLYDLRHSGNFLSWRGKRHDHVVHCRLDRALSNGAWAEDYPASRCIYLCFEGSDHR 60
Query: 231 PLLMRQVVADWGPKP-FRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGS 289
PLL ++ K FR ++ + V+++W + +++EK+ R
Sbjct: 61 PLLTHFDLSKKKKKGVFRYDRRLKNNDEVTALVQEAWNL-----YDTDIVEEKISRCRLE 115
Query: 290 L----RI*NKEVFGDLKTRREKVVQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRY 345
+ R + ++ R+K+ + ++ D +E + L+ E+W+
Sbjct: 116 IVKWSRAKQQSSQKLIEENRQKLEEAMSSQDHNQELLSTINTNLLLAYKAEEEYWK---- 171
Query: 346 QESFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSIVGL-MVDGMWEDEPVRVKAAIHD 404
Q++ WL GD NSG+FH+ + + + DG+ E E + I +
Sbjct: 172 ------QRSRQLWLALGDKNSGYFHAITRGRTVINKFSVIEKEDGVPEYEEAGILNVISE 225
Query: 405 FFQKKFKSVAWLRPKLDGVQFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFN 464
+FQK F + +G + +TI + + + EIK A + K+PGPDGF+
Sbjct: 226 YFQKLFSAN-------EGARAATIKEAIKPFISPEQNPEEIKSACFSIHADKAPGPDGFS 278
Query: 465 FRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGC 524
F Q W + ++V ++ F+ N + I LIPK++ + + D+RPI+L
Sbjct: 279 ASFFQSNWMTVGPNIVLEIQSFFSSSTLQPTINKTHITLIPKIQSLKRMVDYRPIALCTV 338
Query: 525 IYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAK---VKKRPSIF 581
YK++ KL + RL+ ++ ++I E Q FV R D+V++ +E +H K +KR +
Sbjct: 339 FYKIISKLLSRRLQPILQEIISENQSAFVPKRASNDNVLITHEALHYLKSLGAEKRCFMA 398
Query: 582 FKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKG 641
K + KAY+ +EW F++ +M M F WI WI+ C+ + + S L+NGS G E+G
Sbjct: 399 VKTNMSKAYDRIEWDFIKLVMQEMGFHQTWISWILQCITTVSYSFLLNGSAQGAVTPERG 458
Query: 642 LRQG--------ILLRRFFS*LLLKA*MGYSRMQLEL--GNLKVSRL--GQEAILWC 686
LRQG I+ S L KA + S + L + GN +V+ L + I +C
Sbjct: 459 LRQGDPLSPFLFIICSEVLSGLCRKAQLDGSLLGLRVSKGNPRVNHLLFADDTIFFC 515
>At2g28980 putative non-LTR retroelement reverse transcriptase
Length = 1529
Score = 184 bits (466), Expect = 3e-46
Identities = 128/429 (29%), Positives = 210/429 (48%), Gaps = 17/429 (3%)
Query: 233 LMRQVVADWGPKPFRVLNCWLDDPRLFPFVEQSWKG---MKIQGWGAYVLKEKLKGLRGS 289
LM + A G KPF+ +N P+ P VE W + + Y +KLK L+
Sbjct: 535 LMLEAAATGGRKPFKFVNVLTKLPQFLPVVESHWASSAPLYVSTSALYRFSKKLKTLKPH 594
Query: 290 LRI*NKEVFGDLKTR-REKVVQKINLLDVKEEEVGLQP--EELVEMRDLLIEFWRVLRYQ 346
LR KE GDL R RE + LL K+ P E + E ++ + +
Sbjct: 595 LRELGKEKLGDLPKRTREAHI----LLCEKQATTLANPSQETIAEELKAYTDWTHLSELE 650
Query: 347 ESFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSIV---GLMVDGMWEDEPVRVKAA-- 401
E FL QK+ + W+ GD N+ +FH ++ +SI G + + E ++ +A
Sbjct: 651 EGFLKQKSKLHWMNVGDGNNSYFHKAAQVRKMRNSIREIRGPNAETLQTSEEIKGEAERF 710
Query: 402 IHDFFQKKFKSVAWLRPK-LDGVQFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGP 460
++F ++ + + L + + D L EI++ ++ KSPGP
Sbjct: 711 FNEFLNRQSGDFHGISVEDLRNLMSYRCSVTDQNILTREVTGEEIQKVLFAMPNNKSPGP 770
Query: 461 DGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPIS 520
DG+ F + W + D + A++ F+++G P+G NA+ + LIPK + D+RPIS
Sbjct: 771 DGYTSEFFKATWSLTGPDFIAAIQSFFVKGFLPKGLNATILALIPKKDEAIEMKDYRPIS 830
Query: 521 LIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHE-AKVKKRPS 579
+YKV+ K+ A RL+L++ I + Q FV R ++++V++ E++ + K P
Sbjct: 831 CCNVLYKVISKILANRLKLLLPSFILQNQSAFVKERLLMENVLLATELVKDYHKESVTPR 890
Query: 580 IFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHME 639
K+D KA++SV+W FL + +NF + WI C+++AT SV VNG +G F
Sbjct: 891 CAMKIDISKAFDSVQWQFLLNTLEALNFPETFRHWIKLCISTATFSVQVNGELAGFFGSS 950
Query: 640 KGLRQGILL 648
+GLRQG L
Sbjct: 951 RGLRQGCAL 959
>At3g45550 putative protein
Length = 851
Score = 183 bits (464), Expect = 6e-46
Identities = 97/260 (37%), Positives = 149/260 (57%), Gaps = 3/260 (1%)
Query: 426 STITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVRE 485
S++T+ N+EL F + EI +A+ + G K+PGPDG RF ++ WD++ DV+K V+
Sbjct: 35 SSVTNIINSELTQDFRDSEIFEAICQIGDDKAPGPDGLTARFYKQCWDIVGNDVIKEVKL 94
Query: 486 FWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVI 545
F+ N + I +IPK++ PQ L+D+RPI+L +YKV+ K RL+ + ++
Sbjct: 95 FFESSHMKTSVNHTNICMIPKIQNPQTLSDYRPIALCNVLYKVISKCMVNRLKAHLNSIV 154
Query: 546 DERQFTFVGNRNMLDSVVVLNEVMHEAKVKKRPS---IFFKVDYEKAYNSVEWGFLEYMM 602
+ Q F+ R + D+V++ +E+MH KV+KR S + K D KAY+ VEW FLE M
Sbjct: 155 SDSQAAFIPGRIINDNVMIAHEIMHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTM 214
Query: 603 GRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQGILLRRFFS*LLLKA*MG 662
FC KWIGWIM + S SVL+NGSP G +G+RQG L + L
Sbjct: 215 RLFGFCDKWIGWIMAAVKSVHYSVLINGSPHGYISPTRGIRQGDPLSPYLFILCGDILSH 274
Query: 663 YSRMQLELGNLKVSRLGQEA 682
+++ G+++ R+G A
Sbjct: 275 LIKVKASSGDIRGVRIGNGA 294
>At2g41580 putative non-LTR retroelement reverse transcriptase
Length = 1094
Score = 183 bits (464), Expect = 6e-46
Identities = 107/322 (33%), Positives = 172/322 (53%), Gaps = 7/322 (2%)
Query: 344 RYQESFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSIVGLMVDGMWEDEPVRVKAAIH 403
R +E F K+ KW+V GD NS FF +T+ R ++S+ L+ + E R K I
Sbjct: 59 REEEDFWRLKSRDKWMVGGDKNSKFFQATVKANRVSNSLRFLVDENGNEQTVNREKGKIA 118
Query: 404 -DFFQKKFKSV--AWLRPKLDGVQFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGP 460
FF+ F S + + L+G +T+ N +L +E EI +AV+ +PGP
Sbjct: 119 VTFFEDLFSSSYPSSMDSVLEGFN-KRVTEDMNQDLTKKVNEQEIYKAVFSINAESAPGP 177
Query: 461 DGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPIS 520
DGF F QR W ++ ++ + F+ G P N + + LIPK+ P + D RPIS
Sbjct: 178 DGFTALFFQRQWPLVKNQIISDIELFFQTGILPEDWNHTHLCLIPKITKPARMADIRPIS 237
Query: 521 LIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKKRPS- 579
L +YK++ K+ + RL+ + ++ Q FV R + D++++ +E++H + ++ S
Sbjct: 238 LCSVMYKIISKILSARLKKYLPVIVSPTQSAFVAERLVSDNIILAHEIVHNLRTNEKISK 297
Query: 580 --IFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFH 637
+ FK D KAY+ VEW FL+ ++ + F + WI W+M C++S + SVL+NG P G
Sbjct: 298 DFMVFKTDMSKAYDRVEWPFLKGILLALGFNSTWINWMMACVSSVSYSVLINGQPFGHIT 357
Query: 638 MEKGLRQGILLRRFFS*LLLKA 659
+GLRQG L F L +A
Sbjct: 358 PHRGLRQGDPLSPFLFVLCTEA 379
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.339 0.151 0.528
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,333,195
Number of Sequences: 26719
Number of extensions: 1145448
Number of successful extensions: 3361
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 56
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 3109
Number of HSP's gapped (non-prelim): 103
length of query: 1183
length of database: 11,318,596
effective HSP length: 110
effective length of query: 1073
effective length of database: 8,379,506
effective search space: 8991209938
effective search space used: 8991209938
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0058a.1


