

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
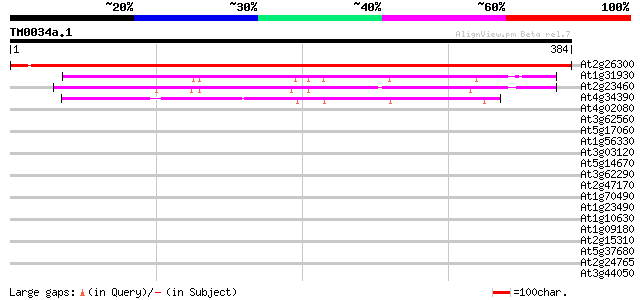
Query= TM0034a.1
(384 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g26300 G protein alpha subunit 1 (GPA1) 671 0.0
At1g31930 hypothetical protein 197 1e-50
At2g23460 extra-large GTP-binding protein (XLG1) 161 5e-40
At4g34390 extra-large G-protein - like 140 2e-33
At4g02080 SAR1/GTP-binding secretory factor 40 0.003
At3g62560 Sar1-like GTP binding protein 40 0.003
At5g17060 ADP-ribosylation factor -like protein 39 0.004
At1g56330 unknown protein 39 0.004
At3g03120 putative ADP-ribosylation factor 39 0.006
At5g14670 ADP-ribosylation factor - like protein 38 0.011
At3g62290 ADP-ribosylation factor-like protein 38 0.011
At2g47170 ADP-ribosylation factor 1 38 0.011
At1g70490 ADP-ribosylation factor 1 like protein 38 0.011
At1g23490 unknown protein (At1g23490) 38 0.011
At1g10630 similar to ADP-ribosylation factor gb|AAD17207; simila... 38 0.011
At1g09180 putative GTP-binding protein, SAR1B 37 0.024
At2g15310 putative ADP-ribosylation factor 35 0.090
At5g37680 ADP-ribosylation factor-like protein (K12B20.15) 34 0.12
At2g24765 ADP-ribosylation factor 3 like protein (At2g24765) 33 0.34
At3g44050 kinesin -like protein 32 0.59
>At2g26300 G protein alpha subunit 1 (GPA1)
Length = 383
Score = 671 bits (1730), Expect = 0.0
Identities = 332/384 (86%), Positives = 354/384 (91%), Gaps = 1/384 (0%)
Query: 1 MGLLCSKNRRYNDADTEENAQTAEIERRIELETKAEKHIQKLLLLGAGESGKSTIFKQIK 60
MGLLCS++R + + DT+EN Q AEIERRIE E KAEKHI+KLLLLGAGESGKSTIFKQIK
Sbjct: 1 MGLLCSRSRHHTE-DTDENTQAAEIERRIEQEAKAEKHIRKLLLLGAGESGKSTIFKQIK 59
Query: 61 LLFQTGFDEAELKSYQPVIHANVYQTIKLLHDGAKELAQNDVDFSKYVISDENKEIGEKL 120
LLFQTGFDE ELKSY PVIHANVYQTIKLLHDG KE AQN+ D +KY++S E+ IGEKL
Sbjct: 60 LLFQTGFDEGELKSYVPVIHANVYQTIKLLHDGTKEFAQNETDSAKYMLSSESIAIGEKL 119
Query: 121 SEIGGRLDYPCLTKELALEIENLWKDAAIQETYARGNELQVPDCTHYFMENLHRLSDANY 180
SEIGGRLDYP LTK++A IE LWKD AIQET ARGNELQVPDCT Y MENL RLSD NY
Sbjct: 120 SEIGGRLDYPRLTKDIAEGIETLWKDPAIQETCARGNELQVPDCTKYLMENLKRLSDINY 179
Query: 181 VPTKDDVLYARVRTTGVVEIQFSPVGENKKSGEVYRLFDVGGQRNERRKWIHLFEGVSAV 240
+PTK+DVLYARVRTTGVVEIQFSPVGENKKSGEVYRLFDVGGQRNERRKWIHLFEGV+AV
Sbjct: 180 IPTKEDVLYARVRTTGVVEIQFSPVGENKKSGEVYRLFDVGGQRNERRKWIHLFEGVTAV 239
Query: 241 IFCAAISEYDQTLFEDENKNRMMETKELFEWVLKQPCFEKTSFMLFLNKFDIFEKKILKV 300
IFCAAISEYDQTLFEDE KNRMMETKELF+WVLKQPCFEKTSFMLFLNKFDIFEKK+L V
Sbjct: 240 IFCAAISEYDQTLFEDEQKNRMMETKELFDWVLKQPCFEKTSFMLFLNKFDIFEKKVLDV 299
Query: 301 PLNVCEWFKDYQPVSTGKQEIEHAYEFVKKKFEESYFQNTAPDRVDRVFKIYRTTALDQK 360
PLNVCEWF+DYQPVS+GKQEIEHAYEFVKKKFEE Y+QNTAPDRVDRVFKIYRTTALDQK
Sbjct: 300 PLNVCEWFRDYQPVSSGKQEIEHAYEFVKKKFEELYYQNTAPDRVDRVFKIYRTTALDQK 359
Query: 361 VVKKTFKLVDETLRRRNLFEAGLL 384
+VKKTFKLVDETLRRRNL EAGLL
Sbjct: 360 LVKKTFKLVDETLRRRNLLEAGLL 383
>At1g31930 hypothetical protein
Length = 848
Score = 197 bits (500), Expect = 1e-50
Identities = 133/394 (33%), Positives = 206/394 (51%), Gaps = 61/394 (15%)
Query: 37 KHIQKLLLLGAGESGKSTIFKQIKLLFQTGFDEAELKSYQPVIHANVYQTIKLLHDGAKE 96
K IQKLLLLG SG STIFKQ K L+ F EL+ + ++ +N+Y+ + +L DG +
Sbjct: 428 KKIQKLLLLGIEGSGTSTIFKQAKFLYGNKFSVEELQDIKLMVQSNMYRYLSILLDGRER 487
Query: 97 LAQNDVDFSKYVISDENKEIGEKLSEIG----------------------------GRLD 128
+ + ++ + + E GE+ ++ G G LD
Sbjct: 488 FEEEALSHTRGLNAVEGDSGGEEANDEGTVTTPQSVYTLNPRLKHFSDWLLDIIATGDLD 547
Query: 129 --YPCLTKELALEIENLWKDAAIQETYARGNELQ-VPDCTHYFMENLHRLSDANYVPTKD 185
+P T+E A +E +WKD AIQ TY R +EL +PD YF+ +S Y P++
Sbjct: 548 AFFPAATREYAPLVEEVWKDPAIQATYRRKDELHFLPDVAEYFLSRAMEVSSNEYEPSER 607
Query: 186 DVLYARVRT--TGVVEIQFS-----PVGENKKSGE--------VYRLFDVGGQ-RNERRK 229
D++YA T G+ ++FS P+ E+ Y+L V + N+ K
Sbjct: 608 DIVYAEGVTQGNGLAFMEFSLSDHSPMSESYPENPDALSSPQPKYQLIRVNAKGMNDSCK 667
Query: 230 WIHLFEGVSAVIFCAAISEYDQTLFEDEN------KNRMMETKELFEWVLKQPCFEKTSF 283
W+ +FE V AVIFC ++S+YDQ E+ +N+M+++KELFE ++K PCF+ T F
Sbjct: 668 WVEMFEDVRAVIFCISLSDYDQINITPESSGTVQYQNKMIQSKELFESMVKHPCFKDTPF 727
Query: 284 MLFLNKFDIFEKKILKVPLNVCEWFKDYQPVSTGK--QEIEH-AYEFVKKKFEESYFQNT 340
+L LNK+D FE+K+ + PL C+WF D+ PV T Q + + AY +V KF+ YF T
Sbjct: 728 ILILNKYDQFEEKLNRAPLTSCDWFSDFCPVRTNNNVQSLAYQAYFYVAMKFKLLYFSIT 787
Query: 341 APDRVDRVFKIYRTTALDQKVVKKTFKLVDETLR 374
++F +++ A D+ V + FK V E L+
Sbjct: 788 G----QKLF-VWQARARDRANVDEGFKYVREVLK 816
>At2g23460 extra-large GTP-binding protein (XLG1)
Length = 888
Score = 161 bits (408), Expect = 5e-40
Identities = 121/396 (30%), Positives = 193/396 (48%), Gaps = 59/396 (14%)
Query: 31 LETKAEKHIQKLLLLGAGESGKSTIFKQIKLLFQ-TGFDEAELKSYQPVIHANVYQTIKL 89
L+ + +QK+LL+G SG STIFKQ K+L++ F E E ++ + +I NVY + +
Sbjct: 475 LDHLEHRTLQKILLVGNSGSGTSTIFKQAKILYKDVPFLEDERENIKVIIQTNVYGYLGM 534
Query: 90 LHDGAKELAQ-------------------------NDVDFSKYVISDENKEIGEKLSEI- 123
L +G + + ND + Y I K + L +
Sbjct: 535 LLEGRERFEEEALALRNTKQCVLENIPADEGDAKSNDKTVTMYSIGPRLKAFSDWLLKTM 594
Query: 124 -GGRLD--YPCLTKELALEIENLWKDAAIQETYARGNELQV-PDCTHYFMENLHRLSDAN 179
G L +P ++E A +E LW+DAAIQ TY R +EL + P YF+E + +
Sbjct: 595 AAGNLGVIFPAASREYAPLVEELWRDAAIQATYKRRSELGLLPSVASYFLERAIDVLTPD 654
Query: 180 YVPTKDDVLYAR--VRTTGVVEIQFS----------PVGENKKSGEVYRLFDVGGQR-NE 226
Y P+ D+LYA ++G+ + FS ++ S Y+L V + E
Sbjct: 655 YEPSDLDILYAEGVTSSSGLACLDFSFPQTASEENLDPSDHHDSLLRYQLIRVPSRGLGE 714
Query: 227 RRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLKQPCFEKTSFMLF 286
KWI +FE V V+F ++S+YDQ ++ N+M+ TK+LFE ++ P FE F+L
Sbjct: 715 NCKWIDMFEDVGMVVFVVSMSDYDQV--SEDGTNKMLLTKKLFESIITHPIFENMDFLLI 772
Query: 287 LNKFDIFEKKILKVPLNVCEWFKDYQPV-------STGKQEI-EHAYEFVKKKFEESYFQ 338
LNK+D+ E+K+ +VPL CEWF+D+ PV + G + + A+ F+ KF+ Y
Sbjct: 773 LNKYDLLEEKVERVPLARCEWFQDFNPVVSRHRGSNNGNPTLGQLAFHFMAVKFKRFYSS 832
Query: 339 NTAPDRVDRVFKIYRTTALDQKVVKKTFKLVDETLR 374
T + + + +LD V + KL E L+
Sbjct: 833 LTG-----KKLFVSSSKSLDPNSVDSSLKLAMEILK 863
>At4g34390 extra-large G-protein - like
Length = 838
Score = 140 bits (352), Expect = 2e-33
Identities = 93/323 (28%), Positives = 166/323 (50%), Gaps = 30/323 (9%)
Query: 36 EKHIQKLLLLGAGESGKSTIFKQIKLLFQTGFDEAELKSYQPVIHANVYQTIKLLHDGAK 95
+K + KLLL+G+ + G +TI+KQ + L+ F + + + +I N+Y + ++ + +
Sbjct: 459 QKMLNKLLLIGSEKGGATTIYKQARSLYNVSFSLEDRERIKFIIQTNLYTYLAMVLEAHE 518
Query: 96 ELAQNDVDFSKYVISDENKEIGEKLSEIGGRLDYPCLTKELALEIENLWKDAAIQETYAR 155
F K + +D++ + K E G +P ++E A + +LW+ AIQ TY R
Sbjct: 519 R-------FEKEMSNDQSSDWVLKEKEDGNLKIFPPSSRENAQTVADLWRVPAIQATYKR 571
Query: 156 GNELQVPDCTHYFMENLHRLSDANYVPTKDDVLYARVRTT--GVVEIQFS-PVGENKKSG 212
+ +P YF+E + +S + Y P+ D+L A ++ G+ + FS P ++S
Sbjct: 572 LRDT-LPRNAVYFLERILEISRSEYDPSDMDILQAEGLSSMEGLSCVDFSFPSTSQEESL 630
Query: 213 EV---------YRLFDVGGQR-NERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENK--N 260
E Y+L + + E K + +FE VIFC ++++Y + + + E N
Sbjct: 631 ESDYQHDTDMKYQLIRLNPRSLGENWKLLEMFEDADLVIFCVSLTDYAENIEDGEGNIVN 690
Query: 261 RMMETKELFEWVLKQPCFEKTSFMLFLNKFDIFEKKILKVPLNVCEWFKDYQPVSTGKQE 320
+M+ TK+LFE ++ P F+L L KFD+ E+KI +VPL CEWF+D+ P+ + Q
Sbjct: 691 KMLATKQLFENMVTHPSLANKRFLLVLTKFDLLEEKIEEVPLRTCEWFEDFNPLISQNQT 750
Query: 321 IEH-------AYEFVKKKFEESY 336
H A+ ++ KF+ Y
Sbjct: 751 SRHNPPMAQRAFHYIGYKFKRLY 773
>At4g02080 SAR1/GTP-binding secretory factor
Length = 193
Score = 39.7 bits (91), Expect = 0.003
Identities = 25/90 (27%), Positives = 41/90 (44%), Gaps = 11/90 (12%)
Query: 204 PVGENKKSGEV-YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRM 262
P E G++ ++ FD+GG + RR W + V AV++ + YD K R
Sbjct: 54 PTSEELSIGKIKFKAFDLGGHQIARRVWKDYYAKVDAVVY--LVDAYD--------KERF 103
Query: 263 METKELFEWVLKQPCFEKTSFMLFLNKFDI 292
E+K+ + +L F++ NK DI
Sbjct: 104 AESKKELDALLSDESLASVPFLILGNKIDI 133
>At3g62560 Sar1-like GTP binding protein
Length = 193
Score = 39.7 bits (91), Expect = 0.003
Identities = 25/90 (27%), Positives = 41/90 (44%), Gaps = 11/90 (12%)
Query: 204 PVGENKKSGEV-YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRM 262
P E G++ ++ FD+GG + RR W + V AV++ + YD K R
Sbjct: 54 PTSEELSIGKIKFKAFDLGGHQIARRVWKDYYAKVDAVVY--LVDAYD--------KERF 103
Query: 263 METKELFEWVLKQPCFEKTSFMLFLNKFDI 292
E+K+ + +L F++ NK DI
Sbjct: 104 AESKKELDALLSDESLANVPFLILGNKIDI 133
>At5g17060 ADP-ribosylation factor -like protein
Length = 192
Score = 39.3 bits (90), Expect = 0.004
Identities = 23/93 (24%), Positives = 44/93 (46%), Gaps = 13/93 (13%)
Query: 214 VYRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVL 273
++ ++DVGGQ R W H F +I+ + + ++ R+ + K+ F+ ++
Sbjct: 62 MFTVWDVGGQEKLRPLWRHYFNNTDGLIY----------VVDSLDRERIGKAKQEFQEII 111
Query: 274 KQPCFEKTSFMLFLNKFDIFEKKILKVPLNVCE 306
K P + ++F NK D+ + P VCE
Sbjct: 112 KDPFMLNSIILVFANKQDM---RGAMSPREVCE 141
>At1g56330 unknown protein
Length = 193
Score = 39.3 bits (90), Expect = 0.004
Identities = 25/90 (27%), Positives = 40/90 (43%), Gaps = 11/90 (12%)
Query: 204 PVGENKKSGEV-YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRM 262
P E G++ ++ FD+GG + RR W + V AV++ + YD K R
Sbjct: 54 PTSEELSIGKIKFKAFDLGGHQIARRVWKDYYAKVDAVVY--LVDAYD--------KERF 103
Query: 263 METKELFEWVLKQPCFEKTSFMLFLNKFDI 292
E+K + +L F++ NK DI
Sbjct: 104 AESKRELDALLSDEALATVPFLILGNKIDI 133
>At3g03120 putative ADP-ribosylation factor
Length = 192
Score = 38.5 bits (88), Expect = 0.006
Identities = 22/93 (23%), Positives = 44/93 (46%), Gaps = 13/93 (13%)
Query: 214 VYRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVL 273
++ ++DVGGQ R W H F +I+ + + ++ R+ + K+ F+ ++
Sbjct: 62 IFTVWDVGGQEKLRPLWRHYFNNTDGLIY----------VVDSLDRERIGKAKQEFQDII 111
Query: 274 KQPCFEKTSFMLFLNKFDIFEKKILKVPLNVCE 306
+ P + ++F NK D+ + P VCE
Sbjct: 112 RDPFMLNSVILVFANKQDM---RGAMSPREVCE 141
>At5g14670 ADP-ribosylation factor - like protein
Length = 188
Score = 37.7 bits (86), Expect = 0.011
Identities = 22/78 (28%), Positives = 38/78 (48%), Gaps = 10/78 (12%)
Query: 215 YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLK 274
+ ++DVGGQ R W H F+ +IF ++ D+ + + +RM+ EL + VL
Sbjct: 63 FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSNDRDRVVEARDELHRMLNEDELRDAVL- 121
Query: 275 QPCFEKTSFMLFLNKFDI 292
++F NK D+
Sbjct: 122 ---------LVFANKQDL 130
>At3g62290 ADP-ribosylation factor-like protein
Length = 181
Score = 37.7 bits (86), Expect = 0.011
Identities = 22/78 (28%), Positives = 38/78 (48%), Gaps = 10/78 (12%)
Query: 215 YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLK 274
+ ++DVGGQ R W H F+ +IF ++ D+ + + +RM+ EL + VL
Sbjct: 63 FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSNDRDRVVEARDELHRMLNEDELRDAVL- 121
Query: 275 QPCFEKTSFMLFLNKFDI 292
++F NK D+
Sbjct: 122 ---------LVFANKQDL 130
>At2g47170 ADP-ribosylation factor 1
Length = 181
Score = 37.7 bits (86), Expect = 0.011
Identities = 22/78 (28%), Positives = 38/78 (48%), Gaps = 10/78 (12%)
Query: 215 YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLK 274
+ ++DVGGQ R W H F+ +IF ++ D+ + + +RM+ EL + VL
Sbjct: 63 FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSNDRDRVVEARDELHRMLNEDELRDAVL- 121
Query: 275 QPCFEKTSFMLFLNKFDI 292
++F NK D+
Sbjct: 122 ---------LVFANKQDL 130
>At1g70490 ADP-ribosylation factor 1 like protein
Length = 181
Score = 37.7 bits (86), Expect = 0.011
Identities = 22/78 (28%), Positives = 38/78 (48%), Gaps = 10/78 (12%)
Query: 215 YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLK 274
+ ++DVGGQ R W H F+ +IF ++ D+ + + +RM+ EL + VL
Sbjct: 63 FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSNDRDRVVEARDELHRMLNEDELRDAVL- 121
Query: 275 QPCFEKTSFMLFLNKFDI 292
++F NK D+
Sbjct: 122 ---------LVFANKQDL 130
>At1g23490 unknown protein (At1g23490)
Length = 181
Score = 37.7 bits (86), Expect = 0.011
Identities = 22/78 (28%), Positives = 38/78 (48%), Gaps = 10/78 (12%)
Query: 215 YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLK 274
+ ++DVGGQ R W H F+ +IF ++ D+ + + +RM+ EL + VL
Sbjct: 63 FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSNDRDRVVEARDELHRMLNEDELRDAVL- 121
Query: 275 QPCFEKTSFMLFLNKFDI 292
++F NK D+
Sbjct: 122 ---------LVFANKQDL 130
>At1g10630 similar to ADP-ribosylation factor gb|AAD17207; similar
to ESTs gb|N65779, gb|AI996946.1, gb|AI725909.1,
gb|T45928, gb|T42202
Length = 181
Score = 37.7 bits (86), Expect = 0.011
Identities = 22/78 (28%), Positives = 38/78 (48%), Gaps = 10/78 (12%)
Query: 215 YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLK 274
+ ++DVGGQ R W H F+ +IF ++ D+ + + +RM+ EL + VL
Sbjct: 63 FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSNDRDRVVEARDELHRMLNEDELRDAVL- 121
Query: 275 QPCFEKTSFMLFLNKFDI 292
++F NK D+
Sbjct: 122 ---------LVFANKQDL 130
>At1g09180 putative GTP-binding protein, SAR1B
Length = 193
Score = 36.6 bits (83), Expect = 0.024
Identities = 23/90 (25%), Positives = 41/90 (45%), Gaps = 11/90 (12%)
Query: 204 PVGENKKSGEV-YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRM 262
P E G++ ++ FD+GG + RR W + V AV++ + YD ++R
Sbjct: 54 PTSEELSIGKINFKAFDLGGHQIARRVWKDCYAKVDAVVY--LVDAYD--------RDRF 103
Query: 263 METKELFEWVLKQPCFEKTSFMLFLNKFDI 292
+E+K + +L ++ NK DI
Sbjct: 104 VESKRELDALLSDEALANVPCLILGNKIDI 133
>At2g15310 putative ADP-ribosylation factor
Length = 205
Score = 34.7 bits (78), Expect = 0.090
Identities = 18/77 (23%), Positives = 33/77 (42%), Gaps = 10/77 (12%)
Query: 215 YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLK 274
+ ++D+GGQ R+ W H F+ +IF + + + R+ E + +L
Sbjct: 63 FTVWDIGGQEKIRKLWRHYFQNAQGLIF----------VVDSSDSERLSEARNELHRILT 112
Query: 275 QPCFEKTSFMLFLNKFD 291
E ++F NK D
Sbjct: 113 DNELEGACVLVFANKQD 129
>At5g37680 ADP-ribosylation factor-like protein (K12B20.15)
Length = 184
Score = 34.3 bits (77), Expect = 0.12
Identities = 21/81 (25%), Positives = 33/81 (39%), Gaps = 4/81 (4%)
Query: 192 VRTTGVVEIQFSPVGENK----KSGEVYRLFDVGGQRNERRKWIHLFEGVSAVIFCAAIS 247
+ T G E VG N K +++D+GGQR R W GVSA+++ +
Sbjct: 39 IATGGYSEDMIPTVGFNMRKVTKGNVTIKIWDLGGQRRFRTMWERYCRGVSAIVYVIDAA 98
Query: 248 EYDQTLFEDENKNRMMETKEL 268
+ D N ++ L
Sbjct: 99 DRDSVPISRSELNDLLTKPSL 119
>At2g24765 ADP-ribosylation factor 3 like protein (At2g24765)
Length = 182
Score = 32.7 bits (73), Expect = 0.34
Identities = 21/78 (26%), Positives = 37/78 (46%), Gaps = 10/78 (12%)
Query: 215 YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLK 274
++++D+GGQ + R W F AVI+ S+ D R+ KE F +L+
Sbjct: 63 FQVWDLGGQTSIRPYWRCYFPNTQAVIYVVDSSDTD----------RIGVAKEEFHAILE 112
Query: 275 QPCFEKTSFMLFLNKFDI 292
+ + ++F NK D+
Sbjct: 113 EDELKGAVVLIFANKQDL 130
>At3g44050 kinesin -like protein
Length = 1229
Score = 32.0 bits (71), Expect = 0.58
Identities = 34/134 (25%), Positives = 60/134 (44%), Gaps = 20/134 (14%)
Query: 9 RRYNDADTEENAQTAEIERRIELETKAEKHIQKLLL-----------LGAGESGKSTIFK 57
RR + D A TAE E ++LE K E I+ L + L SGK +
Sbjct: 519 RREREKDVALQALTAENEASMKLEKKREDEIRGLKMMLKLRDSAIKSLQGVTSGKIPVEA 578
Query: 58 QIK-----LLFQTGFDEAELKSYQ-PVIHANVYQTI--KLLHDGAKELAQNDVDFSKYVI 109
++ L+ + E ++ + Q + A + + + KL+H+ + + D + S
Sbjct: 579 HLQKEKGDLMKEIEEGERDILNQQIQALQAKLLEALDWKLMHESDSSMVKEDGNISNMFC 638
Query: 110 SDENKEIGEKLSEI 123
S++N+E +KLS I
Sbjct: 639 SNQNQE-SKKLSSI 651
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,622,852
Number of Sequences: 26719
Number of extensions: 381411
Number of successful extensions: 1222
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 1176
Number of HSP's gapped (non-prelim): 55
length of query: 384
length of database: 11,318,596
effective HSP length: 101
effective length of query: 283
effective length of database: 8,619,977
effective search space: 2439453491
effective search space used: 2439453491
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0034a.1


