

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
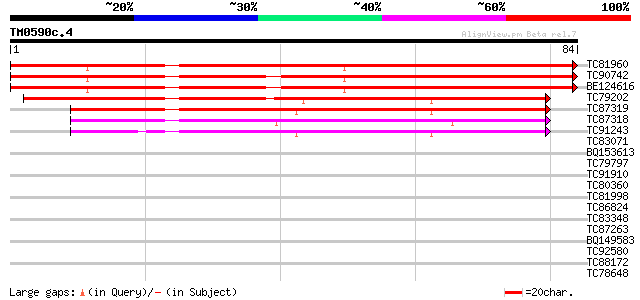
Query= TM0590c.4
(84 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC81960 similar to PIR|S19773|S19773 wound-induced protein - tom... 85 4e-18
TC90742 similar to PIR|S19773|S19773 wound-induced protein - tom... 85 4e-18
BE124616 weakly similar to PIR|T01970|T0197 probable wound-induc... 84 1e-17
TC79202 similar to PIR|T01970|T01970 probable wound-induced prot... 52 3e-08
TC87319 similar to PIR|T01970|T01970 probable wound-induced prot... 52 5e-08
TC87318 similar to PIR|T01970|T01970 probable wound-induced prot... 50 2e-07
TC91243 homologue to GP|6688818|emb|CAB65284.1 putative wound-in... 42 5e-05
TC83071 similar to PIR|T01970|T01970 probable wound-induced prot... 30 0.13
BQ153613 29 0.28
TC79797 28 0.63
TC91910 homologue to GP|6688818|emb|CAB65284.1 putative wound-in... 28 0.82
TC80360 PIR|T01970|T01970 probable wound-induced protein T9A4.6 ... 27 1.1
TC81998 27 1.4
TC86824 homologue to GP|15215820|gb|AAK91455.1 AT4g28240/F26K10_... 27 1.8
TC83348 similar to GP|16588831|gb|AAL26911.1 unknown {Prunus per... 26 2.4
TC87263 similar to GP|9759298|dbj|BAB09804.1 emb|CAB82953.1~gene... 25 6.9
BQ149583 similar to GP|11994595|d receptor-like serine/threonine... 25 6.9
TC92580 similar to GP|22832529|gb|AAF48893.2 CG7274-PA {Drosophi... 25 6.9
TC88172 similar to GP|12003386|gb|AAG43550.1 Avr9/Cf-9 rapidly e... 24 9.1
TC78648 similar to PIR|S51529|S51529 SPF1 protein - sweet potato... 24 9.1
>TC81960 similar to PIR|S19773|S19773 wound-induced protein - tomato
(fragment), partial (63%)
Length = 513
Score = 85.1 bits (209), Expect = 4e-18
Identities = 50/91 (54%), Positives = 62/91 (67%), Gaps = 7/91 (7%)
Frame = +1
Query: 1 MSAATRSLIV----GAVEALKDKLGVYNRRRNYALRSLQQRVVKNNNITPNG---NSSSA 53
MSAATR+ +V GAVEALKD+LGV R NYA RSL Q N +S+S+
Sbjct: 76 MSAATRAWVVASSIGAVEALKDQLGVC--RWNYAFRSLHQHAKNNIRSYTQAKKLSSASS 249
Query: 54 AAIPSKVKRSNQEFMRKVMDLNCWGPSTTKF 84
AA+ +KVKR+ +E M+KVMDLNCWGPST +F
Sbjct: 250 AAVSNKVKRTKEESMKKVMDLNCWGPSTARF 342
>TC90742 similar to PIR|S19773|S19773 wound-induced protein - tomato
(fragment), partial (63%)
Length = 486
Score = 85.1 bits (209), Expect = 4e-18
Identities = 52/93 (55%), Positives = 64/93 (67%), Gaps = 9/93 (9%)
Frame = +3
Query: 1 MSAATRSLIV----GAVEALKDKLGVYNRRRNYALRSLQQRVVKNNNITPNG-----NSS 51
MSAAT++ +V GAVEALKD+LGV R NYA RSL Q NNI +S+
Sbjct: 42 MSAATKAWVVASSIGAVEALKDQLGVC--RWNYAFRSLHQHA--KNNIRSYSQAKKLSSA 209
Query: 52 SAAAIPSKVKRSNQEFMRKVMDLNCWGPSTTKF 84
S+AA+ +KVKRS +E MRKV+DLNCWGPST +F
Sbjct: 210 SSAAVSNKVKRSKEESMRKVIDLNCWGPSTARF 308
>BE124616 weakly similar to PIR|T01970|T0197 probable wound-induced protein
T9A4.6 - Arabidopsis thaliana, partial (63%)
Length = 357
Score = 83.6 bits (205), Expect = 1e-17
Identities = 51/93 (54%), Positives = 63/93 (66%), Gaps = 9/93 (9%)
Frame = +3
Query: 1 MSAATRSLIV----GAVEALKDKLGVYNRRRNYALRSLQQRVVKNNNITPNG-----NSS 51
MSAAT++ +V GAVEALKD+LGV R NY RSL Q NNI +S+
Sbjct: 33 MSAATKAWVVASSIGAVEALKDQLGVC--RWNYVFRSLHQHA--KNNIRSYSQAKKLSSA 200
Query: 52 SAAAIPSKVKRSNQEFMRKVMDLNCWGPSTTKF 84
S+AA+ +KVKRS +E MRKV+DLNCWGPST +F
Sbjct: 201 SSAAVSNKVKRSKEESMRKVIDLNCWGPSTARF 299
>TC79202 similar to PIR|T01970|T01970 probable wound-induced protein T9A4.6
- Arabidopsis thaliana, partial (83%)
Length = 773
Score = 52.4 bits (124), Expect = 3e-08
Identities = 37/86 (43%), Positives = 52/86 (60%), Gaps = 8/86 (9%)
Frame = +2
Query: 3 AATRSLIVGAVEALKDKLGVYNRRRNYALRSLQQRVVKNN----NITPNGNSSSAAAIPS 58
A T + VG VEALKD+LG+ R NYAL+ QQ V KNN + +SSS++ I
Sbjct: 83 AWTVAASVGVVEALKDQLGIC--RWNYALKLAQQHV-KNNVRSFSQAKKLSSSSSSMISK 253
Query: 59 KVK----RSNQEFMRKVMDLNCWGPS 80
++K ++E +R VM L+CWGP+
Sbjct: 254 RLKDDKPNQSEESLRTVMYLSCWGPN 331
>TC87319 similar to PIR|T01970|T01970 probable wound-induced protein T9A4.6
- Arabidopsis thaliana, partial (83%)
Length = 718
Score = 51.6 bits (122), Expect = 5e-08
Identities = 32/77 (41%), Positives = 47/77 (60%), Gaps = 6/77 (7%)
Frame = +1
Query: 10 VGAVEALKDKLGVYNRRRNYALRSLQQRVVKN-NNITPNGNSSSAAAIPSKVK-----RS 63
VG VEALKD+ G+ R NYALR QQ + +I+ N SS++ + +K+K +
Sbjct: 142 VGVVEALKDQAGIC--RWNYALRQAQQHLKNRVRSISQAKNFSSSSFLANKLKDEKKAKQ 315
Query: 64 NQEFMRKVMDLNCWGPS 80
+E +R VM L+CWGP+
Sbjct: 316 AEESLRTVMYLSCWGPN 366
>TC87318 similar to PIR|T01970|T01970 probable wound-induced protein T9A4.6
- Arabidopsis thaliana, partial (83%)
Length = 489
Score = 50.1 bits (118), Expect = 2e-07
Identities = 33/77 (42%), Positives = 46/77 (58%), Gaps = 6/77 (7%)
Frame = +1
Query: 10 VGAVEALKDKLGVYNRRRNYALRSLQQRV---VKNNNITPNGNSSSAAAIPSKVKRSN-- 64
VG VEALKD+ G+ R NYALR QQ + K+ + N +SSS A K ++ +
Sbjct: 139 VGVVEALKDQAGIC--RWNYALRQAQQHLKNRAKSMSQAKNFSSSSFLATKFKDEKKSKQ 312
Query: 65 -QEFMRKVMDLNCWGPS 80
+E +R VM L+CWGP+
Sbjct: 313 AEESLRTVMYLSCWGPN 363
>TC91243 homologue to GP|6688818|emb|CAB65284.1 putative wound-induced
protein {Medicago sativa subsp. x varia}, complete
Length = 656
Score = 41.6 bits (96), Expect = 5e-05
Identities = 31/79 (39%), Positives = 45/79 (56%), Gaps = 8/79 (10%)
Frame = +2
Query: 10 VGAVEALKDKLGVYNRRRNYALRSLQQRVVKN----NNITPNGNSSSAAAIPSKVK---- 61
VG VEALKD+ G+ R N ALRS Q V + + S+S A + S++K
Sbjct: 242 VGVVEALKDQ-GLC--RWNCALRSAQHHVKHHFRSLSQARKVSTSNSYAMVSSRLKEQKA 412
Query: 62 RSNQEFMRKVMDLNCWGPS 80
+ ++E +R VM L+CWGP+
Sbjct: 413 KQSEESLRTVMYLSCWGPN 469
>TC83071 similar to PIR|T01970|T01970 probable wound-induced protein T9A4.6
- Arabidopsis thaliana, partial (21%)
Length = 327
Score = 30.4 bits (67), Expect = 0.13
Identities = 13/38 (34%), Positives = 23/38 (60%)
Frame = +1
Query: 43 NITPNGNSSSAAAIPSKVKRSNQEFMRKVMDLNCWGPS 80
N+ PN +S+ + + K+ +E +R VM L+ WGP+
Sbjct: 166 NMCPNSSSTISKKHSDENKKKAEESLRTVMFLSIWGPN 279
>BQ153613
Length = 562
Score = 29.3 bits (64), Expect = 0.28
Identities = 21/81 (25%), Positives = 40/81 (48%), Gaps = 5/81 (6%)
Frame = +2
Query: 3 AATRSLIVGAVEAL---KDKLGVY--NRRRNYALRSLQQRVVKNNNITPNGNSSSAAAIP 57
AAT ++ G E + K LG NR R ++ ++ +++ + ++ ++ P
Sbjct: 74 AATMAVAQGHTEPIHNCKSALGTIHQNRTRLFSAGGALSALLPLSSVFVS-EATVVSSEP 250
Query: 58 SKVKRSNQEFMRKVMDLNCWG 78
K R + ++KVM +NCWG
Sbjct: 251 KKKLRQTDDSLQKVMYMNCWG 313
>TC79797
Length = 701
Score = 28.1 bits (61), Expect = 0.63
Identities = 13/30 (43%), Positives = 19/30 (63%), Gaps = 3/30 (10%)
Frame = +1
Query: 52 SAAAIPSKVK---RSNQEFMRKVMDLNCWG 78
S A + SKV+ R + ++KVM +NCWG
Sbjct: 229 SDATVSSKVEEKLRQTDDSLQKVMYMNCWG 318
>TC91910 homologue to GP|6688818|emb|CAB65284.1 putative wound-induced
protein {Medicago sativa subsp. x varia}, partial (97%)
Length = 405
Score = 27.7 bits (60), Expect = 0.82
Identities = 12/31 (38%), Positives = 21/31 (67%), Gaps = 4/31 (12%)
Frame = +3
Query: 54 AAIPSKVK----RSNQEFMRKVMDLNCWGPS 80
A + S++K + ++E +R VM L+CWGP+
Sbjct: 246 AMVSSRLKEQEAKRSEESLRTVMYLSCWGPN 338
>TC80360 PIR|T01970|T01970 probable wound-induced protein T9A4.6 -
Arabidopsis thaliana, partial (21%)
Length = 679
Score = 27.3 bits (59), Expect = 1.1
Identities = 17/58 (29%), Positives = 25/58 (42%), Gaps = 17/58 (29%)
Frame = +2
Query: 40 KNNNITPNGNSSSAAAIPSKVK-----------------RSNQEFMRKVMDLNCWGPS 80
+ N I+ + SS PSK K + +E +R VM L+CWGP+
Sbjct: 206 RKNFISSHKGSSRVQHFPSKSKNHVFHQAGANVDAVDRFKKAEESLRTVMYLSCWGPN 379
>TC81998
Length = 697
Score = 26.9 bits (58), Expect = 1.4
Identities = 13/52 (25%), Positives = 28/52 (53%)
Frame = +3
Query: 24 NRRRNYALRSLQQRVVKNNNITPNGNSSSAAAIPSKVKRSNQEFMRKVMDLN 75
N N +L+ +++ NN+ + NSS++ + S K+ + +R ++D N
Sbjct: 21 NNMNNDVKSNLKNKIINNNSFN-SSNSSNSYEVNSGDKQKKTQNLRDLLDTN 173
>TC86824 homologue to GP|15215820|gb|AAK91455.1 AT4g28240/F26K10_120
{Arabidopsis thaliana}, partial (19%)
Length = 491
Score = 26.6 bits (57), Expect = 1.8
Identities = 11/29 (37%), Positives = 19/29 (64%)
Frame = +3
Query: 50 SSSAAAIPSKVKRSNQEFMRKVMDLNCWG 78
S ++ K++R++ +RKVM +NCWG
Sbjct: 216 SDMSSEADEKLRRTDDS-LRKVMYMNCWG 299
>TC83348 similar to GP|16588831|gb|AAL26911.1 unknown {Prunus persica},
partial (27%)
Length = 984
Score = 26.2 bits (56), Expect = 2.4
Identities = 10/22 (45%), Positives = 14/22 (63%)
Frame = +2
Query: 58 SKVKRSNQEFMRKVMDLNCWGP 79
S+ K+ +QEF R + NCW P
Sbjct: 341 SQKKKRSQEFQRVLRS*NCWKP 406
>TC87263 similar to GP|9759298|dbj|BAB09804.1
emb|CAB82953.1~gene_id:MPH15.5~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (65%)
Length = 2229
Score = 24.6 bits (52), Expect = 6.9
Identities = 15/56 (26%), Positives = 27/56 (47%)
Frame = +2
Query: 28 NYALRSLQQRVVKNNNITPNGNSSSAAAIPSKVKRSNQEFMRKVMDLNCWGPSTTK 83
N+ S + +V+KN+N T N +P+K S+ K +DL+ + S +
Sbjct: 605 NHTSNSDETKVMKNSNQTTNATVEGVPIVPNKNLSSDSSL--KGVDLHNYTASLAR 766
>BQ149583 similar to GP|11994595|d receptor-like serine/threonine kinase
{Arabidopsis thaliana}, partial (12%)
Length = 615
Score = 24.6 bits (52), Expect = 6.9
Identities = 9/26 (34%), Positives = 16/26 (60%)
Frame = +1
Query: 46 PNGNSSSAAAIPSKVKRSNQEFMRKV 71
PNG + + SK K+ N+EF+ ++
Sbjct: 400 PNGTLIAVKQLSSKSKQGNREFLNEI 477
>TC92580 similar to GP|22832529|gb|AAF48893.2 CG7274-PA {Drosophila
melanogaster}, partial (2%)
Length = 923
Score = 24.6 bits (52), Expect = 6.9
Identities = 16/48 (33%), Positives = 23/48 (47%), Gaps = 12/48 (25%)
Frame = +3
Query: 35 QQRVVKNNNITPNGNSSSAAA------------IPSKVKRSNQEFMRK 70
+ RVV + I N ++SS A IPSK+K S+Q + K
Sbjct: 729 EHRVVSDEEIEDNRSTSSCEASQIPFKEEEVKSIPSKIKESHQTSVTK 872
>TC88172 similar to GP|12003386|gb|AAG43550.1 Avr9/Cf-9 rapidly elicited
protein 132 {Nicotiana tabacum}, partial (26%)
Length = 1445
Score = 24.3 bits (51), Expect = 9.1
Identities = 12/32 (37%), Positives = 16/32 (49%)
Frame = +3
Query: 51 SSAAAIPSKVKRSNQEFMRKVMDLNCWGPSTT 82
SSA +KR R +M LN W P++T
Sbjct: 921 SSAEGRLKSLKRLLSSSSRGIMSLNPWSPTST 1016
>TC78648 similar to PIR|S51529|S51529 SPF1 protein - sweet potato, partial
(18%)
Length = 990
Score = 24.3 bits (51), Expect = 9.1
Identities = 10/22 (45%), Positives = 16/22 (72%)
Frame = +2
Query: 47 NGNSSSAAAIPSKVKRSNQEFM 68
N NS+ AIP K+ +SNQ+++
Sbjct: 626 NLNSNQLRAIPQKLYQSNQKYI 691
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.126 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,162,276
Number of Sequences: 36976
Number of extensions: 21994
Number of successful extensions: 158
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 148
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 150
length of query: 84
length of database: 9,014,727
effective HSP length: 60
effective length of query: 24
effective length of database: 6,796,167
effective search space: 163108008
effective search space used: 163108008
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 51 (24.3 bits)
Lotus: description of TM0590c.4


