

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0371a.8
(220 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
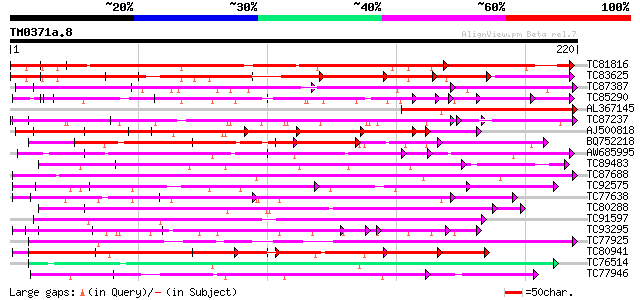
Score E
Sequences producing significant alignments: (bits) Value
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 135 9e-33
TC83625 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 129 7e-31
TC87387 homologue to PIR|T47775|T47775 hypothetical protein F24I... 99 1e-21
TC85290 similar to GP|7767653|gb|AAF69150.1| F27F5.2 {Arabidopsi... 84 4e-17
AL367145 weakly similar to GP|8096269|dbj| KED {Nicotiana tabacu... 83 9e-17
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 83 9e-17
AJ500818 weakly similar to GP|10435304|dbj unnamed protein produ... 71 3e-13
BQ752218 similar to GP|19170914|emb hypothetical protein {Enceph... 70 5e-13
AW685995 similar to PIR|I51618|I516 nucleolar phosphoprotein - A... 70 8e-13
TC89483 similar to GP|18377662|gb|AAL66981.1 unknown protein {Ar... 69 1e-12
TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~un... 67 4e-12
TC92575 weakly similar to GP|21428434|gb|AAM49877.1 LD13350p {Dr... 67 5e-12
TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protei... 66 9e-12
TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein... 66 1e-11
TC91597 similar to GP|21323350|dbj|BAB97978. Hypothetical protei... 64 3e-11
TC93295 weakly similar to GP|5821157|dbj|BAA83720.1 GAMP {Procam... 64 5e-11
TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1 ... 62 1e-10
TC80941 weakly similar to PIR|T39903|T39903 serine-rich protein ... 62 1e-10
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 62 2e-10
TC77946 homologue to GP|10334491|emb|CAC10207. putative splicing... 62 2e-10
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 135 bits (341), Expect = 9e-33
Identities = 78/207 (37%), Positives = 130/207 (62%)
Frame = +3
Query: 13 KEEKTDLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEKDLEDKGEEKKK 72
K E++++ K +K + KE+K + ++D + K VE + EKKEK ++K ++
Sbjct: 63 KMEESNIPIKIDDKSAGDVKEDKVEIEKDL-----EIKSVEKDDEKKEKKDKEKKDKTDV 227
Query: 73 KKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDEGKDGEEKKEKE 132
+GKDK++K+K EK +E +E G+ KK+KEKKKKEKK+KG+ED+ KDGEEKK K+
Sbjct: 228 DEGKDKKDKEKKKKEKKEENVKGEEEDGDEKKDKEKKKKEKKEKGKEDKDKDGEEKKSKK 407
Query: 133 KKKKEKKDKGEEDEGKDGEEKKKDKEKKKEKKDDGKDKTKDVSKLKQKLEKMNGKIEALL 192
K+K+K ++DEG+DG +KKK+K+KK++KK++ + + VS +E+ + +
Sbjct: 408 DKEKKKDKNEDDDEGEDGSKKKKNKDKKEKKKEEDEKEEGKVSVRDIDIEETAKEGKEKK 587
Query: 193 QEKADIERQIKEAEVGSHVVNEKDKDE 219
++K D E + K+ + KDKD+
Sbjct: 588 KKKEDKEEKKKK--------SGKDKDQ 644
Score = 130 bits (327), Expect = 4e-31
Identities = 83/200 (41%), Positives = 120/200 (59%), Gaps = 3/200 (1%)
Frame = +3
Query: 23 SVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEKDLEDKGEEKKKKKGKDKEEKD 82
+V + +S K D+ + K+ + E+E + E K + +D+ +EKK K E+KD
Sbjct: 51 TVSSKMEESNIPIKIDDKSAGDVKEDKVEIEKDLEIKSVEKDDEKKEKKDK-----EKKD 215
Query: 83 KTDAEKGKEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDEGKDGEEKKEKEKKKKEKKDKG 142
KTD ++GK+ KK+KEKKKKEKK++ + E +DG+EKK+KEKKKKEKK+KG
Sbjct: 216 KTDVDEGKD-----------KKDKEKKKKEKKEENVKGEEEDGDEKKDKEKKKKEKKEKG 362
Query: 143 EEDEGKDGEEK--KKDKEKKKEKKDDGKDKTKDVSKLKQKLEKMNGKIEALLQEKADIE- 199
+ED+ KDGEEK KKDKEKKK+K +D D+ +D SK K+ +K K E +E+ +
Sbjct: 363 KEDKDKDGEEKKSKKDKEKKKDKNED-DDEGEDGSKKKKNKDKKEKKKEEDEKEEGKVSV 539
Query: 200 RQIKEAEVGSHVVNEKDKDE 219
R I E +K K E
Sbjct: 540 RDIDIEETAKEGKEKKKKKE 599
Score = 126 bits (316), Expect = 7e-30
Identities = 87/196 (44%), Positives = 126/196 (63%), Gaps = 26/196 (13%)
Frame = +3
Query: 1 MEDSR-QIKVED-----IKEEKT----DLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEK 50
ME+S IK++D +KE+K DL+ KSVEK+ K KE+K K+ +DK++ + +
Sbjct: 66 MEESNIPIKIDDKSAGDVKEDKVEIEKDLEIKSVEKDDEK-KEKKDKEKKDKTDVDEGKD 242
Query: 51 EVEGEGEKKEKDLED-KGEEKKKKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKK 109
+ + E +KKEK E+ KGEE+ + KDKE+K K EKGKE D++ GE KK K+ K
Sbjct: 243 KKDKEKKKKEKKEENVKGEEEDGDEKKDKEKKKKEKKEKGKE---DKDKDGEEKKSKKDK 413
Query: 110 KKEKKDKGE-EDEGKDGEEKKEKEKKKKEKKDKGEEDEG-------------KDGEEKKK 155
+K KKDK E +DEG+DG +KK+ + KK++KK++ E++EG K+G+EKKK
Sbjct: 414 EK-KKDKNEDDDEGEDGSKKKKNKDKKEKKKEEDEKEEGKVSVRDIDIEETAKEGKEKKK 590
Query: 156 DKEKKKE-KKDDGKDK 170
KE K+E KK GKDK
Sbjct: 591 KKEDKEEKKKKSGKDK 638
>TC83625 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (9%)
Length = 908
Score = 129 bits (325), Expect = 7e-31
Identities = 75/133 (56%), Positives = 95/133 (71%), Gaps = 4/133 (3%)
Frame = +2
Query: 38 KDEDKSNKKKKEKEVEGEGEKKEKDLEDKGEEK--KKKKGKDKEEKDKTDAEKGKEKEDD 95
K +DKS KE +VE EKDLE K EK +KK+ KDKE+KDKTD ++GK+K+D
Sbjct: 482 KIDDKSAGDVKEDKVE-----IEKDLEIKSVEKDDEKKEKKDKEKKDKTDVDEGKDKKD- 643
Query: 96 QEAGGEVKKEKEKKKKEKKDKGEEDEGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEK-- 153
KEKKKKEKK++ + E +DG+EKK+KEKKKKEKK+KG+ED+ KDGEEK
Sbjct: 644 ----------KEKKKKEKKEENVKGEEEDGDEKKDKEKKKKEKKEKGKEDKDKDGEEKKS 793
Query: 154 KKDKEKKKEKKDD 166
KKDKEKKK+K +D
Sbjct: 794 KKDKEKKKDKNED 832
Score = 109 bits (272), Expect = 9e-25
Identities = 60/135 (44%), Positives = 91/135 (66%)
Frame = +2
Query: 13 KEEKTDLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEKDLEDKGEEKKK 72
K E++++ K +K + KE+K + ++D + K VE + EKKEK ++K ++
Sbjct: 455 KMEESNIPIKIDDKSAGDVKEDKVEIEKDL-----EIKSVEKDDEKKEKKDKEKKDKTDV 619
Query: 73 KKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDEGKDGEEKKEKE 132
+GKDK++K+K EK +E +E G+ KK+KEKKKKEKK+KG+ED+ KDGEEKK K
Sbjct: 620 DEGKDKKDKEKKKKEKKEENVKGEEEDGDEKKDKEKKKKEKKEKGKEDKDKDGEEKKSK- 796
Query: 133 KKKKEKKDKGEEDEG 147
K K++KKDK E+D+G
Sbjct: 797 KDKEKKKDKNEDDDG 841
Score = 93.6 bits (231), Expect = 5e-20
Identities = 60/139 (43%), Positives = 84/139 (60%), Gaps = 2/139 (1%)
Frame = +2
Query: 51 EVEGEGEKKEKDLEDKGEEKKKKKGK-DKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKK 109
++ + +E ++ K ++K K DK E +K K EK+D++ KEKK
Sbjct: 437 DITVSSKMEESNIPIKIDDKSAGDVKEDKVEIEKDLEIKSVEKDDEK---------KEKK 589
Query: 110 KKEKKDKGEEDEGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEKKKEKKDDGK- 168
KEKKDK + DEGKD KK+KEKKKKEKK++ + E +DG+EKK ++KKKEKK+ GK
Sbjct: 590 DKEKKDKTDVDEGKD---KKDKEKKKKEKKEENVKGEEEDGDEKKDKEKKKKEKKEKGKE 760
Query: 169 DKTKDVSKLKQKLEKMNGK 187
DK KD + K K +K K
Sbjct: 761 DKDKDGEEKKSKKDKEKKK 817
Score = 82.4 bits (202), Expect = 1e-16
Identities = 57/133 (42%), Positives = 83/133 (61%), Gaps = 11/133 (8%)
Frame = +2
Query: 1 MEDSR-QIKVED-----IKEEKT----DLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEK 50
ME+S IK++D +KE+K DL+ KSVEK+ K KE+K K+ +DK++ + +
Sbjct: 458 MEESNIPIKIDDKSAGDVKEDKVEIEKDLEIKSVEKDDEK-KEKKDKEKKDKTDVDEGKD 634
Query: 51 EVEGEGEKKEKDLED-KGEEKKKKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKK 109
+ + E +KKEK E+ KGEE+ + KDKE+K K EKGKE +D + KK+KEK
Sbjct: 635 KKDKEKKKKEKKEENVKGEEEDGDEKKDKEKKKKEKKEKGKEDKDKDGEEKKSKKDKEK- 811
Query: 110 KKEKKDKGEEDEG 122
KKDK E+D+G
Sbjct: 812 ---KKDKNEDDDG 841
Score = 70.9 bits (172), Expect = 4e-13
Identities = 46/125 (36%), Positives = 75/125 (59%)
Frame = +2
Query: 95 DQEAGGEVKKEKEKKKKEKKDKGEEDEGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKK 154
D ++ G+VK++K + +K+ + K E KD E+K EKK KEKKDK + DEGKD ++K+
Sbjct: 488 DDKSAGDVKEDKVEIEKDLEIKSVE---KDDEKK---EKKDKEKKDKTDVDEGKDKKDKE 649
Query: 155 KDKEKKKEKKDDGKDKTKDVSKLKQKLEKMNGKIEALLQEKADIERQIKEAEVGSHVVNE 214
K K++KKE+ G+++ D K K+K +K E + K D ++ +E + +
Sbjct: 650 KKKKEKKEENVKGEEEDGDEKKDKEKKKK-----EKKEKGKEDKDKDGEEKKSKKDKEKK 814
Query: 215 KDKDE 219
KDK+E
Sbjct: 815 KDKNE 829
Score = 26.9 bits (58), Expect = 6.2
Identities = 12/77 (15%), Positives = 45/77 (57%), Gaps = 5/77 (6%)
Frame = +3
Query: 103 KKEKEKKKKEKKDKGEEDEGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEKK-- 160
+++K+++K+ + K + + +++++++++EKK K + ++ + ++ K +
Sbjct: 651 RRKKKRRKRMLRVKKKMVMKRRIRRRRKRKRRRREKKTKTRMVKRRNLRKTRRRKRTRMK 830
Query: 161 ---KEKKDDGKDKTKDV 174
+++K++GK +D+
Sbjct: 831 TMMEDEKEEGKVSVRDI 881
>TC87387 homologue to PIR|T47775|T47775 hypothetical protein F24I3.230 -
Arabidopsis thaliana, partial (14%)
Length = 1074
Score = 99.4 bits (246), Expect = 1e-21
Identities = 63/184 (34%), Positives = 99/184 (53%), Gaps = 20/184 (10%)
Frame = +3
Query: 10 EDIKEEKTDLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEK-------- 61
ED+K+E D + + V+K+ + ++ KK + D KK K E +GE+K K
Sbjct: 186 EDVKKEVADEEGEKVKKDDGEGRKRKKHESADSPAPAKKSKVAEVDGEEKVKTKKVDDAA 365
Query: 62 -DLEDKGEEKKKKKGKDKE------EKDKTDAEKGKE-KEDDQEAGGEVKKEKEKKKKEK 113
++ED +EKKKKK KDKE +++K + EK K+ KE ++ EV+K +KKKK K
Sbjct: 366 VEVEDDKKEKKKKKKKDKENGAAASDEEKVEKEKKKKHKEKGEDGSPEVEKSDKKKKKHK 545
Query: 114 KDKGEEDEGKDGEEKKEKEKKKKEKKDKGEEDEGKD----GEEKKKDKEKKKEKKDDGKD 169
+ + G + +K +KKK+KKDK +D D +E D+ +KK KK KD
Sbjct: 546 ------ETSEVGSPEVDKSEKKKKKKDKEAKDNAADISNGNDESNADRSEKKHKKKKNKD 707
Query: 170 KTKD 173
++
Sbjct: 708 AQEE 719
Score = 68.6 bits (166), Expect = 2e-12
Identities = 50/130 (38%), Positives = 68/130 (51%), Gaps = 13/130 (10%)
Frame = +3
Query: 3 DSRQIKVEDIKEEKTDLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEK---EVEGEGEKK 59
D ++VED K+EK K K E + S EEK +K++ K +K+K E EVE +KK
Sbjct: 354 DDAAVEVEDDKKEKKKKKKKDKENGAAASDEEKVEKEKKKKHKEKGEDGSPEVEKSDKKK 533
Query: 60 EKDLE---------DKGEEKKKKKGKDKEEKDKT-DAEKGKEKEDDQEAGGEVKKEKEKK 109
+K E DK E+KKKKK DKE KD D G ++ + + EK+ K
Sbjct: 534 KKHKETSEVGSPEVDKSEKKKKKK--DKEAKDNAADISNGNDESNAD------RSEKKHK 689
Query: 110 KKEKKDKGEE 119
KK+ KD EE
Sbjct: 690 KKKNKDAQEE 719
Score = 67.8 bits (164), Expect = 3e-12
Identities = 58/178 (32%), Positives = 94/178 (52%), Gaps = 5/178 (2%)
Frame = +3
Query: 48 KEKEVEGEGEKKEKDLEDKGEEKKKKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKE 107
K++ + EGEK +KD GE +K+KK + + A+K K E D E E
Sbjct: 195 KKEVADEEGEKVKKD---DGEGRKRKKHESADSP--APAKKSKVAEVDGE---------E 332
Query: 108 KKKKEKKDKGEEDEGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEKKKEKKDDG 167
K K +K D D + E+ K KEKKKK+KKDK E+ +E+K +KEKKK+ K+ G
Sbjct: 333 KVKTKKVD----DAAVEVEDDK-KEKKKKKKKDK--ENGAAASDEEKVEKEKKKKHKEKG 491
Query: 168 KDKTKDVSK--LKQKLEKMNGKIEALLQEKADIERQIKEAEV---GSHVVNEKDKDEA 220
+D + +V K K+K K ++ + +K++ +++ K+ E + + N D+ A
Sbjct: 492 EDGSPEVEKSDKKKKKHKETSEVGSPEVDKSEKKKKKKDKEAKDNAADISNGNDESNA 665
>TC85290 similar to GP|7767653|gb|AAF69150.1| F27F5.2 {Arabidopsis
thaliana}, partial (32%)
Length = 1558
Score = 84.0 bits (206), Expect = 4e-17
Identities = 47/165 (28%), Positives = 94/165 (56%), Gaps = 11/165 (6%)
Frame = +3
Query: 14 EEKTDLKNKSVEKE---------SPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEKDLE 64
+E + N+S KE K+KE+++K++E+K+ KKEKE E + ++KEK+ +
Sbjct: 651 QEYISIGNESYSKEIFEEYITYLKEKAKEKERKREEEKA---KKEKEREEKEKRKEKEKK 821
Query: 65 DKGEEKKKKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKEKK--DKGEEDEG 122
+K E++K+K K++ +KD++D++ ++ D E KKEK+K++K ++ +D
Sbjct: 822 EKEREREKEKSKERHKKDESDSD-NQDMTDGHGYREEKKKEKDKERKHRRRHQSSMDDVD 998
Query: 123 KDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEKKKEKKDDG 167
+ +EK+E K ++ D+ + + + E + K+ K+D G
Sbjct: 999 SEKDEKEESRKSRRHGSDRKKSRKHANSPESDNESRHKRHKRDHG 1133
Score = 72.8 bits (177), Expect = 1e-13
Identities = 41/120 (34%), Positives = 72/120 (59%), Gaps = 4/120 (3%)
Frame = +3
Query: 57 EKKEKDLEDKGEEKKKKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKEKKDK 116
++K K+ E K EE+K KK K++EEK+K ++ KEKE ++E +K KE+ KK++ D
Sbjct: 720 KEKAKEKERKREEEKAKKEKEREEKEKRKEKEKKEKEREREK----EKSKERHKKDESDS 887
Query: 117 GEED----EGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEKKKEKKDDGKDKTK 172
+D G E+KKEK+K++K + + + D + +K +KE+ ++ + G D+ K
Sbjct: 888 DNQDMTDGHGYREEKKKEKDKERKHR--RRHQSSMDDVDSEKDEKEESRKSRRHGSDRKK 1061
Score = 72.4 bits (176), Expect = 1e-13
Identities = 38/147 (25%), Positives = 77/147 (51%), Gaps = 1/147 (0%)
Frame = +3
Query: 13 KEEKTDLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEKDLED-KGEEKK 71
+EE+ K K E++ + ++EKK+K+ ++ +K KE+ + E + +D+ D G ++
Sbjct: 750 REEEKAKKEKEREEKEKRKEKEKKEKEREREKEKSKERHKKDESDSDNQDMTDGHGYREE 929
Query: 72 KKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDEGKDGEEKKEK 131
KKK KDKE K + + + DD ++ + K+E K ++ D+ + + + E +
Sbjct: 930 KKKEKDKERKHR---RRHQSSMDDVDSEKDEKEESRKSRRHGSDRKKSRKHANSPESDNE 1100
Query: 132 EKKKKEKKDKGEEDEGKDGEEKKKDKE 158
+ K+ K+D G+ E+ +D E
Sbjct: 1101SRHKRHKRDHGDGSRRSGWNEELEDGE 1181
Score = 62.4 bits (150), Expect = 1e-10
Identities = 53/179 (29%), Positives = 95/179 (52%), Gaps = 13/179 (7%)
Frame = +3
Query: 18 DLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEKDLEDKGEEKKKKKGKD 77
+L ++ EKE ++K+ ++ D D +N K++ +E + ++ G +
Sbjct: 501 ELLERAKEKEEKEAKKRQRLAD-DFTNLLYTLKDIITSSTWEECKALFEDTQEYISIGNE 677
Query: 78 KEEKDKTDA------EKGKEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDEGKDGEEKKEK 131
K+ + EK KEKE +E E K +KEK ++E+K+K +E E K+ E ++EK
Sbjct: 678 SYSKEIFEEYITYLKEKAKEKERKRE---EEKAKKEK-EREEKEKRKEKEKKEKEREREK 845
Query: 132 EK-KKKEKKDKGEED--EGKDG----EEKKKDKEKKKEKKDDGKDKTKDVSKLKQKLEK 183
EK K++ KKD+ + D + DG EEKKK+K+K+++ + + DV K + E+
Sbjct: 846 EKSKERHKKDESDSDNQDMTDGHGYREEKKKEKDKERKHRRRHQSSMDDVDSEKDEKEE 1022
Score = 54.3 bits (129), Expect = 4e-08
Identities = 62/235 (26%), Positives = 111/235 (46%), Gaps = 17/235 (7%)
Frame = +3
Query: 2 EDSRQIKVEDIKEEKTDLKNKSVEKE--SPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKK 59
ED IK + +K K + SV ++ S S+E K + + K E+ +E EK+
Sbjct: 354 EDKTLIK-DILKSGKITVATTSVFEDFKSAVSEEATCKTISEINLKLLFEELLERAKEKE 530
Query: 60 EKDLEDKGEEKKKKKGKD---------KEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKK 110
EK E KK+++ D K+ + E+ K +D + + E K+
Sbjct: 531 EK------EAKKRQRLADDFTNLLYTLKDIITSSTWEECKALFEDTQEYISIGNESYSKE 692
Query: 111 KEK------KDKGEEDEGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEKKKEKK 164
+ K+K +E E K EEK +KEK+++EK+ + E+++ + E++K+K K++ KK
Sbjct: 693 IFEEYITYLKEKAKEKERKREEEKAKKEKEREEKEKRKEKEKKEKEREREKEKSKERHKK 872
Query: 165 DDGKDKTKDVSKLKQKLEKMNGKIEALLQEKADIERQIKEAEVGSHVVNEKDKDE 219
D+ +D++ +G E +EK D ER+ + S + +KDE
Sbjct: 873 DESDSDNQDMT-------DGHGYREEKKKEK-DKERKHRRRHQSSMDDVDSEKDE 1013
Score = 52.8 bits (125), Expect = 1e-07
Identities = 37/109 (33%), Positives = 59/109 (53%), Gaps = 5/109 (4%)
Frame = +3
Query: 101 EVKKEKEKKKKEKKDKGEEDEGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEKK 160
E KEKE+K++E+K K E++ E+KEK K+KEKK+K E E +EK K++ KK
Sbjct: 723 EKAKEKERKREEEKAKKEKER-----EEKEKR-KEKEKKEKERERE----KEKSKERHKK 872
Query: 161 KEKKDDGKDKT-----KDVSKLKQKLEKMNGKIEALLQEKADIERQIKE 204
E D +D T ++ K ++ E+ + + + D E+ KE
Sbjct: 873 DESDSDNQDMTDGHGYREEKKKEKDKERKHRRRHQSSMDDVDSEKDEKE 1019
>AL367145 weakly similar to GP|8096269|dbj| KED {Nicotiana tabacum}, partial
(8%)
Length = 284
Score = 82.8 bits (203), Expect = 9e-17
Identities = 44/70 (62%), Positives = 55/70 (77%), Gaps = 2/70 (2%)
Frame = -3
Query: 153 KKKDKEKKKEKKDD--GKDKTKDVSKLKQKLEKMNGKIEALLQEKADIERQIKEAEVGSH 210
+KK K E+++ GKDK+ DVSKLKQKLEK+NGKIEALL++KADIE+QI+EAE +
Sbjct: 252 RKKKTRTKVERRES*LGKDKSNDVSKLKQKLEKINGKIEALLEQKADIEKQIREAEDEGY 73
Query: 211 VVNEKDKDEA 220
VV EK KD A
Sbjct: 72 VVVEKAKDAA 43
Score = 28.5 bits (62), Expect = 2.1
Identities = 11/17 (64%), Positives = 15/17 (87%)
Frame = -2
Query: 32 KEEKKKKDEDKSNKKKK 48
KE+KKK+D+DK KK+K
Sbjct: 262 KEKKKKEDKDKGGKKRK 212
Score = 27.7 bits (60), Expect = 3.6
Identities = 12/15 (80%), Positives = 13/15 (86%)
Frame = -2
Query: 128 KKEKEKKKKEKKDKG 142
K+ KEKKKKE KDKG
Sbjct: 271 KEVKEKKKKEDKDKG 227
Score = 27.7 bits (60), Expect = 3.6
Identities = 12/15 (80%), Positives = 13/15 (86%)
Frame = -2
Query: 103 KKEKEKKKKEKKDKG 117
K+ KEKKKKE KDKG
Sbjct: 271 KEVKEKKKKEDKDKG 227
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 82.8 bits (203), Expect = 9e-17
Identities = 57/234 (24%), Positives = 124/234 (52%), Gaps = 14/234 (5%)
Frame = +1
Query: 1 MEDSRQIKVEDIKEEKTDLKN-KSVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKK 59
+E+ E K+E ++K+ + +++E+ ++K+E+K + E++ NK +++ + E E +K
Sbjct: 124 LENEENKDEEKSKQENEEIKDGEKIQQENEENKDEEKSQQENEENKDEEKSQQENELKKN 303
Query: 60 EKDLEDKGEEKKKKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEK-EKKKKEKKDKGE 118
E ++ GE ++K ++ EE +T++ K KE E+ + G + K++ E +++ K E
Sbjct: 304 EGGEKETGEITEEKSKQENEETSETNS-KDKENEESNQNGSDAKEQVGENHEQDSKQGTE 480
Query: 119 EDEGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEKK-----------KEKKDDG 167
E G +G EK+E + K K+D +++ +DGE+ + +E+ K +
Sbjct: 481 ETNGTEGGEKEEHD---KIKEDTSSDNQVQDGEKNNEAREENYSGDNASSAVVDNKSQES 651
Query: 168 KDKTKDVSKLKQKLEKMNGKIEALLQEKADIERQIKEAEVGSHVV-NEKDKDEA 220
+KT++ K+K E E Q+ ++ + ++ + + NE +KD+A
Sbjct: 652 SNKTEEQFDKKEKNE-----FELESQKNSNETTESTDSTITQNSQGNESEKDQA 798
Score = 75.1 bits (183), Expect = 2e-14
Identities = 40/139 (28%), Positives = 83/139 (58%), Gaps = 3/139 (2%)
Frame = +1
Query: 40 EDKSNKKKKEKEVEGEGEKKEKDLEDKGEEKKKKKGKDKEEKDKTDAEKGKEKEDDQEAG 99
E+KS+ + +E + GE +++ LE+ E+ K+ G+ EEK + E+ K++E ++
Sbjct: 1 EEKSHLENEESKETGESSEEKSHLEN---EESKETGESSEEKSHLENEENKDEEKSKQEN 171
Query: 100 GEVKKEKEKKKKEKKDKGEEDEGKDGEEKKEKEKKKKE---KKDKGEEDEGKDGEEKKKD 156
E+K ++ +++ +++K EE ++ EE K++EK ++E KK++G E E + E+K
Sbjct: 172 EEIKDGEKIQQENEENKDEEKSQQENEENKDEEKSQQENELKKNEGGEKETGEITEEKSK 351
Query: 157 KEKKKEKKDDGKDKTKDVS 175
+E ++ + + KDK + S
Sbjct: 352 QENEETSETNSKDKENEES 408
Score = 67.0 bits (162), Expect = 5e-12
Identities = 51/211 (24%), Positives = 98/211 (46%), Gaps = 27/211 (12%)
Frame = +1
Query: 2 EDSRQIKVEDIKEEKTDLK-NKSVEKESPKSKEEKKKKDED---KSNKKKKEKEVEGE-- 55
+++ + K E+ +++ +LK N+ EKE+ + EEK K++ + ++N K KE E +
Sbjct: 241 QENEENKDEEKSQQENELKKNEGGEKETGEITEEKSKQENEETSETNSKDKENEESNQNG 420
Query: 56 -------GEKKEKDLEDKGEEKKKKKGKDKEEKDK-----------TDAEKGKEKEDDQE 97
GE E+D + EE +G +KEE DK D EK E ++
Sbjct: 421 SDAKEQVGENHEQDSKQGTEETNGTEGGEKEEHDKIKEDTSSDNQVQDGEKNNEAREENY 600
Query: 98 AGGEVKKE-KEKKKKEKKDKGEE--DEGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKK 154
+G + K +E +K EE D+ + E + E +K E + + ++ + +
Sbjct: 601 SGDNASSAVVDNKSQESSNKTEEQFDKKEKNEFELESQKNSNETTESTDSTITQNSQGNE 780
Query: 155 KDKEKKKEKKDDGKDKTKDVSKLKQKLEKMN 185
+K++ + + D K + + KQ+ E+ N
Sbjct: 781 SEKDQAQTENDTPKGSASESDEQKQEQEQNN 873
Score = 43.5 bits (101), Expect = 6e-05
Identities = 33/187 (17%), Positives = 64/187 (33%), Gaps = 13/187 (6%)
Frame = +1
Query: 2 EDSRQIKVEDIKEEKTDLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEK 61
+D+ KV+ T + K E S +SK E + D S + + K
Sbjct: 1198 KDAASNKVQLTTTSDTSSEQKKDESSSAESKSESSQNDNANSGQSNTTSDESANDNKDSS 1377
Query: 62 DLEDKGEEKKKKKGKDKEEKDKT--DAEKGKEKEDDQEAGGEVKKEKEKKKKEKKDKGEE 119
+ E + + D+ D++ + D + + + + + K E
Sbjct: 1378 QVTTSSENSAEGNSNTENNSDENQNDSKNNENTNDSGNTSNDANVNENQNENAAQTKTSE 1557
Query: 120 DEGKDGEEKKEKEKKKKEK-----------KDKGEEDEGKDGEEKKKDKEKKKEKKDDGK 168
+EG E E +K+ E D+G D ++K+ + + +G+
Sbjct: 1558 NEGDAQNESVESKKENNESAHKDVDNNSNSNDQGSSDTSVTQDDKESRVDLGTLPESNGE 1737
Query: 169 DKTKDVS 175
DVS
Sbjct: 1738 SHHNDVS 1758
Score = 42.7 bits (99), Expect = 1e-04
Identities = 39/199 (19%), Positives = 70/199 (34%), Gaps = 33/199 (16%)
Frame = +1
Query: 8 KVEDIKEEKTDLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGE------------ 55
K + +EE N S KS+E K +E K+K E E+E +
Sbjct: 571 KNNEAREENYSGDNASSAVVDNKSQESSNKTEEQFDKKEKNEFELESQKNSNETTESTDS 750
Query: 56 -------GEKKEKDLEDKGEEKKKKKGKDKEEKDKTDAEKGKEKEDDQ-----EAGGEVK 103
G + EKD + K + +E+ + + K+D Q G
Sbjct: 751 TITQNSQGNESEKDQAQTENDTPKGSASESDEQKQEQEQNNTTKDDVQTTDTSSQNGNDT 930
Query: 104 KEKEKKKKEKKDKGEEDEGKDGEEKKEKEKKKKEKKDK---------GEEDEGKDGEEKK 154
EK+ + E + +ED ++ K D+ E +G +
Sbjct: 931 TEKQNETSEDANSKKEDSSALNTTPNNEDSKSGVAGDQADSTTTTSSSETQDGNTNHGEY 1110
Query: 155 KDKEKKKEKKDDGKDKTKD 173
KD + +K+ G++ T++
Sbjct: 1111KDTTNENPEKNSGQEGTQE 1167
>AJ500818 weakly similar to GP|10435304|dbj unnamed protein product {Homo
sapiens}, partial (12%)
Length = 457
Score = 71.2 bits (173), Expect = 3e-13
Identities = 32/108 (29%), Positives = 72/108 (66%)
Frame = -3
Query: 56 GEKKEKDLEDKGEEKKKKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKEKKD 115
G K++ + E G ++ ++K +++K EK +E++ +E E K+++EK+++EK+
Sbjct: 326 GYKRDAEAEAYGYKRDADPVEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQ 147
Query: 116 KGEEDEGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEKKKEK 163
+ + E K EEK+++EK+++EK+ + + E K EEK++++++++EK
Sbjct: 146 EEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRLEEKRQEEKRQEEK 3
Score = 68.6 bits (166), Expect = 2e-12
Identities = 36/113 (31%), Positives = 75/113 (65%), Gaps = 1/113 (0%)
Frame = -3
Query: 47 KKEKEVEGEGEKKEKD-LEDKGEEKKKKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKE 105
K++ E E G K++ D +E+K +E+K+++ K +EEK + E++ +E E K++
Sbjct: 320 KRDAEAEAYGYKRDADPVEEKRQEEKRQEEKRQEEKRQ-------EEKRQEEKRQEEKRQ 162
Query: 106 KEKKKKEKKDKGEEDEGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKE 158
+EK+++EK+ + + E K EEK+++EK+++EK+ + + E K EEK+++++
Sbjct: 161 EEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRLEEKRQEEKRQEEK 3
Score = 61.6 bits (148), Expect = 2e-10
Identities = 35/132 (26%), Positives = 78/132 (58%), Gaps = 4/132 (3%)
Frame = -3
Query: 56 GEKKEKDLEDKGEEKKKKKGKDKEEKD----KTDAEKGKEKEDDQEAGGEVKKEKEKKKK 111
G K++ + + G ++ K E + K DA+ +EK ++ K+++EK+++
Sbjct: 380 GYKRDAEADAYGYKRDAYGYKRDAEAEAYGYKRDADPVEEKRQEE------KRQEEKRQE 219
Query: 112 EKKDKGEEDEGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEKKKEKKDDGKDKT 171
EK+ + + E K EEK+++EK+++EK+ + + E K EEK++++++++EK+ + K
Sbjct: 218 EKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRLE 39
Query: 172 KDVSKLKQKLEK 183
+ + K++ EK
Sbjct: 38 EKRQEEKRQEEK 3
Score = 57.4 bits (137), Expect = 4e-09
Identities = 33/109 (30%), Positives = 66/109 (60%)
Frame = -3
Query: 30 KSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEKDLEDKGEEKKKKKGKDKEEKDKTDAEKG 89
K +EEK+++++ + K+++EK E EK E+K +E+K+++ K +EEK +
Sbjct: 260 KRQEEKRQEEKRQEEKRQEEKRQE------EKRQEEKRQEEKRQEEKRQEEKRQ------ 117
Query: 90 KEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDEGKDGEEKKEKEKKKKEK 138
E K+++EK+++EK+ + + E K EEK+++EK+++EK
Sbjct: 116 -----------EEKRQEEKRQEEKRQEEKRQEEKRLEEKRQEEKRQEEK 3
Score = 52.0 bits (123), Expect = 2e-07
Identities = 28/104 (26%), Positives = 65/104 (61%)
Frame = -3
Query: 10 EDIKEEKTDLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEKDLEDKGEE 69
+ ++E++ + K + +++ K +EEK+++++ + K+++EK E EK E+K +E
Sbjct: 275 DPVEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQE------EKRQEEKRQE 114
Query: 70 KKKKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKEK 113
+K+++ K +E EK +E++ +E E K+++EK+++EK
Sbjct: 113 EKRQEEKRQE-------EKRQEEKRQEEKRLEEKRQEEKRQEEK 3
Score = 48.9 bits (115), Expect = 2e-06
Identities = 26/94 (27%), Positives = 61/94 (64%), Gaps = 3/94 (3%)
Frame = -3
Query: 3 DSRQIKVEDIKEEKTDLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEKD 62
D+ ++ + +E++ + K + +++ K +EEK+++++ + K+++EK E E ++EK
Sbjct: 281 DADPVEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQE-EKRQEEKR 105
Query: 63 LEDKGEEKKKKKGKDKEEK---DKTDAEKGKEKE 93
E+K +E+K+++ K +EEK +K EK +E++
Sbjct: 104 QEEKRQEEKRQEEKRQEEKRLEEKRQEEKRQEEK 3
>BQ752218 similar to GP|19170914|emb hypothetical protein {Encephalitozoon
cuniculi}, partial (5%)
Length = 637
Score = 70.5 bits (171), Expect = 5e-13
Identities = 47/108 (43%), Positives = 63/108 (57%), Gaps = 11/108 (10%)
Frame = -2
Query: 72 KKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDEGKDGEEKKEK 131
KK+ K KEEK K +AEK K+K+ +EA + KK +E K+ K++ +E E K EEK++
Sbjct: 633 KKRPKKKEEKAKKEAEK-KKKKAREEAAKKAKKAREAAYKKAKEEKKEAEKKAKEEKRQA 457
Query: 132 EKK---------KKEKKDKGEEDEGKDGEE--KKKDKEKKKEKKDDGK 168
EKK KK KKDK E+D KD E ++D E KE KD K
Sbjct: 456 EKKIKEEKKEAEKKAKKDKKEQDR-KDAESALSRQDTEDSKESKDGSK 316
Score = 68.9 bits (167), Expect = 1e-12
Identities = 42/111 (37%), Positives = 68/111 (60%)
Frame = -2
Query: 26 KESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEKDLEDKGEEKKKKKGKDKEEKDKTD 85
K+ PK KEEK KK+ +K KKKK +E + KK ++ K +++KK+ + K +++K
Sbjct: 633 KKRPKKKEEKAKKEAEK--KKKKAREEAAKKAKKAREAAYKKAKEEKKEAEKKAKEEKRQ 460
Query: 86 AEKGKEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDEGKDGEEKKEKEKKKK 136
AEK K KE+ +EA E K +K+KK++++KD +D E+ KE + K
Sbjct: 459 AEK-KIKEEKKEA--EKKAKKDKKEQDRKDAESALSRQDTEDSKESKDGSK 316
Score = 58.9 bits (141), Expect = 1e-09
Identities = 39/109 (35%), Positives = 63/109 (57%), Gaps = 4/109 (3%)
Frame = -2
Query: 8 KVEDIKEEKTDLKNKSVEKESPKSKEEK----KKKDEDKSNKKKKEKEVEGEGEKKEKDL 63
K E K+E K K+ E+ + K+K+ + KK E+K +KK KE + + EKK K
Sbjct: 615 KEEKAKKEAEKKKKKAREEAAKKAKKAREAAYKKAKEEKKEAEKKAKEEKRQAEKKIK-- 442
Query: 64 EDKGEEKKKKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKE 112
E+K E +KK K KDK+E+D+ DAE ++D +++ K+ K+ K +
Sbjct: 441 EEKKEAEKKAK-KDKKEQDRKDAESALSRQDTEDS----KESKDGSKPQ 310
Score = 44.7 bits (104), Expect = 3e-05
Identities = 39/118 (33%), Positives = 57/118 (48%), Gaps = 12/118 (10%)
Frame = -2
Query: 104 KEKEKKKKEKKDKGEEDEGKDGEEKKEKEKKKK-------EKKDKGEEDEGKDGEEKKKD 156
K++ KKK+EK KKE EKKKK +K K E K +E+KK+
Sbjct: 633 KKRPKKKEEKA-------------KKEAEKKKKKAREEAAKKAKKAREAAYKKAKEEKKE 493
Query: 157 KEKK-KEKKDDGKDKTKDVSKLKQKLEKMNGKIEALLQEKADIERQ----IKEAEVGS 209
EKK KE+K + K K+ K +K K + K + ++ + RQ KE++ GS
Sbjct: 492 AEKKAKEEKRQAEKKIKEEKKEAEKKAKKDKKEQDRKDAESALSRQDTEDSKESKDGS 319
Score = 39.7 bits (91), Expect = 0.001
Identities = 30/90 (33%), Positives = 47/90 (51%)
Frame = -2
Query: 130 EKEKKKKEKKDKGEEDEGKDGEEKKKDKEKKKEKKDDGKDKTKDVSKLKQKLEKMNGKIE 189
+K KKKE+K K K+ E+KKK ++ KK K ++ + K K EK + +
Sbjct: 633 KKRPKKKEEKAK------KEAEKKKKKAREEAAKK---AKKAREAAYKKAKEEKKEAEKK 481
Query: 190 ALLQEKADIERQIKEAEVGSHVVNEKDKDE 219
A +EK E++IKE + + +KDK E
Sbjct: 480 AK-EEKRQAEKKIKEEKKEAEKKAKKDKKE 394
Score = 35.4 bits (80), Expect = 0.017
Identities = 21/80 (26%), Positives = 44/80 (54%), Gaps = 2/80 (2%)
Frame = -1
Query: 75 GKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKE--KKDKGEEDEGKDGEEKKEKE 132
G+++ EK+ + +G KE+++ G K+ +E + + GEE K+G+ ++E
Sbjct: 637 GQEEAEKEGGEG*EGGRKEEEEGQGRSCKEGQEGQGGSL*EGQGGEEGGRKEGQGREEAG 458
Query: 133 KKKKEKKDKGEEDEGKDGEE 152
K+ + ++G EG++G E
Sbjct: 457 *KEDQGGEEGGRKEGQEG*E 398
>AW685995 similar to PIR|I51618|I516 nucleolar phosphoprotein - African
clawed frog, partial (5%)
Length = 516
Score = 69.7 bits (169), Expect = 8e-13
Identities = 43/138 (31%), Positives = 70/138 (50%), Gaps = 3/138 (2%)
Frame = +3
Query: 30 KSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEKDLEDKGEEKKKKKGKD---KEEKDKTDA 86
KSK KK K E+ KK EK + +G+K K E+K +++KKK+ K + E+D + +
Sbjct: 27 KSKTSKKVKAEEP---KKVEKVEKTKGKKAAKAEEEKKKQQKKKEEKPAPMEVEEDSSSS 197
Query: 87 EKGKEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDEGKDGEEKKEKEKKKKEKKDKGEEDE 146
E+ ED+ A +K KKE + E + E++EK KK K + +E
Sbjct: 198 EESSSSEDEAPAKKAAPAKKAAAKKESSSEEESSSSESESEEEEKPAKKAAAKKESSSEE 377
Query: 147 GKDGEEKKKDKEKKKEKK 164
E++ D+E+K E +
Sbjct: 378 ESSSSEEESDEEEKNEAR 431
Score = 66.6 bits (161), Expect = 7e-12
Identities = 42/151 (27%), Positives = 79/151 (51%)
Frame = +3
Query: 4 SRQIKVEDIKEEKTDLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEKDL 63
S+++K E+ K K + K+ K++ K++EEKKK + K +K EVE + E+
Sbjct: 39 SKKVKAEEPK--KVEKVEKTKGKKAAKAEEEKKK-QQKKKEEKPAPMEVEEDSSSSEESS 209
Query: 64 EDKGEEKKKKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDEGK 123
+ E KK K+ K ++ +E+ E+ E +++ KK KK+ E+E
Sbjct: 210 SSEDEAPAKKAAPAKKAAAKKES-SSEEESSSSESESEEEEKPAKKAAAKKESSSEEESS 386
Query: 124 DGEEKKEKEKKKKEKKDKGEEDEGKDGEEKK 154
EE+ + E++K E + + E++E + G E++
Sbjct: 387 SSEEESD-EEEKNEARRR*EQEEARRGGERR 476
Score = 44.3 bits (103), Expect = 4e-05
Identities = 37/126 (29%), Positives = 61/126 (48%), Gaps = 7/126 (5%)
Frame = +3
Query: 101 EVKKEKEKK-KKEKKDKGEEDEGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEK 159
+VK E+ KK +K +K KG++ K EEKK+++KKK+EK E +E E+ E
Sbjct: 45 KVKAEEPKKVEKVEKTKGKK-AAKAEEEKKKQQKKKEEKPAPMEVEEDSSSSEESSSSED 221
Query: 160 KKEKKDDGKDKTKDVSKLKQKLEKMNGKIEALLQE------KADIERQIKEAEVGSHVVN 213
+ K K K +K + E+ + E+ +E KA +++ E S
Sbjct: 222 EAPAKKAAPAK-KAAAKKESSSEEESSSSESESEEEEKPAKKAAAKKESSSEEESSSSEE 398
Query: 214 EKDKDE 219
E D++E
Sbjct: 399 ESDEEE 416
>TC89483 similar to GP|18377662|gb|AAL66981.1 unknown protein {Arabidopsis
thaliana}, partial (35%)
Length = 1711
Score = 69.3 bits (168), Expect = 1e-12
Identities = 43/178 (24%), Positives = 99/178 (55%), Gaps = 12/178 (6%)
Frame = +1
Query: 12 IKEEKTDLKNKSVEK---ESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEKDLEDKGE 68
++E +T K + E E+P + ++K ++ D+ + K+EKE E ++E + + +
Sbjct: 52 MRERQTGDKKPTSEHDRPETPDRRHDRKSRERDRERELKREKERVLERYEREAERDRIRK 231
Query: 69 EKKKKKGKDKEEKD---KTDAEKGKEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDEGKDG 125
E+++K+ ++ E+ + + +E+E ++E E +KEK++++K +K+ ++E DG
Sbjct: 232 EREQKRRIEEVERQFELQLKEWEYREREKEKERQYEKEKEKDRERKRRKEILYDEEDDDG 411
Query: 126 EEKKE------KEKKKKEKKDKGEEDEGKDGEEKKKDKEKKKEKKDDGKDKTKDVSKL 177
+ +K +EK+ K ++K ++ K EE++ + KK+ ++D + +D KL
Sbjct: 412 DSRKRWRXNAIEEKRNKRLREKEDDLADKQKEEEEIAEAKKRTEEDQQLKRQRDALKL 585
Score = 62.0 bits (149), Expect = 2e-10
Identities = 46/205 (22%), Positives = 102/205 (49%), Gaps = 29/205 (14%)
Frame = +1
Query: 42 KSNKKKKEKEVEGEGEKKEKDLEDKGE-----EKKKKKGKDKEEKDKTDAEKGKEKEDDQ 96
+ N+ K++ G+KK D+ E +K + +D+E + K + E+ E+ + +
Sbjct: 28 RMNRVKRKMRERQTGDKKPTSEHDRPETPDRRHDRKSRERDRERELKREKERVLERYERE 207
Query: 97 EAGGEVKKEKEKKKK---------------EKKDKGEEDEGKDGEEKKEKEKKKKEKKDK 141
++KE+E+K++ E +++ +E E + E++KEK++++K +K+
Sbjct: 208 AERDRIRKEREQKRRIEEVERQFELQLKEWEYREREKEKE-RQYEKEKEKDRERKRRKEI 384
Query: 142 GEEDEGKDGEEKK---------KDKEKKKEKKDDGKDKTKDVSKLKQKLEKMNGKIEALL 192
++E DG+ +K K ++ +EK+DD DK K+ ++ + K + + L
Sbjct: 385 LYDEEDDDGDSRKRWRXNAIEEKRNKRLREKEDDLADKQKEEEEIAE--AKKRTEEDQQL 558
Query: 193 QEKADIERQIKEAEVGSHVVNEKDK 217
+ + D + + E H+VN +D+
Sbjct: 559 KRQRDALKLLTE-----HIVNGRDQ 618
>TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~unknown
protein {Arabidopsis thaliana}, partial (46%)
Length = 2077
Score = 67.4 bits (163), Expect = 4e-12
Identities = 59/230 (25%), Positives = 109/230 (46%), Gaps = 11/230 (4%)
Frame = +2
Query: 2 EDSRQIKVEDIKEEKTDLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEK 61
E+ Q KV +K + K K VE E +S + ++ D+D + + EK+ G + E
Sbjct: 302 EEIAQFKVPLDVGKKLE-KKKRVELEEEESDDFDEELDDDDDDFEGVEKKKAKGGSEGED 478
Query: 62 DLEDKGEEKKKKKGKDKEEKDKTDAEKGKEKED-----DQEAGGEVKKEKEKKKKEKKDK 116
D E++ +E ++ + +E+D+ + KG ED D+ K+E +K E++D+
Sbjct: 479 DFEEEDDEDEEGSEDEDDEEDEKEKVKGGGIEDKFLKIDELTEYLEKEEDNYEKGEERDE 658
Query: 117 GEEDEGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEK-----KKEKKDDGKDKT 171
+ED +D E +K E + ++ D +++E ++ E+ + + K EK KD+
Sbjct: 659 ADEDSEEDDELEKAGEFEMDDEDDDDDDEEDEEAEDMGNVRYEDFFGGKNEKGSKRKDQL 838
Query: 172 KDVSKLKQKLEKMNGKIE-ALLQEKADIERQIKEAEVGSHVVNEKDKDEA 220
+VS + +E K A EK + IK++ G K+ D A
Sbjct: 839 LEVSGDEDDMESTKQKKRTASTYEKQRGKNSIKDSADGES*YRAKNLDYA 988
>TC92575 weakly similar to GP|21428434|gb|AAM49877.1 LD13350p {Drosophila
melanogaster}, partial (3%)
Length = 557
Score = 67.0 bits (162), Expect = 5e-12
Identities = 51/186 (27%), Positives = 98/186 (52%), Gaps = 4/186 (2%)
Frame = -1
Query: 32 KEEKKKKDEDKSNKKKKEKEVEGEGEKKEKDLEDKGEEKKKKKGK---DKEEKDKTDAEK 88
+EEKK + E+ K++K+V+ E +E +D G E K + KEE++ +A+K
Sbjct: 557 EEEKKAEGEEL----KEDKKVDAGDEIEEDKKDDNGFEDDKLSEETELSKEERESVEAKK 390
Query: 89 GKEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDEGKDGEEKKEKEKKKKEKKDKGEEDEGK 148
+ D+E+ E K E +K+K +KD+ ++ + D + +E K +K K +G +GK
Sbjct: 389 PELDAMDEESILEGKDEGSEKEKSQKDREDDKDKVDNKSNEENIKVEKRLKKRG---KGK 219
Query: 149 -DGEEKKKDKEKKKEKKDDGKDKTKDVSKLKQKLEKMNGKIEALLQEKADIERQIKEAEV 207
+GE KK ++ K+ +D + K + + K + + E++ K + + +++KEAE
Sbjct: 218 VNGENVKKKMKELKKIEDIPEAKDESIEKERSRDEEVGDKEKVGGENVKKKMKELKEAEP 39
Query: 208 GSHVVN 213
+ N
Sbjct: 38 TTPTTN 21
Score = 65.9 bits (159), Expect = 1e-11
Identities = 48/187 (25%), Positives = 95/187 (50%), Gaps = 9/187 (4%)
Frame = -1
Query: 2 EDSRQIKVEDIKEEKTDLKNKSVEK--------ESPKSKEEKKKKDEDKSNKKKKEKEVE 53
E+ ++ + E++KE+K +E+ E K EE + E++ + + K+ E++
Sbjct: 557 EEEKKAEGEELKEDKKVDAGDEIEEDKKDDNGFEDDKLSEETELSKEERESVEAKKPELD 378
Query: 54 GEGEKKEKDLEDKGEEKKKKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKEK 113
E+ + +D+G EK+K + +++KDK D + +E +K EK KK+ K
Sbjct: 377 AMDEESILEGKDEGSEKEKSQKDREDDKDKVDNKSNEE---------NIKVEKRLKKRGK 225
Query: 114 KDKGEEDEGK-DGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEKKKEKKDDGKDKTK 172
GK +GE K+K K+ K+ +D E + +E+ +D+E ++K G++ K
Sbjct: 224 --------GKVNGENVKKKMKELKKIEDIPEAKDESIEKERSRDEEVGDKEKVGGENVKK 69
Query: 173 DVSKLKQ 179
+ +LK+
Sbjct: 68 KMKELKE 48
Score = 45.8 bits (107), Expect = 1e-05
Identities = 37/118 (31%), Positives = 64/118 (53%), Gaps = 8/118 (6%)
Frame = -1
Query: 11 DIKEEKTDLKNK---SVEKESPKSKEEKKKKDEDKSNKK--KKEKEVEGEGEKKEKDLED 65
D +E++ L+ K S +++S K +E+ K K ++KSN++ K EK ++ G+ K
Sbjct: 380 DAMDEESILEGKDEGSEKEKSQKDREDDKDKVDNKSNEENIKVEKRLKKRGKGKVN---- 213
Query: 66 KGEEKKKKKGKDKEEKDKTDAEK---GKEKEDDQEAGGEVKKEKEKKKKEKKDKGEED 120
GE KKK + K+ +D +A+ KE+ D+E G + K E KK+ K+ E +
Sbjct: 212 -GENVKKKMKELKKIEDIPEAKDESIEKERSRDEEVGDKEKVGGENVKKKMKELKEAE 42
>TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protein {Oryza
sativa (japonica cultivar-group)}, partial (62%)
Length = 2345
Score = 66.2 bits (160), Expect = 9e-12
Identities = 46/174 (26%), Positives = 82/174 (46%), Gaps = 9/174 (5%)
Frame = +1
Query: 9 VEDIKEEKTDLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEK-DLEDKG 67
V ++ E ++KS KE + +E+ + D SN ++ E E K D
Sbjct: 424 VVPVQNEDVPQESKSEVKEQTEVREQVS--ETDNSNARQFEDNPGDLPEDATKGDSNVSS 597
Query: 68 EEKKKKKGKDKEEKDKTDAEKGKEKEDD------QEAGGEVK-KEKEKKKKEKKDKGEED 120
EEK ++ +K +D ++GK+ ED+ + G E KE E + EKKD+ EE+
Sbjct: 598 EEKSEENSTEKSSEDTKTEDEGKKTEDEGSNTENNKDGEEASTKESESDESEKKDESEEN 777
Query: 121 EGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEKKKEKKDD-GKDKTKD 173
D +E ++K E D E++ + + K+ D+ ++ DD KD++ +
Sbjct: 778 NKSDSDESEKKSSDSNETTDSNVEEKVEQSQNKESDENASEKNTDDNAKDQSSN 939
Score = 52.4 bits (124), Expect = 1e-07
Identities = 31/151 (20%), Positives = 68/151 (44%), Gaps = 12/151 (7%)
Frame = +1
Query: 59 KEKDLEDKGEEKKKKKGKDKEEKDKTDAEKGKEKED------------DQEAGGEVKKEK 106
+ +D+ + + + K++ + +E+ +TD ++ ED D E K E+
Sbjct: 436 QNEDVPQESKSEVKEQTEVREQVSETDNSNARQFEDNPGDLPEDATKGDSNVSSEEKSEE 615
Query: 107 EKKKKEKKDKGEEDEGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEKKKEKKDD 166
+K +D EDEGK E++ + K+ ++ ++ D E +KKD+ ++ K D
Sbjct: 616 NSTEKSSEDTKTEDEGKKTEDEGSNTENNKDGEEASTKESESD-ESEKKDESEENNKSDS 792
Query: 167 GKDKTKDVSKLKQKLEKMNGKIEALLQEKAD 197
+ + K + + K+E +++D
Sbjct: 793 DESEKKSSDSNETTDSNVEEKVEQSQNKESD 885
Score = 40.0 bits (92), Expect = 7e-04
Identities = 30/111 (27%), Positives = 51/111 (45%), Gaps = 16/111 (14%)
Frame = +1
Query: 2 EDSRQIKVEDIKEEKTDLK----------NKSVEKESPKSKE--EKKKKDEDKSNKK--- 46
E+S + ED K E K NK E+ S K E E +KKDE + N K
Sbjct: 613 ENSTEKSSEDTKTEDEGKKTEDEGSNTENNKDGEEASTKESESDESEKKDESEENNKSDS 792
Query: 47 -KKEKEVEGEGEKKEKDLEDKGEEKKKKKGKDKEEKDKTDAEKGKEKEDDQ 96
+ EK+ E + ++E+K E+ + K+ + + TD + K++ ++
Sbjct: 793 DESEKKSSDSNETTDSNVEEKVEQSQNKESDENASEKNTD-DNAKDQSSNE 942
>TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein T28I19.100
- Arabidopsis thaliana, partial (14%)
Length = 1460
Score = 65.9 bits (159), Expect = 1e-11
Identities = 44/186 (23%), Positives = 91/186 (48%), Gaps = 8/186 (4%)
Frame = +3
Query: 12 IKEEKTDLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEKDLEDKGEEKK 71
+K + DL VE + + EE+++ + N + K + E + E +E + + EE +
Sbjct: 339 LKLGRKDLHPGKVEADKNEGHEEEEEDEHIVYNMQNKREHDEQQQEGEEGNKHETEEESE 518
Query: 72 KKKGKDKEEKDKTDAEKGKEKEDDQEA--------GGEVKKEKEKKKKEKKDKGEEDEGK 123
+ +EE+D+ + + G E +++ E+ GG+V+ ++ +K + D EDE
Sbjct: 519 DNVHERREEQDEEENKHGAEVQEENESKSEEVEDEGGDVEIDENDHEKSEADNDREDEVV 698
Query: 124 DGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEKKKEKKDDGKDKTKDVSKLKQKLEK 183
D EEK ++E+ E +++ +EDE K G + + + +E+ D + V+ +
Sbjct: 699 D-EEKDKEEEGDDETENEDKEDEEKGGLVENHENHEAREEHYKADDASSAVAHDTHETST 875
Query: 184 MNGKIE 189
G +E
Sbjct: 876 ETGNLE 893
Score = 63.9 bits (154), Expect = 5e-11
Identities = 50/179 (27%), Positives = 87/179 (47%), Gaps = 8/179 (4%)
Frame = +3
Query: 30 KSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEKDLEDKGEEKKKKKGKDKEEKDK---TDA 86
K +KKK+ + K E + + KDL E K +G ++EE+D+ +
Sbjct: 261 KHNHDKKKEFDKNDTKLPIRTETDQILKLGRKDLHPGKVEADKNEGHEEEEEDEHIVYNM 440
Query: 87 EKGKEKEDDQEAG-----GEVKKEKEKKKKEKKDKGEEDEGKDGEEKKEKEKKKKEKKDK 141
+ +E ++ Q+ G E ++E E E++++ +E+E K G E +E+ + K E+
Sbjct: 441 QNKREHDEQQQEGEEGNKHETEEESEDNVHERREEQDEEENKHGAEVQEENESKSEE--- 611
Query: 142 GEEDEGKDGEEKKKDKEKKKEKKDDGKDKTKDVSKLKQKLEKMNGKIEALLQEKADIER 200
EDEG D E + D E K E +D +D+ D K K++ G E ++K D E+
Sbjct: 612 -VEDEGGDVEIDENDHE-KSEADNDREDEVVDEEKDKEE----EGDDETENEDKEDEEK 770
>TC91597 similar to GP|21323350|dbj|BAB97978. Hypothetical protein
{Corynebacterium glutamicum ATCC 13032}, partial (5%)
Length = 718
Score = 64.3 bits (155), Expect = 3e-11
Identities = 50/183 (27%), Positives = 91/183 (49%), Gaps = 7/183 (3%)
Frame = +2
Query: 10 EDIKEEKTDLKNKSVEKESP----KSKEEKKKKDEDKSNKKKKEKEVEGEGE---KKEKD 62
+D K K D+ ++ E P K +E +K E+K K +EK VE + + K K
Sbjct: 155 KDTKGRKVDVVAEAPELGPPAALTKETKESVEKQEEKPATKLEEKPVEKQEKPIKKPAKK 334
Query: 63 LEDKGEEKKKKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDEG 122
ED+ EK +K ++ EK + +K +K++D+ +++EKK EK +K +E
Sbjct: 335 QEDRPVEKHEKPVEEPIEKQEKPIKKPAKKQEDRPI-----EKQEKKPVEKHEKKPVEEP 499
Query: 123 KDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEKKKEKKDDGKDKTKDVSKLKQKLE 182
EKK EK +K K E+ + + E+++K+ ++ EK K + K ++ +E
Sbjct: 500 IAKSEKKSAEKLEKPIKKPVEKQDHRSVEKQEKNPVEEHEKTPVEKHEKKPAETQEKPVE 679
Query: 183 KMN 185
++
Sbjct: 680 NLS 688
>TC93295 weakly similar to GP|5821157|dbj|BAA83720.1 GAMP {Procambarus
clarkii}, partial (11%)
Length = 504
Score = 63.9 bits (154), Expect = 5e-11
Identities = 46/146 (31%), Positives = 75/146 (50%), Gaps = 7/146 (4%)
Frame = +3
Query: 33 EEKKKKDED---KSNKKKKEKEVEGEGEKKEKDLEDKGEEKKKKKGKDKEEKDKTDAEKG 89
E+ K D D ++++KK + GE + E + ++ K K ++ TD K
Sbjct: 18 EDAKPGDADAKTNTDERKKYEVKPGEVKTGEGTTDVNMQDAKSGDADAKTDERNTDEGKP 197
Query: 90 KEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDEGKDGEEK---KEKEKKKKEKKDK-GEED 145
E + D+ E K E+ K + D+ DE E K K E+K+KE K+K G+E
Sbjct: 198 DEGKTDEGKTDEAKPEEAKPDEANPDEANPDEANPNEIKLDVKMDERKEKEGKEKEGKEK 377
Query: 146 EGKDGEEKKKDKEKKKEKKDDGKDKT 171
EGK+ EEKK++ E+ +E+K++ D T
Sbjct: 378 EGKEKEEKKEE*EE*EERKENLND*T 455
Score = 59.7 bits (143), Expect = 9e-10
Identities = 45/144 (31%), Positives = 76/144 (52%), Gaps = 10/144 (6%)
Frame = +3
Query: 7 IKVEDIKEEKTDLKNKSVEKESPKSKEEKKKKDE---DKSNKKKKEKEVEGEGEKKEKDL 63
+ +ED K D K + E++ + K + K E D + + K + + + +++ D
Sbjct: 9 VNMEDAKPGDADAKTNTDERKKYEVKPGEVKTGEGTTDVNMQDAKSGDADAKTDERNTD- 185
Query: 64 EDKGEEKKKKKGKDKE---EKDKTDAEKGKEKEDDQEAGGEVK---KEKEKKKKEKKDK- 116
E K +E K +GK E E+ K D E D+ E+K K E+K+KE K+K
Sbjct: 186 EGKPDEGKTDEGKTDEAKPEEAKPDEANPDEANPDEANPNEIKLDVKMDERKEKEGKEKE 365
Query: 117 GEEDEGKDGEEKKEKEKKKKEKKD 140
G+E EGK+ EEKKE+ ++ +E+K+
Sbjct: 366 GKEKEGKEKEEKKEE*EE*EERKE 437
Score = 54.7 bits (130), Expect = 3e-08
Identities = 41/132 (31%), Positives = 66/132 (49%), Gaps = 8/132 (6%)
Frame = +3
Query: 60 EKDLEDKGEEKKK---KKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKEKKDK 116
+ D + +E+KK K G+ K + TD K D +A K ++ + K D+
Sbjct: 36 DADAKTNTDERKKYEVKPGEVKTGEGTTDVNMQDAKSGDADA----KTDERNTDEGKPDE 203
Query: 117 GEEDEGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEK---KKDKEKKKE-KKDDGKDKT- 171
G+ DEGK E K E+ K + D+ DE E K K D+ K+KE K+ +GK+K
Sbjct: 204 GKTDEGKTDEAKPEEAKPDEANPDEANPDEANPNEIKLDVKMDERKEKEGKEKEGKEKEG 383
Query: 172 KDVSKLKQKLEK 183
K+ + K++ E+
Sbjct: 384 KEKEEKKEE*EE 419
Score = 49.3 bits (116), Expect = 1e-06
Identities = 39/129 (30%), Positives = 63/129 (48%)
Frame = +3
Query: 2 EDSRQIKVEDIKEEKTDLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEK 61
E + + ++D K D K E+ + + K ++ K DE K+++ K E+ E E
Sbjct: 108 EGTTDVNMQDAKSGDADAKTD--ERNTDEGKPDEGKTDEGKTDEAKPEEAKPDEANPDEA 281
Query: 62 DLEDKGEEKKKKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDE 121
+ ++ + K K E K+K +GKEKE KEK+ KEK++K EE E
Sbjct: 282 NPDEANPNEIKLDVKMDERKEK----EGKEKEG-----------KEKEGKEKEEKKEE*E 416
Query: 122 GKDGEEKKE 130
+ EE+KE
Sbjct: 417 --E*EERKE 437
Score = 48.5 bits (114), Expect = 2e-06
Identities = 38/140 (27%), Positives = 72/140 (51%), Gaps = 7/140 (5%)
Frame = +1
Query: 12 IKEEKTDLKNKSVEKESPKSKEEKK------KKDEDKSNKKKKEKEVE-GEGEKKEKDLE 64
+ +++ +K K+++ K K K+ K +K+ KE+ E + KEK ++
Sbjct: 40 LMQKQIRMKEKNMK*NQVK*KRVKEQRM*ICKMQNQVMLMQKRMKEIRMKENQMKEKRMK 219
Query: 65 DKGEEKKKKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDEGKD 124
+K ++ ++K + +T ++ + K+ + K KEKK+KEKK KG+ K
Sbjct: 220 EKQMKQNRRK---QNRMKQTRTKQTRTKQTRMK*NWM*KWMKEKKRKEKKRKGK----KR 378
Query: 125 GEEKKEKEKKKKEKKDKGEE 144
E K++K KK K+ K KG++
Sbjct: 379 KERKRKKRKKNKKNKKKGKK 438
Score = 37.7 bits (86), Expect = 0.003
Identities = 28/122 (22%), Positives = 53/122 (42%), Gaps = 8/122 (6%)
Frame = +3
Query: 93 EDDQEAGGEVKKEKEKKKKEKKDKGEEDEGK--------DGEEKKEKEKKKKEKKDKGEE 144
ED + + K +++KK + GE G+ D + K + D+G+
Sbjct: 18 EDAKPGDADAKTNTDERKKYEVKPGEVKTGEGTTDVNMQDAKSGDADAKTDERNTDEGKP 197
Query: 145 DEGKDGEEKKKDKEKKKEKKDDGKDKTKDVSKLKQKLEKMNGKIEALLQEKADIERQIKE 204
DEGK E K + + ++ K D+ + + K++ K++ +EK E++ KE
Sbjct: 198 DEGKTDEGKTDEAKPEEAKPDEANPDEANPDEANPNEIKLDVKMDE-RKEKEGKEKEGKE 374
Query: 205 AE 206
E
Sbjct: 375 KE 380
>TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1
{Arabidopsis thaliana}, partial (47%)
Length = 1538
Score = 62.4 bits (150), Expect = 1e-10
Identities = 46/215 (21%), Positives = 101/215 (46%), Gaps = 2/215 (0%)
Frame = +3
Query: 8 KVEDIKEEKTDLKNKSVEKESPKSKE--EKKKKDEDKSNKKKKEKEVEGEGEKKEKDLED 65
++ D+K++K + E ES +SKE + +K KS KK +E + E E E +
Sbjct: 498 EIIDVKKDKEIKQKAKSESESEESKESDSELRKKRRKSYKKSRESDSESESESEV----- 662
Query: 66 KGEEKKKKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDEGKDG 125
E++K++K + E +D++ E+ED +K + ++K ++
Sbjct: 663 --EDRKRRKSRKYSE---SDSDTNSEEED--------RKRRRRRKSSRR----------- 770
Query: 126 EEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEKKKEKKDDGKDKTKDVSKLKQKLEKMN 185
K ++K++ D+ E D+ ++ + +++ K + K ++ VS+ +++E +
Sbjct: 771 RRKSSRKKRRCSDSDESETDDESGYDDSGRKRKRSKRSRKSKKKSSEPVSEGSEEIELGS 950
Query: 186 GKIEALLQEKADIERQIKEAEVGSHVVNEKDKDEA 220
A + E+ D + K AE+ + + K+ E+
Sbjct: 951 DSAVAKINEEIDDVDEEKMAEMNAEALKLKELFES 1055
>TC80941 weakly similar to PIR|T39903|T39903 serine-rich protein - fission
yeast (Schizosaccharomyces pombe), partial (15%)
Length = 557
Score = 62.4 bits (150), Expect = 1e-10
Identities = 33/97 (34%), Positives = 63/97 (64%)
Frame = +2
Query: 72 KKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDEGKDGEEKKEK 131
K+ +DKEEK+K +AEK E +++ G ++EK + E+K++G+++E K EE +EK
Sbjct: 89 KETTEDKEEKEKIEAEKEAEGSIEEKEEG---NDEEKTEVEEKEEGDDNE-KSEEETEEK 256
Query: 132 EKKKKEKKDKGEEDEGKDGEEKKKDKEKKKEKKDDGK 168
E+ +++K + E +E ++ +E DK K+++K + K
Sbjct: 257 EEGDEKEKTEAETEEEEEADEDTIDKSKEEDKAEGSK 367
Score = 57.8 bits (138), Expect = 3e-09
Identities = 30/99 (30%), Positives = 62/99 (62%), Gaps = 1/99 (1%)
Frame = +2
Query: 8 KVEDIKEEKTDLKNKSVEKESPKSKEEKKK-KDEDKSNKKKKEKEVEGEGEKKEKDLEDK 66
+V++ E+K + + EKE+ S EEK++ DE+K+ ++KE+ + E ++E + +++
Sbjct: 83 EVKETTEDKEEKEKIEAEKEAEGSIEEKEEGNDEEKTEVEEKEEGDDNEKSEEETEEKEE 262
Query: 67 GEEKKKKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKE 105
G+EK+K + + +EE++ + K KE+D+ G + KE
Sbjct: 263 GDEKEKTEAETEEEEEADEDTIDKSKEEDKAEGSKGGKE 379
Score = 54.3 bits (129), Expect = 4e-08
Identities = 38/114 (33%), Positives = 63/114 (54%)
Frame = +2
Query: 34 EKKKKDEDKSNKKKKEKEVEGEGEKKEKDLEDKGEEKKKKKGKDKEEKDKTDAEKGKEKE 93
E K+ EDK K+K E E E EG +EK+ +G +++K + ++KEE D D EK +E+
Sbjct: 83 EVKETTEDKEEKEKIEAEKEAEGSIEEKE---EGNDEEKTEVEEKEEGD--DNEKSEEET 247
Query: 94 DDQEAGGEVKKEKEKKKKEKKDKGEEDEGKDGEEKKEKEKKKKEKKDKGEEDEG 147
+++E G ++K+K E E ++ EE E K +++DK E +G
Sbjct: 248 EEKEEG------------DEKEK-TEAETEEEEEADEDTIDKSKEEDKAEGSKG 370
Score = 52.4 bits (124), Expect = 1e-07
Identities = 35/92 (38%), Positives = 58/92 (63%), Gaps = 6/92 (6%)
Frame = +2
Query: 101 EVKKEKEKKKKEKKDKG---EEDEGKDGEEKKEKEKKKK---EKKDKGEEDEGKDGEEKK 154
E K+EKEK + EK+ +G E++EG D EEK E E+K++ +K + E +E ++G+EK+
Sbjct: 101 EDKEEKEKIEAEKEAEGSIEEKEEGND-EEKTEVEEKEEGDDNEKSEEETEEKEEGDEKE 277
Query: 155 KDKEKKKEKKDDGKDKTKDVSKLKQKLEKMNG 186
K E + E++++ + T D SK + K E G
Sbjct: 278 K-TEAETEEEEEADEDTIDKSKEEDKAEGSKG 370
Score = 47.4 bits (111), Expect = 4e-06
Identities = 27/88 (30%), Positives = 48/88 (53%)
Frame = +2
Query: 2 EDSRQIKVEDIKEEKTDLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEK 61
E+ +I+ E E + K + ++E + +E+++ D +KS ++ +EKE E EK E
Sbjct: 110 EEKEKIEAEKEAEGSIEEKEEGNDEEKTEVEEKEEGDDNEKSEEETEEKEEGDEKEKTEA 289
Query: 62 DLEDKGEEKKKKKGKDKEEKDKTDAEKG 89
+ E++ E + K KEE DK + KG
Sbjct: 290 ETEEEEEADEDTIDKSKEE-DKAEGSKG 370
Score = 39.7 bits (91), Expect = 0.001
Identities = 24/89 (26%), Positives = 50/89 (55%)
Frame = +2
Query: 118 EEDEGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEKKKEKKDDGKDKTKDVSKL 177
E E + +E+KEK + +KE +G +E ++G +++K + ++KE+ DD + ++ +
Sbjct: 83 EVKETTEDKEEKEKIEAEKEA--EGSIEEKEEGNDEEKTEVEEKEEGDDNEKSEEETEEK 256
Query: 178 KQKLEKMNGKIEALLQEKADIERQIKEAE 206
++ EK + E +E+AD + K E
Sbjct: 257 EEGDEKEKTEAETEEEEEADEDTIDKSKE 343
Score = 33.9 bits (76), Expect = 0.050
Identities = 30/142 (21%), Positives = 61/142 (42%), Gaps = 10/142 (7%)
Frame = +3
Query: 13 KEEKTDLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEKDLEDKGEEKK- 71
K+++ L+ K K K ++KK N KK+++ + K + L+ + ++
Sbjct: 138 KKQRVALRRKKRVMMRRKLK*RRRKKVMTMRNLKKRQRR--RKRAMKRRKLKQRRRRRRR 311
Query: 72 ---------KKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDEG 122
KKK + + K + + + +E +DD E + ++K + + EED+
Sbjct: 312 PTRILLINQKKKIRPRVVKVEKNDNEVEEVKDDMEVDSVKEPVEDKNDDDDIQEMEEDDK 491
Query: 123 KDGEEKKEKEKKKKEKKDKGEE 144
E + EK E K+ E+
Sbjct: 492 NVAESENEKMDVDAEVKETTED 557
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 61.6 bits (148), Expect = 2e-10
Identities = 52/219 (23%), Positives = 86/219 (38%), Gaps = 13/219 (5%)
Frame = +3
Query: 8 KVEDIKEEKTDLKNKSVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEKDLEDKG 67
KV+ +EE D + S E + P +K K + KK + E E ++D ED+
Sbjct: 513 KVDTSEEE--DSEESSDEDKKPAAKAVPSKNGSAPA--KKAASDEEDTDESSDEDEEDEK 680
Query: 68 EEKKKKKGKD----KEEKDKTDAEKGKEKEDDQEAGGEVKKEKEKKKKEKKDKGEEDEGK 123
K K+ ++ D +++ E D+++ K K KK DE
Sbjct: 681 PAAKAVPSKNGSVPAKKADTESSDEDSESSDEEDKKPAAKASKNVSAPTKKAASSSDEES 860
Query: 124 DGEEKKEKEKK---------KKEKKDKGEEDEGKDGEEKKKDKEKKKEKKDDGKDKTKDV 174
D E ++++ K KK KKD + D+ D +DK+ KK+D + KD
Sbjct: 861 DEESDEDEDAKPVSKPAAVAKKSKKDSSDSDDEDDDSSSDEDKKPVAAKKEDKMNVDKDG 1040
Query: 175 SKLKQKLEKMNGKIEALLQEKADIERQIKEAEVGSHVVN 213
S Q E+ + Q+K + + G N
Sbjct: 1041SDSDQSEEESEDEPSKTPQKKIKDVEMVDAGKSGKKAPN 1157
>TC77946 homologue to GP|10334491|emb|CAC10207. putative splicing factor
{Cicer arietinum}, complete
Length = 2061
Score = 61.6 bits (148), Expect = 2e-10
Identities = 39/166 (23%), Positives = 86/166 (51%), Gaps = 1/166 (0%)
Frame = +3
Query: 41 DKSNKKKKEKEVEGEGEKKEKDLEDKGEEKKKKKGKDKEEKD-KTDAEKGKEKEDDQEAG 99
++ N+ + + E EK +E++ E+K K K+ + D K++ K DD G
Sbjct: 66 ERENRNRI*QWTSTSTEYLEKTVEEQPEDKDSSKKKNTDITDIKSERTYRKHGIDDDLDG 245
Query: 100 GEVKKEKEKKKKEKKDKGEEDEGKDGEEKKEKEKKKKEKKDKGEEDEGKDGEEKKKDKEK 159
GE ++ K + + ++ ++D +D + ++++E +++ E +DGE ++KE+
Sbjct: 246 GEDRRSKRSRGGDDENGSKKDRDRDRDRERDREHRERSSGRHRE----RDGERGSREKER 413
Query: 160 KKEKKDDGKDKTKDVSKLKQKLEKMNGKIEALLQEKADIERQIKEA 205
+K+ + G+DK +D + ++K + K +E+ D ER+ + A
Sbjct: 414 EKDVERGGRDKERD--REREKERERRDKEREKERERRDKEREKERA 545
Score = 60.8 bits (146), Expect = 4e-10
Identities = 39/159 (24%), Positives = 87/159 (54%), Gaps = 4/159 (2%)
Frame = +3
Query: 9 VEDIKEEKTDLKNK-SVEKESPKSKEEKKKKDEDKSNKKKKEKEVEGEGEKKEKDLEDKG 67
+ DIK E+T K+ + + + + K+ + D N KK+++ +++D E
Sbjct: 183 ITDIKSERTYRKHGIDDDLDGGEDRRSKRSRGGDDENGSKKDRD-------RDRDRERDR 341
Query: 68 EEKKKKKGKDKEEKDKTDAEKGKEKEDDQEAGGEVKKE--KEKKKKEKKDKGEEDEGKDG 125
E +++ G+ +E +D + KE+E D E GG K+ + +K++E++DK E E +
Sbjct: 342 EHRERSSGRHRE-RDGERGSREKEREKDVERGGRDKERDREREKERERRDKEREKERERR 518
Query: 126 EEKKEKEKKKKEKKDKGE-EDEGKDGEEKKKDKEKKKEK 163
++++EKE+ + K+ + E ++E + E+++ +K++
Sbjct: 519 DKEREKERA*ERKRGERETKEEYESS*ERQRRARFRKQR 635
Score = 38.1 bits (87), Expect = 0.003
Identities = 21/84 (25%), Positives = 46/84 (54%), Gaps = 2/84 (2%)
Frame = +1
Query: 89 GKEKEDDQEAGGEVKKEKEKKKKE-KKDKGEEDEGKDGEEKKEKEKKKKEKKDKGEED-E 146
G+EK + GE++K K ++E ++++ E + E+++ + + ++D+GE D E
Sbjct: 457 GREK---RRGRGEIRKGKRNGREEIRRERRSVPEREKEEKERPRRSMSRPERDRGERDFE 627
Query: 147 GKDGEEKKKDKEKKKEKKDDGKDK 170
+DG + KE + + D +D+
Sbjct: 628 NRDGRRFRDKKENVEPEADPERDQ 699
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.287 0.118 0.285
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,760,171
Number of Sequences: 36976
Number of extensions: 49523
Number of successful extensions: 4592
Number of sequences better than 10.0: 744
Number of HSP's better than 10.0 without gapping: 1616
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3246
length of query: 220
length of database: 9,014,727
effective HSP length: 92
effective length of query: 128
effective length of database: 5,612,935
effective search space: 718455680
effective search space used: 718455680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 45 (21.7 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0371a.8


