

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0371a.1
(409 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
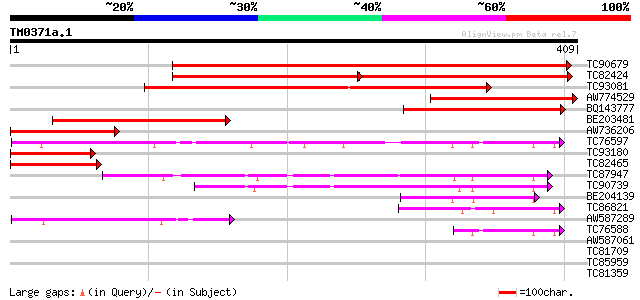
Score E
Sequences producing significant alignments: (bits) Value
TC90679 similar to PIR|T10521|T10521 beta-glucosidase (EC 3.2.1.... 433 e-122
TC82424 similar to PIR|T51283|T51283 glucan 1 3-beta-glucosidase... 236 e-117
TC93081 similar to PIR|T45636|T45636 beta-D-glucan exohydrolase-... 249 2e-66
AW774529 similar to GP|12227502|emb unnamed protein product {Zea... 197 5e-51
BQ143777 similar to PIR|T10521|T10 beta-glucosidase (EC 3.2.1.21... 190 8e-49
BE203481 similar to GP|20259685|gb beta-D-glucan exohydrolase {T... 187 8e-48
AW736206 similar to PIR|T51283|T51 glucan 1 3-beta-glucosidase (... 114 9e-26
TC76597 similar to GP|9759419|dbj|BAB09906.1 xylosidase {Arabido... 91 1e-18
TC93180 similar to PIR|T51283|T51283 glucan 1 3-beta-glucosidase... 89 3e-18
TC82465 similar to PIR|T45637|T45637 beta-D-glucan exohydrolase-... 79 4e-15
TC87947 similar to GP|10178060|dbj|BAB11424. beta-xylosidase {Ar... 72 3e-13
TC90739 similar to GP|10178060|dbj|BAB11424. beta-xylosidase {Ar... 61 7e-10
BE204139 similar to PIR|T52390|T52 beta-1 4-xylosidase [imported... 50 2e-06
TC86821 weakly similar to GP|18086336|gb|AAL57631.1 At1g78060/F2... 49 3e-06
AW587289 similar to GP|14194121|gb At1g02640/T14P4_11 {Arabidops... 48 8e-06
TC76588 similar to GP|9759419|dbj|BAB09906.1 xylosidase {Arabido... 42 4e-04
AW587061 similar to PIR|T49983|T49 beta-xylosidase-like protein ... 38 0.008
TC81709 similar to PIR|T05867|T05867 hypothetical protein T29A15... 30 1.7
TC85959 homologue to PIR|S57471|S57471 GTP-binding protein GTP6 ... 28 4.9
TC81359 weakly similar to GP|5091614|gb|AAD39602.1| Contains a P... 28 6.5
>TC90679 similar to PIR|T10521|T10521 beta-glucosidase (EC 3.2.1.21) -
common nasturtium, partial (43%)
Length = 1017
Score = 433 bits (1114), Expect = e-122
Identities = 208/288 (72%), Positives = 240/288 (83%)
Frame = +3
Query: 118 MIPKFFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGYLGIQ 177
M+P FTEFI+DLT VKN +IP+SRIDDAV RIL VKF MG+FENP AD SL+ LG +
Sbjct: 6 MVPYNFTEFIDDLTFQVKNNIIPISRIDDAVARILRVKFTMGLFENPLADLSLINQLGSK 185
Query: 178 KHRKLAREAVRKSLVLLKNGISAEKPILPLTKKVPKILVAGTHADNLGYQCGGWTIEWQG 237
+HR+LAREAVRKSLVLLKNG A KP+LPL KK K+LVAG+HADNLG QCGGWTI WQG
Sbjct: 186 EHRELAREAVRKSLVLLKNGKYANKPLLPLPKKASKVLVAGSHADNLGNQCGGWTITWQG 365
Query: 238 VSGNNHLEGTTILTAIKNTVDPETTVIYKENPDTEFVESNGFSYAIVVVGEHPYAEMEGD 297
+SG++ GTTIL IK TVDP T V+Y ENPD F++SN FSYAIV+VGE PYAE GD
Sbjct: 366 LSGSDLTTGTTILDGIKQTVDPATEVVYNENPDANFIKSNKFSYAIVIVGEKPYAETFGD 545
Query: 298 SMNLTIPSPGPETITNVCEAITCVVIIISGRPLVIEPYLALIDALVAGWLPGSEGQGVAD 357
S+NLTI PGP TITNVC +I CVV++++GRP+VI+PYL+ IDALVA WLPG+EGQGVAD
Sbjct: 546 SLNLTIAEPGPSTITNVCGSIQCVVVLVTGRPVVIQPYLSKIDALVAAWLPGTEGQGVAD 725
Query: 358 VLYGDYGFTGKLPRTWFKSVDQLPMNVGDPHYDPLFPFGFGLSTQPTK 405
VLYGD+ FTGKL RTWFK+VDQLPMNVGD HYDPLFPFGFGL+T TK
Sbjct: 726 VLYGDFEFTGKLARTWFKTVDQLPMNVGDKHYDPLFPFGFGLTTNLTK 869
>TC82424 similar to PIR|T51283|T51283 glucan 1 3-beta-glucosidase (EC
3.2.1.58) [imported] - common tobacco, partial (45%)
Length = 962
Score = 236 bits (603), Expect(2) = e-117
Identities = 110/154 (71%), Positives = 127/154 (82%)
Frame = +3
Query: 253 IKNTVDPETTVIYKENPDTEFVESNGFSYAIVVVGEHPYAEMEGDSMNLTIPSPGPETIT 312
IKN VD ET V Y+ NP ++V+SN FS+AIV+VGE PYAE GDS+NLTI G TI
Sbjct: 408 IKNAVDKETKVFYEGNPSLDYVKSNDFSHAIVIVGETPYAETNGDSLNLTISGQGYGTIN 587
Query: 313 NVCEAITCVVIIISGRPLVIEPYLALIDALVAGWLPGSEGQGVADVLYGDYGFTGKLPRT 372
NVC + CVV++ISGRP+VI+PYL I+ LVAGWLPGSEG GVADVL+GDYGFTGKLPRT
Sbjct: 588 NVCGVVKCVVVLISGRPIVIQPYLEKIEGLVAGWLPGSEGSGVADVLFGDYGFTGKLPRT 767
Query: 373 WFKSVDQLPMNVGDPHYDPLFPFGFGLSTQPTKA 406
WFK+VDQLPMNVGD HYDPLFPFGFGL+T+ KA
Sbjct: 768 WFKTVDQLPMNVGDSHYDPLFPFGFGLTTEAHKA 869
Score = 205 bits (521), Expect(2) = e-117
Identities = 102/137 (74%), Positives = 115/137 (83%)
Frame = +2
Query: 118 MIPKFFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGYLGIQ 177
MIP +TEFI+ LT+LVK +IPMSRIDDAVKRIL VKF+MG+FENP ADYSL LG Q
Sbjct: 2 MIPHNYTEFIDGLTLLVKKNVIPMSRIDDAVKRILRVKFVMGLFENPLADYSLADQLGSQ 181
Query: 178 KHRKLAREAVRKSLVLLKNGISAEKPILPLTKKVPKILVAGTHADNLGYQCGGWTIEWQG 237
+HR+LAREAVRKSLVLLKNG + +KPILPL KK KILVAG+HADNLGYQCGGWTI+WQG
Sbjct: 182 EHRELAREAVRKSLVLLKNGENVDKPILPLPKKASKILVAGSHADNLGYQCGGWTIQWQG 361
Query: 238 VSGNNHLEGTTILTAIK 254
SGNN GT IL+A K
Sbjct: 362 QSGNNITTGTPILSAYK 412
>TC93081 similar to PIR|T45636|T45636 beta-D-glucan exohydrolase-like
protein - Arabidopsis thaliana, partial (39%)
Length = 788
Score = 249 bits (635), Expect = 2e-66
Identities = 135/252 (53%), Positives = 173/252 (68%), Gaps = 2/252 (0%)
Frame = +2
Query: 98 PH-ANFTYSVEAGVSAGIDMFMIPKFFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKF 156
PH +++ Y + + V+AGIDM M+ + FI +LT L+++ +PMSRIDDAV+RIL VKF
Sbjct: 35 PHGSDYRYCISSAVNAGIDMVMVAVRYKLFIEELTSLIESGEVPMSRIDDAVERILRVKF 214
Query: 157 MMGIFENPFADYSLVGYLGIQKHRKLAREAVRKSLVLLKNGISAEKPILPLTKKVPKILV 216
G+FE P +D SL+ +G + HR LAREAVRKSLVLLKNG +P LPL K +ILV
Sbjct: 215 AAGLFEFPLSDRSLLDIVGCKPHRDLAREAVRKSLVLLKNGKDISEPFLPLNKNAKRILV 394
Query: 217 AGTHADNLGYQCGGWTIEWQGVSGNNHLEGTTILTAIKNTVDPETTVIYKENPDTEFVES 276
GTHADNLG+QCGGWT W G SG + G+TIL A+K V ET VIY++ P + +E
Sbjct: 395 TGTHADNLGFQCGGWTKTWYGASGRITV-GSTILDAVKAAVGAETQVIYEKYPSQDTIER 571
Query: 277 NGFSYAIVVVGEHPYAEMEGDSMNLTIPSPGPETITNVCEAITCVVIIISGRPLVIEPYL 336
N FS+AIV VGE PYAE GD+ LTIP G + I+ V + +VI+ISG LV+EP L
Sbjct: 572 NEFSFAIVAVGEAPYAESLGDNSELTIPFRGTDIISLVADKFPTLVILISG*ALVLEPRL 751
Query: 337 AL-IDALVAGWL 347
+ IDAL A WL
Sbjct: 752 LVKIDALGAAWL 787
>AW774529 similar to GP|12227502|emb unnamed protein product {Zea mays},
partial (16%)
Length = 450
Score = 197 bits (502), Expect = 5e-51
Identities = 90/106 (84%), Positives = 100/106 (93%)
Frame = -3
Query: 304 PSPGPETITNVCEAITCVVIIISGRPLVIEPYLALIDALVAGWLPGSEGQGVADVLYGDY 363
P+PG E ITNVC A+ CVVIIISGRPLVIEPY+ LIDA+VAGWLPGSEGQGVADVL+G+Y
Sbjct: 448 PNPGREIITNVCGAMKCVVIIISGRPLVIEPYVGLIDAVVAGWLPGSEGQGVADVLFGEY 269
Query: 364 GFTGKLPRTWFKSVDQLPMNVGDPHYDPLFPFGFGLSTQPTKAIYS 409
GFTGKLPRTWFKSVDQLPMNVGDPHYDP+FPFGFGL+T+P+K IYS
Sbjct: 268 GFTGKLPRTWFKSVDQLPMNVGDPHYDPVFPFGFGLTTKPSKPIYS 131
>BQ143777 similar to PIR|T10521|T10 beta-glucosidase (EC 3.2.1.21) - common
nasturtium, partial (17%)
Length = 750
Score = 190 bits (483), Expect = 8e-49
Identities = 87/117 (74%), Positives = 98/117 (83%)
Frame = +1
Query: 285 VVGEHPYAEMEGDSMNLTIPSPGPETITNVCEAITCVVIIISGRPLVIEPYLALIDALVA 344
VVGE PYAE GDS+NLTI G ETI NVC + CVV++++GRP+ I+PYL ID LVA
Sbjct: 10 VVGETPYAETNGDSLNLTISGNGTETINNVCGRVKCVVVLVTGRPVAIQPYLNKIDGLVA 189
Query: 345 GWLPGSEGQGVADVLYGDYGFTGKLPRTWFKSVDQLPMNVGDPHYDPLFPFGFGLST 401
WLPGSEG GVADVL+GDYGFTGKL RTWFK+VDQLPMNVGD HYDPLFPFGFGLS+
Sbjct: 190 AWLPGSEGTGVADVLFGDYGFTGKLARTWFKTVDQLPMNVGDSHYDPLFPFGFGLSS 360
>BE203481 similar to GP|20259685|gb beta-D-glucan exohydrolase {Triticum
aestivum}, partial (21%)
Length = 399
Score = 187 bits (474), Expect = 8e-48
Identities = 86/128 (67%), Positives = 106/128 (82%)
Frame = +3
Query: 32 LMRVHMPGYFSSISKGVATIMVSYSSWNGIKMHTNHDMITGFLKNTLHFKGFVISDFEGI 91
LM +HMP Y SI KGV+T+M SYSSWNG+KMH N D+ITG+LKNTL FKGFVISD++GI
Sbjct: 15 LMSLHMPAYIDSIIKGVSTVMASYSSWNGVKMHANRDLITGYLKNTLKFKGFVISDWQGI 194
Query: 92 DRITSPPHANFTYSVEAGVSAGIDMFMIPKFFTEFINDLTILVKNKLIPMSRIDDAVKRI 151
D+IT+PP +N+TYSV+A + AG+DM M+P F +FI DLT+LVKN +IPM RIDDAV+RI
Sbjct: 195 DKITTPPGSNYTYSVQASIEAGVDMVMVPYEFEDFIKDLTLLVKNNIIPMDRIDDAVERI 374
Query: 152 LWVKFMMG 159
L KF MG
Sbjct: 375 LVGKFTMG 398
>AW736206 similar to PIR|T51283|T51 glucan 1 3-beta-glucosidase (EC 3.2.1.58)
[imported] - common tobacco, partial (15%)
Length = 620
Score = 114 bits (284), Expect = 9e-26
Identities = 52/79 (65%), Positives = 63/79 (78%)
Frame = +1
Query: 1 EKVIACAKHYVGDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISKGVATIMVSYSSWNG 60
+KV ACAKH+VGDGGT GI+ +NTV R L +HM Y++SI+K V TIMVSYSSWNG
Sbjct: 382 KKVAACAKHFVGDGGTTEGINEDNTVATRHELFSIHMQAYYNSITKVVLTIMVSYSSWNG 561
Query: 61 IKMHTNHDMITGFLKNTLH 79
KMH+N D++TGFLKNTLH
Sbjct: 562 EKMHSNRDLVTGFLKNTLH 618
>TC76597 similar to GP|9759419|dbj|BAB09906.1 xylosidase {Arabidopsis
thaliana}, partial (77%)
Length = 2713
Score = 90.5 bits (223), Expect = 1e-18
Identities = 107/441 (24%), Positives = 189/441 (42%), Gaps = 42/441 (9%)
Frame = +3
Query: 2 KVIACAKHYVG-DGGTINGID--GNNTVIDRDGLMRVHMPGYFSSISKG-VATIMVSYSS 57
KV AC KHY D NG+D N + + L + + + + G VA++M SY+
Sbjct: 636 KVAACCKHYTAYDLDNWNGVDRFHFNAKVSKQDLADTYDVPFKACVRDGKVASVMCSYNQ 815
Query: 58 WNGIKMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFT--YSVEAGVSAGID 115
NG + +++ ++ G+++SD + + + H T + A + AG+D
Sbjct: 816 VNGKPTCADPELLRNTIRGEWGLNGYIVSDCDSVGVLYDNQHYTRTPEQAAAAAIKAGLD 995
Query: 116 MFMIPKFFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGY-- 173
+ P F D I K LI + ++ A+ ++ V+ +G+F+ Y +G
Sbjct: 996 LDCGP--FLALHTDGAI--KQGLISENDLNLALANLITVQMRLGMFDGDAQPYGNLGTRD 1163
Query: 174 LGIQKHRKLAREAVRKSLVLLKNGISAEKPILPLTKKV-----PKILVAGTHADNLGYQC 228
+ + H +A EA R+ +VLL+N +A P+ P + P V T N
Sbjct: 1164 VCLPSHNDVALEAARQGIVLLQNKGNA-LPLSPTRYRTVGVIGPNSDVTVTMIGNYAGIA 1340
Query: 229 GGWTIEWQGVS---GNNHLEGTTILTAIKNTVDPETTVIYKENPDTEFVESNGFSYAIVV 285
G+T QG++ H G + N + + + ++ T ++V
Sbjct: 1341 CGYTTPLQGIARYVKTIHQAGCKDVGCGGNQLFGLSEQVARQADAT-----------VLV 1487
Query: 286 VGEHPYAEME-GDSMNLTIPSPGPETITNVCEAI--TCVVIIISGRPLVI-----EPYLA 337
+G E E D L +P E ++ V A +++++SG P+ + +P ++
Sbjct: 1488 MGLDQSIEAEFRDRTGLLLPGHQQELVSRVARAARGPVILVLMSGGPIDVTFAKNDPKIS 1667
Query: 338 LIDALVAGWLPGSEGQGVADVLYGDYGFTGKLPRTWFKS--VDQLPMNVGDPHYDP---- 391
I L G+ S G +ADV++G +G+LP TW+ V ++PM D +P
Sbjct: 1668 AI--LWVGYPGQSGGTAIADVIFGRTNPSGRLPNTWYPQDYVRKVPMTNMDMRANPATGY 1841
Query: 392 ------------LFPFGFGLS 400
+FPFG GLS
Sbjct: 1842 PGRTYRFYKGPVVFPFGHGLS 1904
>TC93180 similar to PIR|T51283|T51283 glucan 1 3-beta-glucosidase (EC
3.2.1.58) [imported] - common tobacco, partial (41%)
Length = 798
Score = 89.0 bits (219), Expect = 3e-18
Identities = 40/62 (64%), Positives = 49/62 (78%)
Frame = +1
Query: 1 EKVIACAKHYVGDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISKGVATIMVSYSSWNG 60
+KV ACAKH+VGDGGT GI+ +NTV +R L +HMP Y++SI KGV T+MVSYSSWNG
Sbjct: 595 KKVAACAKHFVGDGGTTKGINEDNTVANRHELFSIHMPAYYNSIIKGVLTVMVSYSSWNG 774
Query: 61 IK 62
K
Sbjct: 775 EK 780
>TC82465 similar to PIR|T45637|T45637 beta-D-glucan exohydrolase-like
protein - Arabidopsis thaliana, partial (42%)
Length = 854
Score = 78.6 bits (192), Expect = 4e-15
Identities = 33/66 (50%), Positives = 48/66 (72%)
Frame = +3
Query: 1 EKVIACAKHYVGDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISKGVATIMVSYSSWNG 60
+ VIACAKH+ GDGGT G++ +T++ + L ++HM Y IS+GV+TIM SY+SWNG
Sbjct: 657 KNVIACAKHFGGDGGTHKGVNEGDTILSYEDLEKIHMAPYLDCISQGVSTIMASYTSWNG 836
Query: 61 IKMHTN 66
K+H +
Sbjct: 837 RKLHAD 854
>TC87947 similar to GP|10178060|dbj|BAB11424. beta-xylosidase {Arabidopsis
thaliana}, partial (55%)
Length = 1662
Score = 72.4 bits (176), Expect = 3e-13
Identities = 80/342 (23%), Positives = 152/342 (44%), Gaps = 18/342 (5%)
Frame = +2
Query: 68 DMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEAG--VSAGIDMFMIPKFFTE 125
D++ G ++ G+++SD + ++ + H T A + +G+D+
Sbjct: 11 DLLKGVIRGKWKLNGYIVSDCDSVEVLFKDQHYTKTPEEAAAKTILSGLDLDC-----GS 175
Query: 126 FINDLT-ILVKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGYLGIQ-----KH 179
++ T VK +L+ + I++AV +G F+ + G LG + ++
Sbjct: 176 YLGQYTGRAVKQRLVDEASINNAVSNNFATLMRLGFFDGDPSKQPY-GNLGPKDVCTPEN 352
Query: 180 RKLAREAVRKSLVLLKNGISAEKPILPLTKKVPKIL-VAGTHADNLGYQCGGWTIEWQGV 238
++LAREA R+ +VLLKN + LPL+ K K L V G +A+ G + E
Sbjct: 353 QELAREAARQGIVLLKNSPGS----LPLSSKAIKSLAVIGPNANATRVMIGNY--EGIPC 514
Query: 239 SGNNHLEGTTILTAIKNTVD-PETTVIYKENPDTEFVESNGFSYAIVVVGEHPYAEMEG- 296
+ L+G T P+ + D + ++ + I+VVG + E E
Sbjct: 515 KYTSPLQGLTAFVPTSYAPGCPDVQCANAQIDDAAKIAASADA-TIIVVGANLAIEAESL 691
Query: 297 DSMNLTIPSPGPETITNVCEAIT--CVVIIISGRPLVI---EPYLALIDALVAGWLPGSE 351
D +N+ +P + + V +++I+SG + + + + L G+ +
Sbjct: 692 DRVNILLPGQQQQLVNEVANVSKGPVILVIMSGGGMDVSFAKTNDKITSILWVGYPGEAG 871
Query: 352 GQGVADVLYGDYGFTGKLPRTWFKS--VDQLPMNVGDPHYDP 391
G +ADV++G Y +G+LP TW+ V+++PM + DP
Sbjct: 872 GAAIADVIFGSYNPSGRLPMTWYPQSYVEKIPMTNMNMRSDP 997
>TC90739 similar to GP|10178060|dbj|BAB11424. beta-xylosidase {Arabidopsis
thaliana}, partial (39%)
Length = 1017
Score = 61.2 bits (147), Expect = 7e-10
Identities = 71/272 (26%), Positives = 117/272 (42%), Gaps = 14/272 (5%)
Frame = +2
Query: 134 VKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGYLG-----IQKHRKLAREAVR 188
VK LI + I++AV +G F+ + + G LG +++LAREA R
Sbjct: 74 VKQGLIGEASINNAVYNNFATLMRLGFFDRDPSKQAY-GNLGPKDVCTSANQELAREAAR 250
Query: 189 KSLVLLKNGISAEKPILPLTKKVPKIL-VAGTHADNLGYQCGGWTIEWQGVSGNNHLEGT 247
+ +VLLKN + LPL K K L V G +A+ G + E + L+G
Sbjct: 251 QGIVLLKNCAGS----LPLNAKAIKSLAVIGPNANATRAMIGNY--EGIPCKYTSPLQGL 412
Query: 248 TILTAIKNTVD-PETTVIYKENPDTEFVESNGFSYAIVVVGEHPYAEMEGDSMNLTIPSP 306
T L P+ D + + ++ + IVV D +N+ +P
Sbjct: 413 TALVPTSFAAGCPDVQCTNAALDDAKKIAASADATVIVVGANLAIEAQSHDRVNILLPGQ 592
Query: 307 GPETITNVCEAITCVVI--IISGRPLVI---EPYLALIDALVAGWLPGSEGQGVADVLYG 361
+ +T V VI I+SG + + + + L G+ + G +ADV++G
Sbjct: 593 QQQLVTEVANVAKGPVILAIMSGGGMDVSFAKTNKKITSILWVGYPGETGGAAIADVIFG 772
Query: 362 DYGFTGKLPRTWFKS--VDQLPMNVGDPHYDP 391
+ +G+LP TW+ VD++PM + DP
Sbjct: 773 YHNPSGRLPMTWYPQSYVDKVPMTNMNMRPDP 868
>BE204139 similar to PIR|T52390|T52 beta-1 4-xylosidase [imported] -
Arabidopsis thaliana, partial (20%)
Length = 601
Score = 50.1 bits (118), Expect = 2e-06
Identities = 35/108 (32%), Positives = 59/108 (54%), Gaps = 8/108 (7%)
Frame = +1
Query: 283 IVVVGEHPYAEMEG-DSMNLTIPSPGPETITNVCEAI--TCVVIIISGRPLVIE--PYLA 337
++VVG E EG D +NLT+P + + +V A T +++I++ P+ I ++
Sbjct: 190 VLVVGLDQSIEAEGLDRVNLTLPGFQEKLVKDVAAATKGTLILVIMAAGPIDISFTKSVS 369
Query: 338 LIDALVAGWLPGSEG-QGVADVLYGDYGFTGKLPRTWFKS--VDQLPM 382
I ++ PG +G +A V++GDY G+ P TW+ VDQ+PM
Sbjct: 370 NIGGILWVGYPGQDGGNAIAQVIFGDYNPGGRSPFTWYPQSYVDQVPM 513
>TC86821 weakly similar to GP|18086336|gb|AAL57631.1 At1g78060/F28K19_32
{Arabidopsis thaliana}, partial (27%)
Length = 1145
Score = 49.3 bits (116), Expect = 3e-06
Identities = 44/145 (30%), Positives = 68/145 (46%), Gaps = 25/145 (17%)
Frame = +1
Query: 281 YAIVVVGEHPYAEMEG-DSMNLTIPSPGPETITNVCEAITCVVIII--SGRPLVIEPYLA 337
Y ++V+G E EG D +L +P E I +V +A VI++ G P+ I A
Sbjct: 214 YVVLVMGLDQSQESEGHDRDDLELPGKQQELINSVAKASKRPVILVLFCGGPVDIS--FA 387
Query: 338 LIDALVAGWL----PGS-EGQGVADVLYGDYGFTGKLPRTWF-KSVDQLPMNVGDPHYDP 391
+D + G + PG G+ +A V++GDY G+LP TW+ K ++PM DP
Sbjct: 388 KVDDKIGGIMWAGYPGELGGRALAQVVFGDYNPGGRLPMTWYPKDFIKIPMTDMRMRADP 567
Query: 392 ----------------LFPFGFGLS 400
++ FG+GLS
Sbjct: 568 SSGYPGRTYRFYTGPKVYEFGYGLS 642
>AW587289 similar to GP|14194121|gb At1g02640/T14P4_11 {Arabidopsis
thaliana}, partial (26%)
Length = 625
Score = 47.8 bits (112), Expect = 8e-06
Identities = 39/167 (23%), Positives = 78/167 (46%), Gaps = 6/167 (3%)
Frame = +2
Query: 2 KVIACAKHYVG-DGGTINGIDGN--NTVIDRDGLMRVHMPGYFSSISKG-VATIMVSYSS 57
KV AC KH+ D NG+D N ++ + + + + +G VA++M SY+
Sbjct: 83 KVAACCKHFTAYDVDNWNGVDRFHFNALVSKQDIEDTFDVPFRMCVKEGKVASVMCSYNQ 262
Query: 58 WNGIKMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEA--GVSAGID 115
NG+ + +++ ++ G+++SD + + + + H T A + AG+D
Sbjct: 263 VNGVPTCADPNLLKKTVRGVWGLDGYIVSDCDSVGVLYNSQHYTSTPEEAAADAIKAGLD 442
Query: 116 MFMIPKFFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFE 162
+ P F D VK L+ + +++A+ L V+ +G+F+
Sbjct: 443 LDCGP-FLGVHTQD---AVKKGLLTEADVNNALVNTLKVQMRLGMFD 571
>TC76588 similar to GP|9759419|dbj|BAB09906.1 xylosidase {Arabidopsis
thaliana}, partial (14%)
Length = 830
Score = 42.0 bits (97), Expect = 4e-04
Identities = 30/103 (29%), Positives = 51/103 (49%), Gaps = 23/103 (22%)
Frame = +1
Query: 321 VVIIISGRPLVI-----EPYLALIDALVAGWLPGSEGQGVADVLYGDYGFTGKLPRTWFK 375
+++++SG P+ + +P ++ I L G+ S G +ADV++G +G+LP TW+
Sbjct: 487 ILVLMSGGPIDVTFAKNDPKISAI--LWVGYPGQSGGTAIADVIFGRTNPSGRLPNTWYP 660
Query: 376 S--VDQLPMNVGDPHYDP----------------LFPFGFGLS 400
V ++PM D +P +FPFG GLS
Sbjct: 661 QDYVRKVPMTNMDMRANPATGYPGRTYRFYKGPVVFPFGHGLS 789
>AW587061 similar to PIR|T49983|T49 beta-xylosidase-like protein -
Arabidopsis thaliana, partial (22%)
Length = 611
Score = 37.7 bits (86), Expect = 0.008
Identities = 28/78 (35%), Positives = 41/78 (51%), Gaps = 5/78 (6%)
Frame = +1
Query: 138 LIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGYLGIQ-----KHRKLAREAVRKSLV 192
L+ +D A+ + V+ +G+F N + G LG Q +H+KLA EA R+ +V
Sbjct: 16 LVKEEDLDRALFNLFSVQMRLGLF-NGDPEKGKFGKLGPQDVCTPEHKKLALEAARQGIV 192
Query: 193 LLKNGISAEKPILPLTKK 210
LLKN + LPL KK
Sbjct: 193 LLKN----DNKFLPLDKK 234
>TC81709 similar to PIR|T05867|T05867 hypothetical protein T29A15.110 -
Arabidopsis thaliana, partial (72%)
Length = 1383
Score = 30.0 bits (66), Expect = 1.7
Identities = 24/95 (25%), Positives = 38/95 (39%)
Frame = +3
Query: 169 SLVGYLGIQKHRKLAREAVRKSLVLLKNGISAEKPILPLTKKVPKILVAGTHADNLGYQC 228
SL LG+ H +L EA + L +E+ I L K + V D
Sbjct: 819 SLNAQLGLSSHLQLQLEAANRDL------FDSEREIQRLRKAIADHCVGYVPHDKSSTGT 980
Query: 229 GGWTIEWQGVSGNNHLEGTTILTAIKNTVDPETTV 263
W+++ + + N HL+G L + T D E +
Sbjct: 981 A-WSVDTRNGNPNGHLDGDNNLDTHEKTKDEEERI 1082
>TC85959 homologue to PIR|S57471|S57471 GTP-binding protein GTP6 - garden
pea, complete
Length = 886
Score = 28.5 bits (62), Expect = 4.9
Identities = 18/60 (30%), Positives = 31/60 (51%)
Frame = -3
Query: 261 TTVIYKENPDTEFVESNGFSYAIVVVGEHPYAEMEGDSMNLTIPSPGPETITNVCEAITC 320
+ VIYK+N V S+ S ++ G PY ++ DS+++ + P E N C+ +C
Sbjct: 365 SNVIYKQNTHGSTVVSSCDSSKPLLTGCVPY--LQFDSLSIKLNCPYFEVNANGCDKTSC 192
>TC81359 weakly similar to GP|5091614|gb|AAD39602.1| Contains a PF|00561
alpha/beta hydrolase fold domain. {Arabidopsis
thaliana}, partial (9%)
Length = 436
Score = 28.1 bits (61), Expect = 6.5
Identities = 10/28 (35%), Positives = 20/28 (70%)
Frame = +1
Query: 53 VSYSSWNGIKMHTNHDMITGFLKNTLHF 80
++Y +N +++H D++TGF +T+HF
Sbjct: 196 LTYFLYNFLEIHFLQDLLTGFSGSTVHF 279
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.140 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,816,721
Number of Sequences: 36976
Number of extensions: 176342
Number of successful extensions: 825
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 817
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 822
length of query: 409
length of database: 9,014,727
effective HSP length: 98
effective length of query: 311
effective length of database: 5,391,079
effective search space: 1676625569
effective search space used: 1676625569
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0371a.1


