

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0369a.2
(1602 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
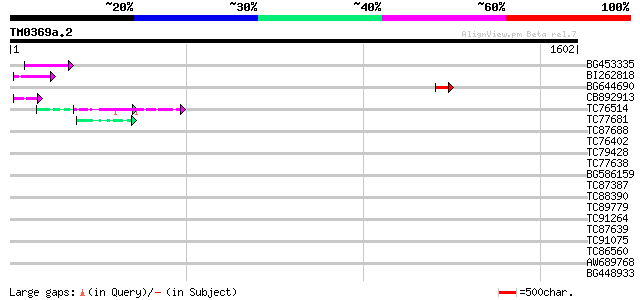
Score E
Sequences producing significant alignments: (bits) Value
BG453335 weakly similar to GP|18071369|g putative gag-pol polypr... 56 1e-07
BI262818 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 55 2e-07
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 49 2e-05
CB892913 similar to GP|1769897|emb lectin receptor kinase {Arabi... 45 2e-04
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 45 2e-04
TC77681 similar to GP|22597156|gb|AAN03465.1 nucleolar histone d... 44 7e-04
TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~un... 41 0.003
TC76402 weakly similar to GP|18086561|gb|AAL57705.1 At2g01100/F2... 41 0.003
TC79428 similar to PIR|T09661|T09661 ascorbate oxidase promoter-... 40 0.006
TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protei... 40 0.007
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 39 0.016
TC87387 homologue to PIR|T47775|T47775 hypothetical protein F24I... 38 0.028
TC88390 similar to GP|13272457|gb|AAK17167.1 unknown protein {Ar... 38 0.028
TC89779 GP|12835923|dbj|BAB23419. data source:MGD source key:MG... 37 0.047
TC91264 weakly similar to PIR|T27543|T27543 hypothetical protein... 37 0.081
TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.... 36 0.14
TC91075 similar to PIR|T48025|T48025 hypothetical protein T12C14... 35 0.18
TC86560 similar to GP|8843737|dbj|BAA97285.1 myosin heavy chain-... 35 0.18
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 35 0.23
BG448933 similar to PIR|T09648|T09 nucleolin homolog nuM1 - alfa... 34 0.52
>BG453335 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (11%)
Length = 660
Score = 55.8 bits (133), Expect = 1e-07
Identities = 37/140 (26%), Positives = 75/140 (53%)
Frame = +1
Query: 41 KKLYSQHHKARAILLSAISYEEYQKITDREFAKGIFDSLKMSHEGNKKVKESKALSLIQK 100
K+L + K ++ ++ +++I+ +K + L+ HEG+ KV++ K SL +K
Sbjct: 112 KELKKKDCKELFLIQ*SLDEGNFKRISKSTRSKEASNILEKYHEGDDKVEQIKLQSLRRK 291
Query: 101 YESFIMEPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRIIRCLPESWMPLVTSIELT 160
++ ME + I + S+ +V I+ + T + V +I+R L + +V +I +
Sbjct: 292 FKLMQMEDEKKIADYISKLINVVNQIKAYGEVVTDQQIVEKIMRTLSPRFDFIVVAIHES 471
Query: 161 RDVENMSLEELISILKCHEL 180
+DV+ + +EEL S L+ H+L
Sbjct: 472 KDVKTLKIEELKSSLEAHKL 531
>BI262818 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (24%)
Length = 615
Score = 55.5 bits (132), Expect = 2e-07
Identities = 34/119 (28%), Positives = 68/119 (56%)
Frame = +1
Query: 11 DLWDIIVDGYERPVDTDGKKIPRSEMTADQKKLYSQHHKARAILLSAISYEEYQKITDRE 70
DLW+ + + YE +D P + K+ ++ KARA L +A+S E + +I +
Sbjct: 244 DLWEAVEEDYEVLPLSDN---PTMAQIKNHKERKTRKSKARATLFAAVSEEIFTRIMTIK 414
Query: 71 FAKGIFDSLKMSHEGNKKVKESKALSLIQKYESFIMEPNESIEEMFSRFQLLVAGIRPL 129
A I++ LK +EG+++++ +AL+LI+++E M+ +E+I+E ++ + +R L
Sbjct: 415 SAFEIWNFLKTEYEGDERIRGMQALNLIREFEMQKMKESETIKEYANKLISIANKVRLL 591
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 48.9 bits (115), Expect = 2e-05
Identities = 28/53 (52%), Positives = 35/53 (65%)
Frame = -3
Query: 1202 RWT*RVPS*MVISQRKSMFINPQVLKMKRNQTMCSN*RNHSTV*SKLPEHGMR 1254
+W *RV M IS+R+ + N LKM+R Q MCS+* H V*SKL EHGM+
Sbjct: 276 KWM*RVHLLMEISKRRCLSSNLLDLKMQRYQIMCSD*IRHYMV*SKLQEHGMK 118
>CB892913 similar to GP|1769897|emb lectin receptor kinase {Arabidopsis
thaliana}, partial (35%)
Length = 837
Score = 45.4 bits (106), Expect = 2e-04
Identities = 23/81 (28%), Positives = 44/81 (53%)
Frame = +3
Query: 11 DLWDIIVDGYERPVDTDGKKIPRSEMTADQKKLYSQHHKARAILLSAISYEEYQKITDRE 70
D+W+++ GY+ D D + + D +K + KA ++ A+ +E++KI++
Sbjct: 195 DVWEVVEKGYKESQDEDSLTKAQRDTLKDSRK---RDKKALFLIYQALDEDEFEKISNAT 365
Query: 71 FAKGIFDSLKMSHEGNKKVKE 91
AK ++ LK S +G KVK+
Sbjct: 366 SAKEAWEKLKTSCQGEDKVKK 428
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 45.4 bits (106), Expect = 2e-04
Identities = 75/340 (22%), Positives = 140/340 (41%), Gaps = 23/340 (6%)
Frame = +3
Query: 181 KRSEMQDLKKKSIALKSKSEK--AKVEKSKALQAEEEESEEASEDSDEDELTLISKRLNR 238
KR +++K S A K K E+ AK + K ++ EE SEE+SE EDE ++
Sbjct: 186 KRQAEEEVKAVS-AKKQKVEEVAAKQKALKVVKKEESSSEESSES--EDEQPVV------ 338
Query: 239 IWKHRQSKFKGSGRAKGKYESSGQKKSSIKEV---TCFECKESGHYKSDCPKLKKDKK-- 293
+A + + KK ++K+ T E +S SD ++KK
Sbjct: 339 -------------KAPAPSKKTPAKKGNVKKAQPETTSEESDSDSSSSDEEEVKKPVSKA 479
Query: 294 -PKKHFKTKKSLMVTFDESESEDVDSDGEVQGLMAIVKDKG-AESKEAVDSDSESEGDPN 351
P K+ + T +E +SE+ + + A+ G A +K+A + +++ +
Sbjct: 480 VPSKNGSAPAKKVDTSEEEDSEESSDEDKKPAAKAVPSKNGSAPAKKAASDEEDTDESSD 659
Query: 352 SDDENE-------------VFASFSTSELKHALSDIMDKYNSLLSTHKKLKKNLSAVSKT 398
D+E+E V A + +E S+ D+ + K KN+SA +K
Sbjct: 660 EDEEDEKPAAKAVPSKNGSVPAKKADTESSDEDSESSDEEDK--KPAAKASKNVSAPTKK 833
Query: 399 PSEHEKIISDLKNENHALVNSNSVLK-NQIAKLEEIVACDASDCRNESKYEKSFQRFLAK 457
+ SD +++ ++ V K +AK + + D+ D ++S ++ + AK
Sbjct: 834 AASSSDEESDEESDEDE--DAKPVSKPAAVAKKSKKDSSDSDDEDDDSSSDEDKKPVAAK 1007
Query: 458 SVDRSLMASMIYGVSRNGMHGIGYSKPIRNEPSVSKAKSL 497
D+ V ++G + +EPS + K +
Sbjct: 1008KEDK-------MNVDKDGSDSDQSEEESEDEPSKTPQKKI 1106
Score = 44.3 bits (103), Expect = 4e-04
Identities = 69/302 (22%), Positives = 116/302 (37%), Gaps = 18/302 (5%)
Frame = +3
Query: 77 DSLKMSHEGNKKVKE----SKALSLIQKYESFIMEPNESIEEMFSRFQLLVAGIRPLNKS 132
+ +K +KV+E KAL +++K ES E +ES +E Q +V P K+
Sbjct: 201 EEVKAVSAKKQKVEEVAAKQKALKVVKKEESSSEESSESEDE-----QPVVKAPAPSKKT 365
Query: 133 YTTKDHVIRIIRCLPESWMPLVTSIELTRDVENMSLEELISILKCHELKRSEMQDLKKKS 192
K +V + P TS E D + EE+ KK
Sbjct: 366 PAKKGNVKKA--------QPETTSEESDSDSSSSDEEEV------------------KKP 467
Query: 193 IALKSKSEKAKVEKSKALQAEEEESEEASEDSDEDELTLISKRLNRIWKHRQSKFKGSGR 252
++ S+ K +EEE+SEE+S++ + + + N +++
Sbjct: 468 VSKAVPSKNGSAPAKKVDTSEEEDSEESSDEDKKPAAKAVPSK-NGSAPAKKAASDEEDT 644
Query: 253 AKGKYESSGQKKSSIKEVT----CFECKESGHYKSDCPKLKKDKKPKK---------HFK 299
+ E +K + K V K++ SD D++ KK
Sbjct: 645 DESSDEDEEDEKPAAKAVPSKNGSVPAKKADTESSDEDSESSDEEDKKPAAKASKNVSAP 824
Query: 300 TKKSLMVTFDESESE-DVDSDGEVQGLMAIVKDKGAESKEAVDSDSESEGDPNSDDENEV 358
TKK+ + +ES+ E D D D + A V K K++ DSD E + + +D+ V
Sbjct: 825 TKKAASSSDEESDEESDEDEDAKPVSKPAAVAKK--SKKDSSDSDDEDDDSSSDEDKKPV 998
Query: 359 FA 360
A
Sbjct: 999 AA 1004
>TC77681 similar to GP|22597156|gb|AAN03465.1 nucleolar histone deacetylase
HD2-P39 {Glycine max}, partial (47%)
Length = 1233
Score = 43.5 bits (101), Expect = 7e-04
Identities = 48/173 (27%), Positives = 70/173 (39%), Gaps = 5/173 (2%)
Frame = +2
Query: 190 KKSIALKSKSEKAKVEKS--KALQAEEEESEEASE-DSDEDELTLISKRLNRIWKHRQSK 246
+K L S+ A V KA + ++SEE E DS+++++ LI+ N
Sbjct: 293 EKEFELSHNSKAASVHFCGYKAYYEDNDDSEEEGETDSEDEDIPLITTEQN--------- 445
Query: 247 FKGSGRAKGKYESSGQKKSSIKEVTCFECKESGHYKSDCPKLKKDKKPKKHFKTKKSLMV 306
G+ + K E E K S K+D K+ P K+ T K + V
Sbjct: 446 ----GKPEPKAE---------------ELKVSEPKKADA----KNAAPAKNAATAKHVKV 556
Query: 307 T--FDESESEDVDSDGEVQGLMAIVKDKGAESKEAVDSDSESEGDPNSDDENE 357
DE +D +SD E+ + E DSDSE E D + DDE E
Sbjct: 557 VDPKDEDSDDDDESDDEI--------GSSDDEMENADSDSEDEDDSDEDDEEE 691
>TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~unknown
protein {Arabidopsis thaliana}, partial (46%)
Length = 2077
Score = 41.2 bits (95), Expect = 0.003
Identities = 45/202 (22%), Positives = 86/202 (42%), Gaps = 28/202 (13%)
Frame = +2
Query: 203 KVEKSKALQAEEEESEEASE--DSDEDELTLISKRLNRIWKHRQSKFK------------ 248
K+EK K ++ EEEES++ E D D+D+ + K+ + + F+
Sbjct: 344 KLEKKKRVELEEEESDDFDEELDDDDDDFEGVEKKKAKGGSEGEDDFEEEDDEDEEGSED 523
Query: 249 --GSGRAKGKYESSG--QKKSSIKEVTCFECKESGHYKSDCPKLKKDKKPKKHFKTKKSL 304
K K + G K I E+T + KE +Y+ + + D+ ++ + +K+
Sbjct: 524 EDDEEDEKEKVKGGGIEDKFLKIDELTEYLEKEEDNYEKGEERDEADEDSEEDDELEKAG 703
Query: 305 MVTFDESESEDVDSDGEVQGLMAIVK---------DKGAESKEAVDSDSESEGDPNSDDE 355
D+ + +D D + E M V+ +KG++ K+ + S E D S +
Sbjct: 704 EFEMDDEDDDDDDEEDEEAEDMGNVRYEDFFGGKNEKGSKRKDQLLEVSGDEDDMESTKQ 883
Query: 356 NEVFAS-FSTSELKHALSDIMD 376
+ AS + K+++ D D
Sbjct: 884 KKRTASTYEKQRGKNSIKDSAD 949
>TC76402 weakly similar to GP|18086561|gb|AAL57705.1 At2g01100/F23H14.7
{Arabidopsis thaliana}, partial (43%)
Length = 1449
Score = 41.2 bits (95), Expect = 0.003
Identities = 37/145 (25%), Positives = 67/145 (45%), Gaps = 2/145 (1%)
Frame = +3
Query: 176 KCHELKRSEMQDLKKKSIALKSKSE-KAKVEKSKALQAEEEESEEASEDSD-EDELTLIS 233
K E K + ++ +K A K+K++ +AK +K K + ++ + D D +DE S
Sbjct: 708 KIMERKEAPLKWEQKLEAAAKAKADAEAKEKKLKTAKHKKRSGSDTDSDRDSDDERKRAS 887
Query: 234 KRLNRIWKHRQSKFKGSGRAKGKYESSGQKKSSIKEVTCFECKESGHYKSDCPKLKKDKK 293
KR +R KHR+ SG + + E S + K+ + ES + SD + +++
Sbjct: 888 KRSHR--KHRKHSHIDSGDHEKRKEKSSKWKTKKRS------SESSDFSSDESESSSEEE 1043
Query: 294 PKKHFKTKKSLMVTFDESESEDVDS 318
++ K +K + S S DS
Sbjct: 1044KRRKKKQRKKIRDQDSRSNSSGSDS 1118
>TC79428 similar to PIR|T09661|T09661 ascorbate oxidase promoter-binding
protein AOBP - winter squash, partial (27%)
Length = 997
Score = 40.4 bits (93), Expect = 0.006
Identities = 57/237 (24%), Positives = 90/237 (37%), Gaps = 21/237 (8%)
Frame = +1
Query: 291 DKKPKKHFKTKKSLMVTFDES-----ESEDVDSDGEVQGLMAIVKDKGAESKEAVDSDSE 345
D K KK +TK + F + E E + DGE A D +E DS+SE
Sbjct: 100 DLKKKKMIETKDPEIKLFGKKILFPGEGEALMIDGEENVSPAAAMDV----EEERDSESE 267
Query: 346 SEGDPNSDDENEVFASFSTSELKHA-----LSDIMDKYNSLLSTHKKLKKNLSAVSKTPS 400
+E D + E + A T E K A ++I + NS + + KTPS
Sbjct: 268 NEDD-EEETEKDPEADKDTEEKKEADPPPDAAEIKNNNNSAATLPE-------GNPKTPS 423
Query: 401 EHEKIISDLKNENHALVNSNSVLKNQIAKLEEIVACDASDCRNE-----SKYEKSFQRFL 455
E+ ++ N+N + + K ++++ C + + + Y + R+
Sbjct: 424 IDEETSKSENEQSETTANNNDTQEKTLKKPDKLLLCPRCNSADTKFCYYNNYNVNQPRYF 603
Query: 456 AKSVDRSLMASMIYGVSRNGMHGIGYSKPIRNEPS------VSKAKSLYECFVPSGT 506
K+ R A G RN G G K N S +S+A P+GT
Sbjct: 604 CKACQRYWTAG---GTMRNVPVGAGRRKNKNNSSSHYRHITISEALDAARIISPNGT 765
>TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protein {Oryza
sativa (japonica cultivar-group)}, partial (62%)
Length = 2345
Score = 40.0 bits (92), Expect = 0.007
Identities = 45/210 (21%), Positives = 97/210 (45%), Gaps = 3/210 (1%)
Frame = +1
Query: 183 SEMQDLKKKSIALKSKSE-KAKVEKSKALQAEEEESEEASEDSDEDELTLISKRLNRIWK 241
S + ++ + + +SKSE K + E + + + + ED+ D +K + +
Sbjct: 418 SSVVPVQNEDVPQESKSEVKEQTEVREQVSETDNSNARQFEDNPGDLPEDATKGDSNVSS 597
Query: 242 HRQSKFKGSGRAKGKYESSGQKKSSIKEVTCFECKESGHYKSDCPKLKKDKKPKKHFKTK 301
+S+ + ++ ++ + K + E + E + G S K + + +K +++
Sbjct: 598 EEKSEENSTEKSSEDTKTEDEGKKTEDEGSNTENNKDGEEAST--KESESDESEKKDESE 771
Query: 302 KSLMVTFDESESEDVDSDGEVQGLMAIVKDKGAESKEAVDSDSESEGDPNSDDE--NEVF 359
++ DESE + DS+ E K + +++KE+ ++ SE D N+ D+ NEVF
Sbjct: 772 ENNKSDSDESEKKSSDSN-ETTDSNVEEKVEQSQNKESDENASEKNTDDNAKDQSSNEVF 948
Query: 360 ASFSTSELKHALSDIMDKYNSLLSTHKKLK 389
S + SEL + + +++ S K+ K
Sbjct: 949 PSGAQSELLNETTTQTGSFSTQCSRVKE*K 1038
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 38.9 bits (89), Expect = 0.016
Identities = 29/109 (26%), Positives = 51/109 (46%)
Frame = +2
Query: 1439 CIRKHQSTSFQVIVMLIMLEIEQREKVLLEIVNFWEAI*SHGQARGNQPLHYQLQRQNIS 1498
CIR + S++ ++IM I EKVLL++ + SHG + N+ L Y+ +QN+
Sbjct: 335 CIRGMEVRSWRHTQIVIMQVI*MIEKVLLDMYLCLVQVQSHGHLKSNRWLLYRQLKQNLL 514
Query: 1499 QQQYAALRCSG*NISWRIIRSLRAISQFIVITLLQSR*ARILSCIQGQS 1547
Q + ++ G R + +L + + T Q + C G+S
Sbjct: 515 QLPFVLVKVFGCEEFSRNLVTLNLEASLCIATTTQQSSFQRTRCFMGRS 661
>TC87387 homologue to PIR|T47775|T47775 hypothetical protein F24I3.230 -
Arabidopsis thaliana, partial (14%)
Length = 1074
Score = 38.1 bits (87), Expect = 0.028
Identities = 44/180 (24%), Positives = 78/180 (42%), Gaps = 1/180 (0%)
Frame = +3
Query: 179 ELKRSEMQDLKKKSIALKSKSEKAKVEKSKALQAEEEESEEASEDSDEDELTLISKRLNR 238
++K+ + + K+K +S A +KSK + + EE + + D K+
Sbjct: 225 KVKKDDGEGRKRKKH--ESADSPAPAKKSKVAEVDGEEKVKTKKVDDAAVEVEDDKKEK- 395
Query: 239 IWKHRQSKFKGSGRAKGKYESSGQKKSSIKEVTCFECKESGHYKSDCPKLKK-DKKPKKH 297
K ++ K K +G A E ++K + KE G S P+++K DKK KKH
Sbjct: 396 --KKKKKKDKENGAAASDEEKVEKEKKK-------KHKEKGEDGS--PEVEKSDKKKKKH 542
Query: 298 FKTKKSLMVTFDESESEDVDSDGEVQGLMAIVKDKGAESKEAVDSDSESEGDPNSDDENE 357
+T + D+SE + D E + A + + G + A S+ + + N D + E
Sbjct: 543 KETSEVGSPEVDKSEKKKKKKDKEAKDNAADISN-GNDESNADRSEKKHKKKKNKDAQEE 719
>TC88390 similar to GP|13272457|gb|AAK17167.1 unknown protein {Arabidopsis
thaliana}, partial (11%)
Length = 976
Score = 38.1 bits (87), Expect = 0.028
Identities = 56/250 (22%), Positives = 95/250 (37%), Gaps = 17/250 (6%)
Frame = +2
Query: 181 KRSEMQDLKKKSIALKSKSE-----------KAKVEKSKALQAEEEESEEASEDSDEDEL 229
K+ E Q L ++++ SE K K+E K E++ E ED D+DE
Sbjct: 53 KKRETQSLTMQTVSTSPLSEVSRKFENDFFSKLKIEDEKIENENEDKHEVEEEDEDDDEF 232
Query: 230 TLISKRLNRIWKHRQSKFKGSGRAKGKYESSGQKKSSIKEVTCFECKESGHYKSDCPKLK 289
T + + KGS + GQ I+ + F ++ H+ D
Sbjct: 233 TFVF-----------ADPKGSPISADDVFDKGQ----IRPIFPFFGQDL-HFTDDYDNES 364
Query: 290 KDKKPKKHFKTKKSLMVTFDESESEDVDSDGEVQGLMAIVKDKGA-ESKEAVDSDSESEG 348
++ P + F + S S+ S V+G K A +S S
Sbjct: 365 GNRSPVRKFFVE-------SPSSSKTATSSAAVEGTYCEWNPKKAVKSNSTGSSKLWKLR 523
Query: 349 DP---NSDDENEVFASFSTSELKHALSDIMDKYNSLLSTHKKLKKNLSAVSKTPSEHEK- 404
DP ++ D + F + S+++ S + N ++ K +K +A PS HEK
Sbjct: 524 DPKLRSNSDGKDAFVFLNPSKVEKTSSSAGETKNDVVKKVKMVKGKKAA--PAPSAHEKH 697
Query: 405 -IISDLKNEN 413
++S + EN
Sbjct: 698 YVMSKARKEN 727
>TC89779 GP|12835923|dbj|BAB23419. data source:MGD source key:MGI:1333811
evidence:ISS~methyl-CpG binding domain protein
1~putative, partial (2%)
Length = 824
Score = 37.4 bits (85), Expect = 0.047
Identities = 50/202 (24%), Positives = 87/202 (42%), Gaps = 7/202 (3%)
Frame = +1
Query: 160 TRDVENMSLEELISILKCHELKRSEMQDLKKKSIALKSK---SEKAKVEKSKALQAE--E 214
T+ ++ SLE H+LK S KKK + K S+ AK K A +++ +
Sbjct: 10 TKKIKRSSLET-----SGHKLKGSSGSPSKKKKKIVNGKQSPSKPAKPRKKYASKSDFYQ 174
Query: 215 EESEEASEDSDEDELTLISKRLNRIWKHRQSKFKGSGRAKGKYESSGQKKSSIKEVTCFE 274
E+++E SE S+ +E T+ISK + SG S ++ +++ +
Sbjct: 175 EQAKETSEISNPEE-TMISK----------ADETDSG-------GSEEELTAVHNEITKK 300
Query: 275 CKESGHYKSDCPKLKKDKKPKK--HFKTKKSLMVTFDESESEDVDSDGEVQGLMAIVKDK 332
K+S P+ K+ KK K H + ++E SED +S + V +
Sbjct: 301 GKKSNKKVRSVPRGKRLKKTKNFHHIEESDDDKRDYNERISEDRESVPQYSSEEKKVDES 480
Query: 333 GAESKEAVDSDSESEGDPNSDD 354
+ +SESEG+ ++ D
Sbjct: 481 SERESVHGEEESESEGEQDNSD 546
>TC91264 weakly similar to PIR|T27543|T27543 hypothetical protein ZC395.10 -
Caenorhabditis elegans, partial (24%)
Length = 767
Score = 36.6 bits (83), Expect = 0.081
Identities = 42/157 (26%), Positives = 61/157 (38%), Gaps = 2/157 (1%)
Frame = +2
Query: 264 KSSIKEVTCFECKESGHYKSDC-PKLKKDKKPKKHFKTKKSLMVTFDESESEDVDSDGEV 322
K S F E K D P+L+KDKK KT S DE E ED +
Sbjct: 179 KRSTTPRNIFLVLEKAEQKQDYWPRLQKDKKKLPFLKTDFSKWK--DEDEDEDEGGPDPI 352
Query: 323 QGL-MAIVKDKGAESKEAVDSDSESEGDPNSDDENEVFASFSTSELKHALSDIMDKYNSL 381
G+ + + +G++S DSD E G DD ++ K S+I ++ S
Sbjct: 353 GGMDFSNLMGEGSDS-ALDDSDEEMPGLETVDDTTKI-----NDSTKEESSEISEESKSE 514
Query: 382 LSTHKKLKKNLSAVSKTPSEHEKIISDLKNENHALVN 418
K +K + + K EK I + E +N
Sbjct: 515 EKEIKSEEKEIKSEEKEIKSEEKEIKSEEKEK*NKIN 625
>TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.2 (TTS-2.2)
{Homo sapiens}, partial (2%)
Length = 1522
Score = 35.8 bits (81), Expect = 0.14
Identities = 18/61 (29%), Positives = 28/61 (45%)
Frame = -1
Query: 242 HRQSKFKGSGRAKGKYESSGQKKSSIKEVTCFECKESGHYKSDCPKLKKDKKPKKHFKTK 301
H+Q + +GS + E + + ++E C+ C E GH +C KK KK K
Sbjct: 400 HKQRRRRGSSSSVSSAEGAPPPRRDMRE--CYSCHERGHIARNCTNTSAAKKKKKGPKVA 227
Query: 302 K 302
K
Sbjct: 226 K 224
>TC91075 similar to PIR|T48025|T48025 hypothetical protein T12C14.30 -
Arabidopsis thaliana, partial (29%)
Length = 686
Score = 35.4 bits (80), Expect = 0.18
Identities = 30/121 (24%), Positives = 51/121 (41%), Gaps = 6/121 (4%)
Frame = +2
Query: 205 EKSKALQAEEEESEEASEDSDEDEL-TLISKRLNRIWKHRQSKFKGSGRAKGKYESSGQK 263
E++K+ + S+E E D + + T R ++W+ AK K K
Sbjct: 218 ERAKSNKRSNAGSDEEDEVRDPNAIPTGFISRDAKVWE-----------AKSKATERNWK 364
Query: 264 KSSIKEVTCFECKESGHYKSDCPK-LKKDKKPKKHFK----TKKSLMVTFDESESEDVDS 318
K +E+ C C ESGH+ CP L + K ++ F+ +S+ F E ++
Sbjct: 365 KRKEEEMICKLCGESGHFTQGCPSTLGANNKSQELFQRIPARDRSVRALFTEKVISKIER 544
Query: 319 D 319
D
Sbjct: 545 D 547
>TC86560 similar to GP|8843737|dbj|BAA97285.1 myosin heavy chain-like
{Arabidopsis thaliana}, partial (28%)
Length = 2450
Score = 35.4 bits (80), Expect = 0.18
Identities = 25/80 (31%), Positives = 43/80 (53%), Gaps = 5/80 (6%)
Frame = +2
Query: 179 ELKRSEMQDLKKKSIALKSKSEKAKV-----EKSKALQAEEEESEEASEDSDEDELTLIS 233
E R E++D+K K+ LK ++E K+ EK EE E+ +A+E S +++T+++
Sbjct: 1499 EEARREVEDMKNKTDELKKEAEATKLALEEAEKKLKEATEEAEAAKAAEASAIEQITVLT 1678
Query: 234 KRLNRIWKHRQSKFKGSGRA 253
+R + R S SG A
Sbjct: 1679 ERTS---ARRASTSSESGAA 1729
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 35.0 bits (79), Expect = 0.23
Identities = 22/48 (45%), Positives = 30/48 (61%)
Frame = +2
Query: 1207 VPS*MVISQRKSMFINPQVLKMKRNQTMCSN*RNHSTV*SKLPEHGMR 1254
+P *MV +RK + +N + LK+ N C++* NHS *SK HGMR
Sbjct: 395 MPF*MVFFKRKCICLNLRDLKLLIN-LWCAS*TNHSMA*SKHHVHGMR 535
>BG448933 similar to PIR|T09648|T09 nucleolin homolog nuM1 - alfalfa, partial
(29%)
Length = 663
Score = 33.9 bits (76), Expect = 0.52
Identities = 39/177 (22%), Positives = 73/177 (41%), Gaps = 2/177 (1%)
Frame = +2
Query: 181 KRSEMQDLKKKSIALKSKSEK--AKVEKSKALQAEEEESEEASEDSDEDELTLISKRLNR 238
KR +++K S A K K E+ AK + K ++ EE SEE+SE EDE ++
Sbjct: 164 KRQAEEEVKAVS-AKKQKVEEVAAKQKALKVVKKEESSSEESSES--EDEQPVV------ 316
Query: 239 IWKHRQSKFKGSGRAKGKYESSGQKKSSIKEVTCFECKESGHYKSDCPKLKKDKKPKKHF 298
+A + + KK ++K+ E S ++ KKP
Sbjct: 317 -------------KAPAPSKKTPAKKGNVKKAQPETTSEESDSDSSSSDEEEVKKPVSKA 457
Query: 299 KTKKSLMVTFDESESEDVDSDGEVQGLMAIVKDKGAESKEAVDSDSESEGDPNSDDE 355
K+ + ++ D D + + + K+ A +K+A + +++ + D+E
Sbjct: 458 VPSKNGSAPAKKVDTSDEDKKPAAKAVPS--KNGSAPAKKAASDEEDTDESSDEDEE 622
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.355 0.156 0.545
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,465,432
Number of Sequences: 36976
Number of extensions: 683613
Number of successful extensions: 8302
Number of sequences better than 10.0: 82
Number of HSP's better than 10.0 without gapping: 2661
Number of HSP's successfully gapped in prelim test: 422
Number of HSP's that attempted gapping in prelim test: 5381
Number of HSP's gapped (non-prelim): 3538
length of query: 1602
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1493
effective length of database: 4,984,343
effective search space: 7441624099
effective search space used: 7441624099
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.6 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0369a.2


