

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0346.7
(449 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
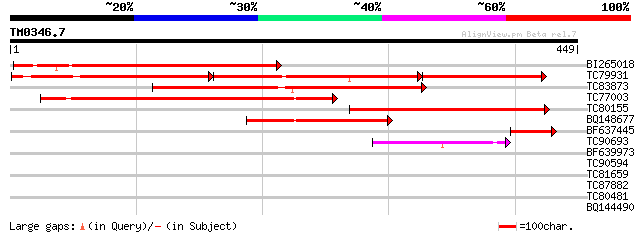
Score E
Sequences producing significant alignments: (bits) Value
BI265018 similar to PIR|T07821|T07 Ca2+/H+-exchanging protein - ... 325 3e-89
TC79931 similar to GP|4512263|dbj|BAA75232.1 H+/Ca2+ exchanger 2... 127 5e-86
TC83873 similar to PIR|T07821|T07821 Ca2+/H+-exchanging protein ... 299 1e-81
TC77003 similar to PIR|T07821|T07821 Ca2+/H+-exchanging protein ... 287 7e-78
TC80155 similar to PIR|T07821|T07821 Ca2+/H+-exchanging protein ... 259 2e-69
BQ148677 weakly similar to PIR|T07821|T07 Ca2+/H+-exchanging pro... 123 2e-28
BF637445 similar to PIR|T07821|T078 Ca2+/H+-exchanging protein -... 65 4e-11
TC90693 similar to GP|20466169|gb|AAM20402.1 unknown protein {Ar... 44 1e-04
BF639973 similar to GP|17065002|gb Unknown protein {Arabidopsis ... 30 1.9
TC90594 similar to GP|10178077|dbj|BAB11496. gene_id:K19B1.7~unk... 30 2.5
TC81659 weakly similar to GP|14596093|gb|AAK68774.1 putative pro... 30 2.5
TC87882 homologue to GP|4589852|dbj|BAA76902.1 cycloartenol synt... 28 5.5
TC80481 similar to GP|22022540|gb|AAM83228.1 At1g14620/T5E21_15 ... 28 7.2
BQ144490 28 7.2
>BI265018 similar to PIR|T07821|T07 Ca2+/H+-exchanging protein - mung bean,
partial (35%)
Length = 661
Score = 325 bits (832), Expect = 3e-89
Identities = 172/214 (80%), Positives = 189/214 (87%), Gaps = 2/214 (0%)
Frame = +2
Query: 4 SVVSPSPSMNEDLESNTTNEETSHENLSRSMV--KRSDSVLVTNNINARFQMLRIFMTNL 61
S P +MNEDLE+N N ET N+S + + K+SD VLVTNN+ RFQ+LR MTN+
Sbjct: 32 STSRPFRTMNEDLENN--NVETVQHNISSNSIVRKKSDMVLVTNNV--RFQILRNVMTNM 199
Query: 62 RVVLLGTKLVVLFPAVPLAVAADFYKFGRPWIFAFSLLGLAPLAERVSFLTEQIAYFTGP 121
+ V+LGTKLVVLFPAVPLAVAADFY GRPWIFA SLLGLAPLAERVSFLTEQIAYFTGP
Sbjct: 200 KEVMLGTKLVVLFPAVPLAVAADFYSLGRPWIFALSLLGLAPLAERVSFLTEQIAYFTGP 379
Query: 122 TVGGLLNATCGNATEMIIAILALRKNKVNVVKFSLLGSVLSNLLLVLGSSLLCGGLANLK 181
TVGGLLNATCGNATEMIIAILAL +NK++VVKFSLLGS+LSNLLLVLGSSLLCGGLANLK
Sbjct: 380 TVGGLLNATCGNATEMIIAILALHQNKIHVVKFSLLGSILSNLLLVLGSSLLCGGLANLK 559
Query: 182 REQRYDRKQADVNSLLLLLGLLCHLLPLLFKYSL 215
REQRYDRKQADVNSLLLLLGLLCHLLPL+FKY+L
Sbjct: 560 REQRYDRKQADVNSLLLLLGLLCHLLPLMFKYAL 661
>TC79931 similar to GP|4512263|dbj|BAA75232.1 H+/Ca2+ exchanger 2 {Ipomoea
nil}, partial (76%)
Length = 1975
Score = 127 bits (319), Expect(3) = 5e-86
Identities = 58/98 (59%), Positives = 79/98 (80%)
Frame = +2
Query: 328 AEHAGSIIFAFKNKLDISLGVAMGSATQISMFVVPLSVIVAWIMGIKMDLDFNLLETGCL 387
AEHA +I+FA K+KLDI++GVA+GS+TQISMFV+P V+V W MG +MDL+F L ET L
Sbjct: 1400 AEHASAIMFAVKDKLDITIGVAVGSSTQISMFVIPFCVVVGWCMGKEMDLNFQLFETATL 1579
Query: 388 AFAIIVTAFTLQDGTSHYMKGVVLTLCYLVIAACFFVN 425
++V AF +Q+GTS+Y KG++L LCYL++AA FFV+
Sbjct: 1580 FITVLVVAFMMQEGTSNYFKGLMLILCYLIVAASFFVH 1693
Score = 117 bits (293), Expect(3) = 5e-86
Identities = 67/162 (41%), Positives = 101/162 (61%), Gaps = 2/162 (1%)
Frame = +3
Query: 2 PVSVVSPSPSMNEDLESNTTNEETSHE-NLSRSMVKRSDSVLVTNNINARFQMLRIFMTN 60
P S SPS + +NT++ H + S + +S S N+ R + +
Sbjct: 441 PFSSTSPSST-----SANTSSSVKPHSFHFDSSSLPKSSSPSFFNHSRTR-----LISKS 590
Query: 61 LRVVLLGTKLVVLFPAVPLAVAADFYKFGRP-WIFAFSLLGLAPLAERVSFLTEQIAYFT 119
+ +VL+ K+ +L P P A+ ++ G+ W+F F+LLG+APLAER+ + TEQ+A++T
Sbjct: 591 IYIVLIKAKINILLPFGPFAIFLHYFSGGKHVWVFFFALLGIAPLAERLGYATEQLAFYT 770
Query: 120 GPTVGGLLNATCGNATEMIIAILALRKNKVNVVKFSLLGSVL 161
GPT+GGLLNAT GNATEMII+I AL+ + + VV+ SLLGS+L
Sbjct: 771 GPTLGGLLNATFGNATEMIISIYALKSDMIRVVQQSLLGSIL 896
Score = 113 bits (282), Expect(3) = 5e-86
Identities = 68/169 (40%), Positives = 106/169 (62%), Gaps = 3/169 (1%)
Frame = +1
Query: 162 SNLLLVLGSSLLCGGLANLKREQRYD-RKQADVNSLLLLLGLLCHLLPLLFKYSLAGGDY 220
SN+LLVLG + GG+ +L+ R+ R VNS LLL+ ++ L P + ++ + +
Sbjct: 898 SNMLLVLGCAFFTGGIVSLQ*TFRFSIRHLPVVNSGLLLMAVMGILFPAVLHFTHS--EV 1071
Query: 221 SIATSTLQLSRASSIVMLLAYVAYIFFQLKTHRRLFDAQEVIIDIEE--DDEEKAVIGFW 278
+ S L LSR SS +ML+AY +Y++FQL++ + + + +D E D+EE+ +
Sbjct: 1072 HLGKSVLSLSRFSSCIMLVAYASYLYFQLRSQQNFYAPVDQEVDTSENSDEEEELELTQS 1251
Query: 279 SAFTWLVGMTLVISLLSEYVVGTIEAASDSWGISVSFISIILLPIVGNA 327
A WL +T+ +S+LS Y+V IE AS+S +SV+FIS+ILLPIVGNA
Sbjct: 1252 EAIIWLAILTVWVSVLSGYLVDAIEGASESLNMSVAFISVILLPIVGNA 1398
>TC83873 similar to PIR|T07821|T07821 Ca2+/H+-exchanging protein - mung
bean, partial (47%)
Length = 693
Score = 299 bits (766), Expect = 1e-81
Identities = 152/220 (69%), Positives = 188/220 (85%), Gaps = 3/220 (1%)
Frame = +1
Query: 114 QIAYFTGPTVGGLLNATCGNATEMIIAILALRKNKVNVVKFSLLGSVLSNLLLVLGSSLL 173
QI+Y+TGPTVGGLLNATCGNATE+IIAI AL NK++VVK+SLLGS+LSNLLLVLG+SLL
Sbjct: 46 QISYYTGPTVGGLLNATCGNATELIIAIFALSNNKIDVVKYSLLGSILSNLLLVLGTSLL 225
Query: 174 CGGLANLKREQRYDRKQADVNSLLLLLGLLCHLLPLLFKYSLAGGDYSI---ATSTLQLS 230
CGG+ANL+ EQ+YDR+QAD+N L+LL+ L CHL+PLLF+Y GD S+ A S+LQ S
Sbjct: 226 CGGIANLRAEQKYDRRQADINLLMLLVALSCHLVPLLFQY----GDASLAVNANSSLQFS 393
Query: 231 RASSIVMLLAYVAYIFFQLKTHRRLFDAQEVIIDIEEDDEEKAVIGFWSAFTWLVGMTLV 290
RA+SIVM++AY AY+FFQL THR+LF+AQ+ + E+ E+AVIGFWS F WLV MT+
Sbjct: 394 RAASIVMVIAYFAYLFFQLWTHRQLFEAQDEDDEDCENASEEAVIGFWSGFAWLVIMTVF 573
Query: 291 ISLLSEYVVGTIEAASDSWGISVSFISIILLPIVGNAAEH 330
I++LSEYVV TI+ ASDSWG+SVSF+SII+LPIVGNAAEH
Sbjct: 574 IAILSEYVVDTIQDASDSWGLSVSFLSIIMLPIVGNAAEH 693
>TC77003 similar to PIR|T07821|T07821 Ca2+/H+-exchanging protein - mung
bean, partial (56%)
Length = 907
Score = 287 bits (734), Expect = 7e-78
Identities = 151/235 (64%), Positives = 184/235 (78%)
Frame = +2
Query: 25 TSHENLSRSMVKRSDSVLVTNNINARFQMLRIFMTNLRVVLLGTKLVVLFPAVPLAVAAD 84
T+H S S+ K+SD LV+ R LR + N++ V+LGTKL +LFPA+P A+ A
Sbjct: 215 TAHGMSSSSLRKKSDHALVSR---IRCASLRNVLVNIQEVILGTKLSILFPAIPAAIVAH 385
Query: 85 FYKFGRPWIFAFSLLGLAPLAERVSFLTEQIAYFTGPTVGGLLNATCGNATEMIIAILAL 144
+ GRPW+F SLLGL PLAERVSFLTEQIA+FTGPTVGGLLNATCGN TE+IIAI AL
Sbjct: 386 YLGLGRPWVFILSLLGLTPLAERVSFLTEQIAFFTGPTVGGLLNATCGNITELIIAIFAL 565
Query: 145 RKNKVNVVKFSLLGSVLSNLLLVLGSSLLCGGLANLKREQRYDRKQADVNSLLLLLGLLC 204
N++ VVK+SLLGS+LSNLLLVLG+SLLCGG+ANL EQ+YDR+QADVNSL+LLL LLC
Sbjct: 566 SSNQIAVVKYSLLGSILSNLLLVLGTSLLCGGIANLGVEQKYDRRQADVNSLMLLLALLC 745
Query: 205 HLLPLLFKYSLAGGDYSIATSTLQLSRASSIVMLLAYVAYIFFQLKTHRRLFDAQ 259
HLLPLLF +S A + ++ S L LSR++SIVML AY AY+ FQL THR+LF A+
Sbjct: 746 HLLPLLFTFSAASPELTVEPS-LYLSRSASIVMLAAYFAYLIFQLWTHRQLF*AE 907
>TC80155 similar to PIR|T07821|T07821 Ca2+/H+-exchanging protein - mung
bean, partial (35%)
Length = 850
Score = 259 bits (661), Expect = 2e-69
Identities = 129/158 (81%), Positives = 143/158 (89%)
Frame = +1
Query: 270 EEKAVIGFWSAFTWLVGMTLVISLLSEYVVGTIEAASDSWGISVSFISIILLPIVGNAAE 329
EEKAVIGF S+F WL GMT+ I+LLSEYVV TI+ ASDSWG+SVSF+SIILLPIVGNAAE
Sbjct: 7 EEKAVIGF*SSFGWLAGMTVFIALLSEYVVDTIQDASDSWGLSVSFLSIILLPIVGNAAE 186
Query: 330 HAGSIIFAFKNKLDISLGVAMGSATQISMFVVPLSVIVAWIMGIKMDLDFNLLETGCLAF 389
HAG+IIFAFKNKLDISLGVA+GSATQISMF VPL VIVAWIMG+KMDL+FNLLETG LA
Sbjct: 187 HAGAIIFAFKNKLDISLGVALGSATQISMFAVPLCVIVAWIMGVKMDLNFNLLETGSLAL 366
Query: 390 AIIVTAFTLQDGTSHYMKGVVLTLCYLVIAACFFVNKT 427
AIIVT FTLQDGTSHY+KG++L LCY VI ACFFV +T
Sbjct: 367 AIIVTGFTLQDGTSHYLKGLILLLCYFVIGACFFVQRT 480
>BQ148677 weakly similar to PIR|T07821|T07 Ca2+/H+-exchanging protein - mung
bean, partial (25%)
Length = 554
Score = 123 bits (308), Expect = 2e-28
Identities = 65/116 (56%), Positives = 83/116 (71%)
Frame = +2
Query: 188 RKQADVNSLLLLLGLLCHLLPLLFKYSLAGGDYSIATSTLQLSRASSIVMLLAYVAYIFF 247
+KQADVNSL+LLL CHLLPLLF +S A + ++ S L LSR++ I ML AY AY+ F
Sbjct: 158 QKQADVNSLMLLLAXXCHLLPLLFTFSAASPELTVEPS-LYLSRSAXIXMLAAYFAYLIF 334
Query: 248 QLKTHRRLFDAQEVIIDIEEDDEEKAVIGFWSAFTWLVGMTLVISLLSEYVVGTIE 303
QL THR+LF+A++ + EE+AVIGFWS+F WL GMT I+L S YVV TI+
Sbjct: 335 QLWTHRQLFEAEDEGEGEXNEAEEEAVIGFWSSFGWLAGMTXFIALXSXYVVDTIQ 502
>BF637445 similar to PIR|T07821|T078 Ca2+/H+-exchanging protein - mung bean,
partial (6%)
Length = 453
Score = 65.5 bits (158), Expect = 4e-11
Identities = 28/37 (75%), Positives = 34/37 (91%)
Frame = +3
Query: 397 TLQDGTSHYMKGVVLTLCYLVIAACFFVNKTAQINQH 433
+LQDGTSHY+KGV+LTLCY+VI+ACFFV KT QIN +
Sbjct: 6 SLQDGTSHYLKGVILTLCYIVISACFFVLKTPQINHY 116
>TC90693 similar to GP|20466169|gb|AAM20402.1 unknown protein {Arabidopsis
thaliana}, partial (24%)
Length = 789
Score = 43.9 bits (102), Expect = 1e-04
Identities = 27/112 (24%), Positives = 57/112 (50%), Gaps = 3/112 (2%)
Frame = +1
Query: 288 TLVISLLSEYVVGTIEAASDSWGISVSFISIILLPIVGNAAEHAGSIIFAFKNK---LDI 344
+L+ + ++ +V ++ SD+ I FIS I LP+ N++E +IIFA ++K +
Sbjct: 373 SLIAAAFADPLVDVVDNFSDATSIPAFFISFIFLPLATNSSEAVSAIIFASRDKRQTASL 552
Query: 345 SLGVAMGSATQISMFVVPLSVIVAWIMGIKMDLDFNLLETGCLAFAIIVTAF 396
+ G+ T ++ + + + + ++ G+ D +L L I++ AF
Sbjct: 553 TFSEIYGAVTMNNVLCLSVFLALVYVRGLTWDFSSEVLV--ILIVCIVMGAF 702
>BF639973 similar to GP|17065002|gb Unknown protein {Arabidopsis thaliana},
partial (22%)
Length = 679
Score = 30.0 bits (66), Expect = 1.9
Identities = 28/105 (26%), Positives = 48/105 (45%), Gaps = 6/105 (5%)
Frame = +2
Query: 113 EQIAYFTGPTVGGLLNATCGNATEMIIAILALRKNKVNVVKF---SLLGSVLSNLLLVLG 169
++I FTGP +G L CG +I + + + + + ++ L L + L
Sbjct: 323 KEIVLFTGPAIGLWL---CGPLMSLIDTAVVGQGSSIELAALGPATVFCDYLGYLFMFLS 493
Query: 170 ---SSLLCGGLANLKREQRYDRKQADVNSLLLLLGLLCHLLPLLF 211
S+++ LA RE+ S+LL +GL+C L+ LLF
Sbjct: 494 IATSNMVATALAKQDREEVQHHI-----SVLLFIGLVCGLVMLLF 613
>TC90594 similar to GP|10178077|dbj|BAB11496. gene_id:K19B1.7~unknown
protein {Arabidopsis thaliana}, partial (11%)
Length = 603
Score = 29.6 bits (65), Expect = 2.5
Identities = 19/81 (23%), Positives = 40/81 (48%), Gaps = 1/81 (1%)
Frame = +3
Query: 196 LLLLLGLLCHLLPLLFKYSLAGGDYSIATSTLQLSRASSIVMLLAYVAYIFFQLKTHRRL 255
+ ++L +L H LPL+ DYS + L R++ IV+ + ++ ++ HRR
Sbjct: 294 IFMVLLILRHTLPLIIS---GNKDYSFPLFMVLLFRSAGIVVPIYFMVRAMALIQRHRRQ 464
Query: 256 F-DAQEVIIDIEEDDEEKAVI 275
+ ++ +D+ E+A +
Sbjct: 465 HREHPSALVSSSDDEIEEAAL 527
>TC81659 weakly similar to GP|14596093|gb|AAK68774.1 putative protein
{Arabidopsis thaliana}, partial (36%)
Length = 684
Score = 29.6 bits (65), Expect = 2.5
Identities = 15/55 (27%), Positives = 31/55 (56%)
Frame = +2
Query: 6 VSPSPSMNEDLESNTTNEETSHENLSRSMVKRSDSVLVTNNINARFQMLRIFMTN 60
VS +++E L ++ S+ +L S++KR ++ L++NN + + R F +N
Sbjct: 14 VSLKQNISESLCPLKRHQSMSYRSLFLSLIKRRNTGLISNNYHHNHRSARFFTSN 178
>TC87882 homologue to GP|4589852|dbj|BAA76902.1 cycloartenol synthase
{Glycyrrhiza glabra}, partial (51%)
Length = 1408
Score = 28.5 bits (62), Expect = 5.5
Identities = 22/81 (27%), Positives = 41/81 (50%), Gaps = 1/81 (1%)
Frame = +2
Query: 205 HLLPLLFKYSLAGGDYSIATSTLQLSRASSIVMLLAYVA-YIFFQLKTHRRLFDAQEVII 263
H+ + +S A + I+ T + +A V+LL+ +A I + +RL+DA VII
Sbjct: 293 HISKGAWPFSTADHGWPISDCTAEGLKA---VLLLSKIAPEIVGEPLDAKRLYDAVNVII 463
Query: 264 DIEEDDEEKAVIGFWSAFTWL 284
++ +D A +++WL
Sbjct: 464 SLQNEDGGLATYELTRSYSWL 526
>TC80481 similar to GP|22022540|gb|AAM83228.1 At1g14620/T5E21_15
{Arabidopsis thaliana}, partial (88%)
Length = 1010
Score = 28.1 bits (61), Expect = 7.2
Identities = 16/43 (37%), Positives = 26/43 (60%), Gaps = 1/43 (2%)
Frame = -2
Query: 202 LLCH-LLPLLFKYSLAGGDYSIATSTLQLSRASSIVMLLAYVA 243
L CH L+ +LFK S G YSI +S + +++S + M ++A
Sbjct: 784 LHCHQLMIILFKNSACSGKYSINSSLVTQTKSSPLPMSNLFLA 656
>BQ144490
Length = 816
Score = 28.1 bits (61), Expect = 7.2
Identities = 17/60 (28%), Positives = 28/60 (46%), Gaps = 12/60 (20%)
Frame = +2
Query: 398 LQDGTSHYMKGVVLTLCYLVIAAC------------FFVNKTAQINQHPRTSFGAKPSFE 445
+ G +++K V LTLCY V+AA ++ + + HP S + PSF+
Sbjct: 71 ISPGVHNHLKQVHLTLCYAVVAAAVAAYLNALPEHWWYSYRNCLLGNHPSRSL-SPPSFQ 247
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.139 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,875,131
Number of Sequences: 36976
Number of extensions: 171175
Number of successful extensions: 1260
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 1240
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1251
length of query: 449
length of database: 9,014,727
effective HSP length: 99
effective length of query: 350
effective length of database: 5,354,103
effective search space: 1873936050
effective search space used: 1873936050
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0346.7


