

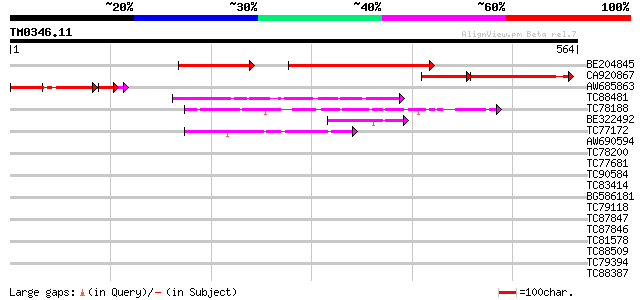
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0346.11
(564 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BE204845 similar to GP|8843796|dbj contains similarity to damage... 259 e-106
CA920867 similar to GP|10798819|dbj putative damage-specific DNA... 154 2e-58
AW685863 similar to GP|10798819|dbj putative damage-specific DNA... 117 2e-35
TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclea... 56 3e-08
TC78188 similar to PIR|C84870|C84870 probable splicing factor [i... 51 1e-06
BE322492 GP|13991907|g receptor for activated protein kinase C R... 43 4e-04
TC77172 similar to GP|20466472|gb|AAM20553.1 putative protein {A... 42 5e-04
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 41 0.001
TC78200 similar to PIR|A84578|A84578 probable WD-40 repeat prote... 41 0.001
TC77681 similar to GP|22597156|gb|AAN03465.1 nucleolar histone d... 39 0.005
TC90584 similar to GP|22136810|gb|AAM91749.1 unknown protein {Ar... 39 0.007
TC83414 weakly similar to GP|17381206|gb|AAL36415.1 unknown prot... 38 0.009
BG586181 similar to GP|4689316|gb| peroxisomal targeting signal ... 38 0.012
TC79118 similar to GP|16323188|gb|AAL15328.1 AT5g67320/K8K14_4 {... 37 0.016
TC87847 similar to GP|15929245|gb|AAH15068.1 Similar to adaptor-... 37 0.020
TC87846 similar to GP|5821151|dbj|BAA83717.1 RNA binding protein... 37 0.020
TC81578 GP|7688063|emb|CAB89693.1 constitutively photomorphogeni... 37 0.026
TC88509 similar to PIR|T01826|T01826 microfibril-associated prot... 36 0.045
TC79394 weakly similar to PIR|T49019|T49019 probable RNA binding... 35 0.059
TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 35 0.059
>BE204845 similar to GP|8843796|dbj contains similarity to damage-specific
DNA binding protein 2~gene_id:MZN1.20 {Arabidopsis
thaliana}, partial (38%)
Length = 657
Score = 259 bits (661), Expect(2) = e-106
Identities = 117/145 (80%), Positives = 134/145 (91%)
Frame = +3
Query: 278 MDINSEKGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCG 337
MDIN EKGLVLVADNFGFLHLVDMRSN++ G+A+LIHKK +KV GIHCNP++PDILLTCG
Sbjct: 222 MDINCEKGLVLVADNFGFLHLVDMRSNHRNGDAVLIHKKGSKVTGIHCNPMQPDILLTCG 401
Query: 338 NDHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKM 397
NDHYAR+WD+RR+E GSS+ L+H RVV+SAYFSP++GNKILTTS DNRLR+WDSIFG M
Sbjct: 402 NDHYARIWDMRRLEAGSSLCSLEHKRVVNSAYFSPITGNKILTTSQDNRLRIWDSIFGNM 581
Query: 398 DSPSREIVHSHDFNRYLTPFKAEWD 422
SPSREIVHSHDFNR+LTPFKAEWD
Sbjct: 582 ASPSREIVHSHDFNRHLTPFKAEWD 656
Score = 145 bits (365), Expect(2) = e-106
Identities = 66/75 (88%), Positives = 70/75 (93%)
Frame = +2
Query: 169 CAVIRYHSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKF 228
CAVI YHSRRIT LEFHPTKNNI+LSGDKKGQLGVWDF +VHEKVVY + HSC+LNNMKF
Sbjct: 2 CAVITYHSRRITSLEFHPTKNNILLSGDKKGQLGVWDFEKVHEKVVYDDKHSCILNNMKF 181
Query: 229 NPTNDCMVYSASSDG 243
NPTNDCMVYSASSDG
Sbjct: 182 NPTNDCMVYSASSDG 226
Score = 31.6 bits (70), Expect = 0.85
Identities = 15/66 (22%), Positives = 34/66 (50%)
Frame = +2
Query: 319 KVVGIHCNPIEPDILLTCGNDHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKI 378
++ + +P + +ILL+ VWD ++ +YD KH+ ++++ F+P + +
Sbjct: 29 RITSLEFHPTKNNILLSGDKKGQLGVWDFEKVHE-KVVYDDKHSCILNNMKFNPTNDCMV 205
Query: 379 LTTSLD 384
+ S D
Sbjct: 206 YSASSD 223
>CA920867 similar to GP|10798819|dbj putative damage-specific DNA binding
protein 2 {Oryza sativa (japonica cultivar-group)},
partial (16%)
Length = 761
Score = 154 bits (389), Expect(2) = 2e-58
Identities = 77/103 (74%), Positives = 89/103 (85%)
Frame = -1
Query: 459 LVAEVMDPNITTISPVNKLHPNDDILASGSSSSLFIWKPKEKSEPVHEKDESKIVVCGRA 518
+VAEVMDPN TTISPVNKLHP DDILA+GSS SLFIWKPKEKSE V EKDES+IVVCG+A
Sbjct: 599 VVAEVMDPNFTTISPVNKLHPRDDILATGSSRSLFIWKPKEKSEMVEEKDESRIVVCGKA 420
Query: 519 EKKRGKKRGADTDESDNEGFNSKLKKSKFQKTSKSQKTKWKLS 561
EKKRGKK+G ++DESD++GF SKLKK KS++T+WKLS
Sbjct: 419 EKKRGKKKGDNSDESDDDGFISKLKK------PKSKQTEWKLS 309
Score = 90.5 bits (223), Expect(2) = 2e-58
Identities = 41/50 (82%), Positives = 45/50 (90%)
Frame = -2
Query: 410 FNRYLTPFKAEWDPKDPSESLAVVGRYISENYNGAALHPIDFIDVSTGKL 459
F +LTPFKAEWDPKDPSESLAV+GRYISEN+ G ALHPIDFID STG+L
Sbjct: 745 FIAHLTPFKAEWDPKDPSESLAVIGRYISENFMGTALHPIDFIDTSTGQL 596
>AW685863 similar to GP|10798819|dbj putative damage-specific DNA binding
protein 2 {Oryza sativa (japonica cultivar-group)},
partial (9%)
Length = 485
Score = 117 bits (294), Expect(2) = 2e-35
Identities = 66/87 (75%), Positives = 69/87 (78%)
Frame = +3
Query: 1 MAPVTRRVPFPKVVIERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEKL 60
MAPVTRR FPKV IERD+DSEQSSS +EEE LE EE LE EEENGV E K EKL
Sbjct: 141 MAPVTRRTSFPKVHIERDSDSEQSSSSDEEEPLE--EEGPLE---EEENGVVENEKIEKL 305
Query: 61 EVGLDANRKGKTPITLPLRKVCKVCKK 87
EVG DANRKGKTPITL LRKVCKVCK+
Sbjct: 306 EVGFDANRKGKTPITLTLRKVCKVCKE 386
Score = 49.7 bits (117), Expect(2) = 2e-35
Identities = 19/20 (95%), Positives = 20/20 (100%)
Frame = +2
Query: 89 GHESGFKGATYIDCPMKPCF 108
GHE+GFKGATYIDCPMKPCF
Sbjct: 392 GHEAGFKGATYIDCPMKPCF 451
Score = 41.6 bits (96), Expect = 8e-04
Identities = 26/87 (29%), Positives = 43/87 (48%), Gaps = 1/87 (1%)
Frame = +1
Query: 33 LEDDEEEALEHEQEEENGVAEIGKNEKLEVGLDANRKGKTPITLPLRKVCKVCKKPGHES 92
++ + + +H ++++ G+ ++ + + L KGK P+ K + KKP S
Sbjct: 223 MKKNHSKKKDHSKKKKMGLLKMKRLRNWKWVLMLIEKGKHPLR*LSEKFVRCVKKPRS*S 402
Query: 93 GFKGATYIDCPMKPCFL-CKTPGHTTL 118
GF G P + F CKTPGHTTL
Sbjct: 403 GF*GCYLY*LPYETLFFFCKTPGHTTL 483
>TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclear
ribonucleoprotein [imported] - Arabidopsis thaliana,
partial (60%)
Length = 1836
Score = 56.2 bits (134), Expect = 3e-08
Identities = 55/232 (23%), Positives = 103/232 (43%), Gaps = 2/232 (0%)
Frame = +1
Query: 163 IPDQVNCAVIRYHSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCL 222
+P+ + ++ H++R T + + P N + + W+ G++
Sbjct: 1036 MPNVKKVSTLKGHTQRATDVAYSPVHKNHLATASADRTAKYWNDQGALLGTFKGHLER-- 1209
Query: 223 LNNMKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINS 282
L + F+P+ + +AS D T D+ET L +G S + +YG+D +
Sbjct: 1210 LARIAFHPSGKYLG-TASYDKTWRLWDVETEEELLLQ-------EGHS--RSVYGLDFHH 1359
Query: 283 EKGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNTK-VVGIHCNPIEPDILLTCGNDHY 341
+ L + D+R TG ++L + + K ++GI +P L T G D+
Sbjct: 1360 DGSLAASCGLDALARVWDLR----TGRSVLALEGHVKSILGISFSP-NGYHLATGGEDNT 1524
Query: 342 ARVWDIRRMETGSSIYDLK-HNRVVSSAYFSPVSGNKILTTSLDNRLRVWDS 392
R+WD+R+ + S+Y + H+ ++S F P G ++T S D +VW S
Sbjct: 1525 CRIWDLRKKK---SLYTIPAHSNLISQVKFEPQEGYFLVTASYDMTAKVWSS 1671
>TC78188 similar to PIR|C84870|C84870 probable splicing factor [imported] -
Arabidopsis thaliana, partial (95%)
Length = 1436
Score = 50.8 bits (120), Expect = 1e-06
Identities = 70/322 (21%), Positives = 136/322 (41%), Gaps = 7/322 (2%)
Frame = +1
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNPTNDC 234
H + ++F+PT +++ SG ++ +W+ + + H + ++ + ++
Sbjct: 310 HQSAVYTMKFNPT-GSVVASGSHDKEIFLWNVHGDCKNFMVLKGHKNAVLDLHWT-SDGT 483
Query: 235 MVYSASSDGTISFTDLETG--ISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVLVADN 292
+ SAS D T+ D ETG I + +L+ P+ LV+ +
Sbjct: 484 QIISASPDKTLRLWDTETGKQIKKMVEHLSYVNSCCPT----------RRGPPLVVSGSD 633
Query: 293 FGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDIRRMET 352
G L DMR +I ++ + + D + T G D+ ++WD+R+ E
Sbjct: 634 DGTAKLWDMRQRG----SIQTFPDKYQITAVSFSDAS-DKIYTGGIDNDVKIWDLRKGEV 798
Query: 353 GSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKMDSPSREIV-----HS 407
++ H +++S SP G+ +LT +D +L +WD + +P V H
Sbjct: 799 TMTLQG--HQDMITSMQLSP-DGSYLLTNGMDCKLCIWDM---RPYAPQNRCVKILEGHQ 960
Query: 408 HDFNRYLTPFKAEWDPKDPSESLAVVGRYISENYNGAALHPIDFIDVSTGKLVAEVMDPN 467
H+F + L K W P D S+ A G++ + D ++ +++ ++ N
Sbjct: 961 HNFEKNL--LKCGWSP-DGSKVTA-----------GSSDRMVYIWDTTSRRILYKLPGHN 1098
Query: 468 ITTISPVNKLHPNDDILASGSS 489
+ V HPN+ I+ S SS
Sbjct: 1099GSVNECV--FHPNEPIVGSCSS 1158
>BE322492 GP|13991907|g receptor for activated protein kinase C RACK1
{Heliothis virescens}, partial (39%)
Length = 452
Score = 42.7 bits (99), Expect = 4e-04
Identities = 28/83 (33%), Positives = 38/83 (45%), Gaps = 3/83 (3%)
Frame = +1
Query: 317 NTKVVGIHCNPIEPDILLTCGNDHYARVWDIRRMETGSSIYDLK---HNRVVSSAYFSPV 373
N V I NP PD++L+ D VW + R ET + + H+ +S S
Sbjct: 91 NGWVTQIATNPKYPDMILSSSRDKTLIVWKLTRDETNYGVPQKRLYGHSHFISDVVLSS- 267
Query: 374 SGNKILTTSLDNRLRVWDSIFGK 396
GN L+ S D LR+WD GK
Sbjct: 268 DGNYALSGSWDKTLRLWDLAAGK 336
>TC77172 similar to GP|20466472|gb|AAM20553.1 putative protein {Arabidopsis
thaliana}, partial (78%)
Length = 4130
Score = 42.4 bits (98), Expect = 5e-04
Identities = 39/178 (21%), Positives = 75/178 (41%), Gaps = 6/178 (3%)
Frame = +3
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVY------GNIHSCLLNNMKF 228
H + LEF+ N++ SG + G++ +WD E + G+ ++ + +
Sbjct: 471 HKGPVRGLEFNSIAPNLLASGAEDGEICIWDLANPLEPTHFPPLKGSGSASQGEVSFLSW 650
Query: 229 NPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVL 288
N ++ S S +GT DL+ S + ++P +G + + + D+ ++ +
Sbjct: 651 NSKVQHILASTSYNGTTVVWDLKKQ-KSVISVVDPVRRRGSA---LQWHPDVATQLAVAS 818
Query: 289 VADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWD 346
D + L DMR+ + H + V+ + P + LLTCG D WD
Sbjct: 819 DEDGSPSIKLWDMRNTMTPVKEFVGHSRG--VIAMSWCPNDSSYLLTCGKDSRTICWD 986
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 41.2 bits (95), Expect = 0.001
Identities = 18/46 (39%), Positives = 29/46 (62%)
Frame = +3
Query: 16 ERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEKLE 61
E D + E+ EEEE+ +D+EEE E E+EEE E G+ ++++
Sbjct: 120 EHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEEDDDEEGEEDEID 257
Score = 35.4 bits (80), Expect = 0.059
Identities = 17/44 (38%), Positives = 25/44 (56%)
Frame = +3
Query: 16 ERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEK 59
E + D E+ +EEE E++EEE EH+ EEE E + E+
Sbjct: 90 EDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEE 221
Score = 33.5 bits (75), Expect = 0.22
Identities = 17/46 (36%), Positives = 26/46 (55%)
Frame = +3
Query: 16 ERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEKLE 61
E + + E+ +EEEE D+EEE E E+EE+ E + E+ E
Sbjct: 69 EGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEE 206
Score = 33.5 bits (75), Expect = 0.22
Identities = 17/56 (30%), Positives = 32/56 (56%)
Frame = +3
Query: 16 ERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEKLEVGLDANRKGK 71
E + + ++ EEE+E E++EEE E E++E + E + E+ E D + +G+
Sbjct: 75 EVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEEDDDEEGE 242
Score = 33.5 bits (75), Expect = 0.22
Identities = 17/39 (43%), Positives = 23/39 (58%)
Frame = +3
Query: 20 DSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNE 58
+ E+ E EEE E DEEE EH++EEE E ++E
Sbjct: 54 NEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDE 170
Score = 32.0 bits (71), Expect = 0.65
Identities = 14/39 (35%), Positives = 24/39 (60%)
Frame = +3
Query: 16 ERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEI 54
E + + ++ EEEEE E++EEE + E+ EE+ + I
Sbjct: 147 EEEEEEDEHDDEEEEEEEEEEEEEEDDDEEGEEDEIDRI 263
Score = 30.0 bits (66), Expect = 2.5
Identities = 15/46 (32%), Positives = 23/46 (49%)
Frame = +3
Query: 16 ERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEKLE 61
E + + E E+E + E+DE + E E+EEE E E+ E
Sbjct: 57 EEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEE 194
Score = 30.0 bits (66), Expect = 2.5
Identities = 15/37 (40%), Positives = 22/37 (58%)
Frame = +3
Query: 12 KVVIERDTDSEQSSSEEEEEALEDDEEEALEHEQEEE 48
KV + + E EEE+E E++E+E E E+EEE
Sbjct: 39 KVEEHNEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEE 149
>TC78200 similar to PIR|A84578|A84578 probable WD-40 repeat protein [imported]
- Arabidopsis thaliana, partial (80%)
Length = 1824
Score = 40.8 bits (94), Expect = 0.001
Identities = 25/99 (25%), Positives = 46/99 (46%), Gaps = 2/99 (2%)
Frame = +1
Query: 300 DMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDIRRMETGSSI--Y 357
D R + HK + V+ N + +L + +D + D+R ++ G S+ +
Sbjct: 934 DTRLGRSPAASFQAHKADVNVLS--WNRLASCMLASGSDDGTISIRDLRLLKEGDSVVAH 1107
Query: 358 DLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGK 396
H ++S +SP + + +S DN+L +WD FGK
Sbjct: 1108 FEYHKHPITSIEWSPHEASSLAVSSADNQLTIWDLFFGK 1224
Score = 40.4 bits (93), Expect = 0.002
Identities = 21/76 (27%), Positives = 34/76 (44%)
Frame = +1
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNPTNDC 234
H+ + L++ PT+ ++ S + +WD H +N + +N C
Sbjct: 844 HTDSVEDLQWSPTEPHVFASCSVDKSIAIWDTRLGRSPAASFQAHKADVNVLSWNRLASC 1023
Query: 235 MVYSASSDGTISFTDL 250
M+ S S DGTIS DL
Sbjct: 1024MLASGSDDGTISIRDL 1071
>TC77681 similar to GP|22597156|gb|AAN03465.1 nucleolar histone deacetylase
HD2-P39 {Glycine max}, partial (47%)
Length = 1233
Score = 38.9 bits (89), Expect = 0.005
Identities = 37/140 (26%), Positives = 58/140 (41%), Gaps = 14/140 (10%)
Frame = +2
Query: 12 KVVIERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEKLEVGLDANRK-- 69
KVV +D DS+ ++E DDE E + + E+E+ E + E +D +K
Sbjct: 551 KVVDPKDEDSDDDDESDDEIGSSDDEMENADSDSEDEDDSDEDDEEETPVKKVDQGKKRP 730
Query: 70 ----GKTPITLPLRKVCKVCKKPGHESGFKGATYIDCPMK----PCFLCKTP----GHTT 117
KTPI+ K K G ++G + P K P KTP G +
Sbjct: 731 NESASKTPISSKKSKNATPEKTDGKKAGHTATPH---PKKGGKTPNSDAKTPKSGGGLSC 901
Query: 118 LTCPHRFSTEHGVVPAPRKK 137
+C F++E G+ + K
Sbjct: 902 SSCSKTFNSETGLTQHSKAK 961
>TC90584 similar to GP|22136810|gb|AAM91749.1 unknown protein {Arabidopsis
thaliana}, partial (18%)
Length = 686
Score = 38.5 bits (88), Expect = 0.007
Identities = 20/54 (37%), Positives = 30/54 (55%)
Frame = -2
Query: 2 APVTRRVPFPKVVIERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIG 55
+P + P P ++ E+S+ EEEA ED+EEE E +EEE+ E+G
Sbjct: 256 SPFSSSSPPPPLLAPLPVSEEESAEPLEEEAAEDEEEEEDEAAEEEEDEEEEVG 95
Score = 36.2 bits (82), Expect = 0.035
Identities = 19/35 (54%), Positives = 25/35 (71%)
Frame = -2
Query: 29 EEEALEDDEEEALEHEQEEENGVAEIGKNEKLEVG 63
EEE+ E EEEA E E+EEE+ AE ++E+ EVG
Sbjct: 199 EEESAEPLEEEAAEDEEEEEDEAAEEEEDEEEEVG 95
>TC83414 weakly similar to GP|17381206|gb|AAL36415.1 unknown protein
{Arabidopsis thaliana}, partial (28%)
Length = 1088
Score = 38.1 bits (87), Expect = 0.009
Identities = 13/40 (32%), Positives = 24/40 (59%)
Frame = +2
Query: 169 CAVIRYHSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVR 208
C + H+ +TC+ F+P +N +SG G++ +W+ VR
Sbjct: 395 CLRVFSHNNYVTCVNFNPVNDNFFISGSIDGKVRIWEVVR 514
Score = 35.8 bits (81), Expect = 0.045
Identities = 16/61 (26%), Positives = 31/61 (50%)
Frame = +2
Query: 333 LLTCGNDHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDS 392
LL+ D R+W + + HN V+ F+PV+ N ++ S+D ++R+W+
Sbjct: 338 LLSSSVDKTVRLWQVG---ISKCLRVFSHNNYVTCVNFNPVNDNFFISGSIDGKVRIWEV 508
Query: 393 I 393
+
Sbjct: 509 V 511
Score = 29.3 bits (64), Expect = 4.2
Identities = 49/250 (19%), Positives = 99/250 (39%), Gaps = 11/250 (4%)
Frame = +2
Query: 192 ILSGDKKGQLGVWDFV---RVHEKVVYGNIHSCLLNNMKFNPTNDCMV-YSASSDGTISF 247
+ SG + G + VW V R +E + N S + K N C+ D +
Sbjct: 26 LASGGEDGIVRVWQVVENERSNELNILDNDPSNIY--FKMNQFTGCVAPLDVDKDKLVKT 199
Query: 248 TDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVLVADNFGFLHLVDMRSNNKT 307
L +S+ + + P+ ++ K L+ +S+ L L GFL + +
Sbjct: 200 EKLRKSSASTCVIIPPKTFR--ILVKPLHEFQGHSDDILDLAWSKSGFLLSSSVDKTVRL 373
Query: 308 -----GNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDIRRMETGSSIYDLKHN 362
+ + N V ++ NP+ + ++ D R+W++ R I D++
Sbjct: 374 WQVGISKCLRVFSHNNYVTCVNFNPVNDNFFISGSIDGKVRIWEVVRCRVVDYI-DIR-- 544
Query: 363 RVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFG--KMDSPSREIVHSHDFNRYLTPFKAE 420
+V++ F P +G + ++ R ++ + KMD+ + +T F +
Sbjct: 545 EIVTAVCFCP-NGQGTIVGTMTGNCRFYNILDNHMKMDAEFSLQGKKKTSGKRITGF--Q 715
Query: 421 WDPKDPSESL 430
+ P DP++ L
Sbjct: 716 FSPSDPTKLL 745
>BG586181 similar to GP|4689316|gb| peroxisomal targeting signal type 2
receptor {Arabidopsis thaliana}, partial (56%)
Length = 804
Score = 37.7 bits (86), Expect = 0.012
Identities = 20/66 (30%), Positives = 32/66 (48%)
Frame = +1
Query: 326 NPIEPDILLTCGNDHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDN 385
NP D+ + D R+WD+R E GS++ H + S ++ I T S+D
Sbjct: 64 NPRHADVFASASGDCTVRIWDVR--EPGSTMILPAHEHEILSCDWNKYDECIIATGSVDK 237
Query: 386 RLRVWD 391
++VWD
Sbjct: 238 SVKVWD 255
Score = 35.4 bits (80), Expect = 0.059
Identities = 20/84 (23%), Positives = 38/84 (44%)
Frame = +1
Query: 308 GNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDIRRMETGSSIYDLKHNRVVSS 367
G+ +++ +++ N + I+ T D +VWD+R ++ + H V
Sbjct: 139 GSTMILPAHEHEILSCDWNKYDECIIATGSVDKSVKVWDVRSYRVPVAVLN-GHGYAVRK 315
Query: 368 AYFSPVSGNKILTTSLDNRLRVWD 391
FSP N +++ S D + VWD
Sbjct: 316 VKFSPHVRNLLVSCSYDMTVCVWD 387
Score = 29.6 bits (65), Expect = 3.2
Identities = 19/79 (24%), Positives = 32/79 (40%)
Frame = +1
Query: 171 VIRYHSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNP 230
++ H I +++ II +G + VWD V N H + +KF+P
Sbjct: 151 ILPAHEHEILSCDWNKYDECIIATGSVDKSVKVWDVRSYRVPVAVLNGHGYAVRKVKFSP 330
Query: 231 TNDCMVYSASSDGTISFTD 249
++ S S D T+ D
Sbjct: 331 HVRNLLVSCSYDMTVCVWD 387
>TC79118 similar to GP|16323188|gb|AAL15328.1 AT5g67320/K8K14_4 {Arabidopsis
thaliana}, partial (44%)
Length = 1098
Score = 37.4 bits (85), Expect = 0.016
Identities = 26/73 (35%), Positives = 36/73 (48%), Gaps = 1/73 (1%)
Frame = +2
Query: 326 NPIEPDILLTCGNDHYARVWDIRRMETGSSIYDLK-HNRVVSSAYFSPVSGNKILTTSLD 384
NP + +L + D ++WDI E G I+ L H V S FSP +G I + SLD
Sbjct: 494 NPNKKLLLASASFDSTVKLWDI---ELGKLIHSLNGHRHPVYSVAFSP-NGEYIASGSLD 661
Query: 385 NRLRVWDSIFGKM 397
L +W GK+
Sbjct: 662 KSLHIWSLKEGKI 700
>TC87847 similar to GP|15929245|gb|AAH15068.1 Similar to adaptor-related
protein complex AP-3 beta 1 subunit {Mus musculus},
partial (2%)
Length = 931
Score = 37.0 bits (84), Expect = 0.020
Identities = 30/91 (32%), Positives = 42/91 (45%)
Frame = +1
Query: 3 PVTRRVPFPKVVIERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEKLEV 62
P + + P KVV E+D DS S EEEE E+D + E E+EE+N + +
Sbjct: 133 PPSTQKPADKVVNEQDEDS----SSEEEEGDEEDGSSS-EDEEEEDNQTQQTPSKDSKPA 297
Query: 63 GLDANRKGKTPITLPLRKVCKVCKKPGHESG 93
N TPI+ P + + G ESG
Sbjct: 298 --SKNPPPSTPISNPKPAESESGSESGSESG 384
>TC87846 similar to GP|5821151|dbj|BAA83717.1 RNA binding protein {Homo
sapiens}, partial (2%)
Length = 550
Score = 37.0 bits (84), Expect = 0.020
Identities = 30/91 (32%), Positives = 42/91 (45%)
Frame = +1
Query: 3 PVTRRVPFPKVVIERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEKLEV 62
P + + P KVV E+D DS S EEEE E+D + E E+EE+N + +
Sbjct: 115 PPSTQKPADKVVNEQDEDS----SSEEEEGDEEDGSSS-EDEEEEDNQTQQTPSKDSKPA 279
Query: 63 GLDANRKGKTPITLPLRKVCKVCKKPGHESG 93
N TPI+ P + + G ESG
Sbjct: 280 --SKNPPPSTPISNPKPAESESGSESGSESG 366
>TC81578 GP|7688063|emb|CAB89693.1 constitutively photomorphogenic 1 protein
{Pisum sativum}, partial (39%)
Length = 842
Score = 36.6 bits (83), Expect = 0.026
Identities = 24/91 (26%), Positives = 44/91 (47%), Gaps = 1/91 (1%)
Frame = +2
Query: 164 PDQVNCAVIRYHSR-RITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCL 222
P +C V+ +R +++CL ++ N I S D +G + VWD V + ++ H
Sbjct: 317 PTDAHCPVVEMTTRSKLSCLSWNKYAKNQIASSDYEGIVTVWD-VTTRKSLMEYEEHEKR 493
Query: 223 LNNMKFNPTNDCMVYSASSDGTISFTDLETG 253
++ F+ T+ M+ S S D + E+G
Sbjct: 494 AWSVDFSRTDPSMLVSGSDDCKVKVLVHESG 586
>TC88509 similar to PIR|T01826|T01826 microfibril-associated protein homolog
T15F16.8 - Arabidopsis thaliana, partial (83%)
Length = 1306
Score = 35.8 bits (81), Expect = 0.045
Identities = 21/58 (36%), Positives = 30/58 (51%), Gaps = 8/58 (13%)
Frame = +1
Query: 16 ERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVA--------EIGKNEKLEVGLD 65
ERDT +E+ E EEEALE+ + +E + E + EI KN +LE +D
Sbjct: 622 ERDTIAERERLEAEEEALEEARKRRMEERRNETRQIVVEEIRKDQEIQKNIELEANID 795
Score = 32.3 bits (72), Expect = 0.50
Identities = 20/56 (35%), Positives = 29/56 (51%), Gaps = 3/56 (5%)
Frame = +1
Query: 2 APVTRRVPFPKVVIERDTDSEQSSSEEEEEALEDDEEEALEHE---QEEENGVAEI 54
A RR + + +RD + EEEEE E++EEE ++E EE GVA +
Sbjct: 430 AMAERRRRIKEKLRQRDQEEALPQEEEEEEEEEEEEEEESDYETDSDEEYTGVAMV 597
Score = 28.5 bits (62), Expect = 7.2
Identities = 15/39 (38%), Positives = 24/39 (61%), Gaps = 1/39 (2%)
Frame = +1
Query: 22 EQSSSEEEEEAL-EDDEEEALEHEQEEENGVAEIGKNEK 59
E+ ++EEAL +++EEE E E+EEE E +E+
Sbjct: 460 EKLRQRDQEEALPQEEEEEEEEEEEEEEESDYETDSDEE 576
>TC79394 weakly similar to PIR|T49019|T49019 probable RNA binding protein -
Arabidopsis thaliana, partial (74%)
Length = 1909
Score = 35.4 bits (80), Expect = 0.059
Identities = 21/46 (45%), Positives = 28/46 (60%), Gaps = 1/46 (2%)
Frame = +2
Query: 12 KVVIERDTDSEQSSSEE-EEEALEDDEEEALEHEQEEENGVAEIGK 56
+V ++ D D E+SS EE E E +E +EE E E+EEE V E K
Sbjct: 233 QVDLDGDNDQEESSEEEVEYEEVEVEEEVEEEEEEEEEEEVEEESK 370
>TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (96%)
Length = 1286
Score = 35.4 bits (80), Expect = 0.059
Identities = 18/55 (32%), Positives = 24/55 (42%), Gaps = 13/55 (23%)
Frame = +1
Query: 81 VCKVCKKPGH------------ESGFKGATYIDCPMKP-CFLCKTPGHTTLTCPH 122
+CK CK+PGH G +C K C+ CK PGH +CP+
Sbjct: 421 LCKNCKRPGHYVRECPNVAVCHNCSLPGHIASECSTKSLCWNCKEPGHMASSCPN 585
Score = 35.0 bits (79), Expect = 0.077
Identities = 18/55 (32%), Positives = 23/55 (41%), Gaps = 1/55 (1%)
Frame = +1
Query: 68 RKGKTPITLPLRKVCKVCKKPGHESGFKGATYIDCPMKP-CFLCKTPGHTTLTCP 121
++G + K C C+K GH + DCP P C LC GH CP
Sbjct: 688 KQGHIAVECTNEKACNNCRKTGHLAR-------DCPNDPICNLCNISGHVARQCP 831
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.134 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,425,203
Number of Sequences: 36976
Number of extensions: 312874
Number of successful extensions: 1989
Number of sequences better than 10.0: 127
Number of HSP's better than 10.0 without gapping: 1536
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1792
length of query: 564
length of database: 9,014,727
effective HSP length: 101
effective length of query: 463
effective length of database: 5,280,151
effective search space: 2444709913
effective search space used: 2444709913
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0346.11


