

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0336.1
(1182 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
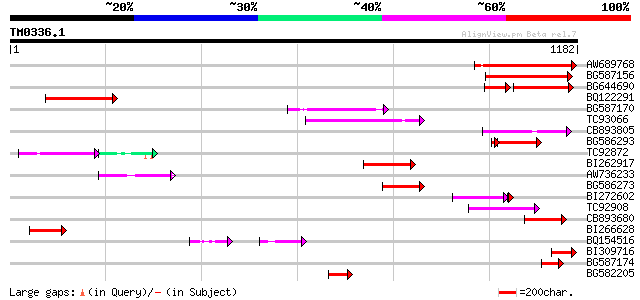
Score E
Sequences producing significant alignments: (bits) Value
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 274 1e-73
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 168 1e-41
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 120 3e-38
BQ122291 147 3e-35
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 140 3e-33
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 137 3e-32
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 124 2e-28
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 94 7e-20
TC92872 65 8e-17
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 80 4e-15
AW736233 GP|21105451|gb small nuclear ribonucleoprotein D1 {Dani... 79 8e-15
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 75 1e-13
BI272602 GP|11602812|db avenaIII {Gallus gallus}, partial (3%) 65 8e-13
TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol po... 72 2e-12
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 69 8e-12
BI266628 67 4e-11
BQ154516 64 3e-10
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 54 4e-07
BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse ... 51 2e-06
BG582205 45 2e-04
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 274 bits (701), Expect = 1e-73
Identities = 135/213 (63%), Positives = 165/213 (77%), Gaps = 1/213 (0%)
Frame = +1
Query: 970 ITRSKNGIVQPRVQPTLLTVQLEPTTVKQALAHPQWLQAMQAEFDALQKNKTWSLVPLPP 1029
ITR+K G ++P+V LT L+ P+WLQAM+ E+ AL NKTW LVPLPP
Sbjct: 1 ITRTKTGKLKPKV---FLTEPCPQDCENLPLSDPRWLQAMKTEYKALIDNKTWDLVPLPP 171
Query: 1030 GREAIGCKWVFRVKHNPDGTLNKYKARLVAKGFHQQPGFDYTETFSPVIKPVTIRVILTL 1089
++AIGCKWV+RVK NPDG++NK+KARLVAKGF Q G DYTETFSPVIKPVTIR+ILT+
Sbjct: 172 HKKAIGCKWVYRVKENPDGSVNKFKARLVAKGFSQTLGCDYTETFSPVIKPVTIRLILTI 351
Query: 1090 ALTNHWTIQQLDVNNAFLNGDLAEEVYMQQPQGFE-HDKKLVCRLHKALYGLKQAPRAWF 1148
A+T W IQQ+D+NNAFLNG L EEVYM QPQGFE +K LVC+L+K+LYGLKQAPRAW+
Sbjct: 352 AITYKWEIQQIDINNAFLNGFLQEEVYMSQPQGFEAANKSLVCKLNKSLYGLKQAPRAWY 531
Query: 1149 EKLTSALVQFGFVASKCDPSLFVLSKKPLVIYV 1181
E LTSA +QFGF S+CDPSL + ++ IY+
Sbjct: 532 EXLTSAQIQFGFTKSRCDPSLLIYNQNGACIYL 630
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 168 bits (426), Expect = 1e-41
Identities = 84/183 (45%), Positives = 121/183 (65%), Gaps = 2/183 (1%)
Frame = -1
Query: 993 PTTVKQALAHPQWLQAMQAEFDALQKNKTWSLVPLPPGREAIGCKWVFRVKHNPDGTLNK 1052
P + ++A+ +W +++ AE A+ KN TW LP G++A+ +W+F +K+ DG++ +
Sbjct: 558 PRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKAVSSRWIFTIKYKADGSIER 379
Query: 1053 YKARLVAKGFHQQPGFDYTETFSPVIKPVTIRVILTLALTNHWTIQQLDVNNAFLNGDLA 1112
K RLVA+GF G DY ETF+PV K TIR++L+LA+ W + Q+DV NAFL G+L
Sbjct: 378 KKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLWQMDVKNAFLQGELE 199
Query: 1113 EEVYMQQPQGFEHDKKL--VCRLHKALYGLKQAPRAWFEKLTSALVQFGFVASKCDPSLF 1170
+EVYM P G EH K V RL KA+YGLKQ+PRAW+ KL++ L GF S+ D +LF
Sbjct: 198 DEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTLNGRGFRKSELDHTLF 19
Query: 1171 VLS 1173
L+
Sbjct: 18 TLT 10
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 120 bits (300), Expect(2) = 3e-38
Identities = 58/128 (45%), Positives = 89/128 (69%), Gaps = 2/128 (1%)
Frame = -2
Query: 1050 LNKYKARLVAKGFHQQPGFDYTETFSPVIKPVTIRVILTLALTNHWTIQQLDVNNAFLNG 1109
+ + K++LV +G++Q+ G DY E FSPV + IR+++ A + + Q+DV +AF+NG
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 1110 DLAEEVYMQQPQGFEHDK--KLVCRLHKALYGLKQAPRAWFEKLTSALVQFGFVASKCDP 1167
DL EEV+++QP GFE + V RL+K LYGLKQAPRAW+E+L+ L++ GF K D
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 1168 SLFVLSKK 1175
+LF+L ++
Sbjct: 64 TLFLLKRE 41
Score = 58.2 bits (139), Expect(2) = 3e-38
Identities = 24/53 (45%), Positives = 33/53 (61%)
Frame = -3
Query: 991 LEPTTVKQALAHPQWLQAMQAEFDALQKNKTWSLVPLPPGREAIGCKWVFRVK 1043
+EP VK+AL W+ +MQ E +++K W LVP P G+ IG +WVFR K
Sbjct: 591 IEPKNVKEALRDADWINSMQEELHQFERSKVWYLVPRPEGKTVIGTRWVFRNK 433
>BQ122291
Length = 487
Score = 147 bits (370), Expect = 3e-35
Identities = 67/150 (44%), Positives = 109/150 (72%)
Frame = -2
Query: 75 RVAGTLNPAYSVWESKDQYLLSWLQSSLSTPVLSRMIGCAHSFQLWEKIHSFFHNQTRAK 134
R AGT NPAY+ WE KD L +W+ S +S +LSR + HS+Q+W++IHS+ Q + +
Sbjct: 477 REAGTRNPAYTEWEDKDSLLCTWILSRISPSLLSRFVLLRHSWQVWDEIHSYCFTQMKTR 298
Query: 135 STQLRTELRSETLGNRTISEFLLQIKSIIDSLFAIGDPISPREHLDVILQGLPSDYEDLI 194
S QLR+ELRS T G+RT+SEF+ +I++I +SL +IGDP+S R+ ++V+L+ LP +++ ++
Sbjct: 297 SRQLRSELRSITKGSRTVSEFIARIRAISESLASIGDPVSHRDLIEVVLEALPEEFDPIV 118
Query: 195 TLISTRFEEHSIDELEALLLAKEARLGKYR 224
++ + E S+DELE+ LL +E+R K++
Sbjct: 117 ASVNAKSEVVSLDELESQLLTQESRKEKFK 28
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 140 bits (353), Expect = 3e-33
Identities = 83/217 (38%), Positives = 126/217 (57%), Gaps = 5/217 (2%)
Frame = -3
Query: 579 SSVCTANLWHFRLGHP---SLNVLRKALQFCKIPVSNKMDVDFCKACVMGKSHRLPSSLS 635
SS+ LWH RLGHP +LN++ + F NK C+AC++GK + +
Sbjct: 605 SSLNKDALWHARLGHPHGRALNLMLPGVVF-----ENKN----CEACILGKHCKNVFPRT 453
Query: 636 TTEYTSPLELVYSDLWGPSPSPSSSGYLYYITFIDAYSRYTWIYLLKAKSEAISAFHQFK 695
+T Y + +L+Y+DLW +PS S + Y++TFID S+YTW+ L+ +K I AF F+
Sbjct: 452 STVYENCFDLIYTDLW-TAPSLSRDNHKYFVTFIDEKSKYTWLTLIPSKDRVIDAFKNFQ 276
Query: 696 ASSELQLNAKIKALQTDWGGEFR--VFPNFLKDFGIHHRLICPHTHHQNGVVERKHRHIV 753
A +AKIK L++D GGE+ F + L GI H+ CP+T QNGV +RK++H++
Sbjct: 275 AYVTNHYHAKIKILRSDNGGEYTSYAFKSHLDHHGILHQTSCPYTPQQNGVAKRKNKHLM 96
Query: 754 DLGLSLLAHASIPLTYWDFAFSTAVYLINRLPTSSLQ 790
++ SL+ A++ STA YLIN +PT L+
Sbjct: 95 EVARSLMFQANV---------STACYLINWIPTKVLR 12
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 137 bits (344), Expect = 3e-32
Identities = 87/258 (33%), Positives = 133/258 (50%), Gaps = 8/258 (3%)
Frame = +1
Query: 616 VDFCKACVM-GKSHRLPSSLSTTEYTSPLELVYSDLWGPSPSPSSSGYLYYITFIDAYSR 674
++FCK + G ++ S +T L+ ++SDLWGPS S G Y +T ID + R
Sbjct: 13 LEFCKHLLFFGNRKKVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPR 192
Query: 675 YTWIYLLKAKSEAISAFHQFKASSELQLNAKIKALQTDWGGEF--RVFPNFLKDFGIHHR 732
W+Y L+ K+E F +++ E Q +K L TD EF F F + GI
Sbjct: 193 KVWVYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARH 372
Query: 733 LICPHTHHQNGVVERKHRHIVDLGLSLLAHASI--PLTYWDFAFSTAVYLINRLPTSSLQ 790
P QNGV ER R +++ +L++A + W A STA +L+NR P S+L
Sbjct: 373 KTIPRNPQQNGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALD 552
Query: 791 FDIPLYKLFKQIPDYTFLKVFGCTCFPLMRPYNSHKLDFRSQECVFLGYSPSHRGYKCLA 850
F +P + DY+ L++FGC + L+ N KL R+ EC+FL Y+ +GY+
Sbjct: 553 FKVPEDIWSGNLVDYSNLRIFGCPAYALV---NDGKLAPRAGECIFLSYASESKGYRLWC 723
Query: 851 S---SGRIYISKDVIFDE 865
S S ++ +S+DV F+E
Sbjct: 724 SDPKSQKLILSRDVTFNE 777
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 124 bits (312), Expect = 2e-28
Identities = 71/194 (36%), Positives = 110/194 (56%), Gaps = 8/194 (4%)
Frame = +3
Query: 986 LLTVQLEPTTVKQALAHPQWLQAMQAEFDALQKNKTWSLVPLPPGREAIGCKWVFRVKHN 1045
+LT+ +PTT ++A+ +W +M E +A ++N TW L L G + IG KW+F+ K N
Sbjct: 21 MLTMTSDPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLN 200
Query: 1046 PDGTLNKYKARLVAKGFHQQPGFDYTETFSPVIKPVTIRVILTLALTNHWTIQQLDVNNA 1105
+G + KYKARLVAKG+ QQ G DYTE F+PV + TIR+++ LA Q+ +
Sbjct: 201 ENGEIEKYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALA-------AQIKRDGV 359
Query: 1106 FLN----GDLAEEVYMQQPQGFEH----DKKLVCRLHKALYGLKQAPRAWFEKLTSALVQ 1157
++ E M++ H + + R+ +ALYGLKQAPRAW+ ++ + +
Sbjct: 360 CIS*M*KAHSCMEN*MRKFLLINHRVM*RRVIS*RVKRALYGLKQAPRAWYSRIEAYFTK 539
Query: 1158 FGFVASKCDPSLFV 1171
GF + +LFV
Sbjct: 540 EGFEKCPYEHTLFV 581
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 94.0 bits (232), Expect(2) = 7e-20
Identities = 46/92 (50%), Positives = 63/92 (68%)
Frame = +2
Query: 1018 KNKTWSLVPLPPGREAIGCKWVFRVKHNPDGTLNKYKARLVAKGFHQQPGFDYTETFSPV 1077
+ +T LV P G + IG +W++++K N DGTL KYKARLVAKG+ +Q G D+ E F+PV
Sbjct: 47 QKQTLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPV 226
Query: 1078 IKPVTIRVILTLALTNHWTIQQLDVNNAFLNG 1109
++ TI ++L LA TN I +DV AFLNG
Sbjct: 227 VRIETI*LLLALAATNGC*IHHIDVKIAFLNG 322
Score = 22.7 bits (47), Expect(2) = 7e-20
Identities = 7/16 (43%), Positives = 13/16 (80%)
Frame = +3
Query: 1005 WLQAMQAEFDALQKNK 1020
W+ AM+AE +++ KN+
Sbjct: 9 WIDAMKAEIESIIKNR 56
>TC92872
Length = 923
Score = 65.5 bits (158), Expect(2) = 8e-17
Identities = 45/171 (26%), Positives = 82/171 (47%), Gaps = 2/171 (1%)
Frame = +1
Query: 18 PHRFKLPSLSTYTISEKLSDRNFLLWKQQVEPIIKAHNL--HVLVQDPEIPPRFVTERDR 75
P K + +I+ KLS +NF WK+QV ++ + H+ D P + T +
Sbjct: 58 PSNTKSSLILNASITFKLSRKNFRAWKRQVTTLLAGIEVMGHI---DGTTPIQTETIINN 228
Query: 76 VAGTLNPAYSVWESKDQYLLSWLQSSLSTPVLSRMIGCAHSFQLWEKIHSFFHNQTRAKS 135
NP Y+ W + DQ +++ L SS++ + LW I S F+N +R+
Sbjct: 229 GVSAPNPDYTKWFTLDQLIINLLLSSMTEADSISFASYETARTLWVAIESQFNNTSRSHV 408
Query: 136 TQLRTELRSETLGNRTISEFLLQIKSIIDSLFAIGDPISPREHLDVILQGL 186
+ +++ T G+++I+++L +KS+ D L I +S + +L GL
Sbjct: 409 MSVTNQIQRCTKGDKSITDYLFSVKSLADELAVIDKSLSDDDITLFVLNGL 561
Score = 40.8 bits (94), Expect(2) = 8e-17
Identities = 38/138 (27%), Positives = 53/138 (37%), Gaps = 16/138 (11%)
Frame = +2
Query: 186 LPSDYEDLITLISTRFEEHSIDELEALLLAKEARLGKYRTPSATGEISVNVALSNPQTPS 245
L +Y D+ I TR + +EL + LLA + L + ++ + V +N
Sbjct: 560 LVPEYNDIAASIRTRQHPFTFEELHSHLLAHDDYLRREAV-----QVDIQVPTAN----- 709
Query: 246 SSAVSTQSASHTTSQGESQDSDPMDSYDSSGHGRGSS----------RGRGARGRGRG-- 293
A HT+ S D + S+G G S RG RGRGRG
Sbjct: 710 -------FAHHTSPNQRSSKDDGILPIPSTGRGNNFSKQSQHHGSNTRGSNYRGRGRGFN 868
Query: 294 ----RSSIQCQLCYKFGH 307
R +CQLC GH
Sbjct: 869 SRGNRPPPRCQLCSLLGH 922
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 80.5 bits (197), Expect = 4e-15
Identities = 41/111 (36%), Positives = 68/111 (60%), Gaps = 1/111 (0%)
Frame = +1
Query: 737 HTHHQNGVVERKHRHIVDLGLSLLAHASIPLTYWDFAFSTAVYLINRLPTSSLQFDIPLY 796
HT QNGV ER +R +++ ++L A + ++W A TA Y+INR P++ + P+
Sbjct: 82 HTPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACYVINRSPSTVIDLKTPM- 258
Query: 797 KLFKQIP-DYTFLKVFGCTCFPLMRPYNSHKLDFRSQECVFLGYSPSHRGY 846
+++K P DY+ L VFGC + + KLD +S++C+FLGY+ + +GY
Sbjct: 259 EMWKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGYADNVKGY 411
>AW736233 GP|21105451|gb small nuclear ribonucleoprotein D1 {Danio rerio},
partial (14%)
Length = 510
Score = 79.3 bits (194), Expect = 8e-15
Identities = 52/167 (31%), Positives = 82/167 (48%), Gaps = 6/167 (3%)
Frame = +1
Query: 185 GLPSDYEDLITLISTRFEEHSIDELEALLLAKEARLGKYRTPSATGEISVNVALSN-PQT 243
GL +Y + +I R + S++E E++L+ +EA+ KYR +S N+A S P
Sbjct: 1 GLLEEYNSFVLVIYIRLDSPSMEEGESILMMQEAQFEKYRQELTNPSVSANLAQSELPSK 180
Query: 244 PSSSAVSTQSASHTTSQGESQDSDPMDSYDSSGHGRGSSRGRGARGRGRGRSS-IQCQLC 302
P G S+ + Y ++G GRG RGRG RGRGR S+ +QCQ+C
Sbjct: 181 P----------------GNSESQEVGTEYYNAGRGRGRGRGRG-RGRGRSNSNRLQCQIC 309
Query: 303 YKFGHDVWQCYHRFNPNFVPQ----KPPQNSPQNSQWNSSPQWHSNS 345
+ HD +C R++ Q PP +S ++ +P W+ +S
Sbjct: 310 ARNNHDAARCCFRYDQASSSQAHHRAPPSEYAASSSYSEAP-WYPDS 447
Score = 40.8 bits (94), Expect = 0.003
Identities = 19/48 (39%), Positives = 25/48 (51%)
Frame = +1
Query: 405 AQAHVAFQTSTSTPQATSPAQSWYPDSGASHHVTSESQNVHQFTPFAG 452
+QAH S ++ WYPDSGASHH+T N+ TP+ G
Sbjct: 367 SQAHHRAPPSEYAASSSYSEAPWYPDSGASHHLTFNPHNLAYRTPYNG 510
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 75.1 bits (183), Expect = 1e-13
Identities = 37/90 (41%), Positives = 58/90 (64%), Gaps = 1/90 (1%)
Frame = -2
Query: 777 AVYLINRLPTSSLQFDIPLYKLFKQIPDYTFLKVFGCTCFPLMRPYNSHKLDFRSQECVF 836
A YLINR+PT L+ P L ++ P T+++VFGC C+ L+ +KL+ RS++ +F
Sbjct: 704 ACYLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMF 525
Query: 837 LGYSPSHRGYKCL-ASSGRIYISKDVIFDE 865
+GYS + +GYKC + R+ +S+DV F E
Sbjct: 524 IGYSTTQKGYKCYDPEARRVLVSRDVKFIE 435
>BI272602 GP|11602812|db avenaIII {Gallus gallus}, partial (3%)
Length = 488
Score = 65.1 bits (157), Expect(2) = 8e-13
Identities = 47/123 (38%), Positives = 63/123 (51%), Gaps = 6/123 (4%)
Frame = +1
Query: 924 QNPSSTSPASAPATSEASAEIPSQVTIPTSSGQVSAPSSSG--NVHPMITRSKNGIVQPR 981
QN +SPAS TS A I + T P + P ++ NV +ITRS+N IV+
Sbjct: 55 QNIQISSPASP--TSNELANITTSPTPPPPPPPTARPHNNHTTNVGGIITRSQNNIVKT- 225
Query: 982 VQPTLLTVQ----LEPTTVKQALAHPQWLQAMQAEFDALQKNKTWSLVPLPPGREAIGCK 1037
++ L V+ +EP+ + QAL W MQ EFDALQ N TW LV + +GCK
Sbjct: 226 IKKLNLHVRPLCPIEPSNITQALRDHDWRSIMQDEFDALQNNNTWDLVSRSFAQNLVGCK 405
Query: 1038 WVF 1040
F
Sbjct: 406 LGF 414
Score = 27.7 bits (60), Expect(2) = 8e-13
Identities = 9/12 (75%), Positives = 11/12 (91%)
Frame = +2
Query: 1038 WVFRVKHNPDGT 1049
WVFR+K NPDG+
Sbjct: 407 WVFRIKRNPDGS 442
>TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol polyprotein
{Oryza sativa}, partial (1%)
Length = 638
Score = 71.6 bits (174), Expect = 2e-12
Identities = 46/147 (31%), Positives = 74/147 (50%)
Frame = +2
Query: 957 VSAPSSSGNVHPMITRSKNGIVQPRVQPTLLTVQLEPTTVKQALAHPQWLQAMQAEFDAL 1016
V+ SSS + H + + R +T +EP QA +WL AM+ E L
Sbjct: 173 VTLNSSSKSYHIFHYVGYSFSAKHRASLAAITSNIEPKNYVQAAQ*QEWLAAMEQEIQVL 352
Query: 1017 QKNKTWSLVPLPPGREAIGCKWVFRVKHNPDGTLNKYKARLVAKGFHQQPGFDYTETFSP 1076
++N T +L PL G++ + C+ V+++ H +G + KYKA+LVAK F Q G D+ +
Sbjct: 353 EENNTSTLEPLREGKKWVDCRPVYKIIHKANGEIEKYKAQLVAKDFVQVEGEDF*D-LCL 529
Query: 1077 VIKPVTIRVILTLALTNHWTIQQLDVN 1103
K R +LT+A + +DV+
Sbjct: 530 SNKDDNCRCLLTIAAAKG*QLHLMDVS 610
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 69.3 bits (168), Expect = 8e-12
Identities = 36/88 (40%), Positives = 54/88 (60%), Gaps = 1/88 (1%)
Frame = -2
Query: 1074 FSPVIKPVTIRVILTLALTNHWTIQQLDVNNAFLNGDLAEEVYMQQPQGFEHD-KKLVCR 1132
F P++K TI +L++ + ++ LDV AFL GDL E++YM QP+GF + K+V +
Sbjct: 554 FVPIVKLNTIMFLLSIVAIENLYLE*LDVKTAFLRGDLVEDIYMHQPEGFS*EVGKMVGK 375
Query: 1133 LHKALYGLKQAPRAWFEKLTSALVQFGF 1160
L K++YGLKQ PR L + + GF
Sbjct: 374 LKKSMYGLKQGPRQCI*SLKALCTRKGF 291
>BI266628
Length = 536
Score = 67.0 bits (162), Expect = 4e-11
Identities = 31/77 (40%), Positives = 47/77 (60%)
Frame = -1
Query: 41 LLWKQQVEPIIKAHNLHVLVQDPEIPPRFVTERDRVAGTLNPAYSVWESKDQYLLSWLQS 100
L WKQQVE +++ V P+IPP ++T+ R AGT PAY+ WE +D L +W+ S
Sbjct: 404 LQWKQQVEGVLRETKTVKFVISPQIPPVYLTDEAREAGTKKPAYTEWEEQDSLLCTWILS 225
Query: 101 SLSTPVLSRMIGCAHSF 117
++ +LSR + HS+
Sbjct: 224 TI*PSLLSRFVLLRHSW 174
>BQ154516
Length = 506
Score = 64.3 bits (155), Expect = 3e-10
Identities = 39/102 (38%), Positives = 54/102 (52%), Gaps = 3/102 (2%)
Frame = +1
Query: 521 SSKVLLQGSVGADGLYKFHEFKLSHLPVSLSAPVQHPVSVHTSVSSSGHAPVEQP---SV 577
S ++LL+G VG+DGLY+F P+Q S+ S+S +A + S
Sbjct: 238 SKQILLEGVVGSDGLYQFK-------------PLQFLPSMSKSLSYCNNATISSVVCNSS 378
Query: 578 VSSVCTANLWHFRLGHPSLNVLRKALQFCKIPVSNKMDVDFC 619
SS + N WH RLGH + NV++ L CKIP NK +DFC
Sbjct: 379 SSSNDSFNKWHCRLGHANPNVVKSILNLCKIPFQNKHVLDFC 504
Score = 55.8 bits (133), Expect = 9e-08
Identities = 34/89 (38%), Positives = 46/89 (51%)
Frame = +3
Query: 375 RYPPAPSGYAPMASSPGVPYRGPAPSPMGYAQAHVAFQTSTSTPQATSPAQSWYPDSGAS 434
R PP S PMA+ ++ P P YA S+ + +P WYPDSGAS
Sbjct: 15 RAPPFNSHMRPMANFAMQGFQAP---PSEYA---------ASSSYSEAP---WYPDSGAS 149
Query: 435 HHVTSESQNVHQFTPFAGQDQLFVGNGQG 463
HH+T N+ TP+ GQ+Q+ + NGQG
Sbjct: 150 HHLTFNPHNLAYRTPYNGQEQVLMDNGQG 236
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 53.9 bits (128), Expect = 4e-07
Identities = 26/56 (46%), Positives = 36/56 (63%), Gaps = 4/56 (7%)
Frame = +2
Query: 1130 VCRLHKALYGLKQAPRAWFEKLTSALVQFGFVASKCDPSLFVLSK----KPLVIYV 1181
VC L K++YGLKQA R W+ KL+ +L+ FG++ S D SLF K L++YV
Sbjct: 20 VCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYV 187
>BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse transcriptase
homolog - rape retrotransposon copia-like (fragment),
partial (84%)
Length = 249
Score = 51.2 bits (121), Expect = 2e-06
Identities = 23/46 (50%), Positives = 33/46 (71%), Gaps = 2/46 (4%)
Frame = -1
Query: 1110 DLAEEVYMQQPQGFEHDKKL--VCRLHKALYGLKQAPRAWFEKLTS 1153
+L E++YM QP+GF K VC+L K+LYGLKQ+PR W+++ S
Sbjct: 249 ELEEKIYMTQPEGFLFPGKEDHVCKLRKSLYGLKQSPRQWYKRFDS 112
>BG582205
Length = 801
Score = 44.7 bits (104), Expect = 2e-04
Identities = 20/51 (39%), Positives = 31/51 (60%)
Frame = +1
Query: 665 YITFIDAYSRYTWIYLLKAKSEAISAFHQFKASSELQLNAKIKALQTDWGG 715
++T +DAYS + WI+ LK KS + +F K +N+ IK++QT GG
Sbjct: 511 FLTCVDAYSPFNWIFPLKLKSHTLQSFKTSKELLNYNINSSIKSVQTV*GG 663
Score = 40.8 bits (94), Expect = 0.003
Identities = 41/137 (29%), Positives = 51/137 (36%), Gaps = 2/137 (1%)
Frame = +3
Query: 617 DFCKACVMGKSHRLPSSLSTTEYTSPLELVYSDLWGPSPSPSSSGYLYYITFIDAYSRYT 676
DFC C +GK+HRLPS PL L S L + +
Sbjct: 387 DFCSFCCLGKAHRLPS-------FHPLYLTLSLLNSFFVICGDYSSRVFFNLCGCLFSF* 545
Query: 677 WIYLLKAKSEAISAFHQFKASSELQLNAKIKALQTDW-GGEFRVFPNFLKDFG-IHHRLI 734
K + + F FK ELQ + GGEFR F +L + G H L
Sbjct: 546 LDISFKTEVSHFTIFQNFKRIVELQYKLLHQVCSNCLRGGEFRPFITYLNNLG*PAHTLT 725
Query: 735 CPHTHHQNGVVERKHRH 751
+ G VERKHRH
Sbjct: 726 T-----KMGSVERKHRH 761
Score = 35.4 bits (80), Expect = 0.13
Identities = 36/119 (30%), Positives = 52/119 (43%), Gaps = 5/119 (4%)
Frame = +2
Query: 635 STTEYTSPLELVYSDLWGPSPSPSSSGYLYYITFIDAYSRYTWIYLLKAKSEAISAFHQF 694
ST YT PLEL++ DLW G L F + + + +++ ++
Sbjct: 440 STVSYTKPLELIFCDLW---------GLLQ*SLF*PVWMPILLLIGYFL*N*SLTLYNL- 589
Query: 695 KASSELQLNAKIKALQTDWGGEFRVFPN-----FLKDFGIHHRLICPHTHHQNGVVERK 748
S+LQ N I + T F++F F F RL CPHTHHQNG+ +K
Sbjct: 590 ---SKLQKNC*I-TI*TPPSSLFKLFEGG*VSPFYYIFE*S-RLTCPHTHHQNGIC*KK 751
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.132 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 45,160,666
Number of Sequences: 36976
Number of extensions: 849519
Number of successful extensions: 15148
Number of sequences better than 10.0: 405
Number of HSP's better than 10.0 without gapping: 7880
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 12830
length of query: 1182
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1075
effective length of database: 5,058,295
effective search space: 5437667125
effective search space used: 5437667125
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0336.1


