

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
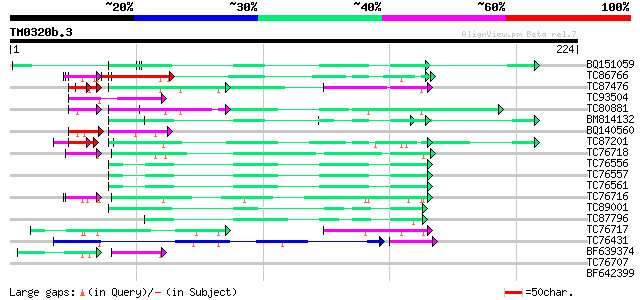
Query= TM0320b.3
(224 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melano... 56 1e-08
TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E... 40 3e-07
TC87476 homologue to PIR|T11622|T11622 extensin class 1 precurso... 38 8e-07
TC93504 weakly similar to PIR|S46286|S46286 RNA-binding protein ... 49 1e-06
TC80881 similar to PIR|T11622|T11622 extensin class 1 precursor ... 47 6e-06
BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein ... 47 8e-06
BQ140560 weakly similar to GP|15128223|db hypothetical protein~s... 39 9e-06
TC87201 similar to PIR|F86388|F86388 hypothetical protein AAG292... 39 1e-05
TC76718 homologue to GP|8132441|gb|AAF73291.1| extensin {Pisum s... 34 6e-05
TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 43 1e-04
TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 43 1e-04
TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 43 1e-04
TC76716 similar to PIR|T10863|T10863 extensin precursor - kidney... 32 3e-04
TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-bi... 41 3e-04
TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear prot... 41 3e-04
TC76717 similar to PIR|T10863|T10863 extensin precursor - kidney... 36 4e-04
TC76431 similar to GP|8132441|gb|AAF73291.1| extensin {Pisum sat... 38 8e-04
BF639374 similar to GP|8132441|gb| extensin {Pisum sativum}, par... 34 9e-04
TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 35 0.010
BF642399 similar to PIR|T10863|T10 extensin precursor - kidney b... 33 0.087
>BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melanogaster},
partial (26%)
Length = 308
Score = 56.2 bits (134), Expect = 1e-08
Identities = 52/170 (30%), Positives = 52/170 (30%)
Frame = +2
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG GGGGGGG GGG GGG
Sbjct: 41 GGGGGGGGGGGGGGGGGGGGGGGGG------------------------GGGGGGGGGGG 148
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGG GGG GGG GGG G GGG
Sbjct: 149 GG---------------------GGGGGGGGGGGGGGGGGGGG--------GGGGGGGGG 241
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G GGGG GGGGGGG G
Sbjct: 242 GGGGGGG------------------------------GGGGGGGGGGGGG 301
Score = 56.2 bits (134), Expect = 1e-08
Identities = 52/170 (30%), Positives = 52/170 (30%)
Frame = +2
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG GGGGGGG GGG GGG
Sbjct: 35 GGGGGGGGGGGGGGGGGGGGGGGGG------------------------GGGGGGGGGGG 142
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGG GGG GGG GGG G GGG
Sbjct: 143 GG---------------------GGGGGGGGGGGGGGGGGGGG--------GGGGGGGGG 235
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G GGGG GGGGGGG G
Sbjct: 236 GGGGGGG------------------------------GGGGGGGGGGGGG 295
Score = 56.2 bits (134), Expect = 1e-08
Identities = 52/170 (30%), Positives = 52/170 (30%)
Frame = +3
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG GGGGGGG GGG GGG
Sbjct: 48 GGGGGGGGGGGGGGGGGGGGGGGGG------------------------GGGGGGGGGGG 155
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGG GGG GGG GGG G GGG
Sbjct: 156 GG---------------------GGGGGGGGGGGGGGGGGGGG--------GGGGGGGGG 248
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G GGGG GGGGGGG G
Sbjct: 249 GGGGGGG------------------------------GGGGGGGGGGGGG 308
Score = 54.7 bits (130), Expect = 3e-08
Identities = 51/165 (30%), Positives = 51/165 (30%)
Frame = +2
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGG 61
GGGGG GG G GGGG GGG GGGG
Sbjct: 38 GGGGGGGG------------------------------GGGGGGGGGGGGGGGGGGGGGG 127
Query: 62 GGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLAT 121
GGG GGG GGG G
Sbjct: 128 GGG------------------------GGGGGGGGGGGGG-------------------- 175
Query: 122 IGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGG 166
G GGG GGG GGG GGG G GGGG GGGG
Sbjct: 176 -GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 307
Score = 50.1 bits (118), Expect = 7e-07
Identities = 48/159 (30%), Positives = 48/159 (30%)
Frame = +3
Query: 51 TVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAV 110
T GGG GGGGGGG GGG GGG G
Sbjct: 30 TPGGGGGGGGGGGG------------------------GGGGGGGGGGGGG--------- 110
Query: 111 VAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVV 170
G GGG GGG GGG GGG G GGGG GGGG
Sbjct: 111 ------------GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG---- 242
Query: 171 VAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GGGGGGG G
Sbjct: 243 --------------------------GGGGGGGGGGGGG 281
Score = 46.2 bits (108), Expect = 1e-05
Identities = 46/157 (29%), Positives = 46/157 (29%)
Frame = +2
Query: 53 EGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVA 112
E G GGGGGGG GGG GGG G
Sbjct: 26 ENPGGGGGGGGG------------------------GGGGGGGGGGGGG----------- 100
Query: 113 AMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVA 172
G GGG GGG GGG GGG G GGGG GGGG
Sbjct: 101 ----------GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG------ 232
Query: 173 AIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GGGGGGG G
Sbjct: 233 ------------------------GGGGGGGGGGGGG 271
Score = 43.9 bits (102), Expect = 5e-05
Identities = 45/158 (28%), Positives = 46/158 (28%)
Frame = +2
Query: 52 VEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVV 111
+ G GGGGGGG GGG GGG G
Sbjct: 17 LSGENPGGGGGGG------------------------GGGGGGGGGGGGG---------- 94
Query: 112 AAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVV 171
G GGG GGG GGG GGG G GGGG GGGG
Sbjct: 95 -----------GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG----- 226
Query: 172 AAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GGGGGGG G
Sbjct: 227 -------------------------GGGGGGGGGGGGG 265
>TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E8.80 -
Arabidopsis thaliana, partial (18%)
Length = 1260
Score = 40.0 bits (92), Expect(2) = 8e-07
Identities = 43/128 (33%), Positives = 49/128 (37%)
Frame = -1
Query: 41 SGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVV 100
+GGGG+ A T +GGG+G GGG *G CG G
Sbjct: 441 TGGGGEG*AYTGDGGGDGDGGG------------------------**GV*CGAGDDGGG 334
Query: 101 GVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGG 160
G +T G GGG GGGDGGG +* GG G GG
Sbjct: 333 G------------------STQGGGGGGGGSTQ--GGGDGGG-----GE*TGGGGECTGG 229
Query: 161 DYGGGGDK 168
GGGGDK
Sbjct: 228 --GGGGDK 211
Score = 38.9 bits (89), Expect(2) = 4e-07
Identities = 41/135 (30%), Positives = 48/135 (35%), Gaps = 8/135 (5%)
Frame = -1
Query: 40 GSGGGGKAVAATVEGGGNGGGG---GGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGR 96
G GGGG + +GGG+GGGG GGG GGG GG +
Sbjct: 321 GGGGGG---GGSTQGGGDGGGGE*TGGGGECT--------------------GGGGGGDK 211
Query: 97 AAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGG--- 153
G + G GGG G GGG GGG
Sbjct: 210 YGYTG----------------GVGE*GGGGGEXGGGGGE*AGGGGGEL***GTGGGGENT 79
Query: 154 --CGVGGGGDYGGGG 166
G GGGG++GGGG
Sbjct: 78 GAGGGGGGGEFGGGG 34
Score = 37.0 bits (84), Expect(2) = 3e-07
Identities = 10/19 (52%), Positives = 11/19 (57%), Gaps = 4/19 (21%)
Frame = -3
Query: 22 IGWWWWWQRWW----LWWW 36
+ WWWW RWW WWW
Sbjct: 184 VRWWWWRXRWWWR*VSWWW 128
Score = 33.9 bits (76), Expect(2) = 3e-07
Identities = 19/30 (63%), Positives = 21/30 (69%), Gaps = 1/30 (3%)
Frame = -1
Query: 37 ***GSGGGGKAVAATVEGGGNG-GGGGGGD 65
***G+GGGG+ A GGG GGGGGGD
Sbjct: 114 ***GTGGGGENTGAGGGGGGGEFGGGGGGD 25
Score = 33.9 bits (76), Expect = 0.051
Identities = 10/18 (55%), Positives = 11/18 (60%), Gaps = 6/18 (33%)
Frame = -3
Query: 24 WWW--WWQRWW----LWW 35
WWW WW RWW +WW
Sbjct: 61 WWW*IWWWRWWRPNTIWW 8
Score = 32.3 bits (72), Expect = 0.15
Identities = 46/151 (30%), Positives = 46/151 (30%), Gaps = 19/151 (12%)
Frame = -1
Query: 34 WWW***GS------------GGGGKAVAATVEGGGNGG--GGGGGDRVVVVVSVVAIEWW 79
WWW * GGGG A GG GGGGGD
Sbjct: 642 WWWSR*KGKRRSSQKNQ*KGGGGGDAYEIPKTGGIGPSYTGGGGGDEYT----------- 496
Query: 80 RRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**G--- 136
G GG A G V A G GGG **G
Sbjct: 495 -----------GGGGEGYAYTGDGGGEVYTGGGG---EG*AYTGDGGGDGDGGG**GV*C 358
Query: 137 --GGDGGGWWWWWWQ*GGGCGVGGGGDYGGG 165
G DGGG GGG GGGD GGG
Sbjct: 357 GAGDDGGGGSTQGGGGGGGGSTQGGGDGGGG 265
Score = 31.6 bits (70), Expect(2) = 4e-07
Identities = 9/23 (39%), Positives = 10/23 (43%), Gaps = 10/23 (43%)
Frame = -3
Query: 24 WWWWWQRWWL----------WWW 36
WWW W WW+ WWW
Sbjct: 403 WWW*WGWWWVIRCLMWRWR*WWW 335
Score = 31.2 bits (69), Expect = 0.33
Identities = 12/27 (44%), Positives = 13/27 (47%), Gaps = 13/27 (48%)
Frame = -3
Query: 24 WWW---------WWQ---RWWLW-WW* 37
WWW WW+ RWW W WW*
Sbjct: 343 WWWRFYTRWRWRWWRFYTRWWRWRWW* 263
Score = 30.4 bits (67), Expect = 0.57
Identities = 20/70 (28%), Positives = 22/70 (30%), Gaps = 5/70 (7%)
Frame = -3
Query: 24 WW---WWWQ--RWWLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEW 78
WW WWW+ W WW ** G + V W
Sbjct: 271 WW*IDWWWR*VYWRRWWG**IWIYGWSR*VR---------------------------WW 173
Query: 79 WRRLWWWWR* 88
W R WWWR*
Sbjct: 172 WWRXRWWWR* 143
Score = 30.4 bits (67), Expect = 0.57
Identities = 42/158 (26%), Positives = 49/158 (30%), Gaps = 31/158 (19%)
Frame = -1
Query: 83 WWWWR*GGG---------CGGGRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA 133
WWW R* G GGG + T + + G GGG +
Sbjct: 642 WWWSR*KGKRRSSQKNQ*KGGGGGDAYEIPKTGGIGP---------SYTGGGGGDEYTGG 490
Query: 134 **GG------GDGGGWWWW--------WWQ*GGGCGVGGG--------GDYGGGGDKVVV 171
GG GDGGG + + GGG G GGG GD GGGG
Sbjct: 489 --GGEGYAYTGDGGGEVYTGGGGEG*AYTGDGGGDGDGGG**GV*CGAGDDGGGGST--- 325
Query: 172 AAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GGGGGGG+ G
Sbjct: 324 -------------------------QGGGGGGGGSTQG 286
Score = 30.4 bits (67), Expect = 0.57
Identities = 11/31 (35%), Positives = 13/31 (41%), Gaps = 15/31 (48%)
Frame = -3
Query: 22 IGWWW---------WWQ------RWWLWWW* 37
+ WWW WW+ RW WWW*
Sbjct: 142 VSWWWRW*AIVVRNWWRW*EYRSRWRRWWW* 50
Score = 29.6 bits (65), Expect = 0.97
Identities = 7/11 (63%), Positives = 7/11 (63%)
Frame = -3
Query: 26 WWWQRWWLWWW 36
WWW WW WW
Sbjct: 61 WWW*IWWWRWW 29
Score = 29.6 bits (65), Expect = 0.97
Identities = 9/17 (52%), Positives = 10/17 (57%)
Frame = -3
Query: 20 VEIGWWWWWQRWWLWWW 36
+ I W WWW W WWW
Sbjct: 421 IGIYW*WWW*--WGWWW 377
Score = 29.3 bits (64), Expect(2) = 8e-07
Identities = 10/25 (40%), Positives = 12/25 (48%), Gaps = 11/25 (44%)
Frame = -3
Query: 23 GWWWW---------WQRWWL--WWW 36
GWWWW W R ++ WWW
Sbjct: 586 GWWWW*CI*DSKDRWYRPFIYRWWW 512
Score = 28.5 bits (62), Expect = 2.2
Identities = 20/81 (24%), Positives = 22/81 (26%), Gaps = 17/81 (20%)
Frame = -3
Query: 24 WWW---WWQR--WWLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEW 78
WWW WW R W W W* ++V W
Sbjct: 175 WWWRXRWWWR*VSWWWRW*----------------------------------AIVVRNW 98
Query: 79 WR--------RLW----WWWR 87
WR R W WWWR
Sbjct: 97 WRW*EYRSRWRRWWW*IWWWR 35
Score = 26.9 bits (58), Expect = 6.3
Identities = 7/13 (53%), Positives = 9/13 (68%), Gaps = 1/13 (7%)
Frame = -3
Query: 25 WWWWQRWWL-WWW 36
WW W+ W + WWW
Sbjct: 286 WWRWRWW*IDWWW 248
>TC87476 homologue to PIR|T11622|T11622 extensin class 1 precursor - cowpea,
partial (48%)
Length = 939
Score = 38.1 bits (87), Expect(2) = 2e-04
Identities = 42/131 (32%), Positives = 48/131 (36%), Gaps = 3/131 (2%)
Frame = -1
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG T GGG+G GGGG L + G G GGG
Sbjct: 336 GEGDGGGGDL*TYGGGGDGDGGGGD-----------------L*TYGGGGEGDGGGGDL* 208
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
+ V GGG G G+GGG + GGG G GGG
Sbjct: 207 I*GGGGEGVG---------------GGGDL*TYGGGGDGEGGGGLL*TYG-GGGDGDGGG 76
Query: 160 GD---YGGGGD 167
GD YGGGG+
Sbjct: 75 GDLYTYGGGGE 43
Score = 35.4 bits (80), Expect(3) = 8e-07
Identities = 25/46 (54%), Positives = 25/46 (54%), Gaps = 3/46 (6%)
Frame = -1
Query: 125 GGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGD---YGGGGD 167
GGG G GDGGG GGG GVGGGGD YGGGGD
Sbjct: 273 GGGDL*TYGGGGEGDGGGGDL*I*G-GGGEGVGGGGDL*TYGGGGD 139
Score = 34.3 bits (77), Expect = 0.039
Identities = 27/60 (45%), Positives = 31/60 (51%), Gaps = 3/60 (5%)
Frame = -3
Query: 136 GGGDGGGWWWWWWQ*GGGCGVGGGGD---YGGGGDKVVVAAIMVVVATPVVTVGRWWRRR 192
G GDGGG + GGG G GGGGD YGGGG+ +V + RWWRRR
Sbjct: 481 GEGDGGGGDL*TYG-GGGEGEGGGGDL*TYGGGGEGDGGRW*LVDIWRR--W*RRWWRRR 311
Score = 32.7 bits (73), Expect(2) = 0.002
Identities = 22/46 (47%), Positives = 26/46 (55%), Gaps = 3/46 (6%)
Frame = -3
Query: 125 GGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGD---YGGGGD 167
GGG + G G+GGG + GGG G GGGGD YGGGG+
Sbjct: 658 GGGDL*I*GGGGEGEGGGGDL*PYG-GGGDGEGGGGDL**YGGGGE 524
Score = 32.3 bits (72), Expect = 0.15
Identities = 17/49 (34%), Positives = 23/49 (46%), Gaps = 15/49 (30%)
Frame = -3
Query: 2 GGGGGNGGRLVLVAVVVT----------VEIGWWWWWQRW-W----LWW 35
GGG G+GGR LV + + + WW W+RW W +WW
Sbjct: 391 GGGEGDGGRW*LVDIWRRW*RRWWRRRFINVWRWWGWRRWRW*LVNIWW 245
Score = 32.3 bits (72), Expect(3) = 8e-07
Identities = 24/68 (35%), Positives = 26/68 (37%), Gaps = 20/68 (29%)
Frame = -3
Query: 40 GSGGGGKAVAATVEGGGNG----------GGGGGGDR----VVVVVSVVAIEWWRR---- 81
G G GG T GGG G GGGG GD +V + WWRR
Sbjct: 481 GEGDGGGGDL*TYGGGGEGEGGGGDL*TYGGGGEGDGGRW*LVDIWRRW*RRWWRRRFIN 302
Query: 82 --LWWWWR 87
WW WR
Sbjct: 301 VWRWWGWR 278
Score = 31.6 bits (70), Expect(2) = 0.10
Identities = 15/20 (75%), Positives = 15/20 (75%), Gaps = 3/20 (15%)
Frame = -1
Query: 151 GGGCGVGGGGD---YGGGGD 167
GGG G GGGGD YGGGGD
Sbjct: 342 GGGEGDGGGGDL*TYGGGGD 283
Score = 30.4 bits (67), Expect = 0.57
Identities = 42/132 (31%), Positives = 47/132 (34%), Gaps = 4/132 (3%)
Frame = -1
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG G GGGGD GGG G G
Sbjct: 330 GDGGGGDL*TY---GGGGDGDGGGGDL*TYG------------------GGGEGDGGGGD 214
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCG-GGSKAVAA**GGGDGGGWWWWWWQ*GGG--CGV 156
+ + V + G G GG + GGGDG G GGG
Sbjct: 213 L*I*GGGGEGVGGGGDL*TYGGGGDGEGGGGLL*TYGGGGDGDG--------GGGDLYTY 58
Query: 157 GGGGDY-GGGGD 167
GGGG+ GGGGD
Sbjct: 57 GGGGEGDGGGGD 22
Score = 30.4 bits (67), Expect(3) = 0.002
Identities = 23/50 (46%), Positives = 25/50 (50%), Gaps = 7/50 (14%)
Frame = -3
Query: 125 GGGSKAVAA**GGGD----GGGWWWWWWQ*GGGCGVGGGGD---YGGGGD 167
GGG +A GGGD GGG G G GGGGD YGGGG+
Sbjct: 538 GGGGEAEG---GGGDL*T*GGG----------GEGDGGGGDL*TYGGGGE 428
Score = 29.6 bits (65), Expect = 0.97
Identities = 19/75 (25%), Positives = 27/75 (35%), Gaps = 7/75 (9%)
Frame = -3
Query: 20 VEIGWWWWWQRWW-----LWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVV 74
V+I WW W+ WW +WW GG G ++ +
Sbjct: 163 VDIWRWW*WRGWWWTLIDIWW---------------------RGGWRWGWW*LIYIWWGR 47
Query: 75 AIEWWRR--LWWWWR 87
WWRR ++ WWR
Sbjct: 46 GG*WWRRRLVYIWWR 2
Score = 29.3 bits (64), Expect = 1.3
Identities = 13/24 (54%), Positives = 13/24 (54%)
Frame = -1
Query: 40 GSGGGGKAVAATVEGGGNGGGGGG 63
G G GG T GGG G GGGG
Sbjct: 96 GDGDGGGGDLYTYGGGGEGDGGGG 25
Score = 27.7 bits (60), Expect = 3.7
Identities = 14/28 (50%), Positives = 16/28 (57%), Gaps = 4/28 (14%)
Frame = -3
Query: 42 GGGGKAVAATVE----GGGNGGGGGGGD 65
GGGG+A + GGG G GGGGD
Sbjct: 538 GGGGEAEGGGGDL*T*GGGGEGDGGGGD 455
Score = 27.3 bits (59), Expect = 4.8
Identities = 13/20 (65%), Positives = 14/20 (70%), Gaps = 3/20 (15%)
Frame = -3
Query: 151 GGGCGVGGGGDY---GGGGD 167
GGG G GGGGD GGGG+
Sbjct: 679 GGGDGDGGGGDL*I*GGGGE 620
Score = 24.6 bits (52), Expect(2) = 0.002
Identities = 7/9 (77%), Positives = 7/9 (77%)
Frame = -2
Query: 79 WRRLWWWWR 87
WRR W WWR
Sbjct: 680 WRR*WRWWR 654
Score = 23.9 bits (50), Expect(3) = 0.002
Identities = 8/12 (66%), Positives = 8/12 (66%), Gaps = 2/12 (16%)
Frame = -2
Query: 78 WWRRLW--WWWR 87
WWRR W W WR
Sbjct: 587 WWRR*WRRWRWR 552
Score = 22.7 bits (47), Expect(2) = 2e-04
Identities = 5/9 (55%), Positives = 6/9 (66%)
Frame = -2
Query: 24 WWWWWQRWW 32
W WW + WW
Sbjct: 395 WRWW*RGWW 369
Score = 21.9 bits (45), Expect(3) = 0.002
Identities = 8/16 (50%), Positives = 10/16 (62%)
Frame = -2
Query: 20 VEIGWWWWWQRWWLWW 35
VE+G W +R W WW
Sbjct: 698 VEVGMIW--RR*WRWW 657
Score = 20.4 bits (41), Expect(3) = 8e-07
Identities = 6/11 (54%), Positives = 7/11 (63%), Gaps = 1/11 (9%)
Frame = -2
Query: 27 WWQR-WWLWWW 36
WW+R W W W
Sbjct: 587 WWRR*WRRWRW 555
Score = 20.0 bits (40), Expect(2) = 0.10
Identities = 6/7 (85%), Positives = 6/7 (85%)
Frame = -3
Query: 186 GRWWRRR 192
G WWRRR
Sbjct: 187 GCWWRRR 167
>TC93504 weakly similar to PIR|S46286|S46286 RNA-binding protein - wood
tobacco, partial (55%)
Length = 576
Score = 49.3 bits (116), Expect = 1e-06
Identities = 19/41 (46%), Positives = 22/41 (53%), Gaps = 2/41 (4%)
Frame = +2
Query: 24 WWWWWQRWWLW--WW***GSGGGGKAVAATVEGGGNGGGGG 62
WWWWW WW W WW + + A + GGGN GGGG
Sbjct: 446 WWWWWL*WWWWKLWW------VVAEIMVAEIMGGGNYGGGG 550
Score = 39.7 bits (91), Expect = 0.001
Identities = 27/73 (36%), Positives = 32/73 (42%), Gaps = 3/73 (4%)
Frame = +3
Query: 136 GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRR---R 192
GGG GGG + GGG G GGGG Y GGG G WWR+ R
Sbjct: 399 GGGYGGGGGY-----GGGGGYGGGGGYSGGGG----------------NYGGWWRKLWWR 515
Query: 193 RRR*SDGGGGGGG 205
+ ++ GG GG
Sbjct: 516 KLWVAETTGGVGG 554
Score = 37.4 bits (85), Expect = 0.005
Identities = 25/64 (39%), Positives = 26/64 (40%), Gaps = 7/64 (10%)
Frame = +3
Query: 40 GSGGGGKAVAATVEGGGNGGGG----GGGDRVVVVVSVVAIEWWRRLWW---WWR*GGGC 92
G GGGG GGG GGG GGG WWR+LWW W G
Sbjct: 390 GFGGGGYGGGGGYGGGGGYGGGGGYSGGGGNYG--------GWWRKLWWRKLWVAETTGG 545
Query: 93 GGGR 96
GGR
Sbjct: 546 VGGR 557
Score = 31.6 bits (70), Expect = 0.25
Identities = 22/65 (33%), Positives = 24/65 (36%), Gaps = 2/65 (3%)
Frame = +3
Query: 152 GGCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGG--GGGANSG 209
GG G GGGG YGGGG GGGG GGG N G
Sbjct: 396 GGGGYGGGGGYGGGGG-----------------------------YGGGGGYSGGGGNYG 488
Query: 210 VSWQQ 214
W++
Sbjct: 489 GWWRK 503
Score = 31.2 bits (69), Expect = 0.33
Identities = 24/66 (36%), Positives = 32/66 (48%)
Frame = +1
Query: 95 GRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGC 154
G AVV VAA ++ A VA +VVV + + GGG+ GG + W + GG
Sbjct: 391 GLVAVVMVAAVAMAAAVAMVVVVAIVVV-------VEIMVGGGGNYGGGNYGWRKLRGGW 549
Query: 155 GVGGGG 160
VG G
Sbjct: 550 VVGTYG 567
>TC80881 similar to PIR|T11622|T11622 extensin class 1 precursor - cowpea,
partial (34%)
Length = 597
Score = 47.0 bits (110), Expect = 6e-06
Identities = 50/148 (33%), Positives = 56/148 (37%), Gaps = 4/148 (2%)
Frame = -3
Query: 52 VEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVV 111
+ GGG G GGGGD L + G G GGG +
Sbjct: 589 IYGGGGEGDGGGGD----------------L*TYGGGGEGEGGGGDLYT*FGGSE----- 473
Query: 112 AAMVVVVLATIGCGGGSKAVAA**GGGDG-GGWWWWWWQ*GGGCGVGGGGD---YGGGGD 167
G GGG GGGDG GG GGG G GGGGD YGGGG+
Sbjct: 472 -----------GDGGGGDL*TYG-GGGDGEGGGGLL*TYGGGGEGDGGGGDL*TYGGGGE 329
Query: 168 KVVVAAIMVVVATPVVTVGRWWRRRRRR 195
VV+ V + RWWR RR R
Sbjct: 328 GRWWRWRFVVIRWVFVDIWRWWRWRRWR 245
Score = 35.8 bits (81), Expect(2) = 2e-04
Identities = 23/61 (37%), Positives = 28/61 (45%), Gaps = 13/61 (21%)
Frame = -3
Query: 40 GSGGGGKAVAATVEGGGNGGG--------GGGGDR-----VVVVVSVVAIEWWRRLWWWW 86
G G GG + T GGG G G GGGG+ VV+ V ++ WR WW W
Sbjct: 430 GDGEGGGGLL*TYGGGGEGDGGGGDL*TYGGGGEGRWWRWRFVVIRWVFVDIWR--WWRW 257
Query: 87 R 87
R
Sbjct: 256 R 254
Score = 30.8 bits (68), Expect = 0.43
Identities = 19/35 (54%), Positives = 21/35 (59%), Gaps = 6/35 (17%)
Frame = -1
Query: 136 GGGDG---GGWWWWWWQ*GGGCGVGGGGD---YGG 164
GGGD GG+ W + GGG G GGGGD YGG
Sbjct: 318 GGGDL***GGYLWTYG--GGGDGEGGGGDL**YGG 220
Score = 30.4 bits (67), Expect = 0.57
Identities = 19/49 (38%), Positives = 24/49 (48%), Gaps = 15/49 (30%)
Frame = -3
Query: 3 GGGGNGG----RLVLVAVVVTVEIGWWWWWQRW-W----LW------WW 36
GGGG G R V++ V V+I WW W+RW W +W WW
Sbjct: 343 GGGGEGRWWRWRFVVIRWVF-VDIWRWWRWRRWRWGFVVIWR*FEIIWW 200
Score = 28.9 bits (63), Expect(2) = 0.014
Identities = 9/21 (42%), Positives = 12/21 (56%), Gaps = 5/21 (23%)
Frame = -2
Query: 20 VEIGWWWWWQRWW-----LWW 35
V+I WW W+ WW +WW
Sbjct: 449 VDIWRWW*WRGWWWTLIDIWW 387
Score = 25.8 bits (55), Expect(2) = 0.014
Identities = 14/30 (46%), Positives = 14/30 (46%), Gaps = 4/30 (13%)
Frame = -1
Query: 40 GSGGGGK----AVAATVEGGGNGGGGGGGD 65
G GGGG GGG G GGGGD
Sbjct: 327 GDGGGGDL***GGYLWTYGGGGDGEGGGGD 238
Score = 25.4 bits (54), Expect(2) = 2e-04
Identities = 8/20 (40%), Positives = 8/20 (40%), Gaps = 7/20 (35%)
Frame = -2
Query: 24 WW-------WWWQRWWLWWW 36
WW W W W WWW
Sbjct: 467 WWRRRLVDIWRWW*WRGWWW 408
>BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein
{Mycobacterium tuberculosis CDC1551}, partial (7%)
Length = 164
Score = 46.6 bits (109), Expect = 8e-06
Identities = 40/113 (35%), Positives = 40/113 (35%)
Frame = +3
Query: 54 GGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAA 113
GGG GGGGGGG GGG GGG A G
Sbjct: 3 GGGRGGGGGGGG-----------------------GGGGGGGGAGGGG------------ 77
Query: 114 MVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGG 166
G GGG GGG GGG GGG G GGGG GGGG
Sbjct: 78 ---------GGGGGG-------GGGGGGG--------GGGGGGGGGGGGGGGG 164
Score = 40.8 bits (94), Expect = 4e-04
Identities = 38/121 (31%), Positives = 38/121 (31%)
Frame = +2
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG GGG GGGGGGG GGG GGGR
Sbjct: 2 GGGAGGGGGGGGGGGGGGGGGGGGG------------------------GGGGGGGRG-- 103
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
GGG GGG GGG GGG G GGG
Sbjct: 104 -------------------------GGG--------GGGGGGG--------GGGGGGGGG 160
Query: 160 G 160
G
Sbjct: 161 G 163
Score = 40.4 bits (93), Expect = 5e-04
Identities = 30/87 (34%), Positives = 30/87 (34%)
Frame = +2
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPV 182
G GGG GGG GGG GGG G GGGG GGGG
Sbjct: 8 GAGGGGGGGGGGGGGGGGGG--------GGGGGGGGGGRGGGGGG--------------- 118
Query: 183 VTVGRWWRRRRRR*SDGGGGGGGANSG 209
GGGGGGG G
Sbjct: 119 ----------------GGGGGGGGGGG 151
Score = 38.9 bits (89), Expect = 0.002
Identities = 24/63 (38%), Positives = 24/63 (38%)
Frame = +1
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGG 61
GGGGG GGR G GGGG A GGG GGGG
Sbjct: 40 GGGGGGGGR-----------------------------GGGGGGGGGGAGGGGGGGGGGG 132
Query: 62 GGG 64
GGG
Sbjct: 133GGG 141
Score = 37.0 bits (84), Expect = 0.006
Identities = 29/87 (33%), Positives = 29/87 (33%)
Frame = +1
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPV 182
G GGG GGG GGG GG G GGGG GGGG
Sbjct: 4 GGGGGGGGGGGGGGGGGGGG--------RGGGGGGGGGGAGGGG---------------- 111
Query: 183 VTVGRWWRRRRRR*SDGGGGGGGANSG 209
GGGGGGG G
Sbjct: 112 ----------------GGGGGGGGGGG 144
Score = 34.3 bits (77), Expect = 0.039
Identities = 27/74 (36%), Positives = 27/74 (36%)
Frame = +1
Query: 136 GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR 195
GGG GGG GGG GGGG GGGG R
Sbjct: 1 GGGGGGG--------GGG---GGGGGGGGGGG---------------------------R 66
Query: 196 *SDGGGGGGGANSG 209
GGGGGGGA G
Sbjct: 67 GGGGGGGGGGAGGG 108
>BQ140560 weakly similar to GP|15128223|db hypothetical protein~similar to
Oryza sativa chromosome 1 P0489A01.2, partial (5%)
Length = 215
Score = 38.9 bits (89), Expect = 0.002
Identities = 11/22 (50%), Positives = 11/22 (50%), Gaps = 9/22 (40%)
Frame = -3
Query: 24 WWWWWQR---------WWLWWW 36
WWWWW R WW WWW
Sbjct: 114 WWWWW*RCST*CRKI*WWWWWW 49
Score = 37.7 bits (86), Expect = 0.004
Identities = 30/85 (35%), Positives = 35/85 (40%)
Frame = -1
Query: 125 GGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPVVT 184
GGG+ A G GG W GGG G+ GGG GGGG + A
Sbjct: 212 GGGAPVDAGRYSXGGGGAPW----NGGGGGGIEGGGGGGGGGRGAPLDA----------- 78
Query: 185 VGRWWRRRRRR*SDGGGGGGGANSG 209
GR+ DGGGGGG +G
Sbjct: 77 -GRY---------DGGGGGGAP*NG 33
Score = 35.8 bits (81), Expect = 0.013
Identities = 10/16 (62%), Positives = 10/16 (62%), Gaps = 3/16 (18%)
Frame = -3
Query: 24 WWWWWQ--RWWLW-WW 36
WWWWW WW W WW
Sbjct: 63 WWWWWSSIEWWGWGWW 16
Score = 33.9 bits (76), Expect(2) = 9e-06
Identities = 11/18 (61%), Positives = 12/18 (66%), Gaps = 4/18 (22%)
Frame = -3
Query: 24 WW-WWW---QRWWLWWW* 37
WW W W +RWW WWW*
Sbjct: 150 WWRWGWH*RRRWWWWWW* 97
Score = 32.7 bits (73), Expect(2) = 0.002
Identities = 15/25 (60%), Positives = 16/25 (64%)
Frame = -1
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGG 64
G GGGG +EGGG GGGGG G
Sbjct: 149 GGGGGG------IEGGGGGGGGGRG 93
Score = 32.0 bits (71), Expect(2) = 9e-06
Identities = 17/34 (50%), Positives = 19/34 (55%), Gaps = 9/34 (26%)
Frame = -1
Query: 40 GSGGGGKAV---AATVEGGG------NGGGGGGG 64
G GGGG+ A +GGG NGGGGGGG
Sbjct: 116 GGGGGGRGAPLDAGRYDGGGGGGAP*NGGGGGGG 15
Score = 30.8 bits (68), Expect = 0.43
Identities = 13/25 (52%), Positives = 15/25 (60%)
Frame = -1
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGG 64
G GGGG A ++ G GGGGGG
Sbjct: 122 GGGGGGGGRGAPLDAGRYDGGGGGG 48
Score = 29.6 bits (65), Expect = 0.97
Identities = 37/106 (34%), Positives = 38/106 (34%), Gaps = 5/106 (4%)
Frame = -1
Query: 42 GGGGKAVAAT--VEGGGNG---GGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGR 96
GGGG V A GGG GGGGGG IE GGG GGGR
Sbjct: 215 GGGGAPVDAGRYSXGGGGAPWNGGGGGG-----------IEGG---------GGGGGGGR 96
Query: 97 AAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGG 142
A + G GGG A * GGG GGG
Sbjct: 95 GAPLDAGRYD----------------GGGGGG---AP*NGGGGGGG 15
Score = 27.7 bits (60), Expect = 3.7
Identities = 16/28 (57%), Positives = 16/28 (57%), Gaps = 2/28 (7%)
Frame = -1
Query: 41 SGGGGKAVAATVEGGG--NGGGGGGGDR 66
S GGG A GGG GGGGGGG R
Sbjct: 179 SXGGGGAPWNGGGGGGIEGGGGGGGGGR 96
Score = 25.0 bits (53), Expect(2) = 0.002
Identities = 6/13 (46%), Positives = 6/13 (46%)
Frame = -3
Query: 24 WWWWWQRWWLWWW 36
W W WW W W
Sbjct: 171 WRWSSMEWWRWGW 133
>TC87201 similar to PIR|F86388|F86388 hypothetical protein AAG29221.1
[imported] - Arabidopsis thaliana, partial (41%)
Length = 991
Score = 38.5 bits (88), Expect(2) = 3e-04
Identities = 58/184 (31%), Positives = 65/184 (34%), Gaps = 16/184 (8%)
Frame = -3
Query: 42 GGGGKAVAATVEGGGNGG----GGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRA 97
GGGG + GGG G GGGGGD L+ + G G GGG
Sbjct: 440 GGGGDS*TY---GGGGGDL*TYGGGGGD----------------LYTYGGGGDGDGGG-- 324
Query: 98 AVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGW--------WWWWWQ 149
G T V T G GGG + GGGDGGG +
Sbjct: 323 ---GDL*TYGGGVY---------TYGGGGGDL*IYG--GGGDGGGGDG*T*GGGGGDLYT 186
Query: 150 *GGGCG----VGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGG 205
GGG G GGGGD GGGGD VV+ + GGGG GG
Sbjct: 185 YGGGGGDL*IYGGGGD-GGGGDG*T*GGARVVICIRI----------------GGGGDGG 57
Query: 206 ANSG 209
G
Sbjct: 56 GGDG 45
Score = 36.2 bits (82), Expect(2) = 1e-05
Identities = 45/133 (33%), Positives = 49/133 (36%), Gaps = 5/133 (3%)
Frame = -3
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG T GG GGGGGD + + GG GGG
Sbjct: 344 GDGDGGGGDL*TYGGGVYTYGGGGGDL*I-----------------YGGGGDGGGGDG*T 216
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGG-----C 154
G L T G GGG + GGGDGGG *GG
Sbjct: 215 *GGGGGD------------LYTYGGGGGDL*IYG--GGGDGGG--GDG*T*GGARVVICI 84
Query: 155 GVGGGGDYGGGGD 167
+GGGGD GGGGD
Sbjct: 83 RIGGGGD-GGGGD 48
Score = 35.8 bits (81), Expect = 0.013
Identities = 44/127 (34%), Positives = 46/127 (35%), Gaps = 13/127 (10%)
Frame = -3
Query: 54 GGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGG-------CGGGRAA-VVGVAAT 105
GGG G GGGGD *GGG CGGG + G
Sbjct: 521 GGGGDGEGGGGDS*T-------------------*GGGGGDL*TYCGGGGDS*TYGGGGG 399
Query: 106 SVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGV--GGGGD-- 161
L T G GGG G GDGGG + GGG GGGGD
Sbjct: 398 D------------L*TYGGGGGDLYTYGGGGDGDGGGGDL*TY--GGGVYTYGGGGGDL* 261
Query: 162 -YGGGGD 167
YGGGGD
Sbjct: 260 IYGGGGD 240
Score = 28.9 bits (63), Expect(2) = 1e-05
Identities = 6/12 (50%), Positives = 9/12 (75%)
Frame = -2
Query: 24 WWWWWQRWWLWW 35
WWW W+ ++WW
Sbjct: 384 WWWRWRLVYIWW 349
Score = 28.5 bits (62), Expect = 2.2
Identities = 14/21 (66%), Positives = 14/21 (66%), Gaps = 4/21 (19%)
Frame = -3
Query: 151 GGGCGVGGGGDY----GGGGD 167
GGG G GGGGD GGGGD
Sbjct: 518 GGGDGEGGGGDS*T*GGGGGD 456
Score = 27.3 bits (59), Expect(2) = 0.45
Identities = 13/33 (39%), Positives = 16/33 (48%), Gaps = 9/33 (27%)
Frame = -3
Query: 40 GSGGGGKA---------VAATVEGGGNGGGGGG 63
G GGGG + + GGG+GGGG G
Sbjct: 143 GDGGGGDG*T*GGARVVICIRIGGGGDGGGGDG 45
Score = 27.3 bits (59), Expect = 4.8
Identities = 10/22 (45%), Positives = 11/22 (49%), Gaps = 6/22 (27%)
Frame = -2
Query: 20 VEIGWWWW-----WQRW-WLWW 35
V+I WW W W W W WW
Sbjct: 393 VDIWWWRWRLVYIWWGW*WRWW 328
Score = 26.6 bits (57), Expect = 8.2
Identities = 22/51 (43%), Positives = 25/51 (48%), Gaps = 4/51 (7%)
Frame = -3
Query: 121 TIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGV---GGGGD-YGGGGD 167
T G GGG + GGD + GGG G+ GGGGD GGGGD
Sbjct: 605 T*GAGGGDL*IY----GGDS*TY-------GGGIGL**YGGGGDGEGGGGD 486
Score = 21.9 bits (45), Expect(2) = 0.45
Identities = 7/16 (43%), Positives = 8/16 (49%), Gaps = 4/16 (25%)
Frame = -2
Query: 24 WWWWWQRW----WLWW 35
W WW +RW W W
Sbjct: 246 W*WWRRRWVNIRWRRW 199
Score = 21.9 bits (45), Expect(2) = 3e-04
Identities = 5/15 (33%), Positives = 8/15 (53%)
Frame = -2
Query: 18 VTVEIGWWWWWQRWW 32
+ + GWW + WW
Sbjct: 609 INIGSGWWGFIDIWW 565
>TC76718 homologue to GP|8132441|gb|AAF73291.1| extensin {Pisum sativum},
partial (63%)
Length = 712
Score = 33.9 bits (76), Expect = 0.051
Identities = 29/98 (29%), Positives = 35/98 (35%), Gaps = 15/98 (15%)
Frame = -2
Query: 5 GGNGGRLVLVAV----VVTVEIGWWWWWQRWWL-WWW***GSGGGGKAVAATVEGGGNGG 59
GG G R VLV V V WW W W L WWW + +
Sbjct: 618 GGGGWRFVLVNWRXR*FVLVNWRWWRWVFIWLLRWWW---------RRLVLVHWWRRGFI 466
Query: 60 GGGGGDRVVVVVS-----VVAIEWWRR-----LWWWWR 87
R V+V+ + + WWRR WWWWR
Sbjct: 465 LVHRWRR*FVLVNWWWRRFILVNWWRR*LVLVNWWWWR 352
Score = 32.3 bits (72), Expect = 0.15
Identities = 22/77 (28%), Positives = 25/77 (31%), Gaps = 8/77 (10%)
Frame = -2
Query: 18 VTVEIGWWWWW------QRWWL--WWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVV 69
V V + W WWW +RW + WWW RVVV
Sbjct: 249 VFVWLLWRWWWGTVMVRRRW*VHRWWW-----------------------------RVVV 157
Query: 70 VVSVVAIEWWRRLWWWW 86
V RLWWWW
Sbjct: 156 V----------RLWWWW 136
Score = 31.6 bits (70), Expect(2) = 6e-05
Identities = 9/16 (56%), Positives = 9/16 (56%), Gaps = 2/16 (12%)
Frame = -1
Query: 23 GWWWWWQRWWL--WWW 36
GWWW W W WWW
Sbjct: 655 GWWW*WVLVWFLGWWW 608
Score = 31.2 bits (69), Expect = 0.33
Identities = 24/73 (32%), Positives = 32/73 (42%), Gaps = 12/73 (16%)
Frame = -2
Query: 27 WWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVV------VSVVAIEWWR 80
WW+R ++ +GGGG +G G GGGG R V+V +V WWR
Sbjct: 684 WWRRGLIFV-----TGGGG-------DGYLYGFLGGGGWRFVLVNWRXR*FVLVNWRWWR 541
Query: 81 RL------WWWWR 87
+ WWW R
Sbjct: 540 WVFIWLLRWWWRR 502
Score = 31.2 bits (69), Expect(2) = 6e-05
Identities = 43/145 (29%), Positives = 51/145 (34%), Gaps = 17/145 (11%)
Frame = -3
Query: 41 SGGGGKAVAATVEGGG-----NGGG------GGGGDRVVVVVSVVAIEWWRRLWWWWR*G 89
+GGGG T GG GGG GGGGD + G
Sbjct: 488 TGGGGDLYLYTGGGGDLYL*TGGGGDLYL*TGGGGDLYL*T------------------G 363
Query: 90 GGCGGGRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ 149
GG G G + + L T GGG +A GG GG W
Sbjct: 362 GGGDGYLYGFFGGGGGDLYLYTGGGGDLYL*T---GGGGEAYLYGFFGGGGGEL*W*G-- 198
Query: 150 *GGGCGVGGGGDY------GGGGDK 168
GGG GGGG++ GGGGD+
Sbjct: 197 -GGGECTGGGGEW***GFGGGGGDE 126
Score = 29.3 bits (64), Expect = 1.3
Identities = 8/15 (53%), Positives = 9/15 (59%), Gaps = 4/15 (26%)
Frame = -2
Query: 26 WWWQ----RWWLWWW 36
WWW+ R W WWW
Sbjct: 177 WWWRVVVVRLWWWWW 133
Score = 26.9 bits (58), Expect = 6.3
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = -2
Query: 17 VVTVEIGWWWWWQ 29
VV V + WWWWW+
Sbjct: 165 VVVVRL-WWWWWR 130
>TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 754
Score = 42.7 bits (99), Expect = 1e-04
Identities = 42/128 (32%), Positives = 42/128 (32%)
Frame = +2
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
GSGGGG GGG GGGG GG GGG GG R
Sbjct: 341 GSGGGG--------GGGRGGGGYGGG-----------------------GGGYGGERRGY 427
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGG GGG G GGG G GGG
Sbjct: 428 GGGGGYG----------------GGGGGGYGERRGGGGGYSRG--------GGGGGYGGG 535
Query: 160 GDYGGGGD 167
G GGGD
Sbjct: 536 GYSRGGGD 559
Score = 37.4 bits (85), Expect = 0.005
Identities = 36/122 (29%), Positives = 39/122 (31%)
Frame = +2
Query: 45 GKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAA 104
G+ + GGGGGG R GGG GGG G
Sbjct: 305 GRNITVNQAQSRGSGGGGGGGRG---------------------GGGYGGGGGGYGGERR 421
Query: 105 TSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGG 164
G GGG GGG GGG+ GG GGGG YGG
Sbjct: 422 ------------------GYGGGGGY-----GGGGGGGYGERRGGGGGYSRGGGGGGYGG 532
Query: 165 GG 166
GG
Sbjct: 533 GG 538
Score = 37.4 bits (85), Expect = 0.005
Identities = 35/96 (36%), Positives = 37/96 (38%), Gaps = 7/96 (7%)
Frame = +2
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCG-----VGGGGDYGGGGDKVVVAAIMVV 177
G GGG + GGG GGG GGG G GGGG YGGGG
Sbjct: 350 GGGGGGRG-----GGGYGGG--------GGGYGGERRGYGGGGGYGGGGG---------- 460
Query: 178 VATPVVTVGRWWRRRRRR*--SDGGGGGGGANSGVS 211
G + RR S GGGGGG G S
Sbjct: 461 --------GGYGERRGGGGGYSRGGGGGGYGGGGYS 544
Score = 34.3 bits (77), Expect = 0.039
Identities = 19/68 (27%), Positives = 20/68 (28%), Gaps = 4/68 (5%)
Frame = +1
Query: 24 WWWWWQRWWLW-WW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWW--R 80
WW W RWW W WW S + WW R
Sbjct: 370 WWRWLWRWWRWIWW---------------------------------RTSWLRWRWWIRR 450
Query: 81 RLWWW-WR 87
R WWW WR
Sbjct: 451 RRWWWIWR 474
Score = 32.7 bits (73), Expect = 0.11
Identities = 14/64 (21%), Positives = 17/64 (25%), Gaps = 1/64 (1%)
Frame = +1
Query: 24 WWWWWQ-RWWLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRL 82
WWW W+ WW W + WW R+
Sbjct: 457 WWWIWRTPWWRRW-------------------------------------LQQRRWWWRI 525
Query: 83 WWWW 86
WWWW
Sbjct: 526 WWWW 537
Score = 32.0 bits (71), Expect = 0.19
Identities = 26/74 (35%), Positives = 27/74 (36%)
Frame = +2
Query: 136 GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR 195
GGG GGG + GGG G GGGG YGG RR
Sbjct: 347 GGGGGGG------RGGGGYG-GGGGGYGG---------------------------ERRG 424
Query: 196 *SDGGGGGGGANSG 209
GGG GGG G
Sbjct: 425 YGGGGGYGGGGGGG 466
Score = 32.0 bits (71), Expect = 0.19
Identities = 8/13 (61%), Positives = 9/13 (68%)
Frame = +1
Query: 24 WWWWWQRWWLWWW 36
WWWW Q+ W WW
Sbjct: 526 WWWWIQQRWR*WW 564
>TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 912
Score = 42.7 bits (99), Expect = 1e-04
Identities = 42/128 (32%), Positives = 42/128 (32%)
Frame = +1
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
GSGGGG GGG GGGG GG GGG GG R
Sbjct: 499 GSGGGG--------GGGRGGGGYGGG-----------------------GGGYGGERRGY 585
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGG GGG G GGG G GGG
Sbjct: 586 GGGGGYG----------------GGGGGGYGERRGGGGGYSRG--------GGGGGYGGG 693
Query: 160 GDYGGGGD 167
G GGGD
Sbjct: 694 GYSRGGGD 717
Score = 37.4 bits (85), Expect = 0.005
Identities = 35/96 (36%), Positives = 37/96 (38%), Gaps = 7/96 (7%)
Frame = +1
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCG-----VGGGGDYGGGGDKVVVAAIMVV 177
G GGG + GGG GGG GGG G GGGG YGGGG
Sbjct: 508 GGGGGGRG-----GGGYGGG--------GGGYGGERRGYGGGGGYGGGGG---------- 618
Query: 178 VATPVVTVGRWWRRRRRR*--SDGGGGGGGANSGVS 211
G + RR S GGGGGG G S
Sbjct: 619 --------GGYGERRGGGGGYSRGGGGGGYGGGGYS 702
Score = 34.3 bits (77), Expect = 0.039
Identities = 19/68 (27%), Positives = 20/68 (28%), Gaps = 4/68 (5%)
Frame = +3
Query: 24 WWWWWQRWWLW-WW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWW--R 80
WW W RWW W WW S + WW R
Sbjct: 528 WWRWLWRWWRWIWW---------------------------------RTSWLRWRWWIRR 608
Query: 81 RLWWW-WR 87
R WWW WR
Sbjct: 609 RRWWWIWR 632
Score = 32.7 bits (73), Expect = 0.11
Identities = 14/64 (21%), Positives = 17/64 (25%), Gaps = 1/64 (1%)
Frame = +3
Query: 24 WWWWWQ-RWWLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRL 82
WWW W+ WW W + WW R+
Sbjct: 615 WWWIWRTPWWRRW-------------------------------------LQQRRWWWRI 683
Query: 83 WWWW 86
WWWW
Sbjct: 684 WWWW 695
Score = 32.0 bits (71), Expect = 0.19
Identities = 26/74 (35%), Positives = 27/74 (36%)
Frame = +1
Query: 136 GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR 195
GGG GGG + GGG G GGGG YGG RR
Sbjct: 505 GGGGGGG------RGGGGYG-GGGGGYGG---------------------------ERRG 582
Query: 196 *SDGGGGGGGANSG 209
GGG GGG G
Sbjct: 583 YGGGGGYGGGGGGG 624
Score = 32.0 bits (71), Expect = 0.19
Identities = 8/13 (61%), Positives = 9/13 (68%)
Frame = +3
Query: 24 WWWWWQRWWLWWW 36
WWWW Q+ W WW
Sbjct: 684 WWWWIQQRWR*WW 722
>TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 1054
Score = 42.7 bits (99), Expect = 1e-04
Identities = 42/128 (32%), Positives = 42/128 (32%)
Frame = +1
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
GSGGGG GGG GGGG GG GGG GG R
Sbjct: 595 GSGGGG--------GGGRGGGGYGGG-----------------------GGGYGGERRGY 681
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGG GGG G GGG G GGG
Sbjct: 682 GGGGGYG----------------GGGGGGYGERRGGGGGYSRG--------GGGGGYGGG 789
Query: 160 GDYGGGGD 167
G GGGD
Sbjct: 790 GYSRGGGD 813
Score = 37.4 bits (85), Expect = 0.005
Identities = 35/96 (36%), Positives = 37/96 (38%), Gaps = 7/96 (7%)
Frame = +1
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCG-----VGGGGDYGGGGDKVVVAAIMVV 177
G GGG + GGG GGG GGG G GGGG YGGGG
Sbjct: 604 GGGGGGRG-----GGGYGGG--------GGGYGGERRGYGGGGGYGGGGG---------- 714
Query: 178 VATPVVTVGRWWRRRRRR*--SDGGGGGGGANSGVS 211
G + RR S GGGGGG G S
Sbjct: 715 --------GGYGERRGGGGGYSRGGGGGGYGGGGYS 798
Score = 34.3 bits (77), Expect = 0.039
Identities = 19/68 (27%), Positives = 20/68 (28%), Gaps = 4/68 (5%)
Frame = +3
Query: 24 WWWWWQRWWLW-WW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWW--R 80
WW W RWW W WW S + WW R
Sbjct: 624 WWRWLWRWWRWIWW---------------------------------RTSWLRWRWWIRR 704
Query: 81 RLWWW-WR 87
R WWW WR
Sbjct: 705 RRWWWIWR 728
Score = 32.7 bits (73), Expect = 0.11
Identities = 14/64 (21%), Positives = 17/64 (25%), Gaps = 1/64 (1%)
Frame = +3
Query: 24 WWWWWQ-RWWLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRL 82
WWW W+ WW W + WW R+
Sbjct: 711 WWWIWRTPWWRRW-------------------------------------LQQRRWWWRI 779
Query: 83 WWWW 86
WWWW
Sbjct: 780 WWWW 791
Score = 32.0 bits (71), Expect = 0.19
Identities = 26/74 (35%), Positives = 27/74 (36%)
Frame = +1
Query: 136 GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR 195
GGG GGG + GGG G GGGG YGG RR
Sbjct: 601 GGGGGGG------RGGGGYG-GGGGGYGG---------------------------ERRG 678
Query: 196 *SDGGGGGGGANSG 209
GGG GGG G
Sbjct: 679 YGGGGGYGGGGGGG 720
Score = 32.0 bits (71), Expect = 0.19
Identities = 8/13 (61%), Positives = 9/13 (68%)
Frame = +3
Query: 24 WWWWWQRWWLWWW 36
WWWW Q+ W WW
Sbjct: 780 WWWWIQQRWR*WW 818
>TC76716 similar to PIR|T10863|T10863 extensin precursor - kidney bean,
partial (48%)
Length = 1275
Score = 31.6 bits (70), Expect(2) = 3e-04
Identities = 9/16 (56%), Positives = 9/16 (56%), Gaps = 2/16 (12%)
Frame = -2
Query: 23 GWWWWWQRWWL--WWW 36
GWWW W W WWW
Sbjct: 704 GWWW*WVLVWFLGWWW 657
Score = 31.6 bits (70), Expect(2) = 0.002
Identities = 24/53 (45%), Positives = 26/53 (48%), Gaps = 10/53 (18%)
Frame = -3
Query: 125 GGGSKAVAA**GGGD-----GGGWWWWWWQ*GGGCGV----GGGGDY-GGGGD 167
GGG GGGD GGG W GGG + GGGG+Y GGGGD
Sbjct: 604 GGGGDGYL*GGGGGDLYLYGGGGEWT-----GGGGDLYL*TGGGGEYTGGGGD 461
Score = 31.6 bits (70), Expect = 0.25
Identities = 15/31 (48%), Positives = 17/31 (54%), Gaps = 3/31 (9%)
Frame = -2
Query: 9 GRLVLVAVVVTVEIGWWWW-W-QRWWL-WWW 36
GRL+LV WWWW W W+L WWW
Sbjct: 212 GRLILV--------NWWWW*WILVWFLGWWW 144
Score = 31.2 bits (69), Expect(2) = 0.022
Identities = 11/25 (44%), Positives = 14/25 (56%), Gaps = 9/25 (36%)
Frame = -2
Query: 20 VEIGWWWW-W----QRWW----LWW 35
V + WWWW W +RWW +WW
Sbjct: 359 VLVNWWWW*WVLIRRRWWRFVLIWW 285
Score = 31.2 bits (69), Expect(2) = 0.010
Identities = 21/76 (27%), Positives = 30/76 (38%), Gaps = 10/76 (13%)
Frame = -2
Query: 22 IGWWWW-W-QRWWL-WWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVS------ 72
+ WWWW W W+L WWW + V G R +V+V+
Sbjct: 452 VNWWWW*WILVWFLGWWW--------WRFVLV-----------NWGRR*LVLVNWWWW*W 330
Query: 73 -VVAIEWWRRLWWWWR 87
++ WWR + WWR
Sbjct: 329 VLIRRRWWRFVLIWWR 282
Score = 30.0 bits (66), Expect(2) = 6e-04
Identities = 12/36 (33%), Positives = 14/36 (38%), Gaps = 21/36 (58%)
Frame = -2
Query: 22 IGWWWW------WQ---------RWWLW------WW 36
+GWWWW W+ RWW W WW
Sbjct: 671 LGWWWWRFVLVNWRRR*FVLVNWRWW*WVLIRRRWW 564
Score = 29.3 bits (64), Expect(2) = 6e-04
Identities = 41/141 (29%), Positives = 47/141 (33%), Gaps = 14/141 (9%)
Frame = -3
Query: 41 SGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVV 100
+GGGG T GGG GGGGD + GGG G +
Sbjct: 529 TGGGGDLYL*T--GGGGEYTGGGGDLYL*T------------------GGGGDGYLYGFL 410
Query: 101 GVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDG----GGWWWWWWQ*GGGCGV 156
G + + GGG GGGDG GG + GGG
Sbjct: 409 GGGGGDLY-------------L*TGGGGDLYL*TGGGGDGYL*GGGGGDLYLYGGGGE*T 269
Query: 157 GGGGDY----------GGGGD 167
GGGGD GGGGD
Sbjct: 268 GGGGDLYL*TGGGE*TGGGGD 206
Score = 28.9 bits (63), Expect(2) = 3e-04
Identities = 47/144 (32%), Positives = 52/144 (35%), Gaps = 17/144 (11%)
Frame = -3
Query: 41 SGGGGKAVAATVEGGGNGG-GGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGG-----CGG 94
+GGGG T GG GGGGGD + W GGG GG
Sbjct: 637 TGGGGDLYL*TGGGGDGYL*GGGGGDLYLYGGG-----------GEWTGGGGDLYL*TGG 491
Query: 95 GRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGD-----GGGWWWWWWQ 149
G G + L T G G G GGGD GGG +
Sbjct: 490 GGEYTGGGGD------------LYL*TGGGGDGYLYGFLGGGGGDLYL*TGGGGDLYL*T 347
Query: 150 *GGGCGV---GGGGD---YGGGGD 167
GGG G GGGGD YGGGG+
Sbjct: 346 GGGGDGYL*GGGGGDLYLYGGGGE 275
Score = 27.3 bits (59), Expect = 4.8
Identities = 10/24 (41%), Positives = 11/24 (45%), Gaps = 8/24 (33%)
Frame = -2
Query: 22 IGWWWWWQRW--------WLWWW* 37
+ WWW RW W WWW*
Sbjct: 245 VNWWW*VDRWRGRLILVNW-WWW* 177
Score = 26.9 bits (58), Expect = 6.3
Identities = 21/49 (42%), Positives = 23/49 (46%), Gaps = 5/49 (10%)
Frame = -3
Query: 124 CGGGSKAVAA**GGGD-----GGGWWWWWWQ*GGGCGVGGGGDYGGGGD 167
CGGG + GGGD GGG + GGG G G GGGGD
Sbjct: 787 CGGGGE*TG---GGGDLYL*TGGGGDLYL*TGGGGDGYLYGFLGGGGGD 650
Score = 25.8 bits (55), Expect(2) = 0.002
Identities = 21/86 (24%), Positives = 24/86 (27%), Gaps = 18/86 (20%)
Frame = -2
Query: 20 VEIGWWWW----WQR------WWL--------WWW***GSGGGGKAVAATVEGGGNGGGG 61
V+I WW W W+R WW WWW*
Sbjct: 797 VDIVWWRW*VNRWRRRFILVNWWRRGLIFVNGWWW*------------------------ 690
Query: 62 GGGDRVVVVVSVVAIEWWRRLWWWWR 87
V+V W WWWWR
Sbjct: 689 ------WVLV-------WFLGWWWWR 651
Score = 23.9 bits (50), Expect(2) = 0.010
Identities = 19/44 (43%), Positives = 22/44 (49%), Gaps = 2/44 (4%)
Frame = -3
Query: 126 GGSKAVAA**GGGD--GGGWWWWWWQ*GGGCGVGGGGDYGGGGD 167
GG + * GGG+ GGG + GGG G G GGGGD
Sbjct: 268 GGGGDLYL*TGGGE*TGGGGDLYL*TGGGGDGYLYGFLGGGGGD 137
Score = 22.7 bits (47), Expect(2) = 0.022
Identities = 14/32 (43%), Positives = 15/32 (46%), Gaps = 7/32 (21%)
Frame = -3
Query: 41 SGGGGKAVAATVEGGGNGGG-------GGGGD 65
+GGGG T G GGG GGGGD
Sbjct: 271 TGGGGDLYL*TGGGE*TGGGGDLYL*TGGGGD 176
>TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-binding
protein {Glycine max}, partial (90%)
Length = 488
Score = 41.2 bits (95), Expect = 3e-04
Identities = 40/126 (31%), Positives = 43/126 (33%)
Frame = +3
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
GSGGGG+ GGG G GGGGG G G GGGR
Sbjct: 291 GSGGGGRGGGGGGYGGGGGYGGGGG------------------------GYGGGGGRRD- 395
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
GG S++ GGG GGG G G GGG
Sbjct: 396 -------------------------GGYSRSGG---GGGYGGGG-------DRGYGGGGG 470
Query: 160 GDYGGG 165
G YGGG
Sbjct: 471 GGYGGG 488
Score = 38.5 bits (88), Expect = 0.002
Identities = 29/87 (33%), Positives = 31/87 (35%)
Frame = +3
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPV 182
G GGG GGG GG + GG GGGG YGGGGD+
Sbjct: 312 GGGGGGYGGGGGYGGGGGGYGGGGGRRDGGYSRSGGGGGYGGGGDR-------------- 449
Query: 183 VTVGRWWRRRRRR*SDGGGGGGGANSG 209
GGGGGGG G
Sbjct: 450 --------------GYGGGGGGGYGGG 488
Score = 38.1 bits (87), Expect = 0.003
Identities = 9/12 (75%), Positives = 10/12 (83%)
Frame = +2
Query: 24 WWWWWQRWWLWW 35
W WWW+ WWLWW
Sbjct: 449 WIWWWRWWWLWW 484
Score = 36.6 bits (83), Expect = 0.008
Identities = 33/88 (37%), Positives = 38/88 (42%), Gaps = 1/88 (1%)
Frame = +3
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPV 182
G GGG + GG GGG+ GGG G GGGG YGGGG +
Sbjct: 291 GSGGGGR-------GGGGGGYGG-----GGGYG-GGGGGYGGGGGR-------------- 389
Query: 183 VTVGRWWRRRRRR*SDGGGG-GGGANSG 209
G + R S GGGG GGG + G
Sbjct: 390 -RDGGYSR------SGGGGGYGGGGDRG 452
Score = 35.0 bits (79), Expect = 0.023
Identities = 27/72 (37%), Positives = 28/72 (38%)
Frame = +3
Query: 138 GDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*S 197
G GGG + GGG G GGGG YGGGG G RR S
Sbjct: 291 GSGGGG-----RGGGGGGYGGGGGYGGGGG----------------GYGGGGGRRDGGYS 407
Query: 198 DGGGGGGGANSG 209
GGGGG G
Sbjct: 408 RSGGGGGYGGGG 443
Score = 34.3 bits (77), Expect = 0.039
Identities = 10/23 (43%), Positives = 10/23 (43%), Gaps = 10/23 (43%)
Frame = +2
Query: 24 WWW----------WWQRWWLWWW 36
WWW WW RWW WW
Sbjct: 416 WWWRLWGRRRSWIWWWRWWWLWW 484
Score = 32.0 bits (71), Expect = 0.19
Identities = 17/77 (22%), Positives = 20/77 (25%), Gaps = 14/77 (18%)
Frame = +2
Query: 25 WWWWQRWWLW-------WW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIE 77
W W +RWW+W WW +
Sbjct: 344 WLWRRRWWIWRRRWSS*WW------------------------------------IQQKR 415
Query: 78 WWRRLW-------WWWR 87
WW RLW WWWR
Sbjct: 416 WWWRLWGRRRSWIWWWR 466
Score = 26.6 bits (57), Expect = 8.2
Identities = 8/23 (34%), Positives = 10/23 (42%), Gaps = 10/23 (43%)
Frame = +2
Query: 24 WWWWWQRW----------WLWWW 36
WW +RW W+WWW
Sbjct: 395 WWIQQKRWWWRLWGRRRSWIWWW 463
>TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear protein.
[strain B95-8 Human herpesvirus 4] {Epstein-barr
virus}, partial (23%)
Length = 431
Score = 41.2 bits (95), Expect = 3e-04
Identities = 40/114 (35%), Positives = 41/114 (35%), Gaps = 2/114 (1%)
Frame = +1
Query: 54 GGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAA 113
GGG GGGGG G GGG G G A VG +
Sbjct: 13 GGGYGGGGGSGG-----------------------GGGGGAGGAHGVGYGSGGGTGG--- 114
Query: 114 MVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGG--GGDYGGG 165
G GGGS GGG GGG GGG G GG GG YGGG
Sbjct: 115 ---------GYGGGSPG-----GGGGGGGS-------GGGGGGGGAHGGAYGGG 213
Score = 37.7 bits (86), Expect = 0.004
Identities = 11/15 (73%), Positives = 11/15 (73%), Gaps = 2/15 (13%)
Frame = +3
Query: 23 GW--WWWWQRWWLWW 35
GW W WW RWWLWW
Sbjct: 123 GWVAWGWWWRWWLWW 167
Score = 37.0 bits (84), Expect = 0.006
Identities = 40/126 (31%), Positives = 43/126 (33%)
Frame = +1
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG GGG+GGGGGGG V + GGG GGG
Sbjct: 13 GGGYGG--------GGGSGGGGGGGAGGAHGVGYGS-------------GGGTGGGYGG- 126
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
+ G GGG GGG GGG GG G G G
Sbjct: 127 --------------------GSPGGGGGGGGSG---GGGGGGGAH------GGAYGGGIG 219
Query: 160 GDYGGG 165
G GGG
Sbjct: 220 GGEGGG 237
Score = 37.0 bits (84), Expect = 0.006
Identities = 18/72 (25%), Positives = 19/72 (26%), Gaps = 8/72 (11%)
Frame = +3
Query: 24 WW--------WWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVA 75
WW WW RWW+W W VA
Sbjct: 66 WWSAWRWIR*WWRNRWWIWGW-------------------------------------VA 134
Query: 76 IEWWRRLWWWWR 87
WW R W WWR
Sbjct: 135 WGWWWRWWLWWR 170
Score = 36.6 bits (83), Expect = 0.008
Identities = 43/163 (26%), Positives = 46/163 (27%)
Frame = +1
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGG 61
GGGGG+GG G G GG GGG GGG
Sbjct: 25 GGGGGSGGG----------------------------GGGGAGGAHGVGYGSGGGTGGGY 120
Query: 62 GGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLAT 121
GGG GGG GGG
Sbjct: 121 GGGSP----------------------GGGGGGG-------------------------- 156
Query: 122 IGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGG 164
G GGG GG GG + GGG G G GG +GG
Sbjct: 157 -GSGGGGGG-----GGAHGGAY-------GGGIGGGEGGGHGG 246
Score = 34.7 bits (78), Expect = 0.030
Identities = 10/25 (40%), Positives = 11/25 (44%), Gaps = 11/25 (44%)
Frame = +3
Query: 23 GWWWWWQRWW-----------LWWW 36
GWWW W WW +WWW
Sbjct: 138 GWWWRWWLWWRRRWWRCAWRSIWWW 212
Score = 33.9 bits (76), Expect = 0.051
Identities = 29/87 (33%), Positives = 30/87 (34%)
Frame = +1
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPV 182
G GGGS GG GGG G G G G GG YGGG
Sbjct: 25 GGGGGS--------GGGGGGGAGGAHGVGYGSGGGTGGGYGGG----------------- 129
Query: 183 VTVGRWWRRRRRR*SDGGGGGGGANSG 209
S GGGGGGG + G
Sbjct: 130 --------------SPGGGGGGGGSGG 168
Score = 30.0 bits (66), Expect = 0.74
Identities = 8/14 (57%), Positives = 10/14 (71%), Gaps = 2/14 (14%)
Frame = +3
Query: 24 WWWWWQ--RWWLWW 35
WWW+W+ RW WW
Sbjct: 204 WWWYWRW*RWRPWW 245
>TC76717 similar to PIR|T10863|T10863 extensin precursor - kidney bean,
partial (56%)
Length = 928
Score = 36.2 bits (82), Expect = 0.010
Identities = 23/62 (37%), Positives = 27/62 (43%), Gaps = 18/62 (29%)
Frame = -1
Query: 22 IGWW--------WWWQR----W--WLWWW***GSGGGGKAVAATVEGGGN----GGGGGG 63
I WW W W+R W W WWW G+ ++V GGG G GGGG
Sbjct: 220 IHWWRR*FVLVNWRWRRSVFVWLLWRWWW---GTVMVRRSVVECTGGGGEW***GFGGGG 50
Query: 64 GD 65
GD
Sbjct: 49 GD 44
Score = 34.7 bits (78), Expect = 0.030
Identities = 20/69 (28%), Positives = 24/69 (33%), Gaps = 7/69 (10%)
Frame = -1
Query: 26 WWWQRW---WLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRR- 81
WWW RW WL+WW R+V+V WWRR
Sbjct: 631 WWWGRWVFIWLFWW--------------------------WWRRLVLV------HWWRRG 548
Query: 82 ---LWWWWR 87
+ WWWR
Sbjct: 547 FILVNWWWR 521
Score = 33.5 bits (75), Expect(2) = 4e-04
Identities = 28/94 (29%), Positives = 35/94 (36%), Gaps = 15/94 (15%)
Frame = -1
Query: 9 GRLVLVAVVVTVEIGWWWW-W-QRWWL-WWW***GSGGGGKAVAATVEGGGNGGGGGGGD 65
GRL+LV WWWW W W+L WWW G+ V
Sbjct: 811 GRLILV--------NWWWW*WILVWFLGWWWWRFVLVNWGRR*LILVNWWW--------- 683
Query: 66 RVVVVVS------VVAIEWWRR------LWWWWR 87
R ++V+ V+ WW R WWWWR
Sbjct: 682 RRFILVNWRRR*FVLVNWWWGRWVFIWLFWWWWR 581
Score = 31.6 bits (70), Expect = 0.25
Identities = 18/89 (20%), Positives = 23/89 (25%), Gaps = 25/89 (28%)
Frame = -1
Query: 24 WWW-------WWQRWWL-------------WWW***GSGGGGKAVAATVEGGGNGGGGGG 63
WWW WW+R ++ WWW
Sbjct: 433 WWWRRLVLVHWWRRGFILVHRWRR*FVLVNWWW--------------------------- 335
Query: 64 GDRVVVVVSVVAIEWWRR-----LWWWWR 87
+ + WWRR WWWWR
Sbjct: 334 -------RRFILVNWWRR*LVLVNWWWWR 269
Score = 29.6 bits (65), Expect(2) = 0.047
Identities = 9/13 (69%), Positives = 9/13 (69%), Gaps = 4/13 (30%)
Frame = -3
Query: 78 WWR----RLWWWW 86
WWR RLWWWW
Sbjct: 89 WWRVVVVRLWWWW 51
Score = 28.9 bits (63), Expect = 1.6
Identities = 10/30 (33%), Positives = 13/30 (43%), Gaps = 11/30 (36%)
Frame = -1
Query: 18 VTVEIGWWWWWQ----RWWL-------WWW 36
V + + WWWW + WW WWW
Sbjct: 613 VFIWLFWWWWRRLVLVHWWRRGFILVNWWW 524
Score = 27.7 bits (60), Expect = 3.7
Identities = 21/104 (20%), Positives = 28/104 (26%), Gaps = 37/104 (35%)
Frame = -1
Query: 20 VEIGWWW-------WWQR------WWLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDR 66
V + WWW WW+R WW W W
Sbjct: 355 VLVNWWWRRFILVNWWRR*LVLVNWWWWRW------------------------------ 266
Query: 67 VVVVV------SVVAIEWWRR-----------------LW-WWW 86
+ + + ++ I WWRR LW WWW
Sbjct: 265 IFIWLLRWWRRRLILIHWWRR*FVLVNWRWRRSVFVWLLWRWWW 134
Score = 27.3 bits (59), Expect = 4.8
Identities = 42/143 (29%), Positives = 49/143 (33%), Gaps = 17/143 (11%)
Frame = -2
Query: 42 GGGGKAVAATVEGGGN------GGG------GGGGDRVVVVVSVVAIEWWRRLWWWWR*G 89
GGGG + GGG+ GGG GGGGD + G
Sbjct: 594 GGGGGDLYLYTGGGGDLYL*TGGGGDLYLYTGGGGDLYL*T------------------G 469
Query: 90 GGCGGGRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGD-----GGGWW 144
GG G +G + + L T GGG GGGD GGG
Sbjct: 468 GGGDGYLYGFLGGGGGDLYLYTGGGGDLYLYT---GGGGDLYL*TGGGGDLYL*TGGGGD 298
Query: 145 WWWWQ*GGGCGVGGGGDYGGGGD 167
+ GGG G G GGGGD
Sbjct: 297 LYL*TGGGGDGYLYGFFGGGGGD 229
Score = 27.3 bits (59), Expect = 4.8
Identities = 10/24 (41%), Positives = 11/24 (45%), Gaps = 8/24 (33%)
Frame = -1
Query: 22 IGWWWWWQRW--------WLWWW* 37
+ WWW RW W WWW*
Sbjct: 844 VNWWW*VDRWRGRLILVNW-WWW* 776
Score = 26.9 bits (58), Expect = 6.3
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = -3
Query: 17 VVTVEIGWWWWWQ 29
VV V + WWWWW+
Sbjct: 80 VVVVRL-WWWWWR 45
Score = 26.6 bits (57), Expect = 8.2
Identities = 6/7 (85%), Positives = 7/7 (99%)
Frame = -3
Query: 144 WWWWWQ* 150
WWWWW+*
Sbjct: 62 WWWWWR* 42
Score = 26.6 bits (57), Expect(2) = 4e-04
Identities = 23/58 (39%), Positives = 25/58 (42%), Gaps = 15/58 (25%)
Frame = -2
Query: 125 GGGSKAVAA**GGGD-----GGGWWWWWWQ*GGG---CGVGGGGDY-------GGGGD 167
GGG GGGD GGG + + GGG GGGGD GGGGD
Sbjct: 591 GGGGDLYLYTGGGGDLYL*TGGGGDLYLYTGGGGDLYL*TGGGGDGYLYGFLGGGGGD 418
Score = 23.9 bits (50), Expect(2) = 2.6
Identities = 19/44 (43%), Positives = 22/44 (49%), Gaps = 2/44 (4%)
Frame = -2
Query: 126 GGSKAVAA**GGGD--GGGWWWWWWQ*GGGCGVGGGGDYGGGGD 167
GG + * GGG+ GGG + GGG G G GGGGD
Sbjct: 867 GGGGDLYL*TGGGE*TGGGGDLYL*TGGGGDGYLYGFLGGGGGD 736
Score = 23.1 bits (48), Expect(2) = 0.047
Identities = 15/36 (41%), Positives = 17/36 (46%), Gaps = 4/36 (11%)
Frame = -2
Query: 41 SGGGGKAVAATVEGGGNGG----GGGGGDRVVVVVS 72
+GGGG+A GGG G GG VVV S
Sbjct: 186 TGGGGEAYLYGFFGGGGGEL*W*GGAWSSAPVVVES 79
Score = 22.7 bits (47), Expect(2) = 2.6
Identities = 6/10 (60%), Positives = 7/10 (70%)
Frame = -1
Query: 78 WWRRLWWWWR 87
WWR + WWR
Sbjct: 910 WWRFVLIWWR 881
>TC76431 similar to GP|8132441|gb|AAF73291.1| extensin {Pisum sativum},
partial (30%)
Length = 890
Score = 37.7 bits (86), Expect = 0.004
Identities = 44/140 (31%), Positives = 50/140 (35%), Gaps = 12/140 (8%)
Frame = -1
Query: 41 SGGGGKAVAATVEGGGN----GGG-----GGGGDRVVVVVSVVAIEW---WRRLWWWWR* 88
+GGGG GGG G G GGGG+ + EW W WWW
Sbjct: 323 TGGGGDEYLYGFFGGGGEW**GLGLT*CTGGGGEEYLYFGG--GGEW*TGWG*G*WWWVG 150
Query: 89 GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWW 148
GGG G + +G GGG *G G G GW W
Sbjct: 149 GGGDG-----------------------YLYGFLGDGGGEW*TG--*GWGCGCGWGW--- 54
Query: 149 Q*GGGCGVGGGGDYGGGGDK 168
GC G G YGGGGD+
Sbjct: 53 ----GC---G*G*YGGGGDE 15
Score = 36.6 bits (83), Expect(2) = 8e-04
Identities = 38/162 (23%), Positives = 44/162 (26%), Gaps = 31/162 (19%)
Frame = -3
Query: 18 VTVEIGWWWWWQRWWLW------WW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVV 71
+ V + WWW W+W WW RVV
Sbjct: 435 IDVTVNWWWRRVINWVWSVGRMNWW-----------------------------RVVNWF 343
Query: 72 -SVVAIEWWRR-------LWWWWR*GGGCGGGRAAVVGVAATS----------------- 106
SV + WW R LWWWWR VVG+ A
Sbjct: 342 WSVRRVNWWWRG*VFVWLLWWWWR----------VVVGLRADVMHWWWW*GILVFWWWW* 193
Query: 107 VVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWW 148
VV V V+VV GWWWW W
Sbjct: 192 VVNWVGVRVMVV-----------------------GWWWWRW 136
Score = 31.2 bits (69), Expect = 0.33
Identities = 23/77 (29%), Positives = 28/77 (35%), Gaps = 3/77 (3%)
Frame = -3
Query: 14 VAVVVTVEIGWWWWWQR---WWLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVV 70
V V + ++ WWWW +W WWW* V V G RV+VV
Sbjct: 270 VVVGLRADVMHWWWW*GILVFW-WWW*----------VVNWV-----------GVRVMVV 157
Query: 71 VSVVAIEWWRRLWWWWR 87
WWWWR
Sbjct: 156 G-----------WWWWR 139
Score = 30.0 bits (66), Expect = 0.74
Identities = 9/15 (60%), Positives = 11/15 (73%)
Frame = -3
Query: 22 IGWWWWWQRWWLWWW 36
+GWWWW RW L W+
Sbjct: 159 VGWWWW--RWILVWF 121
Score = 26.6 bits (57), Expect = 8.2
Identities = 17/37 (45%), Positives = 20/37 (53%), Gaps = 4/37 (10%)
Frame = -1
Query: 135 *GGGDGGGWWWWWWQ*GGGCGVGGG----GDYGGGGD 167
*GG GG W + *GG G GG G +GGGG+
Sbjct: 380 *GG*TGGEW*TGFGV*GG*TGGGGDEYLYGFFGGGGE 270
Score = 22.3 bits (46), Expect(2) = 8e-04
Identities = 10/19 (52%), Positives = 11/19 (57%)
Frame = -2
Query: 151 GGGCGVGGGGDYGGGGDKV 169
GGG GV GGG G +V
Sbjct: 82 GGGVGVDGGGGVDKGDMEV 26
>BF639374 similar to GP|8132441|gb| extensin {Pisum sativum}, partial (45%)
Length = 381
Score = 34.3 bits (77), Expect(2) = 9e-04
Identities = 17/46 (36%), Positives = 18/46 (38%), Gaps = 13/46 (28%)
Frame = -2
Query: 4 GGGNGGRLVLVAVVVTVEIGWWWW------WQR-------WWLWWW 36
GGG G LV +GWWWW W R WW W W
Sbjct: 371 GGGGDGYLVWF-------LGWWWWRFVLVNWGRR*LVLVNWWWW*W 255
Score = 32.3 bits (72), Expect = 0.15
Identities = 22/77 (28%), Positives = 25/77 (31%), Gaps = 8/77 (10%)
Frame = -2
Query: 18 VTVEIGWWWWW------QRWWL--WWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVV 69
V V + W WWW +RW + WWW RVVV
Sbjct: 167 VFVWLLWRWWWGTVMVRRRW*VHRWWW-----------------------------RVVV 75
Query: 70 VVSVVAIEWWRRLWWWW 86
V RLWWWW
Sbjct: 74 V----------RLWWWW 54
Score = 30.4 bits (67), Expect = 0.57
Identities = 21/50 (42%), Positives = 25/50 (50%), Gaps = 6/50 (12%)
Frame = -3
Query: 125 GGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDY------GGGGDK 168
GGG +A GG GG W GGG GGGG++ GGGGD+
Sbjct: 184 GGGGEAYLYGFFGGGGGEL*W*G---GGGECTGGGGEW***GFGGGGGDE 44
Score = 29.3 bits (64), Expect = 1.3
Identities = 8/15 (53%), Positives = 9/15 (59%), Gaps = 4/15 (26%)
Frame = -2
Query: 26 WWWQ----RWWLWWW 36
WWW+ R W WWW
Sbjct: 95 WWWRVVVVRLWWWWW 51
Score = 28.9 bits (63), Expect = 1.6
Identities = 12/27 (44%), Positives = 13/27 (47%)
Frame = -2
Query: 61 GGGGDRVVVVVSVVAIEWWRRLWWWWR 87
GGGGD +V W WWWWR
Sbjct: 371 GGGGDGYLV---------WFLGWWWWR 318
Score = 28.5 bits (62), Expect = 2.2
Identities = 10/17 (58%), Positives = 11/17 (63%), Gaps = 3/17 (17%)
Frame = -2
Query: 136 GGGDGGGWW---WWWWQ 149
GGGDG W WWWW+
Sbjct: 368 GGGDGYLVWFLGWWWWR 318
Score = 26.9 bits (58), Expect = 6.3
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = -2
Query: 17 VVTVEIGWWWWWQ 29
VV V + WWWWW+
Sbjct: 83 VVVVRL-WWWWWR 48
Score = 26.6 bits (57), Expect = 8.2
Identities = 17/54 (31%), Positives = 21/54 (38%), Gaps = 21/54 (38%)
Frame = -2
Query: 3 GGGGNG----------GRLVLV--AVVVTVEIGWWWW---------WQRWWLWW 35
GGGG+G R VLV V + WWWW W+ +WW
Sbjct: 371 GGGGDGYLVWFLGWWWWRFVLVNWGRR*LVLVNWWWW*WVLIRRRWWRFVLIWW 210
Score = 24.6 bits (52), Expect(2) = 9e-04
Identities = 13/27 (48%), Positives = 15/27 (55%), Gaps = 5/27 (18%)
Frame = -3
Query: 41 SGGGGKAVAATVEGGGNG-----GGGG 62
+GGGG+A GGG G GGGG
Sbjct: 187 TGGGGEAYLYGFFGGGGGEL*W*GGGG 107
>TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (87%)
Length = 780
Score = 35.4 bits (80), Expect = 0.018
Identities = 40/130 (30%), Positives = 44/130 (33%), Gaps = 3/130 (2%)
Frame = +1
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GG GGG+GG GGGG GGG GG A
Sbjct: 361 GGGGYNHGGGGYNGGGGHGGHGGGGYN----------------------GGGGHGGHGAA 474
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGD---GGGWWWWWWQ*GGGCGV 156
VA + GGGS GGG GGG + GGG G
Sbjct: 475 ESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNH----GGGSYHHGGG---GYNHGGGGHGG 633
Query: 157 GGGGDYGGGG 166
GGG +GG G
Sbjct: 634 HGGGGHGGHG 663
Score = 33.9 bits (76), Expect = 0.051
Identities = 15/63 (23%), Positives = 16/63 (24%)
Frame = +3
Query: 24 WWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLW 83
WWWW Q+ W W WWRRL
Sbjct: 261 WWWWLQQRWRW----------------------------------------LQPWWRRLQ 320
Query: 84 WWW 86
WWW
Sbjct: 321 WWW 329
Score = 31.2 bits (69), Expect = 0.33
Identities = 34/89 (38%), Positives = 37/89 (41%), Gaps = 15/89 (16%)
Frame = +1
Query: 136 GGG---DGGGWWWWWWQ*GGGCGVGGGGDY-GGGGDKVVVAAIMVVVATPVVT------- 184
GGG GGG+ GGG G GGG Y GGGG AA V V T T
Sbjct: 364 GGGYNHGGGGY-----NGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAK 528
Query: 185 VGRWWRRRRRR*SDGGG----GGGGANSG 209
G + + GGG GGGG N G
Sbjct: 529 YGGGYNHGGGSYNHGGGSYHHGGGGYNHG 615
Score = 30.4 bits (67), Expect(2) = 0.010
Identities = 41/135 (30%), Positives = 45/135 (32%), Gaps = 10/135 (7%)
Frame = +1
Query: 42 GGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGG------- 94
GGGG GGG GGGG + + GGG GG
Sbjct: 304 GGGGYNGGGYNHGGGGYNHGGGG--------------YNHGGGGYNGGGGHGGHGGGGYN 441
Query: 95 GRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGD---GGGWWWWWWQ*G 151
G G A VAV + GGG GGG GGG + G
Sbjct: 442 GGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNH-----GGGSYNHGGG---SYHHGG 597
Query: 152 GGCGVGGGGDYGGGG 166
GG GGGG G GG
Sbjct: 598 GGYNHGGGGHGGHGG 642
Score = 27.7 bits (60), Expect = 3.7
Identities = 15/30 (50%), Positives = 18/30 (60%)
Frame = +1
Query: 137 GGDGGGWWWWWWQ*GGGCGVGGGGDYGGGG 166
GG GGG+ GGG GGG ++GGGG
Sbjct: 157 GGYGGGY-----NHGGGGYNGGGYNHGGGG 231
Score = 26.6 bits (57), Expect = 8.2
Identities = 15/73 (20%), Positives = 17/73 (22%), Gaps = 10/73 (13%)
Frame = +3
Query: 24 WW-----WWWQRWWLW----WW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVV 74
WW WWW WW WW +
Sbjct: 165 WWRLQPRWWWL*WWRL*PRRWW-------------------------------------L 233
Query: 75 AIEW-WRRLWWWW 86
W W + WWWW
Sbjct: 234 QQRWRWLQPWWWW 272
Score = 24.6 bits (52), Expect(2) = 0.010
Identities = 6/13 (46%), Positives = 7/13 (53%)
Frame = +3
Query: 24 WWWWWQRWWLWWW 36
W WW+ WWW
Sbjct: 156 WRLWWRLQPRWWW 194
>BF642399 similar to PIR|T10863|T10 extensin precursor - kidney bean, partial
(33%)
Length = 653
Score = 33.1 bits (74), Expect = 0.087
Identities = 41/138 (29%), Positives = 49/138 (34%), Gaps = 10/138 (7%)
Frame = -2
Query: 41 SGGGGKAVAATVEGGGNGG----GGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGR 96
+GGGG GGG G GGGGD + GGG G
Sbjct: 442 TGGGGDGYLYGFLGGGGGDLYL*TGGGGDLYL*T------------------GGGGDGYL 317
Query: 97 AAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGV 156
G + + L T GGG +A GG GG W GGG
Sbjct: 316 YGFFGGGGGDLYLYTGGGGDLYL*T---GGGGEAYLYGFFGGGGGEL*W*G---GGGECT 155
Query: 157 GGGGDY------GGGGDK 168
GGGG++ GGGGD+
Sbjct: 154 GGGGEW***GFGGGGGDE 101
Score = 32.3 bits (72), Expect = 0.15
Identities = 19/75 (25%), Positives = 22/75 (29%), Gaps = 9/75 (12%)
Frame = -1
Query: 22 IGWWWW-W-QRWWL--WWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIE 77
+ WWWW W W+L WWW V +
Sbjct: 446 VNWWWW*WILVWFLGWWWW-----------------------------------RFVLVN 372
Query: 78 WWRR-----LWWWWR 87
W RR WWWWR
Sbjct: 371 WGRR*LVLVNWWWWR 327
Score = 32.3 bits (72), Expect = 0.15
Identities = 22/77 (28%), Positives = 25/77 (31%), Gaps = 8/77 (10%)
Frame = -1
Query: 18 VTVEIGWWWWW------QRWWL--WWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVV 69
V V + W WWW +RW + WWW RVVV
Sbjct: 224 VFVWLLWRWWWGTVMVRRRW*VHRWWW-----------------------------RVVV 132
Query: 70 VVSVVAIEWWRRLWWWW 86
V RLWWWW
Sbjct: 131 V----------RLWWWW 111
Score = 29.3 bits (64), Expect = 1.3
Identities = 8/15 (53%), Positives = 9/15 (59%), Gaps = 4/15 (26%)
Frame = -1
Query: 26 WWWQ----RWWLWWW 36
WWW+ R W WWW
Sbjct: 152 WWWRVVVVRLWWWWW 108
Score = 28.9 bits (63), Expect = 1.6
Identities = 23/54 (42%), Positives = 25/54 (45%), Gaps = 11/54 (20%)
Frame = -2
Query: 125 GGGSKAVAA**GGGD------GGGWWWWWWQ*GGGCGV----GGGGDY-GGGGD 167
GGG GGGD GG W GGG + GGGG+Y GGGGD
Sbjct: 598 GGGGDGYL*GXGGGDLYLYGXGGEWT------GGGGDLYL*TGGGGEYTGGGGD 455
Score = 27.7 bits (60), Expect = 3.7
Identities = 38/134 (28%), Positives = 48/134 (35%), Gaps = 7/134 (5%)
Frame = -2
Query: 41 SGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVV 100
+GGGG T GGG GGGGD + GGG G +
Sbjct: 523 TGGGGDLYL*T--GGGGEYTGGGGDLYL*T------------------GGGGDGYLYGFL 404
Query: 101 GVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGG---CGVG 157
G + + GGG * GG G G+ + ++ GGG G
Sbjct: 403 GGGGGDLY-------------L*TGGGGDLYL--*TGGGGDGYLYGFFGGGGGDLYLYTG 269
Query: 158 GGGDY----GGGGD 167
GGGD GGGG+
Sbjct: 268 GGGDLYL*TGGGGE 227
Score = 27.7 bits (60), Expect = 3.7
Identities = 16/80 (20%), Positives = 20/80 (25%), Gaps = 17/80 (21%)
Frame = -1
Query: 25 WWWWQ--------------RWWLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVV 70
WWWW+ WW W W
Sbjct: 401 WWWWRFVLVNWGRR*LVLVNWWWWRW---------------------------------- 324
Query: 71 VSVVAIEWWRR---LWWWWR 87
+ + + WWRR L WWR
Sbjct: 323 IFIWLLRWWRRRLILIHWWR 264
Score = 26.9 bits (58), Expect = 6.3
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = -1
Query: 17 VVTVEIGWWWWWQ 29
VV V + WWWWW+
Sbjct: 140 VVVVRL-WWWWWR 105
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.338 0.154 0.636
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,958,051
Number of Sequences: 36976
Number of extensions: 178930
Number of successful extensions: 24201
Number of sequences better than 10.0: 680
Number of HSP's better than 10.0 without gapping: 5338
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 16130
length of query: 224
length of database: 9,014,727
effective HSP length: 93
effective length of query: 131
effective length of database: 5,575,959
effective search space: 730450629
effective search space used: 730450629
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0320b.3


