

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0282.6
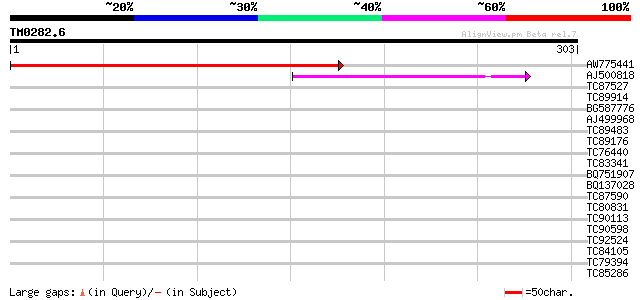
(303 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW775441 homologue to GP|18086381|gb AT3g18790/MVE11_17 {Arabido... 358 2e-99
AJ500818 weakly similar to GP|10435304|dbj unnamed protein produ... 42 2e-04
TC87527 similar to GP|21689681|gb|AAM67462.1 unknown protein {Ar... 36 0.021
TC89914 similar to GP|14335028|gb|AAK59778.1 AT4g37280/C7A10_80 ... 35 0.047
BG587776 weakly similar to PIR|F86242|F86 unknown protein 98896... 34 0.079
AJ499968 weakly similar to GP|23495961|gb| hypothetical protein ... 33 0.10
TC89483 similar to GP|18377662|gb|AAL66981.1 unknown protein {Ar... 32 0.30
TC89176 homologue to GP|13539605|emb|CAC35733. cyclophilin-RNA i... 30 1.1
TC76440 weakly similar to SP|Q9LD55|IF3A_ARATH Eukaryotic transl... 30 1.1
TC83341 weakly similar to GP|8978076|dbj|BAA98104.1 gene_id:K14A... 30 1.5
BQ751907 weakly similar to PIR|T40935|T409 probable utp--glucose... 30 1.5
BQ137028 similar to GP|4914334|gb|A F14N23.20 {Arabidopsis thali... 28 3.3
TC87590 weakly similar to PIR|G96522|G96522 F11A17.16 [imported]... 28 3.3
TC80831 similar to GP|18542939|gb|AAK00428.2 Unknown protein {Or... 28 4.4
TC90113 similar to PIR|C86301|C86301 arginine/serine-rich protei... 28 5.7
TC90598 similar to GP|8953389|emb|CAB96662.1 putative protein {A... 28 5.7
TC92524 similar to GP|19698943|gb|AAL91207.1 unknown protein {Ar... 28 5.7
TC84105 homologue to PIR|C84507|C84507 hypothetical protein At2g... 27 7.4
TC79394 weakly similar to PIR|T49019|T49019 probable RNA binding... 27 7.4
TC85286 similar to GP|3776572|gb|AAC64889.1| ESTs gb|R65052 gb|... 27 7.4
>AW775441 homologue to GP|18086381|gb AT3g18790/MVE11_17 {Arabidopsis
thaliana}, partial (59%)
Length = 665
Score = 358 bits (918), Expect = 2e-99
Identities = 172/178 (96%), Positives = 177/178 (98%)
Frame = +2
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLA+ECRDL+EADKWRQQIMREIGRKVAE
Sbjct: 131 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLASECRDLNEADKWRQQIMREIGRKVAE 310
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNY RHSAKMTDLDGNIVDVPNP
Sbjct: 311 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYARHSAKMTDLDGNIVDVPNP 490
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILE 178
SGRGPGYRYFGAAKKLPGVRELF+KPPELRKR+TRYDIYKRI+AAYYGYRDDEDGILE
Sbjct: 491 SGRGPGYRYFGAAKKLPGVRELFEKPPELRKRKTRYDIYKRINAAYYGYRDDEDGILE 664
>AJ500818 weakly similar to GP|10435304|dbj unnamed protein product {Homo
sapiens}, partial (12%)
Length = 457
Score = 42.4 bits (98), Expect = 2e-04
Identities = 32/127 (25%), Positives = 56/127 (43%)
Frame = -3
Query: 152 RRTRYDIYKRIDAAYYGYRDDEDGILERLEPSAEAAMRREAAEEWYRLDRIRQEARKGVK 211
+R Y + +A YGY+ D G E A R E R + RQE ++ +
Sbjct: 395 KRDAYGYKRDAEADAYGYKRDAYGYKRDAEAEAYGYKRDADPVEEKRQEEKRQEEKRQEE 216
Query: 212 SGEVAEVSAAVREILREEEEDVVEEERMRGREMKERLEENEREFVVHVPLPDEKEIEKMV 271
+ + R+ + +EE EE+R + +E+ +E +R+ +EK E+
Sbjct: 215 KRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQ---EEKRQEEKRQEEKR 45
Query: 272 LEKKKME 278
LE+K+ E
Sbjct: 44 LEEKRQE 24
>TC87527 similar to GP|21689681|gb|AAM67462.1 unknown protein {Arabidopsis
thaliana}, partial (96%)
Length = 1582
Score = 35.8 bits (81), Expect = 0.021
Identities = 30/100 (30%), Positives = 48/100 (48%), Gaps = 1/100 (1%)
Frame = +2
Query: 167 YGYRDDEDGILERLEPSAEAAMRREAAEEWYRLDR-IRQEARKGVKSGEVAEVSAAVREI 225
YG R+ + + AEAA R++A E + DR R++ + + E ++ AA +
Sbjct: 677 YGRRELKMPKKMGVNSKAEAARARKSAAETEKKDRETREKEEQYWREAESSKPRAAKK-- 850
Query: 226 LREEEEDVVEEERMRGREMKERLEENEREFVVHVPLPDEK 265
REEE D E R E++ E E+E + PD+K
Sbjct: 851 -REEEADKKAEAAARRAEVRRLAELEEKELEKMIKKPDKK 967
>TC89914 similar to GP|14335028|gb|AAK59778.1 AT4g37280/C7A10_80
{Arabidopsis thaliana}, partial (79%)
Length = 1291
Score = 34.7 bits (78), Expect = 0.047
Identities = 39/165 (23%), Positives = 73/165 (43%), Gaps = 13/165 (7%)
Frame = +1
Query: 111 DGNIVDVPNPSGRGPGYRYFGAAKKL-----PGVRELFDKPPELRKRRTRYDIYKRIDAA 165
DG DVP+ R + A +K+ P + E + E+RK RY ++
Sbjct: 139 DGPSGDVPSSDSR-----VYSAGEKVLAYHGPRIYEAKVQKAEIRKNEWRYFVH------ 285
Query: 166 YYGYRDDED---GILERLEPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAV 222
Y G+ + D G ++ + E +++ A ++ +Q K VKSG A+V A
Sbjct: 286 YLGWNKNWDEWVGESRLMKHNDENVVKQRALDK-------KQGVDKNVKSGRSAQVKAKS 444
Query: 223 REILREEEEDVVE-EERMRGREMKERLEE----NEREFVVHVPLP 262
+ E+ED+ + + R++ +E+ +E+ F + +P P
Sbjct: 445 SADAKVEKEDIKNIASKGKKRKIDSGVEKGSGNDEKLFKIQIPAP 579
>BG587776 weakly similar to PIR|F86242|F86 unknown protein 98896-95855
[imported] - Arabidopsis thaliana, partial (23%)
Length = 608
Score = 33.9 bits (76), Expect = 0.079
Identities = 27/107 (25%), Positives = 55/107 (51%), Gaps = 1/107 (0%)
Frame = +2
Query: 183 SAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAA-VREILREEEEDVVEEERMRG 241
S+ ++ ++ A E R + ++ ++ + +AE +A V E +R+ ++ + E +R
Sbjct: 203 SSLGSVEQKTAIEKQRKEEEKKRRQQEAELKLIAEETAKRVEEAIRKRVDESLSSEEVRV 382
Query: 242 REMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDL 288
E++ RLEE ++ V +KE E ++E K+ E + EDL
Sbjct: 383 -EIQRRLEEGRKKLNDEVAAQLQKEKEAALIEAKQKEEQARKEKEDL 520
Score = 26.9 bits (58), Expect = 9.7
Identities = 23/109 (21%), Positives = 48/109 (43%), Gaps = 6/109 (5%)
Frame = +2
Query: 176 ILERLEPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEEDVVE 235
I E E A+R+ +E + +R E ++ ++ G ++ +E+E ++E
Sbjct: 299 IAEETAKRVEEAIRKRV-DESLSSEEVRVEIQRRLEEGRKKLNDEVAAQLQKEKEAALIE 475
Query: 236 EERMRGREMKER------LEENEREFVVHVPLPDEKEIEKMVLEKKKME 278
++ + KE+ LEEN R+ E+ + LE+++ E
Sbjct: 476 AKQKEEQARKEKEDLERMLEENRRKI--------EESQRREALEQQRRE 598
>AJ499968 weakly similar to GP|23495961|gb| hypothetical protein {Plasmodium
falciparum 3D7}, partial (19%)
Length = 342
Score = 33.5 bits (75), Expect = 0.10
Identities = 26/103 (25%), Positives = 50/103 (48%), Gaps = 6/103 (5%)
Frame = -3
Query: 200 DRIRQEARKGVKSGEVAEVSAAVREILREEEEDVVEEERMRGREMKERLEENER------ 253
+R R+E + + +A+ REE E + E + +E +ER E+ ER
Sbjct: 331 ERERKEREEREEQERIAKEKEEQERKEREERERLELERIAKEKEERERKEKEEREERERL 152
Query: 254 EFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAK 296
E VV + ++ EK + E+++ LL + V + +E+ ++K
Sbjct: 151 EAVVRIEKERKEAEEKALKEQEEKALLKEGVIDKKEEEEPKSK 23
>TC89483 similar to GP|18377662|gb|AAL66981.1 unknown protein {Arabidopsis
thaliana}, partial (35%)
Length = 1711
Score = 32.0 bits (71), Expect = 0.30
Identities = 35/149 (23%), Positives = 64/149 (42%), Gaps = 18/149 (12%)
Frame = +1
Query: 172 DEDGILERLEPSAEAAMRREAAE-----------------EWYRLDRIRQEARKGVKSGE 214
+++ +LER E AE R+ E EW +R +++ R+ K E
Sbjct: 172 EKERVLERYEREAERDRIRKEREQKRRIEEVERQFELQLKEWEYREREKEKERQYEKEKE 351
Query: 215 VAEVSAAVREILREEEEDVVE-EERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLE 273
+EIL +EE+D + +R R ++E+ + RE L D+++ E+ + E
Sbjct: 352 KDRERKRRKEILYDEEDDDGDSRKRWRXNAIEEKRNKRLRE--KEDDLADKQKEEEEIAE 525
Query: 274 KKKMELLNKYVSEDLMDEQSEAKDMLNIH 302
KK + + + Q +A +L H
Sbjct: 526 AKK-----RTEEDQQLKRQRDALKLLTEH 597
>TC89176 homologue to GP|13539605|emb|CAC35733. cyclophilin-RNA interacting
protein {Paramecium tetraurelia}, partial (3%)
Length = 783
Score = 30.0 bits (66), Expect = 1.1
Identities = 26/90 (28%), Positives = 40/90 (43%), Gaps = 9/90 (10%)
Frame = +2
Query: 223 REILREEEEDVVEEERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNK 282
RE LR E E +R RE + E ERE V + + + I+K EK++ +
Sbjct: 32 RESLRAEWE-----KRFEDREKEREEREKEREKVRRQRVSEWEAIDKENEEKERKRREEE 196
Query: 283 YVSEDLMDEQSEAK---------DMLNIHR 303
+ E ++E+ + DMLN HR
Sbjct: 197 LIRERELEERMNCRRLEWKKRIDDMLNQHR 286
>TC76440 weakly similar to SP|Q9LD55|IF3A_ARATH Eukaryotic translation
initiation factor 3 subunit 10 (eIF-3 theta), partial
(41%)
Length = 1953
Score = 30.0 bits (66), Expect = 1.1
Identities = 26/92 (28%), Positives = 40/92 (43%), Gaps = 29/92 (31%)
Frame = +1
Query: 198 RLDRI-------RQEARKGVKSGEVAEVSAAVREILREEEE------------------- 231
RLDRI +QE K K +V R+ LREEEE
Sbjct: 1033 RLDRISKILLSRKQEREKMRKLKYYLKVEEEKRQKLREEEEAREREEAERKKKEQAERQA 1212
Query: 232 ---DVVEEERMRGREMKERLEENEREFVVHVP 260
++ E+++ R RE++E+ E +RE ++ P
Sbjct: 1213 KLDEIAEKQKQRLREIEEKAEREKREALLGRP 1308
>TC83341 weakly similar to GP|8978076|dbj|BAA98104.1 gene_id:K14A3.4~unknown
protein {Arabidopsis thaliana}, partial (80%)
Length = 1253
Score = 29.6 bits (65), Expect = 1.5
Identities = 14/36 (38%), Positives = 21/36 (57%)
Frame = +1
Query: 228 EEEEDVVEEERMRGREMKERLEENEREFVVHVPLPD 263
+EEE+ EEE E + +E++E +HVP PD
Sbjct: 583 QEEEEEEEEEEEEEEEDDDDVEKDETVEQIHVPQPD 690
>BQ751907 weakly similar to PIR|T40935|T409 probable utp--glucose-1-phosphate
uridylyltransferase - fission yeast (Schizosaccharomyces
pombe), partial (14%)
Length = 558
Score = 29.6 bits (65), Expect = 1.5
Identities = 14/35 (40%), Positives = 19/35 (54%)
Frame = +1
Query: 118 PNPSGRGPGYRYFGAAKKLPGVRELFDKPPELRKR 152
PNP+ R PG +YF +LP + P+ RKR
Sbjct: 64 PNPTARTPGQKYFSRPSRLP-------RLPQWRKR 147
>BQ137028 similar to GP|4914334|gb|A F14N23.20 {Arabidopsis thaliana},
partial (7%)
Length = 687
Score = 28.5 bits (62), Expect = 3.3
Identities = 23/111 (20%), Positives = 52/111 (46%)
Frame = +2
Query: 189 RREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEEDVVEEERMRGREMKERL 248
R+E +E +R+ +EAR E + +E R E + + EER + +K++
Sbjct: 89 RKERRKEIAEKERLEEEARLNDPEEERKRILLEQQEAERIERDRIAFEEREKAWIIKQQQ 268
Query: 249 EENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAKDML 299
+ + E +++++ + LE+ + E L + ED+ D+ +++
Sbjct: 269 QRDLEE--------EQQQLSQRDLEQHESE-LEEEEEEDVDDDDDGRPEII 394
>TC87590 weakly similar to PIR|G96522|G96522 F11A17.16 [imported] -
Arabidopsis thaliana, partial (45%)
Length = 2425
Score = 28.5 bits (62), Expect = 3.3
Identities = 18/52 (34%), Positives = 29/52 (55%), Gaps = 3/52 (5%)
Frame = +2
Query: 204 QEARKGVKSGEVAEVSAAV---REILREEEEDVVEEERMRGREMKERLEENE 252
+ + KG K GEVA+V R + E++ DV E+ +E+ E+LE +E
Sbjct: 566 ESSHKGSKEGEVAKVVVVAPPRRRRIEEDDPDVKEK-----KELLEKLEVSE 706
>TC80831 similar to GP|18542939|gb|AAK00428.2 Unknown protein {Oryza
sativa}, partial (7%)
Length = 1039
Score = 28.1 bits (61), Expect = 4.4
Identities = 23/84 (27%), Positives = 39/84 (46%)
Frame = +2
Query: 205 EARKGVKSGEVAEVSAAVREILREEEEDVVEEERMRGREMKERLEENEREFVVHVPLPDE 264
E R V E+ + ++ + EE+E EEER R KER ++ R+ + E
Sbjct: 71 EERVRVACKEIITLEKQMKLLEEEEKEKREEEERKERRRTKEREKKLRRKERLK---GKE 241
Query: 265 KEIEKMVLEKKKMELLNKYVSEDL 288
K+ EK+ E + ++ E+L
Sbjct: 242 KDREKICSESNDILCTSEISKEEL 313
>TC90113 similar to PIR|C86301|C86301 arginine/serine-rich protein
[imported] - Arabidopsis thaliana, partial (70%)
Length = 1259
Score = 27.7 bits (60), Expect = 5.7
Identities = 14/29 (48%), Positives = 18/29 (61%)
Frame = -1
Query: 226 LREEEEDVVEEERMRGREMKERLEENERE 254
LRE E + E++ R RE +E EE ERE
Sbjct: 110 LREGEREEEEKDLEREREREEEREEEERE 24
>TC90598 similar to GP|8953389|emb|CAB96662.1 putative protein {Arabidopsis
thaliana}, partial (18%)
Length = 602
Score = 27.7 bits (60), Expect = 5.7
Identities = 16/49 (32%), Positives = 29/49 (58%)
Frame = +3
Query: 249 EENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAKD 297
++N ++ + HV +E+E E+ E+ ELL K + EDL ++ +KD
Sbjct: 156 KKNNKKKLTHVTKGNEEEEEEE--EEDAFELLFKKLEEDLKNDDDLSKD 296
>TC92524 similar to GP|19698943|gb|AAL91207.1 unknown protein {Arabidopsis
thaliana}, partial (38%)
Length = 721
Score = 27.7 bits (60), Expect = 5.7
Identities = 29/118 (24%), Positives = 54/118 (45%), Gaps = 5/118 (4%)
Frame = +3
Query: 184 AEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEEDVVEEERMRGRE 243
AE + A E+ RL ++ + K + E S V E ++E E + + + +E
Sbjct: 141 AEQVSQLLATEKNLRL-QLTTDGEKFQQFQEALTKSNDVFETFKQEIEKMAKSMKELKKE 317
Query: 244 MKERLEENEREFVVHVPLPDE-----KEIEKMVLEKKKMELLNKYVSEDLMDEQSEAK 296
+ ++E+ V + L DE K++EK +K+K+E L + + + SE K
Sbjct: 318 NQFLKSKSEKSDVTLIELVDERERLKKQLEKTKNQKEKLESLCRSLQAERKQILSENK 491
>TC84105 homologue to PIR|C84507|C84507 hypothetical protein At2g13370
[imported] - Arabidopsis thaliana, partial (15%)
Length = 834
Score = 27.3 bits (59), Expect = 7.4
Identities = 21/64 (32%), Positives = 32/64 (49%)
Frame = +1
Query: 203 RQEARKGVKSGEVAEVSAAVREILREEEEDVVEEERMRGREMKERLEENEREFVVHVPLP 262
++EA+KG + E+SA ILR E++V+EER K L + E +
Sbjct: 463 KKEAKKGGSFFDKNELSA----ILRFGAEELVKEERNDEESKKRLLSMDIDEILERAEKV 630
Query: 263 DEKE 266
+EKE
Sbjct: 631 EEKE 642
>TC79394 weakly similar to PIR|T49019|T49019 probable RNA binding protein -
Arabidopsis thaliana, partial (74%)
Length = 1909
Score = 27.3 bits (59), Expect = 7.4
Identities = 25/77 (32%), Positives = 32/77 (41%), Gaps = 1/77 (1%)
Frame = +2
Query: 205 EARKGVKSGEVAEVSAAVREILREEEEDVVEEERMRGREMKERLEENEREFVVH-VPLPD 263
E K SGE ++ + EEE EE + +E EE E E PL +
Sbjct: 203 EPEKPTDSGEQVDLDGDNDQEESSEEEVEYEEVEVEEEVEEEEEEEEEEEVEEESKPLDE 382
Query: 264 EKEIEKMVLEKKKMELL 280
E E +K KK ELL
Sbjct: 383 EDEADK----KKHAELL 421
Score = 26.9 bits (58), Expect = 9.7
Identities = 19/63 (30%), Positives = 31/63 (49%)
Frame = +2
Query: 200 DRIRQEARKGVKSGEVAEVSAAVREILREEEEDVVEEERMRGREMKERLEENEREFVVHV 259
D ++E+ + E EV V E EEEE+ VEEE + + E E ++++ +
Sbjct: 251 DNDQEESSEEEVEYEEVEVEEEVEEEEEEEEEEEVEEE---SKPLDEEDEADKKKHAELL 421
Query: 260 PLP 262
LP
Sbjct: 422 ALP 430
>TC85286 similar to GP|3776572|gb|AAC64889.1| ESTs gb|R65052 gb|AA712146
gb|H76533 gb|H76282 gb|AA650771 gb|H76287
gb|AA650887 gb|N37383, partial (77%)
Length = 1947
Score = 27.3 bits (59), Expect = 7.4
Identities = 33/111 (29%), Positives = 49/111 (43%)
Frame = -3
Query: 168 GYRDDEDGILERLEPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILR 227
G ++++G + A +R E E R +R R+ R+ RE R
Sbjct: 910 GCHEEDEGGRVGVRKGASCRIRHEGERERER-ERERERERE--------------RERER 776
Query: 228 EEEEDVVEEERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKME 278
E E + E ER R RE +ER E ERE P+P+ + +VL +K E
Sbjct: 775 ERERER-ERERERERE-RERERERERERERGGPVPN-SSLYCLVLHEKWSE 632
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,684,550
Number of Sequences: 36976
Number of extensions: 75691
Number of successful extensions: 420
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 398
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 409
length of query: 303
length of database: 9,014,727
effective HSP length: 96
effective length of query: 207
effective length of database: 5,465,031
effective search space: 1131261417
effective search space used: 1131261417
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0282.6


