

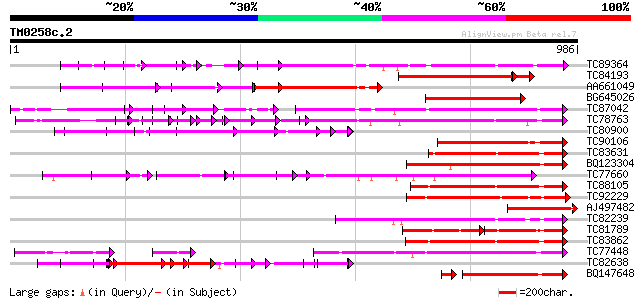
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0258c.2
(986 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89364 weakly similar to PIR|C96745|C96745 hypothetical protein... 356 2e-98
TC84193 homologue to GP|9651941|gb|AAF91322.1| receptor-like pro... 320 1e-87
AA661049 similar to PIR|T50851|T50 receptor protein kinase homol... 284 1e-76
BG645026 similar to GP|9651941|gb receptor-like protein kinase 1... 278 6e-75
TC87042 similar to GP|13605827|gb|AAK32899.1 AT5g48380/MJE7_1 {A... 253 3e-67
TC78763 somatic embryogenesis receptor kinase 1 [Medicago trunca... 247 1e-65
TC80900 weakly similar to PIR|G84652|G84652 probable receptor-li... 237 2e-62
TC90106 similar to GP|3641252|gb|AAC36318.1| leucine-rich recept... 234 9e-62
TC83631 similar to PIR|T10659|T10659 probable serine/threonine-s... 227 1e-59
BQ123304 similar to GP|22093756|d putative receptor protein kina... 226 3e-59
TC77660 similar to GP|22212524|dbj|BAC07504. receptor-like prote... 225 6e-59
TC88105 similar to GP|21593085|gb|AAM65034.1 Putative protein ki... 223 3e-58
TC92229 similar to PIR|B86369|B86369 hypothetical protein AAC980... 222 5e-58
AJ497482 homologue to PIR|T50850|T50 receptor protein kinase hom... 211 1e-54
TC82239 weakly similar to GP|16924042|gb|AAL31654.1 Putative pro... 207 2e-53
TC81789 similar to PIR|B86440|B86440 probable protein kinase [im... 127 2e-53
TC83862 similar to PIR|T05270|T05270 probable serine/threonine-s... 206 3e-53
TC77448 SYMRK; MtSYMRK [Medicago truncatula]; nod region linked ... 205 8e-53
TC82638 weakly similar to PIR|T00712|T00712 protein kinase homol... 142 9e-53
BQ147648 similar to PIR|B86465|B8 probable Protein kinase [impor... 189 2e-52
>TC89364 weakly similar to PIR|C96745|C96745 hypothetical protein T9N14.3
[imported] - Arabidopsis thaliana, partial (45%)
Length = 1903
Score = 356 bits (914), Expect = 2e-98
Identities = 217/566 (38%), Positives = 327/566 (57%), Gaps = 24/566 (4%)
Frame = +1
Query: 431 GVFQLPSVTITELSNNRLNGELPSVIS-GESLGTLTLSNNLFTGKIPAAMKNLRALQSLS 489
GV+ LP+ I +L N +GE+ S I +L + L NN F+GK+P+ + L L+ L
Sbjct: 49 GVWSLPNAKIIDLGFNNFSGEVSSEIGYSTNLSEIVLMNNKFSGKVPSEIGKLVNLEKLY 228
Query: 490 LDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSRNNLAGEVPK 549
L N F G+IP + + L+ +++ N+LTG IP + H + L ++L+ N+L+G +P
Sbjct: 229 LSNNNFSGDIPREIGLLKQLSTLHLEENSLTGVIPKELGHCSRLVDLNLALNSLSGNIPN 408
Query: 550 GMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVPTGGQFLVFNYDKT 609
+ + L+ LNLSRN+++G +PD + M L+++D S N+ +G +P G L+ +K
Sbjct: 409 SVSLMSSLNSLNLSRNKLTGTIPDNLEKM-KLSSVDFSQNSLSGGIPFG--ILIIGGEKA 579
Query: 610 FAGNPNLCFPHRASCPSVLYDSLRKTRAKTARVRAIVIG-----IALATAVLLVAVTVHV 664
F GN LC P +S K K R V + + A+ +H
Sbjct: 580 FVGNKELCVEQ---IPKTSMNSDLKICDKDHGHRRGVFAYKYFLLFFIAVIFAAAIVIHR 750
Query: 665 VRKRRLHR---------AQAWKLTAFQRLEIKAEDVVECLKEENIIGKGGAGIVYRGSMP 715
K R + +Q WK +F +++I A++V L ++N+IG GG G VYR +
Sbjct: 751 CMKNRKEKNLQKGEKEASQKWKQASFHQVDIDADEVSH-LGDDNLIGYGGTGKVYRVKLK 927
Query: 716 N-GTDVAIKRLVGQGSGRNDYGFR---AEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEY 771
G VA+K+L YG + AE+E L KIRH+NI++L +NLL++EY
Sbjct: 928 KTGIVVAVKQL------EKGYGVKILAAEMEILAKIRHKNILKLYACFLKGGSNLLVFEY 1089
Query: 772 MPNGSLGEWLHGAKGGHL---RWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILL 828
MPNG+L + LH + W RYKIA+ A+G+CY+HHDCSP +IHRD+KS+NILL
Sbjct: 1090MPNGNLFQALHREVKDEMVTFDWNQRYKIALGGAKGICYLHHDCSPPVIHRDIKSSNILL 1269
Query: 829 DADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLL 888
DA++EA +ADFG+A+F + S AG++GYIAPE AYT ++ EKSDVYSFGVVLL
Sbjct: 1270DANYEAKIADFGVARFA--EKSQMGYSVFAGTHGYIAPELAYTTEITEKSDVYSFGVVLL 1443
Query: 889 ELIIGRKPV-GEFGDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRL-SGYPLTSVIHMF 946
EL+ GR+PV E+G+ DIV WV +S +D VL+++D R+ S + + +I +
Sbjct: 1444ELVSGREPVEEEYGEAKDIVHWV------MSNLNDRESVLSILDGRVASTHCVEDMIKVL 1605
Query: 947 NIAMMCVKEMGPARPTMREVVHMLTN 972
I + C ++ RPTMR+VV ML +
Sbjct: 1606KIGIKCTTKLPTLRPTMRDVVKMLVD 1683
Score = 104 bits (260), Expect = 1e-22
Identities = 62/169 (36%), Positives = 95/169 (55%)
Frame = +1
Query: 165 EEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKEL 224
E + L K + L N FSG + +L + L N +G+VP + KL L++L
Sbjct: 46 EGVWSLPNAKIIDLGFNNFSGEVSSEIGYSTNLSEIVLMNNKFSGKVPSEIGKLVNLEKL 225
Query: 225 HLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTI 284
+L +N + G IP G ++ L L + +LTG IP LG+ ++L L + +N+L+G I
Sbjct: 226 YLS-NNNFSGDIPREIGLLKQLSTLHLEENSLTGVIPKELGHCSRLVDLNLALNSLSGNI 402
Query: 285 PPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLP 333
P +S M SL SL+LS N LTG IP++ K+K L+ ++F QN G +P
Sbjct: 403 PNSVSLMSSLNSLNLSRNKLTGTIPDNLEKMK-LSSVDFSQNSLSGGIP 546
Score = 98.6 bits (244), Expect = 1e-20
Identities = 67/190 (35%), Positives = 94/190 (49%)
Frame = +1
Query: 120 SLTSLKVLNISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLA 179
SL + K++++ N FSG+ I T L + +N FSG +P EI KL L+ L+L+
Sbjct: 58 SLPNAKIIDLGFNNFSGEVSSEIGYS-TNLSEIVLMNNKFSGKVPSEIGKLVNLEKLYLS 234
Query: 180 GNYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPA 239
N FSG IP + L L L NSLTG +P+ L L +L+L +
Sbjct: 235 NNNFSGDIPREIGLLKQLSTLHLEENSLTGVIPKELGHCSRLVDLNLALN---------- 384
Query: 240 FGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDL 299
+L+G IP S+ ++ L+SL + N LTGTIP L M L S+D
Sbjct: 385 ---------------SLSGNIPNSVSLMSSLNSLNLSRNKLTGTIPDNLEK-MKLSSVDF 516
Query: 300 SINDLTGEIP 309
S N L+G IP
Sbjct: 517 SQNSLSGGIP 546
Score = 95.1 bits (235), Expect = 1e-19
Identities = 63/209 (30%), Positives = 102/209 (48%)
Frame = +1
Query: 198 EFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLT 257
+ + L N+ +G V + L E+ L +N + G +P G + NL L ++N N +
Sbjct: 73 KIIDLGFNNFSGEVSSEIGYSTNLSEIVL-MNNKFSGKVPSEIGKLVNLEKLYLSNNNFS 249
Query: 258 GEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKN 317
G+IP +G L +L +L ++ N+LTG IP EL L+ L+L++N L+G IP S S + +
Sbjct: 250 GDIPREIGLLKQLSTLHLEENSLTGVIPKELGHCSRLVDLNLALNSLSGNIPNSVSLMSS 429
Query: 318 LTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLT 377
L +N +NK G++P NLE +++ +FS +N L+
Sbjct: 430 LNSLNLSRNKLTGTIPD------NLEKMKLSSVDFS-------------------QNSLS 534
Query: 378 GLIPPDLCKSGRLKTFIITDNFFRGPIPK 406
G IP + G K F+ IPK
Sbjct: 535 GGIPFGILIIGGEKAFVGNKELCVEQIPK 621
Score = 90.1 bits (222), Expect = 4e-18
Identities = 54/150 (36%), Positives = 83/150 (55%)
Frame = +1
Query: 88 GHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLFSGQFPGNITVGMT 147
G + EIG L + + N + ++PS++ L +L+ L +S+N FSG P I + +
Sbjct: 106 GEVSSEIGYSTNLSEIVLMNNKFSGKVPSEIGKLVNLEKLYLSNNNFSGDIPREIGL-LK 282
Query: 148 ELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSL 207
+L L +NS +G +P+E+ +L L+LA N SG IP S S SL L L+ N L
Sbjct: 283 QLSTLHLEENSLTGVIPKELGHCSRLVDLNLALNSLSGNIPNSVSLMSSLNSLNLSRNKL 462
Query: 208 TGRVPESLAKLKTLKELHLGYSNAYEGGIP 237
TG +P++L K+K L + N+ GGIP
Sbjct: 463 TGTIPDNLEKMK-LSSVDFS-QNSLSGGIP 546
Score = 89.7 bits (221), Expect = 5e-18
Identities = 58/187 (31%), Positives = 85/187 (45%)
Frame = +1
Query: 290 SMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWE 349
S+ + +DL N+ +GE+ NL+ + NKF G +PS IG L NLE L +
Sbjct: 58 SLPNAKIIDLGFNNFSGEVSSEIGYSTNLSEIVLMNNKFSGKVPSEIGKLVNLEKLYLSN 237
Query: 350 NNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIG 409
NNFS +P +G L K +L T + +N G IPK +G
Sbjct: 238 NNFSGDIPREIG----------------------LLK--QLSTLHLEENSLTGVIPKELG 345
Query: 410 ECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVISGESLGTLTLSNN 469
C L + +A N L G +P V + S+ LS N+L G +P + L ++ S N
Sbjct: 346 HCSRLVDLNLALNSLSGNIPNSVSLMSSLNSLNLSRNKLTGTIPDNLEKMKLSSVDFSQN 525
Query: 470 LFTGKIP 476
+G IP
Sbjct: 526 SLSGGIP 546
>TC84193 homologue to GP|9651941|gb|AAF91322.1| receptor-like protein kinase
1 {Glycine max}, partial (24%)
Length = 748
Score = 320 bits (821), Expect = 1e-87
Identities = 153/208 (73%), Positives = 179/208 (85%), Gaps = 1/208 (0%)
Frame = +2
Query: 677 KLTAFQRLEIKAEDVVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRN-DY 735
++ RL+ +DV++ LKE+NIIGKGGAGIVY+G+MPNG VA+KRL G + D+
Sbjct: 8 EINRIPRLDFTVDDVLDSLKEDNIIGKGGAGIVYKGAMPNGDLVAVKRLPAMSRGSSHDH 187
Query: 736 GFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRY 795
GF AEI+TLG+IRHR+I+RLLG+ SN +TNLL+YEYMPNGSLGE LHG KGGHL W+ RY
Sbjct: 188 GFNAEIQTLGRIRHRHIVRLLGFCSNHETNLLVYEYMPNGSLGEVLHGKKGGHLHWDTRY 367
Query: 796 KIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMS 855
KIAVEAA+GLCY+HHDCSPLI+HRDVKSNNILLD+ FEAHVADFGLAKFL D G S+ MS
Sbjct: 368 KIAVEAAKGLCYLHHDCSPLIVHRDVKSNNILLDSGFEAHVADFGLAKFLQDSGTSECMS 547
Query: 856 SIAGSYGYIAPEYAYTLKVDEKSDVYSF 883
+IAGSYGYIAPEYAYTLKVDEKSDVYSF
Sbjct: 548 AIAGSYGYIAPEYAYTLKVDEKSDVYSF 631
Score = 59.3 bits (142), Expect = 7e-09
Identities = 28/40 (70%), Positives = 32/40 (80%)
Frame = +3
Query: 873 KVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNK 912
K +K+ +FGVVLLEL+ GRKPVGEFGDGVDIV WV K
Sbjct: 600 KSTKKATSTAFGVVLLELVAGRKPVGEFGDGVDIVQWVRK 719
>AA661049 similar to PIR|T50851|T50 receptor protein kinase homolog
[imported] - soybean, partial (19%)
Length = 655
Score = 284 bits (726), Expect = 1e-76
Identities = 147/222 (66%), Positives = 172/222 (77%)
Frame = +3
Query: 426 GPVPPGVFQLPSVTITELSNNRLNGELPSVISGESLGTLTLSNNLFTGKIPAAMKNLRAL 485
GPVPPG FQLPSV I EL NNR NG+LP+ ISG SLG L LSNNLFTG+IPA+MKNLR+L
Sbjct: 3 GPVPPGXFQLPSVQIIELGNNRFNGQLPTEISGNSLGNLALSNNLFTGRIPASMKNLRSL 182
Query: 486 QSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSRNNLAG 545
Q+L LDAN+F+GEIP VF +P+LT++NISGNNLTG IP T+T +SLTAVD SRN L G
Sbjct: 183 QTLLLDANQFLGEIPAEVFALPVLTRINISGNNLTGGIPKTVTQCSSLTAVDFSRNMLTG 362
Query: 546 EVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVPTGGQFLVFN 605
EVPKGMKNL LSI N+S N ISG +PDEIRFMTSLTTLDLS NNFTG VPTGGQFLVFN
Sbjct: 363 EVPKGMKNLKVLSIFNVSHNSISGKIPDEIRFMTSLTTLDLSYNNFTGIVPTGGQFLVFN 542
Query: 606 YDKTFAGNPNLCFPHRASCPSVLYDSLRKTRAKTARVRAIVI 647
D++FAGNP P P+ ++ A+ +A+V+
Sbjct: 543 -DRSFAGNPTYFPP-----PNNMFFIALSFXKNXAKXKAVVL 650
Score = 96.3 bits (238), Expect = 5e-20
Identities = 66/210 (31%), Positives = 100/210 (47%)
Frame = +3
Query: 161 GPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKT 220
GP+P +L ++ + L N F+G +P S SL L L+ N TGR+P S+ L++
Sbjct: 3 GPVPPGXFQLPSVQIIELGNNRFNGQLPTEISG-NSLGNLALSNNLFTGRIPASMKNLRS 179
Query: 221 LKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNL 280
L+ L L + GEIP + L L + + NNL
Sbjct: 180 LQTLLLDANQ-------------------------FLGEIPAEVFALPVLTRINISGNNL 284
Query: 281 TGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLP 340
TG IP ++ SL ++D S N LTGE+P+ LK L++ N N G +P I +
Sbjct: 285 TGGIPKTVTQCSSLTAVDFSRNMLTGEVPKGMKNLKVLSIFNVSHNSISGKIPDEIRFMT 464
Query: 341 NLETLQVWENNFSFVLPHNLGGNGRFLYFD 370
+L TL + NNF+ ++P G+FL F+
Sbjct: 465 SLTTLDLSYNNFTGIVP----TGGQFLVFN 542
Score = 88.6 bits (218), Expect = 1e-17
Identities = 53/174 (30%), Positives = 94/174 (53%)
Frame = +3
Query: 88 GHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLFSGQFPGNITVGMT 147
G +PP L ++ + + N QLP++++ SL L +S+NLF+G+ P ++ +
Sbjct: 3 GPVPPGXFQLPSVQIIELGNNRFNGQLPTEISG-NSLGNLALSNNLFTGRIPASMK-NLR 176
Query: 148 ELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSL 207
L+ L N F G +P E+ L L ++++GN +G IP++ ++ SL + + N L
Sbjct: 177 SLQTLLLDANQFLGEIPAEVFALPVLTRINISGNNLTGGIPKTVTQCSSLTAVDFSRNML 356
Query: 208 TGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIP 261
TG VP+ + LK L ++ + N+ G IP M +L L+++ N TG +P
Sbjct: 357 TGEVPKGMKNLKVLSIFNVSH-NSISGKIPDEIRFMTSLTTLDLSYNNFTGIVP 515
Score = 82.0 bits (201), Expect = 1e-15
Identities = 62/198 (31%), Positives = 93/198 (46%), Gaps = 3/198 (1%)
Frame = +3
Query: 282 GTIPPELSSMMSLMSLDLSINDLTGEIPESFS--KLKNLTLMNFFQNKFRGSLPSFIGDL 339
G +PP + S+ ++L N G++P S L NL L N N F G +P+ + +L
Sbjct: 3 GPVPPGXFQLPSVQIIELGNNRFNGQLPTEISGNSLGNLALSN---NLFTGRIPASMKNL 173
Query: 340 PNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNF 399
+L+TL + N F G IP ++ L I+ N
Sbjct: 174 RSLQTLLLDANQF------------------------LGEIPAEVFALPVLTRINISGNN 281
Query: 400 FRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVIS-G 458
G IPK + +C SLT + + N L G VP G+ L ++I +S+N ++G++P I
Sbjct: 282 LTGGIPKTVTQCSSLTAVDFSRNMLTGEVPKGMKNLKVLSIFNVSHNSISGKIPDEIRFM 461
Query: 459 ESLGTLTLSNNLFTGKIP 476
SL TL LS N FTG +P
Sbjct: 462 TSLTTLDLSYNNFTGIVP 515
Score = 78.2 bits (191), Expect = 1e-14
Identities = 53/174 (30%), Positives = 81/174 (46%)
Frame = +3
Query: 258 GEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKN 317
G +PP L + + + N G +P E+S SL +L LS N TG IP S L++
Sbjct: 3 GPVPPGXFQLPSVQIIELGNNRFNGQLPTEISGN-SLGNLALSNNLFTGRIPASMKNLRS 179
Query: 318 LTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLT 377
L + N+F G +P+ + LP L + + NN + +P + D ++N LT
Sbjct: 180 LQTLLLDANQFLGEIPAEVFALPVLTRINISGNNLTGGIPKTVTQCSSLTAVDFSRNMLT 359
Query: 378 GLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPG 431
G +P + L F ++ N G IP I SLT + ++ N G VP G
Sbjct: 360 GEVPKGMKNLKVLSIFNVSHNSISGKIPDEIRFMTSLTTLDLSYNNFTGIVPTG 521
Score = 38.1 bits (87), Expect = 0.017
Identities = 26/93 (27%), Positives = 48/93 (50%)
Frame = +3
Query: 69 TCDQNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLN 128
T Q + A++ + L G +P + L+ L +S N+++ ++P ++ +TSL L+
Sbjct: 303 TVTQCSSLTAVDFSRNMLTGEVPKGMKNLKVLSIFNVSHNSISGKIPDEIRFMTSLTTLD 482
Query: 129 ISHNLFSGQFPGNITVGMTELEALDAYDNSFSG 161
+S+N F+G P T + L D SF+G
Sbjct: 483 LSYNNFTGIVP-------TGGQFLVFNDRSFAG 560
>BG645026 similar to GP|9651941|gb receptor-like protein kinase 1 {Glycine
max}, partial (16%)
Length = 695
Score = 278 bits (712), Expect = 6e-75
Identities = 131/174 (75%), Positives = 153/174 (87%)
Frame = +3
Query: 723 KRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLH 782
K+L+G G+ +D+GFRAEI+TLG IRHRNI+RLL + SNK+TNLL+YEYM NGSLGE LH
Sbjct: 111 KKLLGFGANNHDHGFRAEIQTLGNIRHRNIVRLLAFCSNKETNLLVYEYMRNGSLGETLH 290
Query: 783 GAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLA 842
G KG L W RYKI++++A+GLCY+HHDCSPLI+HRDVKSNNILL ++FEAHVADFGLA
Sbjct: 291 GKKGAFLSWNFRYKISIDSAKGLCYLHHDCSPLILHRDVKSNNILLSSNFEAHVADFGLA 470
Query: 843 KFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKP 896
KFL D A + MSSIAGSYGYIAPEYAYTL+VDEKSDVYSFGVVLLEL+ GRKP
Sbjct: 471 KFLVDGAAPECMSSIAGSYGYIAPEYAYTLRVDEKSDVYSFGVVLLELLTGRKP 632
>TC87042 similar to GP|13605827|gb|AAK32899.1 AT5g48380/MJE7_1 {Arabidopsis
thaliana}, partial (51%)
Length = 2206
Score = 253 bits (646), Expect = 3e-67
Identities = 170/503 (33%), Positives = 264/503 (51%), Gaps = 29/503 (5%)
Frame = +2
Query: 497 GEIPGGVFEIPMLTKVNISGNNLTGPIPTTI-THRASLTAVDLSRNNLAGEVPKGMKNLM 555
G+ P G+ +T +++S N+L+G IP I T +T++DLS N +GE+P + N
Sbjct: 422 GQFPRGIVNCSSMTGLDLSVNDLSGTIPGDISTLLKFVTSLDLSSNEFSGEIPVSLANCT 601
Query: 556 DLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVPT---GGQFLVFNYDKTFAG 612
L++L LS+N+++G +P + + + T D+S+N TG VP GG+ D +A
Sbjct: 602 YLNVLKLSQNQLTGQIPLLLGTLDRIKTFDVSNNLLTGQVPNFTAGGKV-----DVNYAN 766
Query: 613 NPNLCF-PHRASCPSVLYDSLRKTRAKTARVRAIVIGIALATAVLLVAVTVHVVRK---- 667
N LC P C + +++ TA + +G A+ L VR+
Sbjct: 767 NQGLCGQPSLGVCKATA-----SSKSNTAVIAGAAVGAVTLAALGLGVFMFFFVRRSAYR 931
Query: 668 ------------RRLHRAQAWKLTAFQRL--EIKAEDVVEC---LKEENIIGKGGAGIVY 710
R L + K++ F++ ++K D+++ NIIG G G VY
Sbjct: 932 KKEEDPEGNKWARSLKGTKGIKVSLFEKSISKMKLSDLMKATNNFSNINIIGTGRTGTVY 1111
Query: 711 RGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYE 770
+ ++ +GT +KRL Q S ++ F +E+ TLG ++HRN++ LLG+ K LL+++
Sbjct: 1112KATLEDGTAFMVKRL--QESQHSEKEFMSEMATLGTVKHRNLVPLLGFCVAKKERLLVFK 1285
Query: 771 YMPNGSLGEWLHGAKGG-HLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLD 829
MPNG L + LH A G L W R KIA+ AA+G ++HH C+P IIHR++ S ILLD
Sbjct: 1286NMPNGMLHDQLHPAAGECTLDWPSRLKIAIGAAKGFAWLHHSCNPRIIHRNISSKCILLD 1465
Query: 830 ADFEAHVADFGLAKFL--YDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVL 887
ADFE ++DFGLA+ + D S ++ G +GY+APEY TL K DV+SFG VL
Sbjct: 1466ADFEPKISDFGLARLMNPLDTHLSTFVNGEFGDFGYVAPEYTKTLVATPKGDVFSFGTVL 1645
Query: 888 LELIIGRKPVGEFGDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRLSGYPLTSVIHMFN 947
LEL+ G +P G + + ++ELS S++ L A+ + L+ +
Sbjct: 1646LELVTGERPANVAKAPETFKGNLVEWITELS--SNSKLHDAIDESLLNKGDDNELFQFLK 1819
Query: 948 IAMMCVKEMGPARPTMREVVHML 970
+A CV E+ RPTM EV L
Sbjct: 1820VACNCVTEVPKERPTMFEVYQFL 1888
Score = 73.9 bits (180), Expect = 3e-13
Identities = 59/219 (26%), Positives = 94/219 (41%), Gaps = 4/219 (1%)
Frame = +2
Query: 2 RIRVSYLLVLCFTLIWFRWTVVYSSFSDLDALLKLKESMKGAKAKHHALEDWKFSTSLSA 61
RI SY+ V LI F + Y + +D+ L +K S++ ++ W F+
Sbjct: 167 RILSSYVFVSFLLLISFG--ITYGTETDIFCLKSIKNSIQDPN--NYLTSSWNFNNKTEG 334
Query: 62 H-CSFSGVTC---DQNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSD 117
C F+GV C D+N K+ NL +S L Q P
Sbjct: 335 FICRFNGVECWHPDEN-------------------------KVLNLKLSNMGLKGQFPRG 439
Query: 118 LASLTSLKVLNISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLH 177
+ + +S+ L++S N SG PG+I+ + + +LD N FSG +P + L L
Sbjct: 440 IVNCSSMTGLDLSVNDLSGTIPGDISTLLKFVTSLDLSSNEFSGEIPVSLANCTYLNVLK 619
Query: 178 LAGNYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLA 216
L+ N +G IP ++ ++ N LTG+VP A
Sbjct: 620 LSQNQLTGQIPLLLGTLDRIKTFDVSNNLLTGQVPNFTA 736
Score = 73.6 bits (179), Expect = 4e-13
Identities = 37/115 (32%), Positives = 67/115 (58%), Gaps = 1/115 (0%)
Frame = +2
Query: 249 LEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLM-SLDLSINDLTGE 307
L+++N L G+ P + N + + L + +N+L+GTIP ++S+++ + SLDLS N+ +GE
Sbjct: 395 LKLSNMGLKGQFPRGIVNCSSMTGLDLSVNDLSGTIPGDISTLLKFVTSLDLSSNEFSGE 574
Query: 308 IPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGG 362
IP S + L ++ QN+ G +P +G L ++T V N + +P+ G
Sbjct: 575 IPVSLANCTYLNVLKLSQNQLTGQIPLLLGTLDRIKTFDVSNNLLTGQVPNFTAG 739
Score = 60.1 bits (144), Expect = 4e-09
Identities = 52/200 (26%), Positives = 91/200 (45%), Gaps = 1/200 (0%)
Frame = +2
Query: 269 KLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKL-KNLTLMNFFQNK 327
K+ +L + L G P + + S+ LDLS+NDL+G IP S L K +T ++ N+
Sbjct: 383 KVLNLKLSNMGLKGQFPRGIVNCSSMTGLDLSVNDLSGTIPGDISTLLKFVTSLDLSSNE 562
Query: 328 FRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKS 387
F G +P + + L L++ ++N LTG IP L
Sbjct: 563 FSGEIPVSLANCTYLNVLKL------------------------SQNQLTGQIPLLLGTL 670
Query: 388 GRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNR 447
R+KTF +++N G +P + + ANN G+ PS+ + + + +
Sbjct: 671 DRIKTFDVSNNLLTGQVPNFTAGGK--VDVNYANN-------QGLCGQPSLGVCKATASS 823
Query: 448 LNGELPSVISGESLGTLTLS 467
+ +VI+G ++G +TL+
Sbjct: 824 KSN--TAVIAGAAVGAVTLA 877
Score = 55.1 bits (131), Expect = 1e-07
Identities = 36/111 (32%), Positives = 59/111 (52%), Gaps = 1/111 (0%)
Frame = +2
Query: 200 LGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSM-ENLRLLEMANCNLTG 258
L L+ L G+ P + ++ L L N G IP ++ + + L++++ +G
Sbjct: 395 LKLSNMGLKGQFPRGIVNCSSMTGLDLSV-NDLSGTIPGDISTLLKFVTSLDLSSNEFSG 571
Query: 259 EIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIP 309
EIP SL N T L+ L + N LTG IP L ++ + + D+S N LTG++P
Sbjct: 572 EIPVSLANCTYLNVLKLSQNQLTGQIPLLLGTLDRIKTFDVSNNLLTGQVP 724
Score = 31.2 bits (69), Expect = 2.1
Identities = 16/37 (43%), Positives = 22/37 (59%)
Frame = +2
Query: 560 LNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVP 596
L LS + G P I +S+T LDLS N+ +GT+P
Sbjct: 395 LKLSNMGLKGQFPRGIVNCSSMTGLDLSVNDLSGTIP 505
>TC78763 somatic embryogenesis receptor kinase 1 [Medicago truncatula]
Length = 2737
Score = 247 bits (631), Expect = 1e-65
Identities = 165/495 (33%), Positives = 249/495 (49%), Gaps = 29/495 (5%)
Frame = +3
Query: 505 EIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSR 564
++ L + + NN+TGPIP+ + + +L ++DL N G +P + L L L L+
Sbjct: 699 QLKNLQYLELYSNNITGPIPSDLGNLTNLVSLDLYLNRFNGPIPDSLGKLSKLRFLRLNN 878
Query: 565 NEISGPVPDEIRFMTSLTTLDLSSNNFTGTVPTGGQFLVFNYDKTFAGNPNLCFPHRAS- 623
N + GP+P + +++L LDLS+N +G VP G F +F +FA N NLC P
Sbjct: 879 NSLMGPIPMSLTNISALQVLDLSNNQLSGVVPDNGSFSLFT-PISFANNLNLCGPVTGHP 1055
Query: 624 CPS---------VLYDSLRKTRAKTARVRAIVIGIALATAVLLVAVTVHVVRKRRLHRAQ 674
CP + AI G+A A+L A + RR +
Sbjct: 1056 CPGSPPFSPPPPFVPPPPISAPGSGGATGAIAGGVAAGAALLFAAPAIAFAWWRRRKPQE 1235
Query: 675 AW-----------KLTAFQRLEIKAEDVV-ECLKEENIIGKGGAGIVYRGSMPNGTDVAI 722
+ L +R ++ V + +NI+G+GG G VY+G + +G+ VA+
Sbjct: 1236 FFFDVPAEEDPEVHLGQLKRFSLRELQVATDTFSNKNILGRGGFGKVYKGRLADGSLVAV 1415
Query: 723 KRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLH 782
KRL + + + F+ E+E + HRN++RL G+ LL+Y YM NGS+ L
Sbjct: 1416 KRLKEERTPGGELQFQTEVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLR 1595
Query: 783 GAKGGH--LRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFG 840
L W R +IA+ +ARGL Y+H C P IIHRDVK+ NILLD +FEA V DFG
Sbjct: 1596 ERPPHQEPLDWPTRKRIALGSARGLSYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFG 1775
Query: 841 LAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVG-- 898
LAK L D + +++ G+ G+IAPEY T K EK+DV+ +G++LLELI G++
Sbjct: 1776 LAK-LMDYKDTHVTTAVRGTIGHIAPEYLSTGKSSEKTDVFGYGIMLLELITGQRAFDLA 1952
Query: 899 --EFGDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRL-SGYPLTSVIHMFNIAMMCVKE 955
D V ++ WV + E + +VDP L + Y V + +A++C +
Sbjct: 1953 RLANDDDVMLLDWVKGLLKEKK-------LEMLVDPDLKTNYIEAEVEQLIQVALLCTQG 2111
Query: 956 MGPARPTMREVVHML 970
RP M +VV ML
Sbjct: 2112 SPMDRPKMSDVVRML 2156
Score = 90.9 bits (224), Expect = 2e-18
Identities = 60/205 (29%), Positives = 104/205 (50%)
Frame = +3
Query: 10 VLCFTLIWFRWTVVYSSFSDLDALLKLKESMKGAKAKHHALEDWKFSTSLSAHCSFSGVT 69
+ F L+ + S+ + DAL L+ +++ ++ L+ W +L C++ VT
Sbjct: 456 ICAFFLLLLHPLWLVSANMEGDALHNLRTNLQDP---NNVLQSW--DPTLVNPCTWFHVT 620
Query: 70 CDQNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNI 129
C+ + V+ +++ L G L P++G L+ L+ L + NN+T +PSDL +LT+
Sbjct: 621 CNNDNSVIRVDLGNAALSGTLVPQLGQLKNLQYLELYSNNITGPIPSDLGNLTN------ 782
Query: 130 SHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPE 189
L +LD Y N F+GP+P+ + KL KL++L L N G IP
Sbjct: 783 -------------------LVSLDLYLNRFNGPIPDSLGKLSKLRFLRLNNNSLMGPIPM 905
Query: 190 SYSEFQSLEFLGLNANSLTGRVPES 214
S + +L+ L L+ N L+G VP++
Sbjct: 906 SLTNISALQVLDLSNNQLSGVVPDN 980
Score = 78.6 bits (192), Expect = 1e-14
Identities = 39/111 (35%), Positives = 61/111 (54%)
Frame = +3
Query: 249 LEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEI 308
+++ N L+G + P LG L L L + NN+TG IP +L ++ +L+SLDL +N G I
Sbjct: 648 VDLGNAALSGTLVPQLGQLKNLQYLELYSNNITGPIPSDLGNLTNLVSLDLYLNRFNGPI 827
Query: 309 PESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHN 359
P+S KL L + N G +P + ++ L+ L + N S V+P N
Sbjct: 828 PDSLGKLSKLRFLRLNNNSLMGPIPMSLTNISALQVLDLSNNQLSGVVPDN 980
Score = 74.7 bits (182), Expect = 2e-13
Identities = 39/103 (37%), Positives = 57/103 (54%)
Frame = +3
Query: 231 AYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSS 290
A G + P G ++NL+ LE+ + N+TG IP LGNLT L SL + +N G IP L
Sbjct: 666 ALSGTLVPQLGQLKNLQYLELYSNNITGPIPSDLGNLTNLVSLDLYLNRFNGPIPDSLGK 845
Query: 291 MMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLP 333
+ L L L+ N L G IP S + + L +++ N+ G +P
Sbjct: 846 LSKLRFLRLNNNSLMGPIPMSLTNISALQVLDLSNNQLSGVVP 974
Score = 73.9 bits (180), Expect = 3e-13
Identities = 47/119 (39%), Positives = 68/119 (56%), Gaps = 2/119 (1%)
Frame = +3
Query: 206 SLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLG 265
+L+G + L +LK L+ L L YSN G IP G++ NL L++ G IP SLG
Sbjct: 666 ALSGTLVPQLGQLKNLQYLEL-YSNNITGPIPSDLGNLTNLVSLDLYLNRFNGPIPDSLG 842
Query: 266 NLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPE--SFSKLKNLTLMN 322
L+KL L + N+L G IP L+++ +L LDLS N L+G +P+ SFS ++ N
Sbjct: 843 KLSKLRFLRLNNNSLMGPIPMSLTNISALQVLDLSNNQLSGVVPDNGSFSLFTPISFAN 1019
Score = 72.4 bits (176), Expect = 8e-13
Identities = 38/102 (37%), Positives = 58/102 (56%)
Frame = +3
Query: 184 SGTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSM 243
SGT+ + ++L++L L +N++TG +P L L L L L Y N + G IP + G +
Sbjct: 672 SGTLVPQLGQLKNLQYLELYSNNITGPIPSDLGNLTNLVSLDL-YLNRFNGPIPDSLGKL 848
Query: 244 ENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIP 285
LR L + N +L G IP SL N++ L L + N L+G +P
Sbjct: 849 SKLRFLRLNNNSLMGPIPMSLTNISALQVLDLSNNQLSGVVP 974
Score = 69.3 bits (168), Expect = 7e-12
Identities = 43/137 (31%), Positives = 68/137 (49%), Gaps = 1/137 (0%)
Frame = +3
Query: 293 SLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNF 352
S++ +DL L+G + +LKNL + + N G +PS +G+L NL +L ++ N F
Sbjct: 636 SVIRVDLGNAALSGTLVPQLGQLKNLQYLELYSNNITGPIPSDLGNLTNLVSLDLYLNRF 815
Query: 353 SFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECR 412
+ +P +LG + + + N L G IP L L+ +++N G +P G
Sbjct: 816 NGPIPDSLGKLSKLRFLRLNNNSLMGPIPMSLTNISALQVLDLSNNQLSGVVPDN-GSFS 992
Query: 413 SLTKIRVANNF-LDGPV 428
T I ANN L GPV
Sbjct: 993 LFTPISFANNLNLCGPV 1043
Score = 68.2 bits (165), Expect = 2e-11
Identities = 39/118 (33%), Positives = 62/118 (52%)
Frame = +3
Query: 353 SFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECR 412
S L LG Y ++ N++TG IP DL L + + N F GPIP +G+
Sbjct: 672 SGTLVPQLGQLKNLQYLELYSNNITGPIPSDLGNLTNLVSLDLYLNRFNGPIPDSLGKLS 851
Query: 413 SLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVISGESLGTLTLSNNL 470
L +R+ NN L GP+P + + ++ + +LSNN+L+G +P S ++ +NNL
Sbjct: 852 KLRFLRLNNNSLMGPIPMSLTNISALQVLDLSNNQLSGVVPDNGSFSLFTPISFANNL 1025
Score = 67.8 bits (164), Expect = 2e-11
Identities = 32/102 (31%), Positives = 56/102 (54%)
Frame = +3
Query: 280 LTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDL 339
L+GT+ P+L + +L L+L N++TG IP L NL ++ + N+F G +P +G L
Sbjct: 669 LSGTLVPQLGQLKNLQYLELYSNNITGPIPSDLGNLTNLVSLDLYLNRFNGPIPDSLGKL 848
Query: 340 PNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIP 381
L L++ N+ +P +L D++ N L+G++P
Sbjct: 849 SKLRFLRLNNNSLMGPIPMSLTNISALQVLDLSNNQLSGVVP 974
Score = 57.0 bits (136), Expect = 3e-08
Identities = 42/156 (26%), Positives = 68/156 (42%), Gaps = 2/156 (1%)
Frame = +3
Query: 370 DVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVP 429
D+ L+G + P L + L+ + N GPIP +G +L + + N +GP+P
Sbjct: 651 DLGNAALSGTLVPQLGQLKNLQYLELYSNNITGPIPSDLGNLTNLVSLDLYLNRFNGPIP 830
Query: 430 PGVFQLPSVTITELSNNRLNGELPSVISGESLGTLTLSNNLFTGKIPAAMKNLRALQSLS 489
+ +L + L+NN L G IP ++ N+ ALQ L
Sbjct: 831 DSLGKLSKLRFLRLNNNSL-----------------------MGPIPMSLTNISALQVLD 941
Query: 490 LDANEFIGEIP-GGVFEIPMLTKVNISGN-NLTGPI 523
L N+ G +P G F + T ++ + N NL GP+
Sbjct: 942 LSNNQLSGVVPDNGSFS--LFTPISFANNLNLCGPV 1043
>TC80900 weakly similar to PIR|G84652|G84652 probable receptor-like protein
kinase [imported] - Arabidopsis thaliana, partial (24%)
Length = 1054
Score = 237 bits (604), Expect = 2e-62
Identities = 129/350 (36%), Positives = 203/350 (57%), Gaps = 1/350 (0%)
Frame = +2
Query: 218 LKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQM 277
+K LK ++LGY+N IP G++ +L L + NLTG IP SLGNLT L LF+ +
Sbjct: 11 MKRLKWIYLGYNNLSRE-IPKNIGNLVSLNHLNLVYNNLTGPIPESLGNLTNLQYLFLYL 187
Query: 278 NNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIG 337
N LTG IP + ++ +L+SLDLS N L+GEI L+ L +++ F N F G +P+ I
Sbjct: 188 NKLTGPIPKSIFNLKNLISLDLSDNYLSGEISNLVVNLQKLEILHLFSNNFTGKIPNTIT 367
Query: 338 DLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITD 397
LP+L+ LQ+W N + +P LG + D++ N+LTG IP LC S L I+
Sbjct: 368 SLPHLQVLQLWSNKLTGEIPQTLGIHNNLTILDLSSNNLTGKIPNSLCASKNLHKIILFS 547
Query: 398 NFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGEL-PSVI 456
N +G IPKG+ C++L ++R+ +N L G +P + QLP + + ++S N+ +G++
Sbjct: 548 NSLKGEIPKGLTSCKTLERVRLQDNNLSGKLPLEITQLPQIYLLDISGNKFSGKINDRKW 727
Query: 457 SGESLGTLTLSNNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISG 516
+ SL L L+NN F+G +P + ++ L L N+F G I G +P L ++ ++
Sbjct: 728 NMPSLQMLNLANNNFSGDLPNSFGG-NKVEGLDLSQNQFSGYIQIGFKNLPELVQLKLNN 904
Query: 517 NNLTGPIPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNE 566
NNL G P + L ++DLS N L GE+P+ + + L +L++S N+
Sbjct: 905 NNLFGKFPEELFQCNKLVSLDLSHNRLNGEIPEKLAKMPVLGLLDISENQ 1054
Score = 180 bits (456), Expect = 3e-45
Identities = 117/374 (31%), Positives = 183/374 (48%)
Frame = +2
Query: 168 VKLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLG 227
V +++LK+++L N S IP++ SL L L N+LTG +PESL L L+ L L
Sbjct: 5 VLMKRLKWIYLGYNNLSREIPKNIGNLVSLNHLNLVYNNLTGPIPESLGNLTNLQYLFL- 181
Query: 228 YSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPE 287
Y N G IP + +++NL L++++ L+GEI + NL KL L + NN TG IP
Sbjct: 182 YLNKLTGPIPKSIFNLKNLISLDLSDNYLSGEISNLVVNLQKLEILHLFSNNFTGKIPNT 361
Query: 288 LSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQV 347
++S+ L L L N LTGEIP++ NLT+++ N G +P+ + NL + +
Sbjct: 362 ITSLPHLQVLQLWSNKLTGEIPQTLGIHNNLTILDLSSNNLTGKIPNSLCASKNLHKIIL 541
Query: 348 WENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKG 407
+ N+ L G IP L L+ + DN G +P
Sbjct: 542 FSNS------------------------LKGEIPKGLTSCKTLERVRLQDNNLSGKLPLE 649
Query: 408 IGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVISGESLGTLTLS 467
I + + + ++ N G + + +PS+ + L+NN +G+LP+ G + L LS
Sbjct: 650 ITQLPQIYLLDISGNKFSGKINDRKWNMPSLQMLNLANNNFSGDLPNSFGGNKVEGLDLS 829
Query: 468 NNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTI 527
N F+G I KNL L L L+ N G+ P +F+ L +++S N L G IP +
Sbjct: 830 QNQFSGYIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHNRLNGEIPEKL 1009
Query: 528 THRASLTAVDLSRN 541
L +D+S N
Sbjct: 1010AKMPVLGLLDISEN 1051
Score = 177 bits (450), Expect = 1e-44
Identities = 114/355 (32%), Positives = 177/355 (49%), Gaps = 1/355 (0%)
Frame = +2
Query: 243 MENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSIN 302
M+ L+ + + NL+ EIP ++GNL L+ L + NNLTG IP L ++ +L L L +N
Sbjct: 11 MKRLKWIYLGYNNLSREIPKNIGNLVSLNHLNLVYNNLTGPIPESLGNLTNLQYLFLYLN 190
Query: 303 DLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGG 362
LTG IP+S LKNL ++ N G + + + +L LE L ++ NNF+ +P+ +
Sbjct: 191 KLTGPIPKSIFNLKNLISLDLSDNYLSGEISNLVVNLQKLEILHLFSNNFTGKIPNTITS 370
Query: 363 NGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANN 422
+ N LTG IP L L ++ N G IP + ++L KI + +N
Sbjct: 371 LPHLQVLQLWSNKLTGEIPQTLGIHNNLTILDLSSNNLTGKIPNSLCASKNLHKIILFSN 550
Query: 423 FLDGPVPPGVFQLPSVTITELSNNRLNGELPSVISG-ESLGTLTLSNNLFTGKIPAAMKN 481
L G +P G+ ++ L +N L+G+LP I+ + L +S N F+GKI N
Sbjct: 551 SLKGEIPKGLTSCKTLERVRLQDNNLSGKLPLEITQLPQIYLLDISGNKFSGKINDRKWN 730
Query: 482 LRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSRN 541
+ +LQ L+L N F G++P + GN + G +DLS+N
Sbjct: 731 MPSLQMLNLANNNFSGDLPN-----------SFGGNKVEG--------------LDLSQN 835
Query: 542 NLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVP 596
+G + G KNL +L L L+ N + G P+E+ L +LDLS N G +P
Sbjct: 836 QFSGYIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHNRLNGEIP 1000
Score = 167 bits (423), Expect = 2e-41
Identities = 114/375 (30%), Positives = 185/375 (48%), Gaps = 1/375 (0%)
Frame = +2
Query: 96 LLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLFSGQFPGNITVGMTELEALDAY 155
L+++L+ + + NNL+ ++P ++ +L SL LN+ +N
Sbjct: 8 LMKRLKWIYLGYNNLSREIPKNIGNLVSLNHLNLVYN----------------------- 118
Query: 156 DNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESL 215
+ +GP+PE + L L+YL L N +G IP+S ++L L L+ N L+G + +
Sbjct: 119 --NLTGPIPESLGNLTNLQYLFLYLNKLTGPIPKSIFNLKNLISLDLSDNYLSGEISNLV 292
Query: 216 AKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFV 275
L+ L+ LHL +SN + G IP S+ +L++L++ + LTGEIP +LG L L +
Sbjct: 293 VNLQKLEILHL-FSNNFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGIHNNLTILDL 469
Query: 276 QMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSF 335
NNLTG IP L + +L + L N L GEIP+ + K L + N G LP
Sbjct: 470 SSNNLTGKIPNSLCASKNLHKIILFSNSLKGEIPKGLTSCKTLERVRLQDNNLSGKLPLE 649
Query: 336 IGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFII 395
I LP + L + N FS + ++ N+ +G +P +++ +
Sbjct: 650 ITQLPQIYLLDISGNKFSGKINDRKWNMPSLQMLNLANNNFSGDLPNSF-GGNKVEGLDL 826
Query: 396 TDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSV 455
+ N F G I G L ++++ NN L G P +FQ + +LS+NRLNGE+P
Sbjct: 827 SQNQFSGYIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHNRLNGEIPEK 1006
Query: 456 ISG-ESLGTLTLSNN 469
++ LG L +S N
Sbjct: 1007LAKMPVLGLLDISEN 1051
Score = 162 bits (409), Expect = 8e-40
Identities = 103/322 (31%), Positives = 172/322 (52%), Gaps = 2/322 (0%)
Frame = +2
Query: 79 LNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLFSGQF 138
LN+ L G +P +G L L+ L + +N LT +P + +L +L L++S N SG+
Sbjct: 101 LNLVYNNLTGPIPESLGNLTNLQYLFLYLNKLTGPIPKSIFNLKNLISLDLSDNYLSGEI 280
Query: 139 PGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLE 198
N+ V + +LE L + N+F+G +P I L L+ L L N +G IP++ +L
Sbjct: 281 -SNLVVNLQKLEILHLFSNNFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGIHNNLT 457
Query: 199 FLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTG 258
L L++N+LTG++P SL K L ++ L +SN+ +G IP S + L + + + NL+G
Sbjct: 458 ILDLSSNNLTGKIPNSLCASKNLHKIIL-FSNSLKGEIPKGLTSCKTLERVRLQDNNLSG 634
Query: 259 EIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESF--SKLK 316
++P + L +++ L + N +G I +M SL L+L+ N+ +G++P SF +K++
Sbjct: 635 KLPLEITQLPQIYLLDISGNKFSGKINDRKWNMPSLQMLNLANNNFSGDLPNSFGGNKVE 814
Query: 317 NLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHL 376
L L QN+F G + +LP L L++ NN P L + + D++ N L
Sbjct: 815 GLDLS---QNQFSGYIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHNRL 985
Query: 377 TGLIPPDLCKSGRLKTFIITDN 398
G IP L K L I++N
Sbjct: 986 NGEIPEKLAKMPVLGLLDISEN 1051
Score = 142 bits (357), Expect = 8e-34
Identities = 93/309 (30%), Positives = 149/309 (48%), Gaps = 1/309 (0%)
Frame = +2
Query: 291 MMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWEN 350
M L + L N+L+ EIP++ L +L +N N G +P +G+L NL+ L
Sbjct: 11 MKRLKWIYLGYNNLSREIPKNIGNLVSLNHLNLVYNNLTGPIPESLGNLTNLQYL----- 175
Query: 351 NFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGE 410
FLY N LTG IP + L + ++DN+ G I +
Sbjct: 176 ---------------FLYL----NKLTGPIPKSIFNLKNLISLDLSDNYLSGEISNLVVN 298
Query: 411 CRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVIS-GESLGTLTLSNN 469
+ L + + +N G +P + LP + + +L +N+L GE+P + +L L LS+N
Sbjct: 299 LQKLEILHLFSNNFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGIHNNLTILDLSSN 478
Query: 470 LFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITH 529
TGKIP ++ + L + L +N GEIP G+ L +V + NNL+G +P IT
Sbjct: 479 NLTGKIPNSLCASKNLHKIILFSNSLKGEIPKGLTSCKTLERVRLQDNNLSGKLPLEITQ 658
Query: 530 RASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSN 589
+ +D+S N +G++ N+ L +LNL+ N SG +P+ + LDLS N
Sbjct: 659 LPQIYLLDISGNKFSGKINDRKWNMPSLQMLNLANNNFSGDLPNSFG-GNKVEGLDLSQN 835
Query: 590 NFTGTVPTG 598
F+G + G
Sbjct: 836 QFSGYIQIG 862
Score = 35.0 bits (79), Expect = 0.14
Identities = 23/83 (27%), Positives = 40/83 (47%)
Frame = +2
Query: 75 RVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLF 134
+V L+++ G++ L +L L ++ NNL + P +L L L++SHN
Sbjct: 806 KVEGLDLSQNQFSGYIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHNRL 985
Query: 135 SGQFPGNITVGMTELEALDAYDN 157
+G+ P + M L LD +N
Sbjct: 986 NGEIPEKL-AKMPVLGLLDISEN 1051
>TC90106 similar to GP|3641252|gb|AAC36318.1| leucine-rich receptor-like
protein kinase {Malus x domestica}, partial (25%)
Length = 1241
Score = 234 bits (598), Expect = 9e-62
Identities = 130/228 (57%), Positives = 157/228 (68%), Gaps = 2/228 (0%)
Frame = +1
Query: 745 GKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVEAARG 804
GKI H+NI++L + +D LL+YEYM NGSLG+ LH +KGG L W RYKIAV+AA G
Sbjct: 10 GKIIHKNIVKLWCCCTTRDCQLLVYEYMQNGSLGDLLHSSKGGLLDWPTRYKIAVDAADG 189
Query: 805 LCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGAS-QSMSSIAGSYGY 863
L Y+HHDC P I+HRDVKSNNILLD DF A VADFGLAK + +SMS IAGS GY
Sbjct: 190 LSYLHHDCVPPIVHRDVKSNNILLDGDFGARVADFGLAKVVETTAKGIKSMSIIAGSCGY 369
Query: 864 IAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVG-EFGDGVDIVGWVNKTMSELSQPSD 922
IAPEYAYTLKV+EKSD+YSFGVV+LEL+ GR+PV EFG+ D+V WV T+ D
Sbjct: 370 IAPEYAYTLKVNEKSDIYSFGVVILELVTGRRPVDPEFGE-KDLVKWVCTTL-------D 525
Query: 923 TALVLAVVDPRLSGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 970
V V+D RL + +FNI +MC + RP+MR VV ML
Sbjct: 526 QKGVDHVLDSRLDSCFKEEICKVFNIGLMCTSPLPINRPSMRRVVKML 669
>TC83631 similar to PIR|T10659|T10659 probable serine/threonine-specific
protein kinase (EC 2.7.1.-) T5F17.100 - Arabidopsis
thaliana, partial (24%)
Length = 975
Score = 227 bits (579), Expect = 1e-59
Identities = 117/247 (47%), Positives = 169/247 (68%), Gaps = 5/247 (2%)
Frame = +1
Query: 729 GSGRNDYGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGH 788
GSG + G E+ LG++RHRNI+RLLG++ N +++YE+M NG+LG+ +HG +
Sbjct: 22 GSGDDLVG---EVNLLGRLRHRNIVRLLGFLYNDTDVMIVYEFMVNGNLGDAMHGKQSER 192
Query: 789 LR--WEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLY 846
L W RY IA+ A+GL Y+HHDC P +IHRD+KSNNILLDA+ EA +ADFGLAK +
Sbjct: 193 LLVDWVSRYNIALGIAQGLAYLHHDCHPPVIHRDIKSNNILLDANLEARIADFGLAKMMV 372
Query: 847 DPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVG-EFGDGVD 905
++++S IAGSYGYIAPEY Y+LKVDEK D+YSFG+VLLELI G++P+ +FG+ VD
Sbjct: 373 --RKNETVSMIAGSYGYIAPEYGYSLKVDEKIDIYSFGIVLLELITGKRPIDPDFGESVD 546
Query: 906 IVGWVNKTMSELSQPSDTALVLAVVDPRLSG--YPLTSVIHMFNIAMMCVKEMGPARPTM 963
IVGW+ + + + S +DP + + ++ + IA++C ++ RP+M
Sbjct: 547 IVGWIRRKIDKNSPEE-------ALDPSVGNCKHVQEEMLLVLRIALLCTAKLPKERPSM 705
Query: 964 REVVHML 970
R+V+ ML
Sbjct: 706 RDVIMML 726
>BQ123304 similar to GP|22093756|d putative receptor protein kinase {Oryza
sativa (japonica cultivar-group)}, partial (19%)
Length = 979
Score = 226 bits (577), Expect = 3e-59
Identities = 126/294 (42%), Positives = 179/294 (60%), Gaps = 13/294 (4%)
Frame = +2
Query: 690 DVVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRH 749
D L +E IIG GG+G VYR +P G VA+K++ + F E++TLG+I+H
Sbjct: 14 DATNNLSDEFIIGSGGSGTVYRVELPTGETVAVKKISLKDEYLLHKSFIREVKTLGRIKH 193
Query: 750 RNIMRLLGYVSNKDT----NLLLYEYMPNGSLGEWLHGAK---GGHLRWEMRYKIAVEAA 802
R++++L+G SN+ NLL+YE+M NGS+ +WLHG L W+ R+KIA+ A
Sbjct: 194 RHLVKLVGCCSNRHKGNGCNLLIYEFMENGSVWDWLHGNALKLRRSLDWDTRFKIALGLA 373
Query: 803 RGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYD--PGASQSMSSIAGS 860
+G+ Y+HHDC P IIHRD+KS+NILLD++ +AH+ DFGLAK + + ++S S AGS
Sbjct: 374 QGMEYLHHDCVPKIIHRDIKSSNILLDSNMDAHLGDFGLAKAIVENLDSNTESTSCFAGS 553
Query: 861 YGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPV-GEFGDGVDIVGWVNKTMSELSQ 919
YGYIAPE+ Y+LK EKSDVYS GVVL+EL+ G+ P F VD+V WV ++
Sbjct: 554 YGYIAPEFGYSLKATEKSDVYSMGVVLMELVSGKLPTDAAFRGSVDMVRWVEMRINMKGT 733
Query: 920 PSDTALVLAVVDPRLS---GYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 970
+ +VDP L Y + + IA+ C K RP+ R+V +L
Sbjct: 734 ERE-----ELVDPELKPLLPYEEFAAFQVLEIAIQCTKTTPMKRPSSRQVCDLL 880
>TC77660 similar to GP|22212524|dbj|BAC07504. receptor-like protein kinase
{Nicotiana tabacum}, partial (69%)
Length = 1886
Score = 225 bits (574), Expect = 6e-59
Identities = 154/496 (31%), Positives = 250/496 (50%), Gaps = 44/496 (8%)
Frame = +2
Query: 464 LTLSNNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPI 523
L + +G + + L L+ +SL N+ + +IP + + L +N++ N +G +
Sbjct: 308 LVFKSRKLSGILSPTIGKLTELKEISLSDNKLVDQIPTSIVDCRKLEFLNLANNLFSGEV 487
Query: 524 PTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTT 583
P+ + L +D+S N L+G + ++ +L L+++ N +G VP +R +L
Sbjct: 488 PSEFSSLIRLRFLDISGNKLSGNL-NFLRYFPNLETLSVADNHFTGRVPVSVRSFRNLRH 664
Query: 584 LDLSSNNFTGTVPTGGQFLVFN---------YDKTFAGNPNLCFPHRASCPSVL-----Y 629
+ S N F VP + L + Y N + PHR+ P
Sbjct: 665 FNFSGNRFLEGVPLNQKLLGYEDTDNTAPKRYILAETNNSSQTRPHRSHSPGAAPAPAPA 844
Query: 630 DSLRKTRAKTARVRAIVIG-IALATAVLLVAVTVHVVRKRRL----HRAQAWKLTAFQRL 684
L K + ++ ++G +A A A +L ++ K L + + + L
Sbjct: 845 APLHKHKKSRRKLAGWILGFVAGAFAGILSGFVFSLLFKLALILIKGKGKGSGPAIYSSL 1024
Query: 685 EIKAEDVVECLKEENI-----IGKGGAGIVYRGSMP--NGTDVAIKRLVGQGSGRNDYG- 736
KAED+ KE+ + IG+GG VY+ +P NG +AIK+++ +
Sbjct: 1025IKKAEDLAFLEKEDGLASLEKIGQGGCVEVYKAELPGSNGKMIAIKKIIQPPKDAAELAE 1204
Query: 737 ------------FRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGA 784
++EI+T+G+IRHRN++ LL ++S D + L+YE+M NGSL + LH
Sbjct: 1205EDSKLLHKKMRQIKSEIDTVGQIRHRNLLPLLAHISRPDCHYLVYEFMKNGSLQDMLHKV 1384
Query: 785 KGGH--LRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLA 842
+ G L W R+KIA+ A GL Y+H SP IIHRD+K N+LLD + EA +ADFGLA
Sbjct: 1385ERGEAELDWLARHKIALGIAAGLEYLHTSHSPRIIHRDLKPANVLLDDEMEARIADFGLA 1564
Query: 843 KFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEF-- 900
K + D + S++AG+ GYIAPEY LK ++K D+YSFGV+L L+IG+ P +F
Sbjct: 1565KAMPDAQTHITTSNVAGTVGYIAPEYHQILKFNDKCDIYSFGVMLGVLVIGKLPSDDFFT 1744
Query: 901 -GDGVDIVGWVNKTMS 915
D + +V W+ M+
Sbjct: 1745NTDEMSLVKWMRNVMT 1792
Score = 79.3 bits (194), Expect = 7e-15
Identities = 52/164 (31%), Positives = 81/164 (48%), Gaps = 8/164 (4%)
Frame = +2
Query: 57 TSLSAHCSFSGVTCDQNL--------RVVALNVTLVPLFGHLPPEIGLLEKLENLTISMN 108
T+ + C+ GV C++ L RV L L G L P IG L +L+ +++S N
Sbjct: 218 TTTNNLCNKEGVFCERRLTNNESYALRVTKLVFKSRKLSGILSPTIGKLTELKEISLSDN 397
Query: 109 NLTDQLPSDLASLTSLKVLNISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIV 168
L DQ+P+ + L+ LN+++NLFSG+ P + + L LD N SG L +
Sbjct: 398 KLVDQIPTSIVDCRKLEFLNLANNLFSGEVPSEFS-SLIRLRFLDISGNKLSGNL-NFLR 571
Query: 169 KLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSLTGRVP 212
L+ L +A N+F+G +P S F++L + N VP
Sbjct: 572 YFPNLETLSVADNHFTGRVPVSVRSFRNLRHFNFSGNRFLEGVP 703
Score = 77.0 bits (188), Expect = 3e-14
Identities = 39/125 (31%), Positives = 69/125 (55%)
Frame = +2
Query: 376 LTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQL 435
L+G++ P + K LK ++DN IP I +CR L + +ANN G VP L
Sbjct: 329 LSGILSPTIGKLTELKEISLSDNKLVDQIPTSIVDCRKLEFLNLANNLFSGEVPSEFSSL 508
Query: 436 PSVTITELSNNRLNGELPSVISGESLGTLTLSNNLFTGKIPAAMKNLRALQSLSLDANEF 495
+ ++S N+L+G L + +L TL++++N FTG++P ++++ R L+ + N F
Sbjct: 509 IRLRFLDISGNKLSGNLNFLRYFPNLETLSVADNHFTGRVPVSVRSFRNLRHFNFSGNRF 688
Query: 496 IGEIP 500
+ +P
Sbjct: 689 LEGVP 703
Score = 68.2 bits (165), Expect = 2e-11
Identities = 40/126 (31%), Positives = 67/126 (52%)
Frame = +2
Query: 256 LTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKL 315
L+G + P++G LT+L + + N L IP + L L+L+ N +GE+P FS L
Sbjct: 329 LSGILSPTIGKLTELKEISLSDNKLVDQIPTSIVDCRKLEFLNLANNLFSGEVPSEFSSL 508
Query: 316 KNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNH 375
L ++ NK G+L +F+ PNLETL V +N+F+ +P ++ +F+ + N
Sbjct: 509 IRLRFLDISGNKLSGNL-NFLRYFPNLETLSVADNHFTGRVPVSVRSFRNLRHFNFSGNR 685
Query: 376 LTGLIP 381
+P
Sbjct: 686 FLEGVP 703
Score = 62.0 bits (149), Expect = 1e-09
Identities = 37/106 (34%), Positives = 56/106 (51%), Gaps = 1/106 (0%)
Frame = +2
Query: 143 TVG-MTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLG 201
T+G +TEL+ + DN +P IV KL++L+LA N FSG +P +S L FL
Sbjct: 350 TIGKLTELKEISLSDNKLVDQIPTSIVDCRKLEFLNLANNLFSGEVPSEFSSLIRLRFLD 529
Query: 202 LNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLR 247
++ N L+G + L L+ L + N + G +P + S NLR
Sbjct: 530 ISGNKLSGNL-NFLRYFPNLETLSVA-DNHFTGRVPVSVRSFRNLR 661
Score = 51.2 bits (121), Expect = 2e-06
Identities = 36/137 (26%), Positives = 60/137 (43%), Gaps = 1/137 (0%)
Frame = +2
Query: 389 RLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRL 448
R+ + G + IG+ L +I +++N L +P + + L+NN
Sbjct: 296 RVTKLVFKSRKLSGILSPTIGKLTELKEISLSDNKLVDQIPTSIVDCRKLEFLNLANNLF 475
Query: 449 NGELPSVISG-ESLGTLTLSNNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFEIP 507
+GE+PS S L L +S N +G + ++ L++LS+ N F G +P V
Sbjct: 476 SGEVPSEFSSLIRLRFLDISGNKLSGNL-NFLRYFPNLETLSVADNHFTGRVPVSVRSFR 652
Query: 508 MLTKVNISGNNLTGPIP 524
L N SGN +P
Sbjct: 653 NLRHFNFSGNRFLEGVP 703
>TC88105 similar to GP|21593085|gb|AAM65034.1 Putative protein kinase
{Arabidopsis thaliana}, partial (72%)
Length = 2379
Score = 223 bits (568), Expect = 3e-58
Identities = 121/279 (43%), Positives = 173/279 (61%), Gaps = 5/279 (1%)
Frame = +2
Query: 697 EENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLL 756
++NIIG+GG G+VY+G + NG VAIK+L+ G+ + FR E+E +G +RH+N++RLL
Sbjct: 731 KDNIIGEGGYGVVYQGQLINGNPVAIKKLLNN-LGQAEKEFRVEVEAIGHVRHKNLVRLL 907
Query: 757 GYVSNKDTNLLLYEYMPNGSLGEWLHGA--KGGHLRWEMRYKIAVEAARGLCYMHHDCSP 814
G+ LL+YEY+ NG+L +WLHGA + G+L W+ R KI + A+ L Y+H P
Sbjct: 908 GFCIEGTHRLLIYEYVNNGNLEQWLHGAMRQYGYLTWDARIKILLGTAKALAYLHEAIEP 1087
Query: 815 LIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKV 874
++HRD+KS+NIL+D DF A ++DFGLAK L G S + + G++GY+APEYA + +
Sbjct: 1088 KVVHRDIKSSNILIDDDFNAKISDFGLAKLL-GAGKSHITTRVMGTFGYVAPEYANSGLL 1264
Query: 875 DEKSDVYSFGVVLLELIIGRKPV--GEFGDGVDIVGWVNKTMSELSQPSDTALVLAVVDP 932
+EKSDVYSFGV+LLE I GR PV V++V W+ + VVDP
Sbjct: 1265 NEKSDVYSFGVLLLEAITGRDPVDYNRSAAEVNLVDWLKMMVGNRHAEE-------VVDP 1423
Query: 933 RLSGYPLTSVI-HMFNIAMMCVKEMGPARPTMREVVHML 970
+ P TS + + A+ CV RP M +VV ML
Sbjct: 1424 NIETRPSTSALKRVLLTALRCVDPDSEKRPKMSQVVRML 1540
>TC92229 similar to PIR|B86369|B86369 hypothetical protein AAC98010.1
[imported] - Arabidopsis thaliana, partial (44%)
Length = 1347
Score = 222 bits (566), Expect = 5e-58
Identities = 120/290 (41%), Positives = 178/290 (61%), Gaps = 4/290 (1%)
Frame = +2
Query: 690 DVVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRH 749
++ EN+IG+GG G VY+ MP+G A+K L+ GSG+ + FRAE++T+ ++ H
Sbjct: 179 EITNGFSSENVIGEGGFGRVYKALMPDGRVGALK-LLKAGSGQGEREFRAEVDTISRVHH 355
Query: 750 RNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVEAARGLCYMH 809
R+++ L+GY + +L+YE++PNG+L + LH ++ L W R KIA+ AARGL Y+H
Sbjct: 356 RHLVSLIGYCIAEQQRVLIYEFVPNGNLDQHLHESQWNVLDWPKRMKIAIGAARGLAYLH 535
Query: 810 HDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYA 869
C+P IIHRD+KS+NILLD +EA VADFGLA+ D S + + G++GY+APEYA
Sbjct: 536 EGCNPKIIHRDIKSSNILLDDSYEAQVADFGLARLTDDTNTHVS-TRVMGTFGYMAPEYA 712
Query: 870 YTLKVDEKSDVYSFGVVLLELIIGRKPVG---EFGDGVDIVGWVNKTMSELSQPSDTALV 926
+ K+ ++SDV+SFGVVLLEL+ GRKPV GD +V W + + D +
Sbjct: 713 TSGKLTDRSDVFSFGVVLLELVTGRKPVDPTQPVGD-ESLVEWARPILLRAIETGDFS-- 883
Query: 927 LAVVDPRL-SGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHMLTNPPQ 975
+ DPRL Y + + M A C++ P RP M ++ L + Q
Sbjct: 884 -ELADPRLHRQYIDSEMFRMIEAAAACIRHSAPKRPRMVQIARALDSGDQ 1030
>AJ497482 homologue to PIR|T50850|T50 receptor protein kinase homolog
[imported] - soybean, partial (11%)
Length = 583
Score = 211 bits (536), Expect = 1e-54
Identities = 106/121 (87%), Positives = 112/121 (91%)
Frame = +3
Query: 866 PEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNKTMSELSQPSDTAL 925
PEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVDIVGW+NKT EL QPSD AL
Sbjct: 3 PEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVDIVGWINKTELELYQPSDKAL 182
Query: 926 VLAVVDPRLSGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHMLTNPPQSNTSTQDLIN 985
V AVVDPRL+GYPLTSVI+MFNIAMMCVKEMGPARPTMREVVHMLTNPP S ++ +LIN
Sbjct: 183 VSAVVDPRLNGYPLTSVIYMFNIAMMCVKEMGPARPTMREVVHMLTNPPHS--TSHNLIN 356
Query: 986 L 986
L
Sbjct: 357 L 359
>TC82239 weakly similar to GP|16924042|gb|AAL31654.1 Putative protein kinase
{Oryza sativa}, partial (14%)
Length = 1476
Score = 207 bits (527), Expect = 2e-53
Identities = 142/425 (33%), Positives = 213/425 (49%), Gaps = 21/425 (4%)
Frame = +2
Query: 567 ISGPVPDEIRFMTSLTTLDLSSNNFTGTVPTGGQF-LVFNYDKTFAGNPNLCFPHRASCP 625
+SG VP++I M SL++L+LS N+ G VP G F L ++ + N +LC + P
Sbjct: 5 LSGKVPNQISGMLSLSSLNLSYNHLEGNVPKSGIFKLNSSHALDLSNNQDLCGSFKGLIP 184
Query: 626 SVLYDSLRKTRAKTARVRAIVIGIALATAVLLVAVTVHVV-----RKRRLHRAQAWKLTA 680
+ S + I I +L A+ L V V V+ +K R R ++K+
Sbjct: 185 CNVSSSEPSDGGSNKKKVVIPIVASLGGALFLSLVIVGVILLCYKKKSRTLRKSSFKMPN 364
Query: 681 -----FQRLEIKAEDVVECLKE---ENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGR 732
+ + D++E + IG+G G VY+ + G A+K+L
Sbjct: 365 PFSIWYFNGRVVYSDIIEATNNFDNKYCIGEGAFGNVYKAELKGGQIFAVKKLKCDEENL 544
Query: 733 NDYG---FRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGG-H 788
+ F +E+E + + RHRNI++L G+ L+YEYM GSL + L K
Sbjct: 545 DTESIKTFESEVEAMTETRHRNIVKLYGFCCEGMHTFLVYEYMDRGSLEDMLIDDKRALE 724
Query: 789 LRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDP 848
L W R++I A L YMHHDCSP +IHRD+ S N+LL + EAHV+DFG A+FL
Sbjct: 725 LDWSKRFEIVKGVASALSYMHHDCSPALIHRDISSKNVLLSKNLEAHVSDFGTARFL--K 898
Query: 849 GASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVDIVG 908
S +S AG+YGY APE AYT+ V EK DV+SFGV+ E++ G+ P D+V
Sbjct: 899 PNSPIWTSFAGTYGYAAPELAYTMAVTEKCDVFSFGVLAFEILTGKHP-------SDLVS 1057
Query: 909 WVNKTMSELSQPSDTALVLAVVDPRLSGYP---LTSVIHMFNIAMMCVKEMGPARPTMRE 965
++ + + + ++DPRL P L + + N+A+ C+ +RPTMR
Sbjct: 1058YIQTSNDQKIDFKE------ILDPRLPSPPKNILKELALVANLALSCLHTHPQSRPTMRS 1219
Query: 966 VVHML 970
V L
Sbjct: 1220VAQFL 1234
>TC81789 similar to PIR|B86440|B86440 probable protein kinase [imported] -
Arabidopsis thaliana, partial (54%)
Length = 1189
Score = 127 bits (319), Expect(2) = 2e-53
Identities = 66/146 (45%), Positives = 95/146 (64%), Gaps = 3/146 (2%)
Frame = +2
Query: 684 LEIKAEDVV---ECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAE 740
L ++D++ E L EE+IIG GG G VY+ +M +G A+K++V G + + F E
Sbjct: 119 LPYSSKDIIKKLETLNEEHIIGVGGFGTVYKLAMDDGNVFALKKIVKLNEGFDRF-FERE 295
Query: 741 IETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVE 800
+ LG I+HR ++ L GY ++ + LL+Y+Y+P GSL E LH K L W+ R I +
Sbjct: 296 LAILGSIKHRYLVNLRGYCNSPTSKLLIYDYLPGGSLDEVLH-EKSEQLDWDSRLNIIMG 472
Query: 801 AARGLCYMHHDCSPLIIHRDVKSNNI 826
AA+GL Y+HHDCSP IIHRD+KS+NI
Sbjct: 473 AAKGLAYLHHDCSPRIIHRDIKSSNI 550
Score = 101 bits (252), Expect(2) = 2e-53
Identities = 57/146 (39%), Positives = 89/146 (60%), Gaps = 2/146 (1%)
Frame = +3
Query: 827 LLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVV 886
LLD +A V+DFGLAK L D S + +AG++GY+APEY + + EK+DVYSFGV+
Sbjct: 552 LLDGKLDARVSDFGLAKLLEDE-ESHITTIVAGTFGYLAPEYMQSGRATEKTDVYSFGVL 728
Query: 887 LLELIIGRKPVGE--FGDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRLSGYPLTSVIH 944
LE++ G++P G+++VGW+N ++E ++P + +VDP G + S+
Sbjct: 729 TLEVLSGKRPTDASFIEKGLNVVGWLNFLITE-NRPRE------IVDPLCDGVQVESLDA 887
Query: 945 MFNIAMMCVKEMGPARPTMREVVHML 970
+ ++A+ CV RPTM VV +L
Sbjct: 888 LLSMAIQCVSSNPEDRPTMHRVVQLL 965
>TC83862 similar to PIR|T05270|T05270 probable serine/threonine-specific
protein kinase (EC 2.7.1.-) T4L20.80 - Arabidopsis
thaliana, partial (60%)
Length = 948
Score = 206 bits (525), Expect = 3e-53
Identities = 112/287 (39%), Positives = 173/287 (60%), Gaps = 5/287 (1%)
Frame = +1
Query: 689 EDVVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIR 748
E+ + E ++IG+ G GIVYRG + +G+ VA+K L+ G+ + F+ E+E +GK+R
Sbjct: 31 ENATDGFAEGSVIGERGYGIVYRGILQDGSIVAVKNLLNN-KGQAEKEFKVEVEAIGKVR 207
Query: 749 HRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKG--GHLRWEMRYKIAVEAARGLC 806
H+N++ L+GY + +L+YEY+ NG+L +WLHG G L W++R KIAV A+GL
Sbjct: 208 HKNLVGLVGYCAEGAKRMLVYEYVDNGNLEQWLHGDVGPVSPLTWDIRMKIAVGTAKGLA 387
Query: 807 YMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAP 866
Y+H P ++HRDVKS+NILLD + A V+DFGLAK L G S + + G++GY++P
Sbjct: 388 YLHEGLEPKVVHRDVKSSNILLDKKWHAKVSDFGLAKLL-GSGKSYVTTRVMGTFGYVSP 564
Query: 867 EYAYTLKVDEKSDVYSFGVVLLELIIGRKPV--GEFGDGVDIVGWVNKTMSELSQPSDTA 924
EYA T ++E SDVYSFG++L+EL+ GR P+ +++V W ++
Sbjct: 565 EYASTGMLNEGSDVYSFGILLMELVTGRSPIDYSRAPAEMNLVDWFKGMVASRRGEE--- 735
Query: 925 LVLAVVDPRLSGYPL-TSVIHMFNIAMMCVKEMGPARPTMREVVHML 970
+VDP + P S+ + + C+ RP M ++VHML
Sbjct: 736 ----LVDPLIEIQPSPRSLKRALLVCLRCIDLDANKRPKMGQIVHML 864
>TC77448 SYMRK; MtSYMRK [Medicago truncatula]; nod region linked receptor
kinase [Medicago truncatula]
Length = 3568
Score = 205 bits (521), Expect = 8e-53
Identities = 152/477 (31%), Positives = 240/477 (49%), Gaps = 34/477 (7%)
Frame = +1
Query: 528 THRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTS--LTTLD 585
T + +T +DLS NNL G +P + + +L ILNLS N+ P F S L +LD
Sbjct: 1582 TGSSIITKLDLSSNNLKGAIPSIVTKMTNLQILNLSHNQFDMLFPS---FPPSSLLISLD 1752
Query: 586 LSSNNFTGTVPTGGQFLVFNYDKTFAGNPNLCFPHRASCPSVLYDS-LRKTRAKTARVRA 644
LS N+ +G +P L F NP++ S L ++ + +AK +
Sbjct: 1753 LSYNDLSGWLPESIISLPHLKSLYFGCNPSMSDEDTTKLNSSLINTDYGRCKAKKPKFGQ 1932
Query: 645 IVIGIALATAVLLVAVTVHVVRKRRLHRAQAWKLTAFQRLEIKAEDVVECLKEEN----- 699
+ + A+ + LL+ + V ++ R +R ++ L F + A +++ L ++
Sbjct: 1933 VFVIGAITSGSLLITLAVGILFFCR-YRHKSITLEGFGKTYPMATNIIFSLPSKDDFFIK 2109
Query: 700 ---------------------IIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFR 738
+IG+GG G VYRG++ +G +VA+K + S + F
Sbjct: 2110 SVSVKPFTLEYIEQATEQYKTLIGEGGFGSVYRGTLDDGQEVAVK-VRSSTSTQGTREFD 2286
Query: 739 AEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHG--AKGGHLRWEMRYK 796
E+ L I+H N++ LLGY + D +L+Y +M NGSL + L+G +K L W R
Sbjct: 2287 NELNLLSAIQHENLVPLLGYCNEYDQQILVYPFMSNGSLLDRLYGEASKRKILDWPTRLS 2466
Query: 797 IAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSS 856
IA+ AARGL Y+H +IHRDVKS+NILLD A VADFG +K+ G S
Sbjct: 2467 IALGAARGLAYLHTFPGRSVIHRDVKSSNILLDQSMCAKVADFGFSKYAPQEGDSYVSLE 2646
Query: 857 IAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVD--IVGWVNKTM 914
+ G+ GY+ PEY T ++ EKSDV+SFGVVLLE++ GR+P+ ++ +V W +
Sbjct: 2647 VRGTAGYLDPEYYKTQQLSEKSDVFSFGVVLLEIVSGREPLNIKRPRIEWSLVEWAKPYI 2826
Query: 915 SELSQPSDTALVLAVVDPRL-SGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 970
+ V +VDP + GY ++ + +A+ C++ RP M ++V L
Sbjct: 2827 R-------ASKVDEIVDPGIKGGYHAEALWRVVEVALQCLEPYSTYRPCMVDIVREL 2976
Score = 62.0 bits (149), Expect = 1e-09
Identities = 47/173 (27%), Positives = 80/173 (46%)
Frame = +1
Query: 9 LVLCFTLIWFRWTVVYSSFSDLDALLKLKESMKGAKAKHHALEDWKFSTSLSAHCSFSGV 68
L+ + ++ R + ++ DL+ + K++E + ++ ALE W + + G+
Sbjct: 1393 LLNAYEILQARSWIEETNQKDLEVIQKMREELLLHNQENEALESWSGDPCMIF--PWKGI 1566
Query: 69 TCDQNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLN 128
TCD + ++ KL+ +S NNL +PS + +T+L++LN
Sbjct: 1567 TCDDSTGS------------------SIITKLD---LSSNNLKGAIPSIVTKMTNLQILN 1683
Query: 129 ISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGN 181
+SHN F FP + L +LD N SG LPE I+ L LK L+ N
Sbjct: 1684 LSHNQFDMLFPSFPPSSL--LISLDLSYNDLSGWLPESIISLPHLKSLYFGCN 1836
Score = 44.3 bits (103), Expect = 2e-04
Identities = 28/75 (37%), Positives = 41/75 (54%)
Frame = +1
Query: 249 LEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEI 308
L++++ NL G IP + +T L L + N P S + L+SLDLS NDL+G +
Sbjct: 1606 LDLSSNNLKGAIPSIVTKMTNLQILNLSHNQFDMLFPSFPPSSL-LISLDLSYNDLSGWL 1782
Query: 309 PESFSKLKNLTLMNF 323
PES L +L + F
Sbjct: 1783 PESIISLPHLKSLYF 1827
Score = 38.1 bits (87), Expect = 0.017
Identities = 18/42 (42%), Positives = 25/42 (58%)
Frame = +1
Query: 294 LMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSF 335
+ LDLS N+L G IP +K+ NL ++N N+F PSF
Sbjct: 1597 ITKLDLSSNNLKGAIPSIVTKMTNLQILNLSHNQFDMLFPSF 1722
>TC82638 weakly similar to PIR|T00712|T00712 protein kinase homolog F22O13.7
- Arabidopsis thaliana, partial (27%)
Length = 1074
Score = 142 bits (357), Expect(2) = 9e-53
Identities = 78/185 (42%), Positives = 116/185 (62%), Gaps = 1/185 (0%)
Frame = +2
Query: 174 KYLHLAGNYFSGTIPESYSEFQSLEF-LGLNANSLTGRVPESLAKLKTLKELHLGYSNAY 232
++L L ++F G+IP+S S+F+ E + S + KL +L+ + +GY N +
Sbjct: 506 RHLILEASFFEGSIPKSISKFE*SEVPRSIWE*SHREKSRLRFGKLSSLEYMIIGY-NEF 682
Query: 233 EGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMM 292
EGGIP FG++ L+ L++A N+ GEIP LG L L+++F+ N+ G IP + +M
Sbjct: 683 EGGIPKEFGNLTKLKYLDLAEGNVGGEIPDELGKLKLLNTVFLYKNSFEGKIPTNIGNMT 862
Query: 293 SLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNF 352
SL+ LDLS N L+G IP S+LKNL L+NF +NK G +PS +GDLP LE L++W N+
Sbjct: 863 SLVLLDLSDNMLSGNIPAEISQLKNLQLLNFMRNKLSGPVPSGLGDLPQLEVLELWNNSL 1042
Query: 353 SFVLP 357
S LP
Sbjct: 1043SRPLP 1057
Score = 89.7 bits (221), Expect = 5e-18
Identities = 49/142 (34%), Positives = 78/142 (54%)
Frame = +2
Query: 169 KLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGY 228
KL L+Y+ + N F G IP+ + L++L L ++ G +P+ L KLK L + L Y
Sbjct: 638 KLSSLEYMIIGYNEFEGGIPKEFGNLTKLKYLDLAEGNVGGEIPDELGKLKLLNTVFL-Y 814
Query: 229 SNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPEL 288
N++EG IP G+M +L LL++++ L+G IP + L L L N L+G +P L
Sbjct: 815 KNSFEGKIPTNIGNMTSLVLLDLSDNMLSGNIPAEISQLKNLQLLNFMRNKLSGPVPSGL 994
Query: 289 SSMMSLMSLDLSINDLTGEIPE 310
+ L L+L N L+ +P+
Sbjct: 995 GDLPQLEVLELWNNSLSRPLPK 1060
Score = 88.2 bits (217), Expect = 1e-17
Identities = 62/203 (30%), Positives = 96/203 (46%)
Frame = +2
Query: 394 IITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELP 453
I+ +FF G IPK I + F VP +++ + L +L+
Sbjct: 515 ILEASFFEGSIPKSISK------------FE*SEVPRSIWE*SHREKSRLRFGKLS---- 646
Query: 454 SVISGESLGTLTLSNNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFEIPMLTKVN 513
SL + + N F G IP NL L+ L L GEIP + ++ +L V
Sbjct: 647 ------SLEYMIIGYNEFEGGIPKEFGNLTKLKYLDLAEGNVGGEIPDELGKLKLLNTVF 808
Query: 514 ISGNNLTGPIPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPD 573
+ N+ G IPT I + SL +DLS N L+G +P + L +L +LN RN++SGPVP
Sbjct: 809 LYKNSFEGKIPTNIGNMTSLVLLDLSDNMLSGNIPAEISQLKNLQLLNFMRNKLSGPVPS 988
Query: 574 EIRFMTSLTTLDLSSNNFTGTVP 596
+ + L L+L +N+ + +P
Sbjct: 989 GLGDLPQLEVLELWNNSLSRPLP 1057
Score = 84.7 bits (208), Expect(2) = 9e-53
Identities = 49/133 (36%), Positives = 75/133 (55%)
Frame = +1
Query: 48 HALEDWKFSTSLSAHCSFSGVTCDQNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISM 107
++L DWK + AHC+++GV C+ V LN++ + L G + EI L+ L L +
Sbjct: 130 NSLHDWKDGGAAQAHCNWTGVQCNSAGAVEKLNLSHMNLSGSVSNEIQSLKSLTFLNLCC 309
Query: 108 NNLTDQLPSDLASLTSLKVLNISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEI 167
N L + +LTSLK L++S N F+G FP + +EL L+A N+FSG LPE++
Sbjct: 310 NGFESSLSKHITNLTSLKSLDVSQNFFTGGFPLGLGKA-SELLTLNASSNNFSGFLPEDL 486
Query: 168 VKLEKLKYLHLAG 180
+ L+ L L G
Sbjct: 487 GNISSLETLDLRG 525
Score = 79.7 bits (195), Expect(2) = 2e-24
Identities = 47/165 (28%), Positives = 77/165 (46%)
Frame = +2
Query: 265 GNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFF 324
G L+ L + + N G IP E ++ L LDL+ ++ GEIP+ KLK L + +
Sbjct: 635 GKLSSLEYMIIGYNEFEGGIPKEFGNLTKLKYLDLAEGNVGGEIPDELGKLKLLNTVFLY 814
Query: 325 QNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDL 384
+N F G +P+ IG++ +L L D++ N L+G IP ++
Sbjct: 815 KNSFEGKIPTNIGNMTSLVLL------------------------DLSDNMLSGNIPAEI 922
Query: 385 CKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVP 429
+ L+ N GP+P G+G+ L + + NN L P+P
Sbjct: 923 SQLKNLQLLNFMRNKLSGPVPSGLGDLPQLEVLELWNNSLSRPLP 1057
Score = 79.3 bits (194), Expect = 7e-15
Identities = 51/146 (34%), Positives = 70/146 (47%), Gaps = 4/146 (2%)
Frame = +2
Query: 312 FSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGG----NGRFL 367
F KL +L M N+F G +P G+L L+ L + E N +P LG N FL
Sbjct: 632 FGKLSSLEYMIIGYNEFEGGIPKEFGNLTKLKYLDLAEGNVGGEIPDELGKLKLLNTVFL 811
Query: 368 YFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGP 427
Y KN G IP ++ L ++DN G IP I + ++L + N L GP
Sbjct: 812 Y----KNSFEGKIPTNIGNMTSLVLLDLSDNMLSGNIPAEISQLKNLQLLNFMRNKLSGP 979
Query: 428 VPPGVFQLPSVTITELSNNRLNGELP 453
VP G+ LP + + EL NN L+ LP
Sbjct: 980 VPSGLGDLPQLEVLELWNNSLSRPLP 1057
Score = 79.0 bits (193), Expect = 9e-15
Identities = 46/142 (32%), Positives = 73/142 (51%)
Frame = +2
Query: 146 MTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNAN 205
++ LE + N F G +P+E L KLKYL LA G IP+ + + L + L N
Sbjct: 641 LSSLEYMIIGYNEFEGGIPKEFGNLTKLKYLDLAEGNVGGEIPDELGKLKLLNTVFLYKN 820
Query: 206 SLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLG 265
S G++P ++ + +L L L N G IP ++NL+LL L+G +P LG
Sbjct: 821 SFEGKIPTNIGNMTSLVLLDLS-DNMLSGNIPAEISQLKNLQLLNFMRNKLSGPVPSGLG 997
Query: 266 NLTKLHSLFVQMNNLTGTIPPE 287
+L +L L + N+L+ +P +
Sbjct: 998 DLPQLEVLELWNNSLSRPLPKD 1063
Score = 62.8 bits (151), Expect = 6e-10
Identities = 32/102 (31%), Positives = 59/102 (57%)
Frame = +2
Query: 88 GHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLFSGQFPGNITVGMT 147
G +P E+G L+ L + + N+ ++P+++ ++TSL +L++S N+ SG P I+ +
Sbjct: 758 GEIPDELGKLKLLNTVFLYKNSFEGKIPTNIGNMTSLVLLDLSDNMLSGNIPAEIS-QLK 934
Query: 148 ELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPE 189
L+ L+ N SGP+P + L +L+ L L N S +P+
Sbjct: 935 NLQLLNFMRNKLSGPVPSGLGDLPQLEVLELWNNSLSRPLPK 1060
Score = 54.3 bits (129), Expect = 2e-07
Identities = 31/86 (36%), Positives = 48/86 (55%)
Frame = +1
Query: 511 KVNISGNNLTGPIPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGP 570
K+N+S NL+G + I SLT ++L N + K + NL L L++S+N +G
Sbjct: 220 KLNLSHMNLSGSVSNEIQSLKSLTFLNLCCNGFESSLSKHITNLTSLKSLDVSQNFFTGG 399
Query: 571 VPDEIRFMTSLTTLDLSSNNFTGTVP 596
P + + L TL+ SSNNF+G +P
Sbjct: 400 FPLGLGKASELLTLNASSNNFSGFLP 477
Score = 52.4 bits (124), Expect(2) = 2e-24
Identities = 31/104 (29%), Positives = 49/104 (46%)
Frame = +1
Query: 170 LEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYS 229
+EKL H+ SG++ +SL FL L N + + + L +LK L +
Sbjct: 214 VEKLNLSHMN---LSGSVSNEIQSLKSLTFLNLCCNGFESSLSKHITNLTSLKSLDVS-Q 381
Query: 230 NAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSL 273
N + GG P G L L ++ N +G +P LGN++ L +L
Sbjct: 382 NFFTGGFPLGLGKASELLTLNASSNNFSGFLPEDLGNISSLETL 513
Score = 47.4 bits (111), Expect = 3e-05
Identities = 23/67 (34%), Positives = 36/67 (53%)
Frame = +2
Query: 531 ASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNN 590
+SL + + N G +PK NL L L+L+ + G +PDE+ + L T+ L N+
Sbjct: 644 SSLEYMIIGYNEFEGGIPKEFGNLTKLKYLDLAEGNVGGEIPDELGKLKLLNTVFLYKNS 823
Query: 591 FTGTVPT 597
F G +PT
Sbjct: 824 FEGKIPT 844
Score = 46.2 bits (108), Expect = 6e-05
Identities = 25/63 (39%), Positives = 36/63 (56%)
Frame = +1
Query: 536 VDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTV 595
++LS NL+G V +++L L+ LNL N + I +TSL +LD+S N FTG
Sbjct: 223 LNLSHMNLSGSVSNEIQSLKSLTFLNLCCNGFESSLSKHITNLTSLKSLDVSQNFFTGGF 402
Query: 596 PTG 598
P G
Sbjct: 403 PLG 411
Score = 38.5 bits (88), Expect = 0.013
Identities = 37/170 (21%), Positives = 61/170 (35%)
Frame = +1
Query: 279 NLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGD 338
N TG + +S ++ L+LS +L+G + LK+LT +N N F SL
Sbjct: 178 NWTGV---QCNSAGAVEKLNLSHMNLSGSVSNEIQSLKSLTFLNLCCNGFESSL------ 330
Query: 339 LPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDN 398
H+T L LK+ ++ N
Sbjct: 331 ----------------------------------SKHITNLTS--------LKSLDVSQN 384
Query: 399 FFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRL 448
FF G P G+G+ L + ++N G +P + + S+ +L L
Sbjct: 385 FFTGGFPLGLGKASELLTLNASSNNFSGFLPEDLGNISSLETLDLRGKLL 534
>BQ147648 similar to PIR|B86465|B8 probable Protein kinase [imported] -
Arabidopsis thaliana, partial (19%)
Length = 657
Score = 189 bits (481), Expect(2) = 2e-52
Identities = 97/189 (51%), Positives = 125/189 (65%), Gaps = 6/189 (3%)
Frame = +3
Query: 788 HLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYD 847
+L WE RYKIAV +A+GL Y+HHDC P I+HRDVK NNILLD+ FEA++ADFGLAK +
Sbjct: 108 NLDWETRYKIAVGSAQGLAYLHHDCVPSILHRDVKCNNILLDSKFEAYIADFGLAKLMNS 287
Query: 848 PGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGE---FGDGV 904
P +MS +AGSYGYIAPEY YT+ + EKSDVYS+GVVLLE++ GR V + GDG
Sbjct: 288 PNYHHAMSRVAGSYGYIAPEYGYTMNITEKSDVYSYGVVLLEILSGRSAVEDGQHVGDGQ 467
Query: 905 DIVGWVNKTMSELSQPSDTALVLAVVDPRLSGYP---LTSVIHMFNIAMMCVKEMGPARP 961
IV WV K M+ ++++D +L P + ++ IAM CV RP
Sbjct: 468 HIVEWVKKKMASFEP------AVSILDTKLQSLPNQVVQEMLQTLGIAMFCVNSSPVERP 629
Query: 962 TMREVVHML 970
TM+EVV +L
Sbjct: 630 TMKEVVALL 656
Score = 35.8 bits (81), Expect(2) = 2e-52
Identities = 15/27 (55%), Positives = 20/27 (73%)
Frame = +2
Query: 751 NIMRLLGYVSNKDTNLLLYEYMPNGSL 777
NI+RL+GY SN LLLY ++ NG+L
Sbjct: 2 NIVRLIGYCSNGSVKLLLYNFIQNGNL 82
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.138 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,284,189
Number of Sequences: 36976
Number of extensions: 419416
Number of successful extensions: 6440
Number of sequences better than 10.0: 873
Number of HSP's better than 10.0 without gapping: 3116
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4698
length of query: 986
length of database: 9,014,727
effective HSP length: 105
effective length of query: 881
effective length of database: 5,132,247
effective search space: 4521509607
effective search space used: 4521509607
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0258c.2


