

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0216.15
(500 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
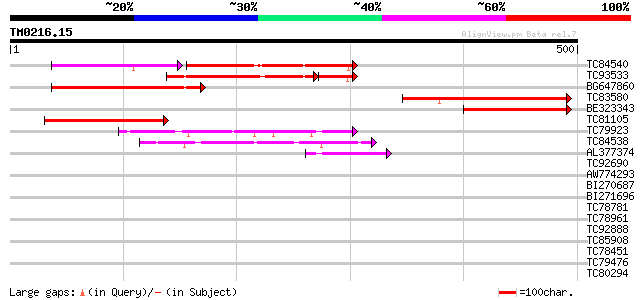
Score E
Sequences producing significant alignments: (bits) Value
TC84540 similar to GP|21539577|gb|AAM53341.1 unknown protein {Ar... 182 3e-70
TC93533 similar to GP|17065464|gb|AAL32886.1 Unknown protein {Ar... 148 1e-45
BG647860 weakly similar to GP|17065464|gb Unknown protein {Arabi... 131 5e-31
TC83580 similar to GP|1666171|emb|CAA70322.1 unknown {Nicotiana ... 126 2e-29
BE323343 similar to GP|1666171|emb unknown {Nicotiana plumbagini... 103 1e-22
TC81105 weakly similar to PIR|F84721|F84721 probable RING zinc f... 92 4e-19
TC79923 similar to PIR|T04748|T04748 hypothetical protein T16H5.... 85 7e-17
TC84538 weakly similar to GP|22296418|dbj|BAC10186. similar to p... 80 2e-15
AL377374 weakly similar to GP|8885575|dbj| emb|CAB75487.1~gene_i... 46 3e-05
TC92690 weakly similar to GP|15292861|gb|AAK92801.1 unknown prot... 40 0.003
AW774293 similar to GP|13881624|gb PPE family protein {Mycobacte... 33 0.25
BI270687 weakly similar to GP|1903347|gb EST gb|ATTS5672 comes f... 32 0.74
BI271696 weakly similar to GP|21593120|gb putative splicing regu... 30 2.8
TC78781 weakly similar to PIR|T02291|T02291 hypothetical protein... 30 2.8
TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Ar... 30 2.8
TC92888 similar to GP|3219304|dbj|BAA28847.1 MUS38 {Neurospora c... 30 2.8
TC85908 similar to GP|11385445|gb|AAG34805.1 glutathione S-trans... 29 3.7
TC78451 similar to GP|9279625|dbj|BAB01083.1 gb|AAF23830.1~gene_... 29 3.7
TC79476 similar to GP|14423560|gb|AAK62462.1 Unknown protein {Ar... 29 3.7
TC80294 similar to PIR|T47806|T47806 hypothetical protein F24G16... 29 3.7
>TC84540 similar to GP|21539577|gb|AAM53341.1 unknown protein {Arabidopsis
thaliana}, partial (38%)
Length = 1027
Score = 182 bits (462), Expect(2) = 3e-70
Identities = 81/152 (53%), Positives = 107/152 (70%), Gaps = 2/152 (1%)
Frame = +3
Query: 157 LTLRCPDPSCDVAVDRDMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVCY 216
L LRCPDP+C AVD+DMI AS K YE++L+ SY++ K +WCPAPGC++ V +
Sbjct: 582 LMLRCPDPACAAAVDQDMIDAFASAEDKKKYERYLVRSYIEVNKKTKWCPAPGCEHAVNF 761
Query: 217 QGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDCETAGRWMKKIDKSTSENGKWILAHTK 276
D G + DV+CLC +SFCW C E++H PVDC+T +W+ K + + SEN WILA+TK
Sbjct: 762 --DAG-DENYDVSCLCSYSFCWNCTEDAHRPVDCDTVSKWILK-NSAESENTNWILAYTK 929
Query: 277 PCPRCKSLIEKNQGFKYMTCK--CGFQFCWVC 306
PCP+CK IEKN+G +MTC C FQFCW+C
Sbjct: 930 PCPKCKRSIEKNRGCMHMTCSAPCRFQFCWLC 1025
Score = 101 bits (252), Expect(2) = 3e-70
Identities = 52/119 (43%), Positives = 69/119 (57%), Gaps = 4/119 (3%)
Frame = +2
Query: 38 NFTILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCEDEEKVQKA 97
NF+IL E +I+ QED I VA+VL IP +A LL H WNV+ V E+W DE+ V++
Sbjct: 215 NFSILKESDIRERQEDDIRSVAAVLSIPPVAASILLRHYNWNVSNVNEAWFADEDGVRRK 394
Query: 98 AGLLNQPKVE----IDWLKDLCQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNINKG 152
GLL +P + K C +CF + KI +A C HP C +CW YI T+IN G
Sbjct: 395 VGLLEKPAYKNPDANKMPKLTCGICFEAYRPSKIHTASCGHPYCSSCWGGYIGTSINDG 571
>TC93533 similar to GP|17065464|gb|AAL32886.1 Unknown protein {Arabidopsis
thaliana}, partial (38%)
Length = 660
Score = 148 bits (373), Expect(2) = 1e-45
Identities = 66/134 (49%), Positives = 87/134 (64%)
Frame = +3
Query: 139 ACWMDYIDTNINKGPSKCLTLRCPDPSCDVAVDRDMIRELASESSKVMYEQFLLWSYVDS 198
+CW YI T+IN GP CL LRCP+P+C A+ +DMI LA K Y ++L SY++
Sbjct: 6 SCWEGYISTSINDGPG-CLMLRCPEPTCGAAIGQDMIDLLACNEDKEKYARYLRRSYIED 182
Query: 199 KAKMRWCPAPGCDYVVCYQGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDCETAGRWMK 258
K +WCPAPGC+Y V + GG + DV+CLC +SFCW C EE+H PVDC T +W+
Sbjct: 183 NKKSKWCPAPGCEYAVTFDAGGG---NYDVSCLCSYSFCWNCTEEAHRPVDCGTVVKWIM 353
Query: 259 KIDKSTSENGKWIL 272
K + + SEN WIL
Sbjct: 354 K-NSAESENMNWIL 392
Score = 53.9 bits (128), Expect(2) = 1e-45
Identities = 21/36 (58%), Positives = 28/36 (77%), Gaps = 2/36 (5%)
Frame = +1
Query: 273 AHTKPCPRCKSLIEKNQGFKYMTC--KCGFQFCWVC 306
A++KPCP+CK IEKNQG ++TC C F+FCW+C
Sbjct: 394 ANSKPCPKCKRPIEKNQGCMHITCTPPCKFEFCWLC 501
>BG647860 weakly similar to GP|17065464|gb Unknown protein {Arabidopsis
thaliana}, partial (23%)
Length = 719
Score = 131 bits (330), Expect = 5e-31
Identities = 65/136 (47%), Positives = 87/136 (63%), Gaps = 1/136 (0%)
Frame = +3
Query: 38 NFTILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCEDEEKVQKA 97
NF IL E +I+ QED I+ +A+VL IP +A LL H WNV+ V ++W DE++V++
Sbjct: 273 NFAILKESDIRDRQEDDISSLATVLSIPPVAASILLRHYNWNVSNVNDAWFADEDRVRRK 452
Query: 98 AGLLNQPKVEIDWLKDL-CQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNINKGPSKC 156
GLL +P E K+L C +CF + KI +A C HP C +CW YI T+IN GP C
Sbjct: 453 VGLLEKPVYENPDAKELTCGICFEAYRPSKIHNASCGHPYCFSCWGGYIGTSINDGPG-C 629
Query: 157 LTLRCPDPSCDVAVDR 172
L LRCPDP+C AVD+
Sbjct: 630 LMLRCPDPACGAAVDQ 677
>TC83580 similar to GP|1666171|emb|CAA70322.1 unknown {Nicotiana
plumbaginifolia}, partial (49%)
Length = 1022
Score = 126 bits (316), Expect = 2e-29
Identities = 68/153 (44%), Positives = 94/153 (60%), Gaps = 4/153 (2%)
Frame = +2
Query: 347 KKCNHYYECWARNDFLSKSILQKLIDLDRKY---LSMSLQKRETYFDFVKKAWQQIAECR 403
++ HYYE WA N + L L + + LS + + E+ F+ +AW QI ECR
Sbjct: 17 ERYTHYYERWASNQSSRQKALADLHQMQTVHMEKLSDTQCQPESQLKFITEAWLQIVECR 196
Query: 404 RILKWTNVYGYFLPKNEKAKKQFSEYIQVEAEVALEKLQQCAKVEETMF-QDNYPEESLN 462
R+LKWT YGY+L ++E AKKQF EY+Q EAE LE+L QCA+ E +F P + N
Sbjct: 197 RVLKWTYAYGYYLAEHEHAKKQFFEYLQGEAESGLERLHQCAEKELQVFLSAEGPSKEFN 376
Query: 463 KFQEKVIHLTDVTKIYFENLLRAFEDGLDKVEA 495
F+ K+ LT VTK +FENL+RA E+GL V++
Sbjct: 377 DFRTKLAGLTSVTKNFFENLVRALENGLCDVDS 475
>BE323343 similar to GP|1666171|emb unknown {Nicotiana plumbaginifolia},
partial (34%)
Length = 413
Score = 103 bits (258), Expect = 1e-22
Identities = 53/96 (55%), Positives = 66/96 (68%), Gaps = 1/96 (1%)
Frame = +3
Query: 401 ECRRILKWTNVYGYFLPKNEKAKKQFSEYIQVEAEVALEKLQQCAKVEETMF-QDNYPEE 459
ECRR+LKWT YGY+LP++E AKKQF EY+Q EAE LE+L QCA+ E F +
Sbjct: 3 ECRRVLKWTYAYGYYLPEHEGAKKQFFEYLQGEAESGLERLHQCAEKELQQFLTAEGQSK 182
Query: 460 SLNKFQEKVIHLTDVTKIYFENLLRAFEDGLDKVEA 495
N F+ K+ LT VTK YFENL+RA E+GL V+A
Sbjct: 183 EFNDFRTKLAGLTSVTKNYFENLVRALENGLSDVDA 290
>TC81105 weakly similar to PIR|F84721|F84721 probable RING zinc finger
protein [imported] - Arabidopsis thaliana, partial (23%)
Length = 683
Score = 92.4 bits (228), Expect = 4e-19
Identities = 47/111 (42%), Positives = 68/111 (60%), Gaps = 1/111 (0%)
Frame = +3
Query: 31 EEEDKRENFTILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCED 90
E +NFTIL E +I+ QED I++VA+VL + +A LL H+ W+V+KV ++W D
Sbjct: 351 ESRRPEQNFTILKESDIRVRQEDDIDRVAAVLSVSRVAASILLRHHNWSVSKVHDAWVAD 530
Query: 91 EEKVQKAAGLLNQPKVEIDWLKDL-CQVCFRSSEKGKILSAGCAHPLCEAC 140
EE+V KA GLL +P V+ +L C +CF + + I A C HP C +C
Sbjct: 531 EERVXKAVGLLEKPVVQHPNTSELTCGICFENYPRSGIGMASCGHPYCFSC 683
>TC79923 similar to PIR|T04748|T04748 hypothetical protein T16H5.30 -
Arabidopsis thaliana, partial (28%)
Length = 1001
Score = 84.7 bits (208), Expect = 7e-17
Identities = 57/227 (25%), Positives = 98/227 (43%), Gaps = 17/227 (7%)
Frame = +2
Query: 97 AAGLLNQPKVEIDWLKDLCQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNINKGPSKC 156
A GL+ P V + L + C +C +++ C+H C C Y D G +C
Sbjct: 224 AMGLVTFP-VNDEKLLENCSICCDDKPVPMMITLKCSHTFCSHCLRSYAD-----GKLQC 385
Query: 157 --LTLRCPDPSCDVAVDRDMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVV 214
+ +RCP P C + + S E+ L + + ++++ +CP P C ++
Sbjct: 386 CQVPIRCPQPGCRYCISTPECKSFLPFISFESLEKALSEANI-AQSERFYCPFPNCSVLL 562
Query: 215 ----CYQGDGGCARDLDVTCL----CYHSFCWRCGEESHSPVDCETAGRWMKKIDKST-- 264
C G + D +C+ C C CG HS + CE ++ ++
Sbjct: 563 DPCECLSAMDGSSSQSDNSCIECPVCQRFICVGCGVPWHSSMSCEEFQSLPEEERDASDI 742
Query: 265 -----SENGKWILAHTKPCPRCKSLIEKNQGFKYMTCKCGFQFCWVC 306
++N +W K C +C+ +IE QG +MTC+CG +FC+ C
Sbjct: 743 TLHRLAQNKRW-----KRCQQCRIMIELTQGCYHMTCRCGHEFCYSC 868
Score = 29.6 bits (65), Expect = 2.8
Identities = 14/44 (31%), Positives = 20/44 (44%), Gaps = 1/44 (2%)
Frame = +2
Query: 228 VTCLCYHSFCWRCGEE-SHSPVDCETAGRWMKKIDKSTSENGKW 270
+TC C H FC+ CG E C+ A + S E+ +W
Sbjct: 830 MTCRCGHEFCYSCGAEYRDGQQTCQCAFWDEDSLTNSLQESEQW 961
>TC84538 weakly similar to GP|22296418|dbj|BAC10186. similar to pre-mRNA
splicing factor ATP-dependent RNA helicase, partial
(11%)
Length = 973
Score = 80.1 bits (196), Expect = 2e-15
Identities = 61/214 (28%), Positives = 87/214 (40%), Gaps = 5/214 (2%)
Frame = +2
Query: 115 CQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNI-NKG--PSKCLTLRCPDPSCDVAVD 171
C +C E G L GC H C C ++ ++ I N+G P C C DP +
Sbjct: 293 CPICLCEVEDGYKLE-GCGHLFCRLCLVEQCESAIKNQGSFPICCAHQGCGDP----ILL 457
Query: 172 RDMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDLDVTCL 231
D L+++ ++ L S R+CP+P C V D A + V
Sbjct: 458 TDFRTLLSNDKLDELFRASLGAFVASSSGTYRFCPSPDCPSVYRV-ADSDTASEPFVCGA 634
Query: 232 CYHSFCWRCGEESHSPVDCETAGRWMKKIDKSTSENGKWILA--HTKPCPRCKSLIEKNQ 289
CY C +C E H + CE R+ + D S +W K C C +IEK
Sbjct: 635 CYSETCTKCHLEYHPYLSCE---RYRELKDDPDSSLKEWCKGKEQVKSCFACGQIIEKID 805
Query: 290 GFKYMTCKCGFQFCWVCFHEQLKCESRSCSPFIR 323
G ++ CKCG CWVC ++ S C +R
Sbjct: 806 GCNHVECKCGKHVCWVCL--EIFTSSDECYDHLR 901
>AL377374 weakly similar to GP|8885575|dbj|
emb|CAB75487.1~gene_id:F15L12.13~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (7%)
Length = 386
Score = 46.2 bits (108), Expect = 3e-05
Identities = 24/75 (32%), Positives = 35/75 (46%)
Frame = +3
Query: 262 KSTSENGKWILAHTKPCPRCKSLIEKNQGFKYMTCKCGFQFCWVCFHEQLKCESRSCSPF 321
KS + W K C +C +IE +G +MTC+CGF+FC+ C E ++ P
Sbjct: 36 KSLASRSLW-----KQCVKCNHMIELAEGCYHMTCRCGFEFCYKCGAEWKDKKATCSCPL 200
Query: 322 IRMHIMEFQGTDGEE 336
+ Q D EE
Sbjct: 201 WEEDNIWLQDRDDEE 245
>TC92690 weakly similar to GP|15292861|gb|AAK92801.1 unknown protein
{Arabidopsis thaliana}, partial (15%)
Length = 629
Score = 39.7 bits (91), Expect = 0.003
Identities = 22/103 (21%), Positives = 42/103 (40%), Gaps = 9/103 (8%)
Frame = +2
Query: 51 QEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCEDEEKVQKAAGLLNQPKVEIDW 110
Q++ + +V +L + A +L ++ W+V K+ E + E + A + Q V +D
Sbjct: 326 QKEDLRRVMDMLSVRQEHARIMLIYHRWDVEKLFEVYVEKGK-----AFMFAQAGVSVDE 490
Query: 111 LKD---------LCQVCFRSSEKGKILSAGCAHPLCEACWMDY 144
D +C++C + C H C CW +
Sbjct: 491 YHDSGSLDSAIVMCEICMDDIPSDEATRMDCGHCFCNTCWTQH 619
>AW774293 similar to GP|13881624|gb PPE family protein {Mycobacterium
tuberculosis CDC1551}, partial (1%)
Length = 597
Score = 33.1 bits (74), Expect = 0.25
Identities = 19/60 (31%), Positives = 26/60 (42%)
Frame = -1
Query: 198 SKAKMRWCPAPGCDYVVCYQGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDCETAGRWM 257
+K M C P C CY C+R +C C FCW+C HS + + G W+
Sbjct: 411 TKCSMPGCCCPKCSLPCCYPK---CSRP---SC-CMSCFCWQCFMGKHSGRNGKQRG*WL 253
>BI270687 weakly similar to GP|1903347|gb EST gb|ATTS5672 comes from this
gene. {Arabidopsis thaliana}, partial (17%)
Length = 665
Score = 31.6 bits (70), Expect = 0.74
Identities = 23/90 (25%), Positives = 44/90 (48%)
Frame = +3
Query: 364 KSILQKLIDLDRKYLSMSLQKRETYFDFVKKAWQQIAECRRILKWTNVYGYFLPKNEKAK 423
K I + ++ L ++ ++ ++R+ Y +KK W IA L N YG+ L K
Sbjct: 102 KDISEFMLHLAQEAEDLASKERQNYSPILKK-WNAIAAALAALTLNNCYGHVL------K 260
Query: 424 KQFSEYIQVEAEVALEKLQQCAKVEETMFQ 453
+ SE + E+ + LQ+ ++E+ + Q
Sbjct: 261 QYLSEIKSITVELII-VLQKAKRLEDILVQ 347
>BI271696 weakly similar to GP|21593120|gb putative splicing regulatory
protein {Arabidopsis thaliana}, partial (66%)
Length = 690
Score = 29.6 bits (65), Expect = 2.8
Identities = 16/36 (44%), Positives = 19/36 (52%), Gaps = 5/36 (13%)
Frame = +1
Query: 287 KNQGF-KYMTCKCGF----QFCWVCFHEQLKCESRS 317
K GF K C CG + CW+C HEQ C +RS
Sbjct: 484 KISGF*KNKVCLCGC*AKGK*CWICNHEQSLCINRS 591
>TC78781 weakly similar to PIR|T02291|T02291 hypothetical protein T13D8.28 -
Arabidopsis thaliana, partial (12%)
Length = 1300
Score = 29.6 bits (65), Expect = 2.8
Identities = 16/54 (29%), Positives = 26/54 (47%)
Frame = +3
Query: 11 SHGLVCGLRERDKNLPNTEEEEEDKRENFTILSELEIKRLQEDAINKVASVLCI 64
S L C L + NL N EEEED + + LS+ + R+ + + + C+
Sbjct: 51 SQSLTCSLFPSNPNLLNMAEEEEDGEDRISSLSDDLLVRILSNLHTRESVSTCV 212
>TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Arabidopsis
thaliana}, partial (71%)
Length = 974
Score = 29.6 bits (65), Expect = 2.8
Identities = 19/71 (26%), Positives = 32/71 (44%)
Frame = +2
Query: 197 DSKAKMRWCPAPGCDYVVCYQGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDCETAGRW 256
DS+ + P PG DG ARD + + C+RCGE H +C+ + +
Sbjct: 311 DSREYLGRGPPPGSGRCFNCGIDGHWARDCKAGD--WKNKCYRCGERGHIEKNCKNSPKK 484
Query: 257 MKKIDKSTSEN 267
+ + +S S +
Sbjct: 485 LSRHARSVSRS 517
>TC92888 similar to GP|3219304|dbj|BAA28847.1 MUS38 {Neurospora crassa},
partial (23%)
Length = 703
Score = 29.6 bits (65), Expect = 2.8
Identities = 12/35 (34%), Positives = 16/35 (45%)
Frame = +3
Query: 230 CLCYHSFCWRCGEESHSPVDCETAGRWMKKIDKST 264
C C S WR G + P+ C R + K D+ T
Sbjct: 486 CACVLSILWRVGRRAALPLGCAARKRCLYKADQGT 590
>TC85908 similar to GP|11385445|gb|AAG34805.1 glutathione S-transferase GST
15 {Glycine max}, partial (65%)
Length = 970
Score = 29.3 bits (64), Expect = 3.7
Identities = 10/18 (55%), Positives = 13/18 (71%)
Frame = +2
Query: 76 NLWNVTKVLESWCEDEEK 93
NLW KV+E WC D++K
Sbjct: 530 NLWERLKVIEDWCLDDKK 583
>TC78451 similar to GP|9279625|dbj|BAB01083.1
gb|AAF23830.1~gene_id:MOJ10.9~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (88%)
Length = 2452
Score = 29.3 bits (64), Expect = 3.7
Identities = 11/27 (40%), Positives = 14/27 (51%)
Frame = -1
Query: 114 LCQVCFRSSEKGKILSAGCAHPLCEAC 140
+C C R+ EK GC P+CE C
Sbjct: 1567 VCANCCRNDEKTDFTVRGCQTPVCEPC 1487
>TC79476 similar to GP|14423560|gb|AAK62462.1 Unknown protein {Arabidopsis
thaliana}, partial (43%)
Length = 956
Score = 29.3 bits (64), Expect = 3.7
Identities = 12/38 (31%), Positives = 22/38 (57%)
Frame = +3
Query: 29 EEEEEDKRENFTILSELEIKRLQEDAINKVASVLCIPD 66
EEEEE++ N + +K+ + +N++AS + PD
Sbjct: 87 EEEEEEETNNLKHRNTKPVKQTLPEVLNRLASTILFPD 200
>TC80294 similar to PIR|T47806|T47806 hypothetical protein F24G16.90 -
Arabidopsis thaliana, partial (22%)
Length = 985
Score = 29.3 bits (64), Expect = 3.7
Identities = 17/52 (32%), Positives = 25/52 (47%)
Frame = +3
Query: 275 TKPCPRCKSLIEKNQGFKYMTCKCGFQFCWVCFHEQLKCESRSCSPFIRMHI 326
++ RC K+ G + C F+ C CFHEQ ++CS R+HI
Sbjct: 567 SRSAERCAECCAKS*GCDFRDWSC-FESC--CFHEQSGLGQQTCSLERRVHI 713
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.136 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,423,881
Number of Sequences: 36976
Number of extensions: 371436
Number of successful extensions: 2329
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 2226
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2304
length of query: 500
length of database: 9,014,727
effective HSP length: 100
effective length of query: 400
effective length of database: 5,317,127
effective search space: 2126850800
effective search space used: 2126850800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0216.15


