

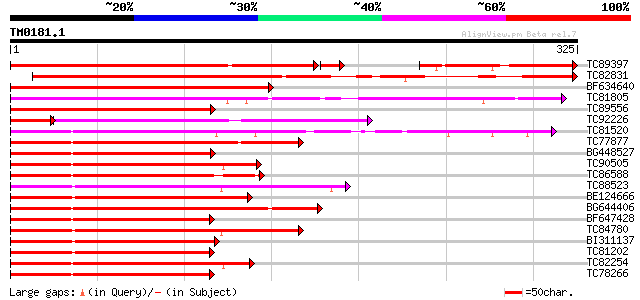
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0181.1
(325 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89397 homologue to GP|2832408|emb|CAA74605.1 R2R3-MYB transcri... 321 2e-88
TC82831 similar to PIR|T51650|T51650 probable transcription fact... 290 7e-79
BF634640 homologue to GP|2832408|emb R2R3-MYB transcription fact... 271 3e-73
TC81805 similar to GP|9758323|dbj|BAB08797.1 Myb-related transcr... 254 2e-68
TC89556 similar to GP|18419456|gb|AAL69334.1 blind {Lycopersicon... 221 4e-58
TC92226 similar to GP|18419456|gb|AAL69334.1 blind {Lycopersicon... 159 8e-51
TC81520 similar to PIR|D86470|D86470 hypothetical protein AAD460... 172 2e-43
TC77877 similar to GP|5139806|dbj|BAA81732.1 GmMYB29A2 {Glycine ... 170 6e-43
BG448527 similar to PIR|T51509|T51 probable transcription factor... 169 1e-42
TC90505 similar to GP|3941528|gb|AAC83640.1| putative transcript... 169 1e-42
TC86588 similar to GP|5139814|dbj|BAA81736.1 GmMYB29B2 {Glycine ... 168 2e-42
TC88523 similar to PIR|T05690|T05690 myb-related transcription f... 167 7e-42
BE124666 similar to PIR|JQ0957|JQ0 myb-related protein 330 - gar... 164 4e-41
BG644406 homologue to PIR|T07398|T07 myb-related transcription f... 164 6e-41
BF647428 homologue to PIR|T51621|T51 myb-like protein [imported]... 162 1e-40
TC84780 homologue to PIR|F85021|F85021 probable transcription fa... 160 8e-40
BI311137 similar to GP|17380966|gb putative transcription factor... 159 1e-39
TC81202 homologue to GP|17380966|gb|AAL36295.1 putative transcri... 156 9e-39
TC82254 similar to GP|22266673|dbj|BAC07543. myb-related transcr... 154 3e-38
TC78266 similar to GP|22266673|dbj|BAC07543. myb-related transcr... 153 8e-38
>TC89397 homologue to GP|2832408|emb|CAA74605.1 R2R3-MYB transcription
factor {Arabidopsis thaliana}, partial (40%)
Length = 1063
Score = 321 bits (822), Expect(2) = 2e-88
Identities = 155/179 (86%), Positives = 170/179 (94%), Gaps = 2/179 (1%)
Frame = +3
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKANVK+GPWSPEEDAKLK+YI+ +GTGGNWIALPQKIGLKRCGKSCRLRWLN
Sbjct: 39 MGRAPCCDKANVKKGPWSPEEDAKLKSYIEQNGTGGNWIALPQKIGLKRCGKSCRLRWLN 218
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNIKHGGFSEEED+IIC+LYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK
Sbjct: 219 YLRPNIKHGGFSEEEDDIICSLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 398
Query: 121 QRKEQQAQARRVSTMKQEIKRETDQNLMVAS-GVNTTHAAFYWPAEYSI-PMPVANASI 177
QRKEQ QARRVS MKQE+KRET+QNLM+A+ GVN+ +A YWPAEYS+ PMPV+N+SI
Sbjct: 399 QRKEQ--QARRVSNMKQEMKRETNQNLMMAALGVNSMSSAPYWPAEYSVHPMPVSNSSI 569
Score = 100 bits (248), Expect = 1e-21
Identities = 61/98 (62%), Positives = 66/98 (67%), Gaps = 8/98 (8%)
Frame = +2
Query: 236 PVAMVSNA--NIGGTTCQQSNVFQGFENFTSDFSELVCVNPQ-----TIDGFYGMESLEM 288
P +MVSN N GG CQ S++FQGFENF D SELVCVN Q TIDGFY +M
Sbjct: 629 PFSMVSNTITNNGGN-CQPSHIFQGFENFPRDLSELVCVNQQQIMDRTIDGFY-----DM 790
Query: 289 SNVSST-NTPSAESTSWGDMSSLVYSPLVSDYFGMQMQ 325
SN ST T S ESTSWGDM+SLVYSPLVSDY G Q
Sbjct: 791 SNGGSTITTTSTESTSWGDMNSLVYSPLVSDYEGCCQQ 904
Score = 22.7 bits (47), Expect(2) = 2e-88
Identities = 9/14 (64%), Positives = 11/14 (78%)
Frame = +1
Query: 179 DYDFHNQTSLKSLL 192
DYD +NQTS SL+
Sbjct: 574 DYDTNNQTSFTSLM 615
>TC82831 similar to PIR|T51650|T51650 probable transcription factor MYB36
[imported] - Arabidopsis thaliana, partial (34%)
Length = 940
Score = 290 bits (741), Expect = 7e-79
Identities = 173/324 (53%), Positives = 197/324 (60%), Gaps = 12/324 (3%)
Frame = +2
Query: 14 RGPWSPEEDAKLKAYIDHHGTGGNWIALPQKI-GLKRCGKSCRLRWLNYLRPNIKHGGFS 72
RGPWSPEED+KLKAYI HGTGGNWIALP+KI GLKRCGKSCRLRWLNYLRPN+KHG FS
Sbjct: 2 RGPWSPEEDSKLKAYIQQHGTGGNWIALPKKIVGLKRCGKSCRLRWLNYLRPNLKHGSFS 181
Query: 73 EEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGKQRKEQQAQARRV 132
EEEDNIIC+LYV+IGSRWSIIAAQLPGRTDNDIKNYWNT+LKKKLL KQRKEQQ+QARRV
Sbjct: 182 EEEDNIICSLYVNIGSRWSIIAAQLPGRTDNDIKNYWNTKLKKKLLTKQRKEQQSQARRV 361
Query: 133 STMKQEIKRETDQNLMVASGVNTTHAAFYWPAEY-SIP-MPVAN-ASIQDYDFHNQTSLK 189
QEIKRE+ ++ + T YWP ++ S P +PV N +SIQ+Y+
Sbjct: 362 FNXNQEIKRESGNSVWPIGFIGQT--PNYWPTQHNSFPNIPVTNPSSIQNYNLKE----- 520
Query: 190 SLLINKQGGVRFSYDHQ-QPYSNAITATANSQDPCDI-------YPASTMNMINPVAMVS 241
V F DHQ QP + A N Q PCDI Y STMN IN
Sbjct: 521 ---------VEFYDDHQLQP----VNANNNCQYPCDIYFQQDQLYCPSTMNSINSAG--- 652
Query: 242 NANIGGTTCQQSNVFQGFENFTSDFSELVCVNPQTIDGFYGMESLEMSNVSSTNTPSAES 301
L VN Q +DG + + S ES
Sbjct: 653 ---------------------------LTYVNQQQLDG--------------SISTSTES 709
Query: 302 TSWGDMSSLVYSPLVSDYFGMQMQ 325
+SWGDMSSL++SPL SDY G Q
Sbjct: 710 SSWGDMSSLIHSPLASDYEGGHQQ 781
>BF634640 homologue to GP|2832408|emb R2R3-MYB transcription factor
{Arabidopsis thaliana}, partial (39%)
Length = 657
Score = 271 bits (693), Expect = 3e-73
Identities = 123/151 (81%), Positives = 138/151 (90%)
Frame = +3
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKANVK+GPWSPEED+KLK+YI+ HGTGGNWIALPQKIGLKRCGKSCRLRWLN
Sbjct: 81 MGRAPCCDKANVKKGPWSPEEDSKLKSYIEQHGTGGNWIALPQKIGLKRCGKSCRLRWLN 260
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPN+KHGGFSEEEDNIIC+LY+SIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK
Sbjct: 261 YLRPNLKHGGFSEEEDNIICSLYISIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 440
Query: 121 QRKEQQAQARRVSTMKQEIKRETDQNLMVAS 151
RK+QQ QAR +K+E++ N +++S
Sbjct: 441 HRKDQQQQARNRGNNGAIVKQESNNNRVMSS 533
>TC81805 similar to GP|9758323|dbj|BAB08797.1 Myb-related transcription
factor-like protein {Arabidopsis thaliana}, partial
(46%)
Length = 1221
Score = 254 bits (650), Expect = 2e-68
Identities = 149/358 (41%), Positives = 199/358 (54%), Gaps = 39/358 (10%)
Frame = +2
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKANVK+GPWSPEEDA LK+YI+ +GTGGNWIALPQKIGLKRCGKSCRLRWLN
Sbjct: 20 MGRAPCCDKANVKKGPWSPEEDATLKSYIETNGTGGNWIALPQKIGLKRCGKSCRLRWLN 199
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNIKHGGF+EEEDNIIC+LY+SIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK
Sbjct: 200 YLRPNIKHGGFTEEEDNIICSLYISIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 379
Query: 121 QRK----------------EQQAQARRVST-----MKQEIKRETDQNLMVASGVNTTHAA 159
+++ ++ + + +S+ ++ ++ ++ QN + S N AA
Sbjct: 380 RKQSNINNCLMNQKDTNGIDENSYSNSLSSSALERLQLHMQLQSLQNPL--SFYNNNPAA 553
Query: 160 FYWPAEYSIPMPVANASIQDYDFHNQTSLKSLLINKQGGVRFSYDHQQPYSNAITATANS 219
WP + + S+Q + +N +++ FS N I N+
Sbjct: 554 LVWPKLHPSQEKMIQISLQ--NSNNNPMMQN---------AFSSPQVDLLENIIPLENNN 700
Query: 220 QDPCDIYPASTMNMINPVAMVSNANIGGTTCQQSNVFQGFENFTSDFSELV--------- 270
+ + + N M S+ G ++S +G + S+ E++
Sbjct: 701 NNSVTFNASGNSSNNNNSIMHSSVAPRGEAVEKSTNNEGIQELESELDEILNNRNIITME 880
Query: 271 ---------CVNPQTIDGFYGMESLEMSNVSSTNTPSAESTSWGDMSSLVYSPLVSDY 319
C +G + SN S +T S S SW ++L+ + DY
Sbjct: 881 DEYRVAEFDCFRDMNNNGSKDQNLIWWSN-DSGDTKSGSSNSWDSTTNLMQEGMFQDY 1051
>TC89556 similar to GP|18419456|gb|AAL69334.1 blind {Lycopersicon
esculentum}, partial (37%)
Length = 990
Score = 221 bits (562), Expect = 4e-58
Identities = 95/118 (80%), Positives = 108/118 (91%)
Frame = +3
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR+PCCDKANVK+GPWSPEEDAKLK YI+ HGTGGNWI LP+K+GL RCGKSCRLRWLN
Sbjct: 18 MGRSPCCDKANVKKGPWSPEEDAKLKEYIEKHGTGGNWITLPKKVGLTRCGKSCRLRWLN 197
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLL 118
YLRPNIKHG FS+ ED IIC L+ SIGSRWSIIA++L GRTDND+KNYWNT+LKKK++
Sbjct: 198 YLRPNIKHGEFSDSEDKIICTLFASIGSRWSIIASKLEGRTDNDVKNYWNTKLKKKIM 371
>TC92226 similar to GP|18419456|gb|AAL69334.1 blind {Lycopersicon
esculentum}, partial (37%)
Length = 819
Score = 159 bits (401), Expect(2) = 8e-51
Identities = 85/184 (46%), Positives = 105/184 (56%), Gaps = 1/184 (0%)
Frame = +1
Query: 26 KAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLNYLRPNIKHGGFSEEEDNIICNLYVS 85
+ YI HGTGGNWI+LPQK GL+RCGKS R RWLNY RPNIKHG S+E+D IIC LY +
Sbjct: 109 REYIQKHGTGGNWISLPQKAGLRRCGKSSR*RWLNY*RPNIKHGECSDEQDRIICTLYAN 288
Query: 86 IGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGKQRKEQQAQARRVSTMKQEIKRETDQ 145
IGSRWSIIAAQ P RTDNDIKNYWNT+L KKL+ + + +
Sbjct: 289 IGSRWSIIAAQ*PRRTDNDIKNYWNTKLNKKLMNLLPQSHN------TVLPYLTSSSFPS 450
Query: 146 NLMVASGVNTTHAAFYWPAEYSIPMPVANASIQDYDFHNQTS-LKSLLINKQGGVRFSYD 204
+ T+ +F E +P +++SI +HNQ S L N + Y
Sbjct: 451 HCTSYFNQTPTNTSFTTSLEQQFSVPSSSSSISLPFYHNQESLLNGFSSNTNTNISMQYQ 630
Query: 205 HQQP 208
Q P
Sbjct: 631 LQNP 642
Score = 59.3 bits (142), Expect(2) = 8e-51
Identities = 24/26 (92%), Positives = 25/26 (95%)
Frame = +3
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLK 26
MGRAPCCDKANVK+GPWSPEED KLK
Sbjct: 33 MGRAPCCDKANVKKGPWSPEEDTKLK 110
>TC81520 similar to PIR|D86470|D86470 hypothetical protein AAD46010.1
[imported] - Arabidopsis thaliana, partial (39%)
Length = 1259
Score = 172 bits (435), Expect = 2e-43
Identities = 119/336 (35%), Positives = 171/336 (50%), Gaps = 23/336 (6%)
Frame = +2
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR+PCCD++ +K+GPW+PEED KL +I HG G +W ALP+ GL RCGKSCRLRW N
Sbjct: 56 MGRSPCCDESGLKKGPWTPEEDQKLVEHIQQHGHG-SWRALPKLAGLNRCGKSCRLRWTN 232
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKL--L 118
YLRP+IK G FS+EE+ I +L+ +G++WS IA LPGRTDN+IKN+WNT LKKKL +
Sbjct: 233 YLRPDIKRGKFSQEEEQTILHLHSILGNKWSAIATHLPGRTDNEIKNFWNTHLKKKLIQM 412
Query: 119 GKQRKEQQAQARRVSTMKQEI----KRETDQNLMVASGVNTTHAAFYWPAEYSIPMPVAN 174
G Q + VST+ + E + +S + HAA ++ +
Sbjct: 413 GFDPMTHQPRTDLVSTIPYLLALANMTEIIDHQNQSSWDDQQHAAMSSLQAEAVHL---- 580
Query: 175 ASIQDYDFHNQTSLKSLLINKQGGVRFSYDHQQPYSNAITATANSQDPCDIYPASTMNMI 234
A +Q + Q+S S+ IN SYD NA+ + P + +T++ +
Sbjct: 581 AKLQCLQYLLQSSNSSININNN-----SYD-----QNAMITNMEQEQPLSL--LNTISNV 724
Query: 235 NPVAMVSNANIGGTTC--------QQSNVFQGFENFTSDFSELVCVNPQ-----TIDGFY 281
++ + + T QS + + FS C+N + TI F
Sbjct: 725 KENTIMDCSQLDSTHAASFSQPLHHQSVLPHFLDPQQVSFSSQSCLNNEQSQGGTITNFA 904
Query: 282 GMESLEMSNVSSTN----TPSAESTSWGDMSSLVYS 313
++ NV S+ P+A S GD SS S
Sbjct: 905 TVDETSWINVPSSAPISVPPNAIGISAGDASSCTSS 1012
>TC77877 similar to GP|5139806|dbj|BAA81732.1 GmMYB29A2 {Glycine max},
partial (78%)
Length = 1156
Score = 170 bits (431), Expect = 6e-43
Identities = 80/168 (47%), Positives = 113/168 (66%)
Frame = +3
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
M RAPCC+K +K+GPW+ EED L +YI+ HG NW ALP+ GL RCGKSCRLRW+N
Sbjct: 66 MVRAPCCEKKGLKKGPWTLEEDEILTSYINKHGHS-NWRALPKHAGLLRCGKSCRLRWIN 242
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YL+P+IK G F+ EE+ I ++ S+G+RWS IAA+LPGRTDN+IKN W+T LKK+LL
Sbjct: 243 YLKPDIKRGNFTNEEEETIIKMHESLGNRWSAIAAKLPGRTDNEIKNVWHTHLKKRLLNT 422
Query: 121 QRKEQQAQARRVSTMKQEIKRETDQNLMVASGVNTTHAAFYWPAEYSI 168
+ + ++ KQ+IKR + + + N T ++ + E ++
Sbjct: 423 NNNQPNSNTKK-RVSKQKIKRSDSNSSTLTTASNCTFSSDFSSQEKNL 563
>BG448527 similar to PIR|T51509|T51 probable transcription factor (MYB9) -
Arabidopsis thaliana, partial (40%)
Length = 633
Score = 169 bits (429), Expect = 1e-42
Identities = 75/118 (63%), Positives = 94/118 (79%)
Frame = +1
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR+PCCD+ VK+GPW+ EED KL YI+ HG G NW L ++ GL RCGKSCRLRW N
Sbjct: 40 MGRSPCCDEIGVKKGPWTQEEDEKLIDYINKHGHG-NWGTLSKRAGLNRCGKSCRLRWTN 216
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLL 118
YLRP+IK G F++EE+ +I NL+ +G++WS IAA LPGRTDN+IKNYWNT ++KKLL
Sbjct: 217 YLRPDIKRGKFTDEEERVIINLHSVLGNKWSKIAAHLPGRTDNEIKNYWNTNIRKKLL 390
>TC90505 similar to GP|3941528|gb|AAC83640.1| putative transcription factor
{Arabidopsis thaliana}, partial (49%)
Length = 984
Score = 169 bits (428), Expect = 1e-42
Identities = 82/154 (53%), Positives = 103/154 (66%), Gaps = 10/154 (6%)
Frame = +1
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR PCCDK VK+GPW+PEED L YI HG G NW A+P K GL RC KSCRLRW N
Sbjct: 157 MGRPPCCDKEGVKKGPWTPEEDIILVTYIQEHGPG-NWRAVPTKTGLSRCSKSCRLRWTN 333
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRP IK G F+E+E+ +I +L +G+RW+ IA+ LP RTDNDIKNYWNT LKKKL
Sbjct: 334 YLRPGIKRGNFTEQEEKMIIHLQDLLGNRWAAIASYLPQRTDNDIKNYWNTHLKKKLKKS 513
Query: 121 Q----------RKEQQAQARRVSTMKQEIKRETD 144
Q +E+ + ++R+ + E + +TD
Sbjct: 514 QTGNNAECINFEEEKFSASQRIPRGQWERRLQTD 615
>TC86588 similar to GP|5139814|dbj|BAA81736.1 GmMYB29B2 {Glycine max},
partial (85%)
Length = 1389
Score = 168 bits (426), Expect = 2e-42
Identities = 83/146 (56%), Positives = 105/146 (71%)
Frame = +2
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
M RAPCC+K +KRGPWS EED L +YI HG G NW ALP+ GL RCGKSCRLRW+N
Sbjct: 107 MVRAPCCEKMGLKRGPWSLEEDEILTSYIQKHGHG-NWRALPKLAGLLRCGKSCRLRWIN 283
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRP+IK G F+ EE+ I L+ +G+RWS IAA+LPGRTDN+IKN W+T LKKKL
Sbjct: 284 YLRPDIKRGNFTNEEEENIIKLHEMLGNRWSAIAAKLPGRTDNEIKNVWHTHLKKKL--- 454
Query: 121 QRKEQQAQARRVSTMKQEIKRETDQN 146
+ ++A++ + K +IKR +D N
Sbjct: 455 --NQTNSEAKKKAISKPKIKR-SDSN 523
>TC88523 similar to PIR|T05690|T05690 myb-related transcription factor MYB4
- Arabidopsis thaliana, partial (54%)
Length = 1292
Score = 167 bits (422), Expect = 7e-42
Identities = 81/204 (39%), Positives = 121/204 (58%), Gaps = 9/204 (4%)
Frame = +1
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR+PCC+KA+ +G W+ EED +L +YI HG G W +LP+ GL RCGKSCRLRW+N
Sbjct: 271 MGRSPCCEKAHTNKGAWTKEEDDRLISYIRAHGEGC-WRSLPKAAGLLRCGKSCRLRWIN 447
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRP++K G F+EEED +I L+ +G++WS+IA +LPGRTDN+IKNYWNT +++KLL +
Sbjct: 448 YLRPDLKRGNFTEEEDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIRRKLLNR 627
Query: 121 ------QRKEQQAQARRVSTMKQEIKRETDQNLMVASGVNTTHAAFYWPAEYSIPMPVAN 174
R + + + ++ + + VA+ +T A P S +
Sbjct: 628 GIDPATHRPLNEVSHSQSQSQTLHLQNQEAVTIAVAASTSTPTATKTLPTTISFASSIKQ 807
Query: 175 ASIQDYDFH---NQTSLKSLLINK 195
+ H N +K L++ +
Sbjct: 808 EQYHHHHHHQEMNTNMVKGLVLER 879
>BE124666 similar to PIR|JQ0957|JQ0 myb-related protein 330 - garden
snapdragon, partial (48%)
Length = 536
Score = 164 bits (415), Expect = 4e-41
Identities = 74/139 (53%), Positives = 101/139 (72%)
Frame = +3
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR+PCC+K + +G W+ EED +L YI HG G W +LP+ GL RCGKSCRLRW+N
Sbjct: 27 MGRSPCCEKEHTNKGAWTKEEDERLINYIKLHGEGC-WRSLPKAAGLLRCGKSCRLRWIN 203
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRP++K G F+E+ED++I NL+ +G++WS+IAA+LPGRTDN+IKNYWNT +K+KL +
Sbjct: 204 YLRPDLKRGNFTEQEDDLIINLHSLLGNKWSLIAARLPGRTDNEIKNYWNTHIKRKLYSR 383
Query: 121 QRKEQQAQARRVSTMKQEI 139
Q ++ ST I
Sbjct: 384 GVDPQTHRSLNDSTTSTTI 440
>BG644406 homologue to PIR|T07398|T07 myb-related transcription factor THM6 -
tomato, partial (42%)
Length = 762
Score = 164 bits (414), Expect = 6e-41
Identities = 80/180 (44%), Positives = 109/180 (60%), Gaps = 1/180 (0%)
Frame = +1
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR PCCDK VK+GPW+PEED L +++ HG G NW +P GL+RC KSCRLRW N
Sbjct: 145 MGRPPCCDKTGVKKGPWTPEEDIMLVSFVQEHGPG-NWRTVPTHTGLRRCSKSCRLRWTN 321
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRP IK G F+++E+ +I L +G++W+ IA+ LP RTDNDIKNYWNT LKKKL
Sbjct: 322 YLRPGIKRGSFTDQEEKMIIQLQALLGNKWAAIASYLPERTDNDIKNYWNTHLKKKLKKL 501
Query: 121 QRKEQQA-QARRVSTMKQEIKRETDQNLMVASGVNTTHAAFYWPAEYSIPMPVANASIQD 179
+ + +ST + + ++ L + +NT AF P+ + +I D
Sbjct: 502 ETSGLYSNDGHCLSTYNSTSRGQWERTLQ--ADINTAKQAFTNALSLDKSSPIPDYTITD 675
>BF647428 homologue to PIR|T51621|T51 myb-like protein [imported] -
Arabidopsis thaliana, partial (44%)
Length = 689
Score = 162 bits (411), Expect = 1e-40
Identities = 74/117 (63%), Positives = 87/117 (74%)
Frame = +2
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
M R PCCDK VK+GPW+PEED L +YI HG G NW A+P GL RC KSCRLRW N
Sbjct: 92 MVRPPCCDKEGVKKGPWTPEEDIILVSYIQEHGPG-NWKAVPTNTGLLRCSKSCRLRWTN 268
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKL 117
YLRP IK G F+++E+ +I +L +G+RW+ IAA LP RTDNDIKNYWNT LKKKL
Sbjct: 269 YLRPGIKRGNFTDQEEKMIIHLQALLGNRWAAIAAYLPQRTDNDIKNYWNTYLKKKL 439
>TC84780 homologue to PIR|F85021|F85021 probable transcription factor
[imported] - Arabidopsis thaliana, partial (39%)
Length = 971
Score = 160 bits (404), Expect = 8e-40
Identities = 81/181 (44%), Positives = 113/181 (61%), Gaps = 13/181 (7%)
Frame = +1
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR CC K +++G WSPEED KL +I +G G W ++P++ GL+RCGKSCRLRW+N
Sbjct: 103 MGRHSCCYKQKLRKGLWSPEEDEKLLNHITKYGHGC-WSSVPKQAGLQRCGKSCRLRWIN 279
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRP++K G FS+EE+N+I L+ +G+RWS IAAQLPGRTDN+IKN WN+ LKKKL K
Sbjct: 280 YLRPDLKRGTFSQEEENLIIELHSVLGNRWSQIAAQLPGRTDNEIKNLWNSCLKKKLRQK 459
Query: 121 -------------QRKEQQAQARRVSTMKQEIKRETDQNLMVASGVNTTHAAFYWPAEYS 167
+ +++ R EI + N++ + + + A Y P + S
Sbjct: 460 GIDPVTHKPLSEVENVDEEEGKTRSQEKTAEISESDELNIVRSESTSKSDAVSYEPKQSS 639
Query: 168 I 168
I
Sbjct: 640 I 642
>BI311137 similar to GP|17380966|gb putative transcription factor
{Arabidopsis thaliana}, partial (35%)
Length = 761
Score = 159 bits (403), Expect = 1e-39
Identities = 74/120 (61%), Positives = 91/120 (75%)
Frame = +1
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR CC K +++G WSPEED KL YI HG G W ++P+ GL+RCGKSCRLRW+N
Sbjct: 148 MGRHSCCYKQKLRKGLWSPEEDEKLLNYITKHGHGC-WSSVPKLAGLQRCGKSCRLRWIN 324
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRP++K G FSE+E+N I L+ +G+RWS IAAQLPGRTDN+IKN WN+ LKKKL K
Sbjct: 325 YLRPDLKRGAFSEQEENSIIELHALLGNRWSQIAAQLPGRTDNEIKNLWNSSLKKKLKQK 504
>TC81202 homologue to GP|17380966|gb|AAL36295.1 putative transcription
factor {Arabidopsis thaliana}, partial (40%)
Length = 1506
Score = 156 bits (395), Expect = 9e-39
Identities = 70/117 (59%), Positives = 91/117 (76%)
Frame = +2
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
+GR CC K +++G WSPEED KL YI HG G W ++P+ GL+RCGKSCRLRW+N
Sbjct: 59 LGRHSCCYKQKLRKGLWSPEEDEKLLNYITKHGHGC-WSSVPKLAGLQRCGKSCRLRWIN 235
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKL 117
YLRP++K G FS++E+N+I L+ +G+RWS IAAQLPGRTDN+IKN WN+ LKK+L
Sbjct: 236 YLRPDLKRGAFSQQEENLIIELHAVLGNRWSQIAAQLPGRTDNEIKNLWNSCLKKRL 406
>TC82254 similar to GP|22266673|dbj|BAC07543. myb-related transcription
factor VlMYBB1-1 {Vitis labrusca x Vitis vinifera},
partial (46%)
Length = 754
Score = 154 bits (390), Expect = 3e-38
Identities = 73/147 (49%), Positives = 101/147 (68%), Gaps = 7/147 (4%)
Frame = +2
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
M R P CDK +++G W+PEED KL AY+ +G NW LP+ GL+RCGKSCRLRWLN
Sbjct: 98 MVRTPYCDKNGLRKGTWTPEEDKKLIAYVTRYGCW-NWRQLPKFAGLERCGKSCRLRWLN 274
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRP+IK G FS EE+ I L+ +G+RW++I+A LPGRTDN+IKNYW+T +KK L+
Sbjct: 275 YLRPDIKRGSFSHEEEETIIKLHEKLGNRWTMISANLPGRTDNEIKNYWHTTIKKTLVKN 454
Query: 121 Q-------RKEQQAQARRVSTMKQEIK 140
+ +K + + ++ TM++ K
Sbjct: 455 KSNTKRGTKKAKDSNSKNHPTMEKPKK 535
>TC78266 similar to GP|22266673|dbj|BAC07543. myb-related transcription
factor VlMYBB1-1 {Vitis labrusca x Vitis vinifera},
partial (46%)
Length = 665
Score = 153 bits (387), Expect = 8e-38
Identities = 68/118 (57%), Positives = 91/118 (76%), Gaps = 1/118 (0%)
Frame = +1
Query: 1 MGRAPCCDK-ANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWL 59
M R P CDK + +++G W+ EED KL AY+ +G NW LP+ GL RCGKSCRLRWL
Sbjct: 85 MARTPSCDKKSGMRKGTWTAEEDRKLIAYVTRYGCW-NWRQLPKFAGLSRCGKSCRLRWL 261
Query: 60 NYLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKL 117
NYLRPNIK G F++EE+ +I ++ ++G+RWS IAA+LPGRTDN++KNYW+T LKK++
Sbjct: 262 NYLRPNIKRGNFTQEEEELIIRMHKNLGNRWSTIAAELPGRTDNEVKNYWHTSLKKRV 435
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.130 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,778,390
Number of Sequences: 36976
Number of extensions: 151503
Number of successful extensions: 909
Number of sequences better than 10.0: 102
Number of HSP's better than 10.0 without gapping: 839
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 855
length of query: 325
length of database: 9,014,727
effective HSP length: 96
effective length of query: 229
effective length of database: 5,465,031
effective search space: 1251492099
effective search space used: 1251492099
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0181.1


