

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
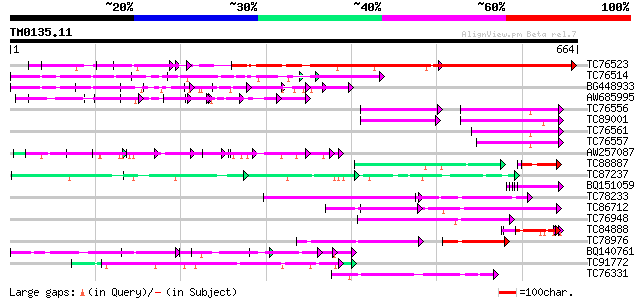
Query= TM0135.11
(664 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 465 e-131
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 326 2e-89
BG448933 similar to PIR|T09648|T09 nucleolin homolog nuM1 - alfa... 150 2e-36
AW685995 similar to PIR|I51618|I516 nucleolar phosphoprotein - A... 89 4e-18
TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 87 2e-17
TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-bi... 86 4e-17
TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 80 2e-15
TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 79 4e-15
AW257087 homologue to GP|6683126|dbj KIAA0339 protein {Homo sapi... 74 2e-13
TC88887 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproli... 49 4e-13
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 72 9e-13
BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melano... 71 2e-12
TC78233 similar to GP|15809838|gb|AAL06847.1 AT5g51120/MWD22_6 {... 70 2e-12
TC86712 similar to GP|17528972|gb|AAL38696.1 putative RNA-bindin... 70 3e-12
TC76948 similar to SP|Q08935|ROC1_NICSY 29 kDa ribonucleoprotein... 69 4e-12
TC84888 similar to GP|21322752|dbj|BAB78536. cold shock protein-... 69 8e-12
TC78976 similar to GP|10178188|dbj|BAB11662. poly(A)-binding pro... 68 1e-11
BQ140761 weakly similar to GP|18076245|emb phosphophoryn {Rattus... 68 1e-11
TC91772 weakly similar to GP|17380998|gb|AAL36311.1 unknown prot... 68 1e-11
TC76331 similar to GP|7673355|gb|AAF66823.1| poly(A)-binding pro... 68 1e-11
>TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (69%)
Length = 1615
Score = 465 bits (1197), Expect = e-131
Identities = 260/411 (63%), Positives = 300/411 (72%), Gaps = 7/411 (1%)
Frame = +3
Query: 260 ADSDSSDSDSSSDEDDDKKKPATKVAKKLPAAAPIKKVEKSESSDDESSESSDEENDSKP 319
A S +SD SDED+D K P +K PAA K + S SDDE +SS +E D KP
Sbjct: 159 ASSSDEESDEESDEDEDAK-PVSK-----PAAVAKKSKKDSSDSDDEDDDSSSDE-DKKP 317
Query: 320 ATTTVAKPSAVKKSASSDSDDEDSSDSDSEVKKDKMDVDESDSSEDSDEEDEKSTKTPQK 379
VA V +S S SDDED K +D D SDS E +E +++ +KTPQK
Sbjct: 318 ----VAAKKEVSESESDSSDDED---------KMNVDKDGSDSDESEEESEDEPSKTPQK 458
Query: 380 KVKDVEMVDAS-SGKNAPKTPAAQTENEGSKTLFVGNLSFSVQRSDVENFFKDCGEVVDV 438
K+KDVEMVDA SGK AP TPA EN GSKTLFVGNLSFSVQRSD+E FF+DCGEVVDV
Sbjct: 459 KIKDVEMVDAGKSGKKAPNTPATPIENSGSKTLFVGNLSFSVQRSDIEKFFQDCGEVVDV 638
Query: 439 RLATDEDGRFKGFGHVEFATAAAAQSALEYNGSELLQRPVRLDLARERGAYTPNSGAANN 498
R ++DE+GRFKGFGHVEFA+A AAQSALE NG ELLQR VRLDLARERGA+TPN+ +N
Sbjct: 639 RFSSDEEGRFKGFGHVEFASAEAAQSALEMNGQELLQRAVRLDLARERGAFTPNNN-SNY 815
Query: 499 SFQKGGRGQSQTVFVRGFDKFQGEDDIKSSLQEHF-GSCGEITRISVPKDFESGNVKGFA 557
S Q GGRGQSQTVFVRGFDK GED+I++ L EHF G+CGE TR+S+PKDFESG KGFA
Sbjct: 816 SAQSGGRGQSQTVFVRGFDKNLGEDEIRAKLMEHFGGTCGEPTRVSIPKDFESGYSKGFA 995
Query: 558 YLDFSDTNSVNKALELHDSDLGGFTLAVDEAKPRDNQGSGGRSGGGRSGGGR-GGRFDSG 616
Y+DF D++S +KALELH+S+L G+ L+VDEAKPRD+QGSGGR GGGRSGGGR GG SG
Sbjct: 996 YMDFKDSDSFSKALELHESELDGYQLSVDEAKPRDSQGSGGRGGGGRSGGGRFGGGGRSG 1175
Query: 617 RGGGRGRFGSGGR--GGDRGGRGGRGG-GGRGGRGTP-LRAEGKKTTFADE 663
GGR GGR G D GGRGGRGG GGRGG P +EGKKTTFAD+
Sbjct: 1176GFGGRSGGRGGGRFGGRDSGGRGGRGGRGGRGGFNKPSFASEGKKTTFADD 1328
Score = 93.2 bits (230), Expect = 3e-19
Identities = 109/412 (26%), Positives = 167/412 (40%), Gaps = 25/412 (6%)
Frame = +3
Query: 122 IAKKPTKVVVPKKEESSSDEDSSSDEDSSSESDDEKPAAKVAVPAKKAPAKNGSVTAPAK 181
+ +KP K+ V + + +S D SDE+S E +D KP +K A AKK+ K
Sbjct: 111 LQQKPLKM*VHPQRKPASSSDEESDEESD-EDEDAKPVSKPAAVAKKS-----------K 254
Query: 182 KAQPAPSSSSESEDDDSSEEDEPSAKQPKKEVAVKPAVQKKKEESSSDDSSSESEDEKPA 241
K SS S+ EDDDSS +++ KK VA KKE S S+ SS+ ED+
Sbjct: 255 K----DSSDSDDEDDDSSSDED------KKPVAA------KKEVSESESDSSDDEDKM-- 380
Query: 242 VKVSAPSKVVPAAVKNGAADSDSSDSDSSSDEDDDKKKPATKVAKKLPAAAPIKKVEKSE 301
D D SDSD S +E +D+ P+ P KK++ E
Sbjct: 381 -----------------NVDKDGSDSDESEEESEDE-----------PSKTPQKKIKDVE 476
Query: 302 SSDDESSESSDEENDSKPATTTVAKPSAVKKSASS--DSDDEDSSDSDSEVKKDKMDVDE 359
D S + P + +K V + S SD E EV + DE
Sbjct: 477 MVDAGKSGKKAPNTPATPIENSGSKTLFVGNLSFSVQRSDIEKFFQDCGEVVDVRFSSDE 656
Query: 360 SDSSEDSDEEDEKSTKTPQKKVK-------------DVEMVDASSGKNAPKTPAAQTENE 406
+ + S + Q ++ D+ + N +AQ+
Sbjct: 657 EGRFKGFGHVEFASAEAAQSALEMNGQELLQRAVRLDLARERGAFTPNNNSNYSAQSGGR 836
Query: 407 G-SKTLFVGNLSFSVQRSDV-----ENFFKDCGEVVDVRLATD-EDGRFKGFGHVEFATA 459
G S+T+FV ++ ++ E+F CGE V + D E G KGF +++F +
Sbjct: 837 GQSQTVFVRGFDKNLGEDEIRAKLMEHFGGTCGEPTRVSIPKDFESGYSKGFAYMDFKDS 1016
Query: 460 AAAQSALEYNGSELLQRPVRLDLARER---GAYTPNSGAANNSFQKGGRGQS 508
+ ALE + SEL + +D A+ R G+ G + + GG G+S
Sbjct: 1017DSFSKALELHESELDGYQLSVDEAKPRDSQGSGGRGGGGRSGGGRFGGGGRS 1172
Score = 56.2 bits (134), Expect = 4e-08
Identities = 53/177 (29%), Positives = 78/177 (43%), Gaps = 14/177 (7%)
Frame = +3
Query: 38 KQVSAKKQKIAEVAAKQKKETKLQKVKKESSSDDDSSSE-----DEKPAPKVAPLKTSVK 92
K ++ +KI + K K + K SSSD++S E D KP K A +
Sbjct: 75 KTLNLLMKKIRNLQQKPLKM*VHPQRKPASSSDEESDEESDEDEDAKPVSKPAAV----- 239
Query: 93 NGTTPAKKAKPAPSSSSSDEDSSDEEEEVIAKKPTKVVVPKKEESSSDEDSSSDED---- 148
AKK+K S S ++D S +E+ KKP V KKE S S+ DSS DED
Sbjct: 240 -----AKKSKKDSSDSDDEDDDSSSDED---KKP---VAAKKEVSESESDSSDDEDKMNV 386
Query: 149 -----SSSESDDEKPAAKVAVPAKKAPAKNGSVTAPAKKAQPAPSSSSESEDDDSSE 200
S ES++E P KK K+ + K + AP++ + ++ S+
Sbjct: 387 DKDGSDSDESEEESEDEPSKTPQKK--IKDVEMVDAGKSGKKAPNTPATPIENSGSK 551
Score = 50.8 bits (120), Expect = 2e-06
Identities = 35/113 (30%), Positives = 53/113 (45%), Gaps = 1/113 (0%)
Frame = +3
Query: 102 KPAPSSSSSDEDSSDEEEEVIAKKPTKVVVPKKEESSSDEDSSSDEDSSSESDDEKPAAK 161
KPA SS ++ SDE+E+ V K ++ SSD D D+DSSS+ D + AAK
Sbjct: 153 KPASSSDEESDEESDEDEDAKPVSKPAAVAKKSKKDSSDSDDE-DDDSSSDEDKKPVAAK 329
Query: 162 VAVPAKKAPAKNGSVTAPAKK-AQPAPSSSSESEDDDSSEEDEPSAKQPKKEV 213
V ++ + + K + S ESED+ S K P+K++
Sbjct: 330 KEVSESESDSSDDEDKMNVDKDGSDSDESEEESEDEPS--------KTPQKKI 464
Score = 48.9 bits (115), Expect = 6e-06
Identities = 49/172 (28%), Positives = 72/172 (41%), Gaps = 1/172 (0%)
Frame = +3
Query: 23 KSAKKAKRAAEDEVEKQVSAKK-QKIAEVAAKQKKETKLQKVKKESSSDDDSSSEDEKPA 81
K A + +++E ++ AK K A VA K KK++ + DD SS ED+KP
Sbjct: 153 KPASSSDEESDEESDEDEDAKPVSKPAAVAKKSKKDSS----DSDDEDDDSSSDEDKKP- 317
Query: 82 PKVAPLKTSVKNGTTPAKKAKPAPSSSSSDEDSSDEEEEVIAKKPTKVVVPKKEESSSDE 141
AKK S S+ DSSD+E+++ K+ S SDE
Sbjct: 318 --------------VAAKK-----EVSESESDSSDDEDKMNV---------DKDGSDSDE 413
Query: 142 DSSSDEDSSSESDDEKPAAKVAVPAKKAPAKNGSVTAPAKKAQPAPSSSSES 193
ED S++ +K V A K+ K AP A P +S S++
Sbjct: 414 SEEESEDEPSKTPQKKIKDVEMVDAGKSGKK-----APNTPATPIENSGSKT 554
Score = 39.3 bits (90), Expect = 0.005
Identities = 26/57 (45%), Positives = 37/57 (64%), Gaps = 5/57 (8%)
Frame = +2
Query: 248 SKVVPAAVKNGA-----ADSDSSDSDSSSDEDDDKKKPATKVAKKLPAAAPIKKVEK 299
+K VP+ KNG+ AD++SSD DS S +++D KKPA K +K + +AP KK K
Sbjct: 8 AKAVPS--KNGSVPAKKADTESSDEDSESSDEED-KKPAAKASKNV--SAPTKKACK 163
Score = 37.7 bits (86), Expect = 0.014
Identities = 22/49 (44%), Positives = 30/49 (60%)
Frame = +2
Query: 166 AKKAPAKNGSVTAPAKKAQPAPSSSSESEDDDSSEEDEPSAKQPKKEVA 214
AK P+KNGSV PAKKA + SS+ + + S EED+ A + K V+
Sbjct: 8 AKAVPSKNGSV--PAKKAD---TESSDEDSESSDEEDKKPAAKASKNVS 139
Score = 37.7 bits (86), Expect = 0.014
Identities = 26/57 (45%), Positives = 32/57 (55%), Gaps = 1/57 (1%)
Frame = +2
Query: 80 PAPKVAPLKTSVKNGTTPAKKAKPAPSSSSSDEDSSDEEEEVIAKKPTK-VVVPKKE 135
PA K P K NG+ PAKKA SS D +SSDEE++ A K +K V P K+
Sbjct: 2 PAAKAVPSK----NGSVPAKKAD--TESSDEDSESSDEEDKKPAAKASKNVSAPTKK 154
Score = 36.2 bits (82), Expect = 0.042
Identities = 21/52 (40%), Positives = 29/52 (55%), Gaps = 8/52 (15%)
Frame = +2
Query: 126 PTKVVVPKKEES----SSDEDSSSDEDSSSESDDEKPAAK----VAVPAKKA 169
P VP K S +D +SS ++ SS+ +D+KPAAK V+ P KKA
Sbjct: 2 PAAKAVPSKNGSVPAKKADTESSDEDSESSDEEDKKPAAKASKNVSAPTKKA 157
Score = 36.2 bits (82), Expect = 0.042
Identities = 24/55 (43%), Positives = 26/55 (46%)
Frame = +2
Query: 280 PATKVAKKLPAAAPIKKVEKSESSDDESSESSDEENDSKPATTTVAKPSAVKKSA 334
PA K + P KK + S DE SESSDEE D KPA SA K A
Sbjct: 2 PAAKAVPSKNGSVPAKKADTESS--DEDSESSDEE-DKKPAAKASKNVSAPTKKA 157
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 326 bits (835), Expect = 2e-89
Identities = 221/447 (49%), Positives = 270/447 (59%), Gaps = 8/447 (1%)
Frame = +3
Query: 1 MPKSSKKSATKVEAAAPVAAAPKSAKKAKRAAEDEVEKQVSAKKQKIAEVAAKQKKETKL 60
MPKSSKKSATKV+AA V + KS KK KR AE+EV K VSAKKQK+ EVAAKQK L
Sbjct: 102 MPKSSKKSATKVDAAPVVVSPVKSGKKGKRQAEEEV-KAVSAKKQKVEEVAAKQK---AL 269
Query: 61 QKVKK-ESSSDDDSSSEDEKPAPKV-AP-LKTSVKNGTTPAKKAKPAPSSSSSDEDSSDE 117
+ VKK ESSS++ S SEDE+P K AP KT K G KKA+P +S SD DSS
Sbjct: 270 KVVKKEESSSEESSESEDEQPVVKAPAPSKKTPAKKGN--VKKAQPETTSEESDSDSSSS 443
Query: 118 EEEVIAKKPTKVVVPKKEESSSDE--DSSSDEDSSSESDDEKPAAKVAVPAKKAPAKNGS 175
+EE + KKP VP K S+ + D+S +EDS SD++K A AK P+KNGS
Sbjct: 444 DEEEV-KKPVSKAVPSKNGSAPAKKVDTSEEEDSEESSDEDKKPA-----AKAVPSKNGS 605
Query: 176 VTAPAKKAQPAPSSSSESEDDDSSEEDEPSAKQPKKEVAVKPAVQKKKEESSSDDSSSES 235
APAKKA + ES D+D E+++P+AK + PA + E S D SS+
Sbjct: 606 --APAKKAASDEEDTDESSDED-EEDEKPAAKAVPSKNGSVPAKKADTESSDEDSESSDE 776
Query: 236 EDEKPAVKVSAPSKVVPAAVKNGAADSDSSDSDSSSDEDDDKKKPATKVAKKLPAAAPIK 295
ED+KPA K SK V A K A+ SD +SD SDED+D KP +K PAA K
Sbjct: 777 EDKKPAAKA---SKNVSAPTKKAASSSD-EESDEESDEDED-AKPVSK-----PAAVAKK 926
Query: 296 KVEKSESSDDESSESSDEENDSKPATTTVAKPSAVKKSASSDSDDEDSSDSDSEVKKDKM 355
+ S SDDE +SS +E+ KP A KK +DKM
Sbjct: 927 SKKDSSDSDDEDDDSSSDED---------KKPVAAKK-------------------EDKM 1022
Query: 356 DVDE--SDSSEDSDEEDEKSTKTPQKKVKDVEMVDA-SSGKNAPKTPAAQTENEGSKTLF 412
+VD+ SDS + +E +++ +KTPQKK+KDVEMVDA SGK AP TPA EN GSKTLF
Sbjct: 1023NVDKDGSDSDQSEEESEDEPSKTPQKKIKDVEMVDAGKSGKKAPNTPATPIENSGSKTLF 1202
Query: 413 VGNLSFSVQRSDVENFFKDCGEVVDVR 439
VGNLSFSVQRSD+E FF+DCGEVVDVR
Sbjct: 1203VGNLSFSVQRSDIEKFFQDCGEVVDVR 1283
Score = 43.5 bits (101), Expect = 3e-04
Identities = 70/217 (32%), Positives = 87/217 (39%), Gaps = 14/217 (6%)
Frame = -1
Query: 143 SSSDEDSSSESDDEKPAAKVAVPAKKAPAKNGSVTAPAKKAQPAPSSSSESEDDDSSEED 202
SSS DSSS+S E+ AA G++T +A A SS SED +SS ED
Sbjct: 886 SSSSSDSSSDSSSEELAAFFV----------GALT--FLEAFAAGFLSSSSEDSESSSED 743
Query: 203 EPSA----KQPKKEVAVKPAVQKKKEESSSDDSSSESEDEKPAVKVSAPSKVVPAAVKNG 258
SA P E A SSS+DSS S E A A + A+ G
Sbjct: 742 SVSAFFAGTLPFLE-GTAFAAGFSSSSSSSEDSSVSSSSE-AAFFAGALPFLDGTALAAG 569
Query: 259 AADSDSSDSDSSSDEDDDKKKPATKVAKKLP---AAAPIKKVEKSESSDDESSESSDEEN 315
S S+SSS E +T A LP A S SS++E SES E
Sbjct: 568 FLSSSEDSSESSSSE------VSTFFAGALPFLDGTALDTGFFTSSSSEEELSESDSSEV 407
Query: 316 DSKPATTTVA-------KPSAVKKSASSDSDDEDSSD 345
S A T + + + S SD EDSS+
Sbjct: 406 VSGWAFFTFPFLAGVFFEGAGAFTTGCSSSDSEDSSE 296
Score = 43.1 bits (100), Expect = 3e-04
Identities = 56/179 (31%), Positives = 69/179 (38%)
Frame = -1
Query: 186 APSSSSESEDDDSSEEDEPSAKQPKKEVAVKPAVQKKKEESSSDDSSSESEDEKPAVKVS 245
A SSSS+S D SSEE A + A SSS+DS S SED A
Sbjct: 889 ASSSSSDSSSDSSSEE---LAAFFVGALTFLEAFAAGFLSSSSEDSESSSEDSVSAFFAG 719
Query: 246 APSKVVPAAVKNGAADSDSSDSDSSSDEDDDKKKPATKVAKKLPAAAPIKKVEKSESSDD 305
+ A G + S SS DSS + A A LP SS +
Sbjct: 718 TLPFLEGTAFAAGFSSSSSSSEDSSVSSSSE----AAFFAGALPFLDGTALAAGFLSSSE 551
Query: 306 ESSESSDEENDSKPATTTVAKPSAVKKSASSDSDDEDSSDSDSEVKKDKMDVDESDSSE 364
+SSESS E +T A + D+ SS S+ E + ESDSSE
Sbjct: 550 DSSESSSSE-----VSTFFAGALPFLDGTALDTGFFTSSSSEEE-------LSESDSSE 410
Score = 36.2 bits (82), Expect = 0.042
Identities = 48/171 (28%), Positives = 65/171 (37%)
Frame = -1
Query: 106 SSSSSDEDSSDEEEEVIAKKPTKVVVPKKEESSSDEDSSSDEDSSSESDDEKPAAKVAVP 165
SSSSSD S EE+ A + E ++ SSS EDS S S+D + A
Sbjct: 886 SSSSSDSSSDSSSEELAAFFVGALTF--LEAFAAGFLSSSSEDSESSSED----SVSAFF 725
Query: 166 AKKAPAKNGSVTAPAKKAQPAPSSSSESEDDDSSEEDEPSAKQPKKEVAVKPAVQKKKEE 225
A P G+ A SSSS S +D S +A +
Sbjct: 724 AGTLPFLEGTAFAAGF------SSSSSSSEDSSVSSSSEAAFFAGALPFLDGTALAAGFL 563
Query: 226 SSSDDSSSESEDEKPAVKVSAPSKVVPAAVKNGAADSDSSDSDSSSDEDDD 276
SSS+DSS S E A + A+ G S SS+ + S + +
Sbjct: 562 SSSEDSSESSSSEVSTFFAGALPFLDGTALDTGFFTSSSSEEELSESDSSE 410
>BG448933 similar to PIR|T09648|T09 nucleolin homolog nuM1 - alfalfa, partial
(29%)
Length = 663
Score = 150 bits (379), Expect = 2e-36
Identities = 109/219 (49%), Positives = 132/219 (59%), Gaps = 3/219 (1%)
Frame = +2
Query: 1 MPKSSKKSATKVEAAAPVAAAPKSAKKAKRAAEDEVEKQVSAKKQKIAEVAAKQKKETKL 60
MPKSSKKSATKV+AA V + KS KK KR AE+EV K VSAKKQK+ EVAAKQK L
Sbjct: 80 MPKSSKKSATKVDAAPVVVSPVKSGKKGKRQAEEEV-KAVSAKKQKVEEVAAKQK---AL 247
Query: 61 QKVKK-ESSSDDDSSSEDEKPAPKV-AP-LKTSVKNGTTPAKKAKPAPSSSSSDEDSSDE 117
+ VKK ESSS++ S SEDE+P K AP KT K G KKA+P +S SD DSS
Sbjct: 248 KVVKKEESSSEESSESEDEQPVVKAPAPSKKTPAKKGN--VKKAQPETTSEESDSDSSSS 421
Query: 118 EEEVIAKKPTKVVVPKKEESSSDEDSSSDEDSSSESDDEKPAAKVAVPAKKAPAKNGSVT 177
+EE + K +K V K + + + +SDE D+KPAAK P+KNGS
Sbjct: 422 DEEEVKKPVSKAVPSKNGSAPAKKVDTSDE-------DKKPAAKA------VPSKNGS-- 556
Query: 178 APAKKAQPAPSSSSESEDDDSSEEDEPSAKQPKKEVAVK 216
APAKKA +S E + D+SS+EDE K K V K
Sbjct: 557 APAKKA-----ASDEEDTDESSDEDEEXXKPAAKAVPSK 658
Score = 101 bits (252), Expect = 8e-22
Identities = 74/203 (36%), Positives = 102/203 (49%), Gaps = 7/203 (3%)
Frame = +2
Query: 157 KPAAKVAVPAKKAPAKNGSVTAPAKKAQPAPSSSSESEDDDSSEEDEPSAKQPKKEVAVK 216
K + K A AP V +P K + + E S++ KQ +EVA K
Sbjct: 86 KSSKKSATKVDAAPV----VVSPVKSGKKGKRQAEEEVKAVSAK------KQKVEEVAAK 235
Query: 217 PAVQK--KKEESSSDDSSSESEDEKPAVKVSAPSKVVPAAVKN-----GAADSDSSDSDS 269
K KKEESSS++SS ESEDE+P VK APSK PA N S+ SDSDS
Sbjct: 236 QKALKVVKKEESSSEESS-ESEDEQPVVKAPAPSKKTPAKKGNVKKAQPETTSEESDSDS 412
Query: 270 SSDEDDDKKKPATKVAKKLPAAAPIKKVEKSESSDDESSESSDEENDSKPATTTVAKPSA 329
SS ++++ KKP +K +AP KKV+ S+ ++++ +N S PA
Sbjct: 413 SSSDEEEVKKPVSKAVPSKNGSAPAKKVDTSDEDKKPAAKAVPSKNGSAPA--------- 565
Query: 330 VKKSASSDSDDEDSSDSDSEVKK 352
KK+AS + D ++SSD D E K
Sbjct: 566 -KKAASDEEDTDESSDEDEEXXK 631
Score = 99.8 bits (247), Expect = 3e-21
Identities = 82/213 (38%), Positives = 109/213 (50%), Gaps = 7/213 (3%)
Frame = +2
Query: 78 EKPAPKV--APLKTS-VKNGTTPAKKAKPAPSSSSSDEDSSDEEEEVIAKKPTKVVVPKK 134
+K A KV AP+ S VK+G ++A+ + S+ + + EEV AK+ VV K
Sbjct: 95 KKSATKVDAAPVVVSPVKSGKKGKRQAEEEVKAVSAKKQ---KVEEVAAKQKALKVVKK- 262
Query: 135 EESSSDEDSSSDEDSSSE----SDDEKPAAKVAVPAKKAPAKNGSVTAPAKKAQPAPSSS 190
E+SSSE S+DE+P K P+KK PAK G+V KKAQ P ++
Sbjct: 263 ------------EESSSEESSESEDEQPVVKAPAPSKKTPAKKGNV----KKAQ--PETT 388
Query: 191 SESEDDDSSEEDEPSAKQPKKEVAVKPAVQKKKEESSSDDSSSESEDEKPAVKVSAPSKV 250
SE D DSS DE K+P V AV K + + + ED+KPA K + PSK
Sbjct: 389 SEESDSDSSSSDEEEVKKP-----VSKAVPSKNGSAPAKKVDTSDEDKKPAAK-AVPSKN 550
Query: 251 VPAAVKNGAADSDSSDSDSSSDEDDDKKKPATK 283
A K A SD D+D SSDED++ KPA K
Sbjct: 551 GSAPAKKAA--SDEEDTDESSDEDEEXXKPAAK 643
Score = 78.6 bits (192), Expect = 7e-15
Identities = 66/206 (32%), Positives = 92/206 (44%), Gaps = 11/206 (5%)
Frame = +2
Query: 130 VVPKKEESSSDEDSSSDEDSSSESDDEKPAAKVAVPAKKAPAKNGSV---TAPAKKAQPA 186
++PK + S+ + ++ S +K + K AK V A K +
Sbjct: 77 IMPKSSKKSATKVDAAPVVVSPVKSGKKGKRQAEEEVKAVSAKKQKVEEVAAKQKALKVV 256
Query: 187 PSSSSESEDDDSSEEDEPSAKQP---KKEVAVKPAVQKKK-----EESSSDDSSSESEDE 238
S SE+ SE+++P K P KK A K V+K + EES SD SSS+ E+
Sbjct: 257 KKEESSSEESSESEDEQPVVKAPAPSKKTPAKKGNVKKAQPETTSEESDSDSSSSDEEEV 436
Query: 239 KPAVKVSAPSKVVPAAVKNGAADSDSSDSDSSSDEDDDKKKPATKVAKKLPAAAPIKKVE 298
K V + PS KNG+A + D +SDED KKPA K +AP KK
Sbjct: 437 KKPVSKAVPS-------KNGSAPAKKVD---TSDED---KKPAAKAVPSKNGSAPAKKAA 577
Query: 299 KSESSDDESSESSDEENDSKPATTTV 324
E DESS+ ++E KPA V
Sbjct: 578 SDEEDTDESSD--EDEEXXKPAAKAV 649
Score = 64.3 bits (155), Expect = 1e-10
Identities = 53/186 (28%), Positives = 82/186 (43%), Gaps = 18/186 (9%)
Frame = +2
Query: 205 SAKQPKKEVAVKPAVQ------KKKEESSSDDSSSESEDEKPAVKVSAPSKVVPAAVKNG 258
S+K+ +V P V KK + + ++ + S ++ +V+A K + K
Sbjct: 89 SSKKSATKVDAAPVVVSPVKSGKKGKRQAEEEVKAVSAKKQKVEEVAAKQKALKVVKKE- 265
Query: 259 AADSDSSDSDSSSDEDDDKKKPATKVAKKLPAA-APIKKVEKSESSD--DESSESSDEEN 315
+S S +S S DE K PA +KK PA +KK + +S+ D S SSDEE
Sbjct: 266 --ESSSEESSESEDEQPVVKAPAP--SKKTPAKKGNVKKAQPETTSEESDSDSSSSDEEE 433
Query: 316 DSKPATTTVAKPSAVKKSASSDSDDED---------SSDSDSEVKKDKMDVDESDSSEDS 366
KP + V + + D+ DED S + + KK D +++D S D
Sbjct: 434 VKKPVSKAVPSKNGSAPAKKVDTSDEDKKPAAKAVPSKNGSAPAKKAASDEEDTDESSDE 613
Query: 367 DEEDEK 372
DEE K
Sbjct: 614 DEEXXK 631
Score = 61.6 bits (148), Expect = 9e-10
Identities = 61/189 (32%), Positives = 87/189 (45%), Gaps = 24/189 (12%)
Frame = +2
Query: 238 EKPAVKVSAPSKVVPAAVKNGAADSDSSDSDSSSDEDDDKKKPATKVAKKLPAAAPIKKV 297
+K A KV A + VV + VK+G ++ + + KK+ +VA K A +K V
Sbjct: 95 KKSATKVDA-APVVVSPVKSGKKGKRQAEEEVKAVSA--KKQKVEEVAAKQKA---LKVV 256
Query: 298 EKSESSDDESSESSDEENDSK---PATTTVAKPSAVKK----SASSDSDDEDSSDSDSEV 350
+K ESS +ESSES DE+ K P+ T AK VKK + S +SD + SS + EV
Sbjct: 257 KKEESSSEESSESEDEQPVVKAPAPSKKTPAKKGNVKKAQPETTSEESDSDSSSSDEEEV 436
Query: 351 KK---------------DKMDVDESDSSEDSDEEDEKSTKTPQKK-VKDVEMVDASSGKN 394
KK K+D + D + K+ P KK D E D SS ++
Sbjct: 437 KKPVSKAVPSKNGSAPAKKVDTSDEDKKPAAKAVPSKNGSAPAKKAASDEEDTDESSDED 616
Query: 395 APKT-PAAQ 402
PAA+
Sbjct: 617 EEXXKPAAK 643
Score = 31.2 bits (69), Expect = 1.3
Identities = 35/141 (24%), Positives = 56/141 (38%), Gaps = 8/141 (5%)
Frame = +2
Query: 278 KKPATKVAKKLPAAAPIKKVEKSESSDDESSESSDEENDSKPATTTVAKPSAVKKSASSD 337
KK ATKV +P+K +K + +E ++ + AK A+K
Sbjct: 95 KKSATKVDAAPVVVSPVKSGKKGKRQAEEEVKAVSAKKQK--VEEVAAKQKALKV----- 253
Query: 338 SDDEDSSDSDSEVKKDKMDVDESDSSEDSDEEDEK--------STKTPQKKVKDVEMVDA 389
VKK+ ES S E S+ EDE+ S KTP KK
Sbjct: 254 ------------VKKE-----ESSSEESSESEDEQPVVKAPAPSKKTPAKK--------G 358
Query: 390 SSGKNAPKTPAAQTENEGSKT 410
+ K P+T + +++++ S +
Sbjct: 359 NVKKAQPETTSEESDSDSSSS 421
>AW685995 similar to PIR|I51618|I516 nucleolar phosphoprotein - African
clawed frog, partial (5%)
Length = 516
Score = 89.4 bits (220), Expect = 4e-18
Identities = 58/150 (38%), Positives = 77/150 (50%)
Frame = +3
Query: 7 KSATKVEAAAPVAAAPKSAKKAKRAAEDEVEKQVSAKKQKIAEVAAKQKKETKLQKVKKE 66
K++ KV+A P K K+AA+ E EK+ KK K++K ++ +
Sbjct: 33 KTSKKVKAEEPKKVEKVEKTKGKKAAKAEEEKKKQQKK--------KEEKPAPMEVEEDS 188
Query: 67 SSSDDDSSSEDEKPAPKVAPLKTSVKNGTTPAKKAKPAPSSSSSDEDSSDEEEEVIAKKP 126
SSS++ SSSEDE PA K AP AKKA SSS +E SS E E +KP
Sbjct: 189 SSSEESSSSEDEAPAKKAAP-----------AKKAAAKKESSSEEESSSSESESEEEEKP 335
Query: 127 TKVVVPKKEESSSDEDSSSDEDSSSESDDE 156
K KKE SS +E SSS+E+S E +E
Sbjct: 336 AKKAAAKKESSSEEESSSSEEESDEEEKNE 425
Score = 72.8 bits (177), Expect = 4e-13
Identities = 50/138 (36%), Positives = 74/138 (53%)
Frame = +3
Query: 181 KKAQPAPSSSSESEDDDSSEEDEPSAKQPKKEVAVKPAVQKKKEESSSDDSSSESEDEKP 240
K +P E + + E K+ +K+ KPA + +E+SSS + SS SEDE P
Sbjct: 51 KAEEPKKVEKVEKTKGKKAAKAEEEKKKQQKKKEEKPAPMEVEEDSSSSEESSSSEDEAP 230
Query: 241 AVKVSAPSKVVPAAVKNGAADSDSSDSDSSSDEDDDKKKPATKVAKKLPAAAPIKKVEKS 300
A K +AP+K AA K +++ +SS S+S S+E++ KPA K A K
Sbjct: 231 A-KKAAPAKKA-AAKKESSSEEESSSSESESEEEE---KPAKKAA------------AKK 359
Query: 301 ESSDDESSESSDEENDSK 318
ESS +E S SS+EE+D +
Sbjct: 360 ESSSEEESSSSEEESDEE 413
Score = 71.2 bits (173), Expect = 1e-12
Identities = 64/194 (32%), Positives = 84/194 (42%), Gaps = 2/194 (1%)
Frame = +3
Query: 23 KSAKKAKRAAEDEVEKQVSAKKQKIAEVAAKQKKETKLQKVKKESSSDDDSSSEDEKPAP 82
K++KK K +VEK K +K AAK ++E K Q+ KKE EKPAP
Sbjct: 33 KTSKKVKAEEPKKVEKVEKTKGKK----AAKAEEEKKKQQKKKE-----------EKPAP 167
Query: 83 KVAPLKTSVKNGTTPAKKAKPAPSSSSSDEDSSDEEEEVIAKK--PTKVVVPKKEESSSD 140
SSS E+SS E+E AKK P K KKE SS +
Sbjct: 168 M-------------------EVEEDSSSSEESSSSEDEAPAKKAAPAKKAAAKKESSSEE 290
Query: 141 EDSSSDEDSSSESDDEKPAAKVAVPAKKAPAKNGSVTAPAKKAQPAPSSSSESEDDDSSE 200
E SSS+ +S E EKPA K A AKK + SS SE++ E
Sbjct: 291 ESSSSESESEEE---EKPAKKAA----------------AKKESSSEEESSSSEEESDEE 413
Query: 201 EDEPSAKQPKKEVA 214
E + ++ ++E A
Sbjct: 414 EKNEARRR*EQEEA 455
Score = 69.3 bits (168), Expect = 4e-12
Identities = 58/149 (38%), Positives = 76/149 (50%)
Frame = +3
Query: 88 KTSVKNGTTPAKKAKPAPSSSSSDEDSSDEEEEVIAKKPTKVVVPKKEESSSDEDSSSDE 147
KTS K KK + + ++EE++ KK + P + E EDSSS E
Sbjct: 33 KTSKKVKAEEPKKVEKVEKTKGKKAAKAEEEKKKQQKKKEEKPAPMEVE----EDSSSSE 200
Query: 148 DSSSESDDEKPAAKVAVPAKKAPAKNGSVTAPAKKAQPAPSSSSESEDDDSSEEDEPSAK 207
+SSS S+DE PA K A PAKKA AK S + SSSSESE SEE+E
Sbjct: 201 ESSS-SEDEAPAKKAA-PAKKAAAKKES-------SSEEESSSSESE----SEEEE---- 329
Query: 208 QPKKEVAVKPAVQKKKEESSSDDSSSESE 236
+P K+ A K ++E SSS++ S E E
Sbjct: 330 KPAKKAAAKKESSSEEESSSSEEESDEEE 416
Score = 63.9 bits (154), Expect = 2e-10
Identities = 48/143 (33%), Positives = 68/143 (46%), Gaps = 4/143 (2%)
Frame = +3
Query: 100 KAKPAPSSSSSDEDSSDEEEEVIAKKPTKVVVPKKEESSSDEDSSS----DEDSSSESDD 155
K+K + + + ++ E+ KK K KK++ E+ + +EDSSS +
Sbjct: 27 KSKTSKKVKAEEPKKVEKVEKTKGKKAAKAEEEKKKQQKKKEEKPAPMEVEEDSSS--SE 200
Query: 156 EKPAAKVAVPAKKAPAKNGSVTAPAKKAQPAPSSSSESEDDDSSEEDEPSAKQPKKEVAV 215
E +++ PAKKA APAKKA SSSE E S E E K KK A
Sbjct: 201 ESSSSEDEAPAKKA--------APAKKAAAKKESSSEEESSSSESESEEEEKPAKKAAA- 353
Query: 216 KPAVQKKKEESSSDDSSSESEDE 238
K+ESSS++ SS SE+E
Sbjct: 354 -------KKESSSEEESSSSEEE 401
Score = 62.0 bits (149), Expect = 7e-10
Identities = 46/150 (30%), Positives = 75/150 (49%), Gaps = 3/150 (2%)
Frame = +3
Query: 206 AKQPKKEVAVKPAVQKKKEESSSDDSSSESEDEKPAVKVSAPSKVVPAAVKNGAADSDSS 265
+K KK A +P + +K E + ++++E+EK + K P V+ DS SS
Sbjct: 30 SKTSKKVKAEEPK-KVEKVEKTKGKKAAKAEEEKKKQQKKKEEKPAPMEVEE---DSSSS 197
Query: 266 DSDSSSDEDDDKKKPATKVAKKLPAAAPIKKV--EKSESSDDESSES-SDEENDSKPATT 322
+ SSS+++ KK AAP KK +K SS++ESS S S+ E + KPA
Sbjct: 198 EESSSSEDEAPAKK-----------AAPAKKAAAKKESSSEEESSSSESESEEEEKPAKK 344
Query: 323 TVAKPSAVKKSASSDSDDEDSSDSDSEVKK 352
AK + + SS S++E + +E ++
Sbjct: 345 AAAKKESSSEEESSSSEEESDEEEKNEARR 434
Score = 60.5 bits (145), Expect = 2e-09
Identities = 47/149 (31%), Positives = 68/149 (45%), Gaps = 3/149 (2%)
Frame = +3
Query: 130 VVPKKEESSSDEDSSSDEDSSSESDDEKPAAKVAVPAKKAPAKNGSVTAPAKKAQPAPSS 189
++ K + S + + E K AAK KK K AP + +
Sbjct: 18 IMGKSKTSKKVKAEEPKKVEKVEKTKGKKAAKAEEEKKKQQKKKEEKPAPMEVEE----D 185
Query: 190 SSESEDDDSSEEDEPSAKQPKKEVAVKPAVQKKKEESSSDDSSSES---EDEKPAVKVSA 246
SS SE+ SSE++ P+ KK K A KK+ S + SSSES E+EKPA K
Sbjct: 186 SSSSEESSSSEDEAPA----KKAAPAKKAAAKKESSSEEESSSSESESEEEEKPAKKA-- 347
Query: 247 PSKVVPAAVKNGAADSDSSDSDSSSDEDD 275
AA K +++ +SS S+ SDE++
Sbjct: 348 ------AAKKESSSEEESSSSEEESDEEE 416
Score = 37.7 bits (86), Expect = 0.014
Identities = 37/124 (29%), Positives = 50/124 (39%), Gaps = 2/124 (1%)
Frame = +3
Query: 4 SSKKSATKVEAAAPVAAAPK--SAKKAKRAAEDEVEKQVSAKKQKIAEVAAKQKKETKLQ 61
S + S+++ EA A AA K +AKK + E
Sbjct: 195 SEESSSSEDEAPAKKAAPAKKAAAKKESSSEE---------------------------- 290
Query: 62 KVKKESSSDDDSSSEDEKPAPKVAPLKTSVKNGTTPAKKAKPAPSSSSSDEDSSDEEEEV 121
ESSS + S E+EKPA K A AKK + SSS E+ SDEEE+
Sbjct: 291 ----ESSSSESESEEEEKPAKKAA------------AKKESSSEEESSSSEEESDEEEKN 422
Query: 122 IAKK 125
A++
Sbjct: 423 EARR 434
Score = 32.0 bits (71), Expect = 0.79
Identities = 33/101 (32%), Positives = 46/101 (44%), Gaps = 1/101 (0%)
Frame = -3
Query: 246 APSKVVPAAVKNGAADSDSSDSDSSSDEDDDKKKPATKVAKKLPAAAPIKKVEKSESSDD 305
+P + + + A SS SDSSSDE D + + +A A S SSD
Sbjct: 469 SPPRRASSCSQRRRASFFSSSSDSSSDELDSSSELDSFLAAAFLAGF-------SSSSDS 311
Query: 306 ESSE-SSDEENDSKPATTTVAKPSAVKKSASSDSDDEDSSD 345
+S E S E DS A +A A + +S S+ EDSS+
Sbjct: 310 DSDELDSSSELDSFLAAAFLA--GAAFLAGASSSELEDSSE 194
Score = 29.3 bits (64), Expect = 5.1
Identities = 30/73 (41%), Positives = 35/73 (47%), Gaps = 2/73 (2%)
Frame = -3
Query: 106 SSSSSDE-DSSDEEEEVIAKKPTKVVVPKKEESSSDEDSSSDE-DSSSESDDEKPAAKVA 163
S SSSDE DSS E + +A SS DS SDE DSSSE D AA +A
Sbjct: 406 SDSSSDELDSSSELDSFLA-------AAFLAGFSSSSDSDSDELDSSSELDSFLAAAFLA 248
Query: 164 VPAKKAPAKNGSV 176
A A A + +
Sbjct: 247 GAAFLAGASSSEL 209
Score = 28.5 bits (62), Expect = 8.7
Identities = 29/87 (33%), Positives = 35/87 (39%), Gaps = 2/87 (2%)
Frame = -3
Query: 226 SSSDDSSSESEDEKPAVKVSAPSKVVPAAVKNGAADSDSSDSD--SSSDEDDDKKKPATK 283
SSS DSSS+ D S + AA G + S SDSD SS E D A
Sbjct: 415 SSSSDSSSDELDSS-----SELDSFLAAAFLAGFSSSSDSDSDELDSSSELDSFLAAAFL 251
Query: 284 VAKKLPAAAPIKKVEKSESSDDESSES 310
A A ++E S + SS S
Sbjct: 250 AGAAFLAGASSSELEDSSELELSSSTS 170
>TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 754
Score = 87.4 bits (215), Expect = 2e-17
Identities = 54/129 (41%), Positives = 73/129 (55%), Gaps = 8/129 (6%)
Frame = +2
Query: 528 SLQEHFGSCGEITRISVPKDFESGNVKGFAYLDFSDTNSVNKALE-LHDSDLGGFTLAVD 586
+L++ F GEI + D E+G +GF ++ F++ S+N A+E ++ DL G + V+
Sbjct: 146 ALEKAFSQYGEIVDSKIINDRETGRSRGFGFVTFANEKSMNDAIEAMNGQDLDGRNITVN 325
Query: 587 EAKPRDNQGSGG--RSGGGRSGGG---RGGRFDSGRGGGRGRFGSGGRGGDRGGRGG--R 639
+A+ R + G GG R GGG GGG G R G GGG G G GG G RGG GG R
Sbjct: 326 QAQSRGSGGGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGGGGGGGYGERRGGGGGYSR 505
Query: 640 GGGGRGGRG 648
GGGG G G
Sbjct: 506 GGGGGGYGG 532
Score = 55.1 bits (131), Expect = 9e-08
Identities = 33/97 (34%), Positives = 49/97 (50%), Gaps = 2/97 (2%)
Frame = +2
Query: 412 FVGNLSFSVQRSDVENFFKDCGEVVDVRLATD-EDGRFKGFGHVEFATAAAAQSALE-YN 469
FVG L+++ +E F GE+VD ++ D E GR +GFG V FA + A+E N
Sbjct: 110 FVGGLAWATDNEALEKAFSQYGEIVDSKIINDRETGRSRGFGFVTFANEKSMNDAIEAMN 289
Query: 470 GSELLQRPVRLDLARERGAYTPNSGAANNSFQKGGRG 506
G +L R + ++ A+ RG+ G GG G
Sbjct: 290 GQDLDGRNITVNQAQSRGSGGGGGGGRGGGGYGGGGG 400
>TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-binding
protein {Glycine max}, partial (90%)
Length = 488
Score = 86.3 bits (212), Expect = 4e-17
Identities = 51/127 (40%), Positives = 70/127 (54%), Gaps = 6/127 (4%)
Frame = +3
Query: 528 SLQEHFGSCGEITRISVPKDFESGNVKGFAYLDFSDTNSVNKALE-LHDSDLGGFTLAVD 586
+L++ F GEIT V D E+G +GF ++ F++ S+ A+E ++ D+ G + V+
Sbjct: 96 ALEQAFSKFGEITDSKVINDRETGRSRGFGFVTFAEEKSMRDAIEEMNGQDIDGRNITVN 275
Query: 587 EAKPRDNQGSGGRSGGGRSGGGRG-GRFDSGRGGGRGR----FGSGGRGGDRGGRGGRGG 641
EA+ R + G G GGG GGG G G G GGG GR + G GG GG G RG
Sbjct: 276 EAQSRGSGGGGRGGGGGGYGGGGGYGGGGGGYGGGGGRRDGGYSRSGGGGGYGGGGDRGY 455
Query: 642 GGRGGRG 648
GG GG G
Sbjct: 456 GGGGGGG 476
Score = 51.6 bits (122), Expect = 1e-06
Identities = 30/95 (31%), Positives = 49/95 (51%), Gaps = 2/95 (2%)
Frame = +3
Query: 412 FVGNLSFSVQRSDVENFFKDCGEVVDVRLATD-EDGRFKGFGHVEFATAAAAQSAL-EYN 469
FVG L+++ +E F GE+ D ++ D E GR +GFG V FA + + A+ E N
Sbjct: 60 FVGGLAWATDSQALEQAFSKFGEITDSKVINDRETGRSRGFGFVTFAEEKSMRDAIEEMN 239
Query: 470 GSELLQRPVRLDLARERGAYTPNSGAANNSFQKGG 504
G ++ R + ++ A+ RG+ G + GG
Sbjct: 240 GQDIDGRNITVNEAQSRGSGGGGRGGGGGGYGGGG 344
>TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 1054
Score = 80.5 bits (197), Expect = 2e-15
Identities = 49/115 (42%), Positives = 66/115 (56%), Gaps = 8/115 (6%)
Frame = +1
Query: 542 ISVPKDFESGNVKGFAYLDFSDTNSVNKALE-LHDSDLGGFTLAVDEAKPRDNQGSGG-- 598
+ + D E+G +GF ++ F++ S+N A+E ++ DL G + V++A+ R + G GG
Sbjct: 442 VQIINDRETGRSRGFGFVTFANEKSMNDAIEAMNGQDLDGRNITVNQAQSRGSGGGGGGG 621
Query: 599 RSGGGRSGGG---RGGRFDSGRGGGRGRFGSGGRGGDRGGRGG--RGGGGRGGRG 648
R GGG GGG G R G GGG G G GG G RGG GG RGGGG G G
Sbjct: 622 RGGGGYGGGGGGYGGERRGYGGGGGYGGGGGGGYGERRGGGGGYSRGGGGGGYGG 786
>TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 912
Score = 79.3 bits (194), Expect = 4e-15
Identities = 49/110 (44%), Positives = 64/110 (57%), Gaps = 8/110 (7%)
Frame = +1
Query: 547 DFESGNVKGFAYLDFSDTNSVNKALE-LHDSDLGGFTLAVDEAKPRDNQGSGG--RSGGG 603
D E+G +GF ++ F++ S+N A+E ++ DL G + V++A+ R + G GG R GGG
Sbjct: 361 DRETGRSRGFGFVTFANEKSMNDAIEAMNGQDLDGRNITVNQAQSRGSGGGGGGGRGGGG 540
Query: 604 RSGGG---RGGRFDSGRGGGRGRFGSGGRGGDRGGRGG--RGGGGRGGRG 648
GGG G R G GGG G G GG G RGG GG RGGGG G G
Sbjct: 541 YGGGGGGYGGERRGYGGGGGYGGGGGGGYGERRGGGGGYSRGGGGGGYGG 690
>AW257087 homologue to GP|6683126|dbj KIAA0339 protein {Homo sapiens},
partial (2%)
Length = 466
Score = 73.6 bits (179), Expect = 2e-13
Identities = 53/155 (34%), Positives = 73/155 (46%), Gaps = 1/155 (0%)
Frame = +1
Query: 137 SSSDEDSSSDEDSSSESDDEKPAAK-VAVPAKKAPAKNGSVTAPAKKAQPAPSSSSESED 195
SSS SSS DS S+S+ E PA K V V K AP S ++ K + S S+S+
Sbjct: 16 SSSSSSSSSSSDSDSDSESEAPAKKAVPVVTKTAPTHAKSASS---KMEVDSDSDSDSDS 186
Query: 196 DDSSEEDEPSAKQPKKEVAVKPAVQKKKEESSSDDSSSESEDEKPAVKVSAPSKVVPAAV 255
SS + K V+ A KKEES SD S S + S+ S
Sbjct: 187 SSSSSTSSSVSSVKKPAVSAAKATPAKKEESDSDSDSDSSSSSSSSSSSSSSS------- 345
Query: 256 KNGAADSDSSDSDSSSDEDDDKKKPATKVAKKLPA 290
++ S SS S SS+ +D+++++ A V KK PA
Sbjct: 346 ---SSSSSSSSSSSSNSDDEEEEEEAKPVVKKTPA 441
Score = 73.6 bits (179), Expect = 2e-13
Identities = 53/138 (38%), Positives = 71/138 (51%), Gaps = 11/138 (7%)
Frame = +1
Query: 226 SSSDDSSSESEDEKPAVK-VSAPSKVVPAAVKNGAA----DSDS-SDSDSSSDEDDDK-- 277
SSS DS S+SE E PA K V +K P K+ ++ DSDS SDSDSSS
Sbjct: 37 SSSSDSDSDSESEAPAKKAVPVVTKTAPTHAKSASSKMEVDSDSDSDSDSSSSSSTSSSV 216
Query: 278 ---KKPATKVAKKLPAAAPIKKVEKSESSDDESSESSDEENDSKPATTTVAKPSAVKKSA 334
KKPA AK PA +K ES D S+SS + S ++++ + S+ S+
Sbjct: 217 SSVKKPAVSAAKATPA-------KKEESDSDSDSDSSSSSSSSSSSSSSSSSSSSSSSSS 375
Query: 335 SSDSDDEDSSDSDSEVKK 352
SS+SDDE+ + V K
Sbjct: 376 SSNSDDEEEEEEAKPVVK 429
Score = 68.2 bits (165), Expect = 1e-11
Identities = 54/166 (32%), Positives = 68/166 (40%), Gaps = 9/166 (5%)
Frame = +1
Query: 98 AKKAKPAPSSSSSDEDSSDEEEEVIAKKPTKVV---VPKKEESSS-----DEDSSSDEDS 149
+ + + SSSSS + SD E E AKK VV P +S+S D DS SD DS
Sbjct: 7 SSSSSSSSSSSSSSDSDSDSESEAPAKKAVPVVTKTAPTHAKSASSKMEVDSDSDSDSDS 186
Query: 150 SSESDDEKPAAKVAVPAKKAPAKNGSVTAPAKKAQPAPSSSSESEDDDSSEEDEPSAKQP 209
SS S + V K PA + + PAKK + S S+S SS S+
Sbjct: 187 SSSSSTSSSVSSV-----KKPAVSAAKATPAKKEESDSDSDSDSSSSSSSSSSSSSSSSS 351
Query: 210 KKEVAVKPAVQKKKEESSSDDSSSESEDEKPAV-KVSAPSKVVPAA 254
SS+ D E E+ KP V K A S P A
Sbjct: 352 SS--------SSSSSSSSNSDDEEEEEEAKPVVKKTPAKSASTPVA 465
Score = 64.7 bits (156), Expect = 1e-10
Identities = 46/137 (33%), Positives = 60/137 (43%), Gaps = 13/137 (9%)
Frame = +1
Query: 257 NGAADSDSSDSDSSSDEDDDKKKPATKVAKKLP--AAAPIKKVEKSESSDDESSESSDEE 314
+ ++ S SSDSDS S+ + KK V K P A + K+E SD +S SS
Sbjct: 22 SSSSSSSSSDSDSDSESEAPAKKAVPVVTKTAPTHAKSASSKMEVDSDSDSDSDSSSSSS 201
Query: 315 NDS------KPATTTVAKPSAVKKSASSDSDDEDSSDSDSEVKKDKMDVDESDSSEDS-- 366
S KPA + A K+ + SDSD + SS S S S SS S
Sbjct: 202 TSSSVSSVKKPAVSAAKATPAKKEESDSDSDSDSSSSSSSSSSSSSSSSSSSSSSSSSSS 381
Query: 367 ---DEEDEKSTKTPQKK 380
DEE+E+ K KK
Sbjct: 382 NSDDEEEEEEAKPVVKK 432
Score = 61.6 bits (148), Expect = 9e-10
Identities = 56/167 (33%), Positives = 69/167 (40%), Gaps = 16/167 (9%)
Frame = +1
Query: 67 SSSDDDSSSEDEKPAPKVAPLKTSVKNGTTPAKKAKP-----APSSSSSDEDSSDEEEEV 121
SSSD DS SE E PA K P+ T K T AK A + S S SD SS
Sbjct: 40 SSSDSDSDSESEAPAKKAVPVVT--KTAPTHAKSASSKMEVDSDSDSDSDSSSSSSTSSS 213
Query: 122 IA--KKPT----KVVVPKKEESSSDEDS-----SSDEDSSSESDDEKPAAKVAVPAKKAP 170
++ KKP K KKEES SD DS SS SSS S
Sbjct: 214 VSSVKKPAVSAAKATPAKKEESDSDSDSDSSSSSSSSSSSSSSS---------------- 345
Query: 171 AKNGSVTAPAKKAQPAPSSSSESEDDDSSEEDEPSAKQPKKEVAVKP 217
+ + + SSSS S+D++ EE +P K+ + A P
Sbjct: 346 ---------SSSSSSSSSSSSNSDDEEEEEEAKPVVKKTPAKSASTP 459
Score = 59.7 bits (143), Expect = 4e-09
Identities = 49/160 (30%), Positives = 65/160 (40%), Gaps = 3/160 (1%)
Frame = +1
Query: 19 AAAPKSAKKAKRAAEDEVEKQVSAKKQKIAEVAAKQKKETKLQKVKKESSSDDDSSSEDE 78
+++ S+ + ++ + E + AKK + V K K E SD DS S D
Sbjct: 13 SSSSSSSSSSSSDSDSDSESEAPAKKA-VPVVTKTAPTHAKSASSKMEVDSDSDSDS-DS 186
Query: 79 KPAPKVAPLKTSVKNGTTPAKKAKPA---PSSSSSDEDSSDEEEEVIAKKPTKVVVPKKE 135
+ + +SVK A KA PA S S SD DSS
Sbjct: 187 SSSSSTSSSVSSVKKPAVSAAKATPAKKEESDSDSDSDSSSSSSS-----------SSSS 333
Query: 136 ESSSDEDSSSDEDSSSESDDEKPAAKVAVPAKKAPAKNGS 175
SSS SSS SSS SDDE+ + KK PAK+ S
Sbjct: 334 SSSSSSSSSSSSSSSSNSDDEEEEEEAKPVVKKTPAKSAS 453
Score = 59.7 bits (143), Expect = 4e-09
Identities = 47/153 (30%), Positives = 65/153 (41%), Gaps = 20/153 (13%)
Frame = +1
Query: 259 AADSDSSDSDSS----SDEDDDKKKPATKVAKKLPAAAPIKKVEKSESSDDESSESSDEE 314
++ S SS S SS SD D + + PA K + AP KS SS E SD +
Sbjct: 4 SSSSSSSSSSSSSSSDSDSDSESEAPAKKAVPVVTKTAPTH--AKSASSKMEVDSDSDSD 177
Query: 315 NDSKPATTTVAKPSAVKKSA----------------SSDSDDEDSSDSDSEVKKDKMDVD 358
+DS +++T + S+VKK A SDSD SS S S
Sbjct: 178 SDSSSSSSTSSSVSSVKKPAVSAAKATPAKKEESDSDSDSDSSSSSSSSSSSSSSSSSSS 357
Query: 359 ESDSSEDSDEEDEKSTKTPQKKVKDVEMVDASS 391
S SS S+ +DE+ + + VK AS+
Sbjct: 358 SSSSSSSSNSDDEEEEEEAKPVVKKTPAKSAST 456
Score = 45.4 bits (106), Expect = 7e-05
Identities = 37/139 (26%), Positives = 56/139 (39%), Gaps = 5/139 (3%)
Frame = +1
Query: 5 SKKSATKVEAAAPVAAAPKSAKKAKRAAEDE-----VEKQVSAKKQKIAEVAAKQKKETK 59
+KK+ V AP A S+K + D S+ + + A K T
Sbjct: 82 AKKAVPVVTKTAPTHAKSASSKMEVDSDSDSDSDSSSSSSTSSSVSSVKKPAVSAAKATP 261
Query: 60 LQKVKKESSSDDDSSSEDEKPAPKVAPLKTSVKNGTTPAKKAKPAPSSSSSDEDSSDEEE 119
+K + +S SD DSSS S + ++ + + + SSSSS +SD+EE
Sbjct: 262 AKKEESDSDSDSDSSS--------------SSSSSSSSSSSSSSSSSSSSSSSSNSDDEE 399
Query: 120 EVIAKKPTKVVVPKKEESS 138
E KP P K S+
Sbjct: 400 EEEEAKPVVKKTPAKSAST 456
Score = 38.9 bits (89), Expect = 0.006
Identities = 37/116 (31%), Positives = 46/116 (38%), Gaps = 21/116 (18%)
Frame = +1
Query: 300 SESSDDESSESSDEENDSK--PATT----TVAKPSAVKKSASSDSD-DEDSSDSD----- 347
S SS D S+S E K P T T AK ++ K SDSD D DSS S
Sbjct: 34 SSSSSDSDSDSESEAPAKKAVPVVTKTAPTHAKSASSKMEVDSDSDSDSDSSSSSSTSSS 213
Query: 348 -SEVKKDKMDV--------DESDSSEDSDEEDEKSTKTPQKKVKDVEMVDASSGKN 394
S VKK + +ESDS DSD S+ + +SS +
Sbjct: 214 VSSVKKPAVSAAKATPAKKEESDSDSDSDSSSSSSSSSSSSSSSSSSSSSSSSSSS 381
Score = 34.7 bits (78), Expect = 0.12
Identities = 22/96 (22%), Positives = 42/96 (42%), Gaps = 2/96 (2%)
Frame = +1
Query: 4 SSKKSATKVEAAAPVAAAPKSAKKAKRAAEDEVEKQVSAKKQKIAEVAAKQKKETKLQKV 63
S S++ ++ V++ K A A +A + E+ S + ++ +
Sbjct: 172 SDSDSSSSSSTSSSVSSVKKPAVSAAKATPAKKEESDSDSDSDSSSSSSSSSSSSSSSSS 351
Query: 64 KKESSSDDDSSSEDEKPAPKVAPL--KTSVKNGTTP 97
SSS S+S+DE+ + P+ KT K+ +TP
Sbjct: 352 SSSSSSSSSSNSDDEEEEEEAKPVVKKTPAKSASTP 459
Score = 28.5 bits (62), Expect = 8.7
Identities = 14/44 (31%), Positives = 24/44 (53%)
Frame = -3
Query: 188 SSSSESEDDDSSEEDEPSAKQPKKEVAVKPAVQKKKEESSSDDS 231
SSSS DD+ +EDE + ++ + +++ + ES SD S
Sbjct: 401 SSSSSLLDDEDEDEDEDEDEDEDEDEDDEELLEESESESESDSS 270
>TC88887 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproline-rich
glycoprotein DZ-HRGP {Volvox carteri f. nagariensis},
partial (18%)
Length = 1681
Score = 49.3 bits (116), Expect(2) = 4e-13
Identities = 30/49 (61%), Positives = 31/49 (63%), Gaps = 2/49 (4%)
Frame = -3
Query: 600 SGGGRSGGGRGGRFDSGRGGGRGRFGSG-GRGGDRGGRGGRGGG-GRGG 646
+GG GGRGG GG RG FG G GRGG GGRGG GGG GRGG
Sbjct: 728 AGGPMGFGGRGGF-----GGMRGGFGGGYGRGGFNGGRGGFGGGYGRGG 597
Score = 48.1 bits (113), Expect = 1e-05
Identities = 30/54 (55%), Positives = 32/54 (58%), Gaps = 2/54 (3%)
Frame = -3
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRG--GRGGRGGGGRGG 646
G+GG G G GG G R G G GRG F +GGRGG G GRGG GGG GG
Sbjct: 731 GAGGPMGFGGRGGFGGMRGGFGGGYGRGGF-NGGRGGFGGGYGRGGFAGGGGGG 573
Score = 43.5 bits (101), Expect(2) = 4e-13
Identities = 49/204 (24%), Positives = 81/204 (39%), Gaps = 28/204 (13%)
Frame = -2
Query: 405 NEGSKTLFVGNLSFSVQRSDVENFFKDCGEVVDVRLATDEDGRFKGFGHVEFATAAAAQS 464
++ + ++VGNLS+ V+ +++F + GEV+ + +G KG G VE+AT AQ+
Sbjct: 1371 SQQDRRVYVGNLSYDVKWHHLKDFMRQAGEVLFADVLLLPNGMSKGCGIVEYATREQAQN 1192
Query: 465 AL-EYNGSELLQRPVRLDLARE-----------RGAYTPNSGAANNSFQKGG-------- 504
A+ + L+ R V + RE RG + G GG
Sbjct: 1191 AVATLSNQNLMGRLVYVREDREAEPRFIGATANRGGFGGGPGGPGGPGGPGGMHPGGFGG 1012
Query: 505 -------RGQSQTVFVRGFDKFQGEDDIKSSLQEHFGSCGEITRISVPKDFESGNVKGFA 557
G S+ +FV G D+K ++ + G + R V G KG
Sbjct: 1011 GGPGSYSTGGSRQIFVANLPYTVGWQDLKDLFRQAARN-GVVIRADVHLG-PDGRPKGSG 838
Query: 558 YLDFSDTNSVNKAL-ELHDSDLGG 580
+ F + A+ + H D G
Sbjct: 837 IVVFENPEDARNAIQQFHGYDWQG 766
Score = 28.5 bits (62), Expect = 8.7
Identities = 27/90 (30%), Positives = 40/90 (44%), Gaps = 11/90 (12%)
Frame = -2
Query: 550 SGNVKGFAYLDFSDTNSVNKALE-LHDSDLGGFTLAVDE---AKPR-----DNQGS--GG 598
+G KG ++++ A+ L + +L G + V E A+PR N+G GG
Sbjct: 1251 NGMSKGCGIVEYATREQAQNAVATLSNQNLMGRLVYVREDREAEPRFIGATANRGGFGGG 1072
Query: 599 RSGGGRSGGGRGGRFDSGRGGGRGRFGSGG 628
G G GG G GGG G + +GG
Sbjct: 1071 PGGPGGPGGPGGMHPGGFGGGGPGSYSTGG 982
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 71.6 bits (174), Expect = 9e-13
Identities = 84/430 (19%), Positives = 165/430 (37%), Gaps = 23/430 (5%)
Frame = +1
Query: 3 KSSKKSATKVEAAAPVAAAPKSAKKAKRAAEDEVEKQVSAKKQKIAEVAAKQKKETKLQK 62
KS +++ E + +S + A E E KQ E + E +
Sbjct: 343 KSKQENEETSETNSKDKENEESNQNGSDAKEQVGENHEQDSKQGTEETNGTEGGEKEEHD 522
Query: 63 VKKESSSDDDSSSEDEKPAPKVAPLKTSVKNGTTPAKKAKPAPSSSSSDEDSSDEEEEVI 122
KE +S D+ + EK + A A + S E S+ EE+
Sbjct: 523 KIKEDTSSDNQVQDGEK--------NNEAREENYSGDNASSAVVDNKSQESSNKTEEQFD 678
Query: 123 AKKPTKVVVPKKEESSSDEDS-------------SSDEDSSSESDDEKPAAKVAVPAKKA 169
K+ + + ++ S+ +S S + + +E+D K +A + K+
Sbjct: 679 KKEKNEFELESQKNSNETTESTDSTITQNSQGNESEKDQAQTENDTPKGSASESDEQKQE 858
Query: 170 PAKNGSVTAPAKKAQPAPSSSSE--------SEDDDSSEEDEPSAKQPKKEVAVKPAVQK 221
+N + + + + ++ SED +S +ED + K V
Sbjct: 859 QEQNNTTKDDVQTTDTSSQNGNDTTEKQNETSEDANSKKEDSSALNTTPNNEDSKSGVAG 1038
Query: 222 KKEESSSDDSSSESEDEKPAVKVSAPSKVVPAAVKNGAADSDSSDSDSSSDEDDDKKKPA 281
+ +S++ SSSE++D + + + +S SSS+ D+K +
Sbjct: 1039DQADSTTTTSSSETQDGN--TNHGEYKDTTNENPEKNSGQEGTQESGSSSNTFDNKDAAS 1212
Query: 282 TKVAKKLPAAAPIKKVEKSESSDDESSESSDEENDSKPATTTVAKPSA--VKKSASSDSD 339
KV + ++ + SS + SESS +N + + T + SA K S+ +
Sbjct: 1213NKVQLTTTSDTSSEQKKDESSSAESKSESSQNDNANSGQSNTTSDESANDNKDSSQVTTS 1392
Query: 340 DEDSSDSDSEVKKDKMDVDESDSSEDSDEEDEKSTKTPQKKVKDVEMVDASSGKNAPKTP 399
E+S++ +S + + D +++DS + + D +T V + + +A+ K +
Sbjct: 1393SENSAEGNSNTENNS-DENQNDSKNNENTNDSGNTSN-DANVNENQNENAAQTKTSENEG 1566
Query: 400 AAQTENEGSK 409
AQ E+ SK
Sbjct: 1567DAQNESVESK 1596
Score = 54.3 bits (129), Expect = 1e-07
Identities = 101/499 (20%), Positives = 180/499 (35%), Gaps = 35/499 (7%)
Frame = +1
Query: 134 KEESSSDEDSSSDEDSSSESDDEKPAAKVAVPAKKAPAKNGSVTAPAKKAQPAPSSSSES 193
+ E S + SS+E S E+++ K + SS +S
Sbjct: 19 ENEESKETGESSEEKSHLENEESKETGE--------------------------SSEEKS 120
Query: 194 EDDDSSEEDEPSAKQPKKEVAVKPAVQKKKEESSSDDSSSESEDEKPAVKVSAPSKVVPA 253
++ +DE +KQ +E+ +Q++ EE+ ++ S + +E K S+
Sbjct: 121 HLENEENKDEEKSKQENEEIKDGEKIQQENEENKDEEKSQQENEEN---KDEEKSQQENE 291
Query: 254 AVKNGAADSDSSDSDSSSDEDDDKKKPATKVAKKLPAAAPIKKVEKSESSDDESSESSDE 313
KN + ++ + + ++++ T K + SD + +
Sbjct: 292 LKKNEGGEKETGEITEEKSKQENEETSETNSKDKE------NEESNQNGSDAKEQVGENH 453
Query: 314 ENDSKPATTTV--------AKPSAVKKSASSDSDDEDSSDSDSEVKKDKMDVDESDSSED 365
E DSK T + +K+ SSD+ +D + ++E +++ D + S+
Sbjct: 454 EQDSKQGTEETNGTEGGEKEEHDKIKEDTSSDNQVQD-GEKNNEAREENYSGDNASSAVV 630
Query: 366 SDEEDEKSTKTPQ---KKVKD-------------VEMVDA----SSGKNAPKTPAAQTEN 405
++ E S KT + KK K+ E D+ +S N + AQTEN
Sbjct: 631 DNKSQESSNKTEEQFDKKEKNEFELESQKNSNETTESTDSTITQNSQGNESEKDQAQTEN 810
Query: 406 EGSKTLFVGNLSFS---VQRSDVENFFKDCGEVVDVRLATDEDGRFKGFGHVEFATAAAA 462
+ K G+ S S Q + N KD + D D K + T+ A
Sbjct: 811 DTPK----GSASESDEQKQEQEQNNTTKDDVQTTDTSSQNGNDTTEK-----QNETSEDA 963
Query: 463 QSALEYNGSELLQRPVRLD----LARERGAYTPNSGAANNSFQKGGRGQSQTVFVRGFDK 518
S E + S L P D +A ++ T + ++ G+ +
Sbjct: 964 NSKKE-DSSALNTTPNNEDSKSGVAGDQADSTTTTSSSETQDGNTNHGE--------YKD 1116
Query: 519 FQGEDDIKSSLQEHFGSCGEITRISVPKDFESGNVKGFAYLDFSDTNSVNKALELHDSDL 578
E+ K+S QE G + KD S V+ SDT+S K E ++
Sbjct: 1117TTNENPEKNSGQEGTQESGSSSNTFDNKDAASNKVQ---LTTTSDTSSEQKKDESSSAE- 1284
Query: 579 GGFTLAVDEAKPRDNQGSG 597
+ E+ DN SG
Sbjct: 1285-----SKSESSQNDNANSG 1326
Score = 45.1 bits (105), Expect = 9e-05
Identities = 47/285 (16%), Positives = 112/285 (38%), Gaps = 8/285 (2%)
Frame = +1
Query: 3 KSSKKSATKVEAAAPVAAAPKSAKKAKRAAEDEVEKQVSAKKQKIAEVAAK--QKKETKL 60
++S+ + +K E ++ + P + A D+ + + + + + K+T
Sbjct: 946 ETSEDANSKKEDSSALNTTPNNEDSKSGVAGDQADSTTTTSSSETQDGNTNHGEYKDTTN 1125
Query: 61 QKVKKESS------SDDDSSSEDEKPAPKVAPLKTSVKNGTTPAKKAKPAPSSSSSDEDS 114
+ +K S S S++ D K A T+ + ++ KK + + + S S+
Sbjct: 1126 ENPEKNSGQEGTQESGSSSNTFDNKDAASNKVQLTTTSDTSSEQKKDESSSAESKSESSQ 1305
Query: 115 SDEEEEVIAKKPTKVVVPKKEESSSDEDSSSDEDSSSESDDEKPAAKVAVPAKKAPAKNG 174
+D + ++SDE ++ ++DSS + + A+
Sbjct: 1306 NDNANS------------GQSNTTSDESANDNKDSSQ-----------VTTSSENSAEGN 1416
Query: 175 SVTAPAKKAQPAPSSSSESEDDDSSEEDEPSAKQPKKEVAVKPAVQKKKEESSSDDSSSE 234
S T S ++E+ +D + ++ + + + E A + + + ++ ++ S+
Sbjct: 1417 SNTENNSDENQNDSKNNENTNDSGNTSNDANVNENQNENAAQTKTSENEGDAQNESVESK 1596
Query: 235 SEDEKPAVKVSAPSKVVPAAVKNGAADSDSSDSDSSSDEDDDKKK 279
E+ + A K V N + +D SD+S +DD + +
Sbjct: 1597 KENNESAHK----------DVDNNSNSNDQGSSDTSVTQDDKESR 1701
>BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melanogaster},
partial (26%)
Length = 308
Score = 70.9 bits (172), Expect = 2e-12
Identities = 35/60 (58%), Positives = 35/60 (58%)
Frame = +1
Query: 589 KPRDNQGSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
KPR G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 28 KPRGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 207
Score = 68.2 bits (165), Expect = 1e-11
Identities = 33/57 (57%), Positives = 34/57 (58%)
Frame = +2
Query: 592 DNQGSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
+N G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 26 ENPGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 196
Score = 67.8 bits (164), Expect = 1e-11
Identities = 34/67 (50%), Positives = 36/67 (52%)
Frame = +2
Query: 582 TLAVDEAKPRDNQGSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGG 641
T+ + P G GG GGG GGG GG G GGG G G GG GG GG GG GG
Sbjct: 8 TIPLSGENPGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 187
Query: 642 GGRGGRG 648
GG GG G
Sbjct: 188 GGGGGGG 208
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +3
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 54 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 215
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +3
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 72 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 233
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +3
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 57 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 218
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +2
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 50 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 211
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +1
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 61 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 222
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +3
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 66 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 227
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +1
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 67 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 228
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +2
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 107 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 268
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +1
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 73 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 234
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +3
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 78 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 239
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +2
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 80 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 241
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +2
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 86 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 247
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +1
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 133 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 294
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +2
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 89 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 250
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +3
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 93 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 254
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +2
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 95 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 256
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +1
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 97 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 258
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +2
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 101 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 262
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +2
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 104 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 265
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +3
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 132 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 293
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +1
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 109 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 270
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +1
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 112 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 273
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +3
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 117 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 278
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +3
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 120 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 281
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +2
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 122 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 283
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +3
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 126 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 287
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +3
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 129 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 290
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +3
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 84 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 245
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +1
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 58 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 219
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +1
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 136 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 297
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +1
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 139 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 300
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +3
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 144 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 305
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/54 (59%), Positives = 32/54 (59%)
Frame = +1
Query: 595 GSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRGGRG 648
G GG GGG GGG GG G GGG G G GG GG GG GG GGGG GG G
Sbjct: 145 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 306
Score = 64.3 bits (155), Expect = 1e-10
Identities = 33/63 (52%), Positives = 34/63 (53%)
Frame = +1
Query: 586 DEAKPRDNQGSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGGGRG 645
D P + GG GGG GGG GG G GGG G G GG GG GG GG GGGG G
Sbjct: 4 DNYPPFRGKPRGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 183
Query: 646 GRG 648
G G
Sbjct: 184 GGG 192
>TC78233 similar to GP|15809838|gb|AAL06847.1 AT5g51120/MWD22_6 {Arabidopsis
thaliana}, partial (66%)
Length = 1183
Score = 70.5 bits (171), Expect = 2e-12
Identities = 45/188 (23%), Positives = 87/188 (45%), Gaps = 1/188 (0%)
Frame = +3
Query: 298 EKSESSDDESSESSDEENDSKPATTTVAKPSAVKKSASSDSD-DEDSSDSDSEVKKDKMD 356
EK E +E + ++ + V + D+D D + + D E+ +
Sbjct: 48 EK*EKDKEEKKKEEEDRIAMEQEEHEVYGADIPDEEVDMDADIDAEHQEGDEELASNHTT 227
Query: 357 VDESDSSEDSDEEDEKSTKTPQKKVKDVEMVDASSGKNAPKTPAAQTENEGSKTLFVGNL 416
+ D + E +E+++ + + K + + A A+ E +++++VGN+
Sbjct: 228 KELEDMKKRLKEIEEEASALREMQAKVEKEMGAVQDPAGSSVSQAEKEEVDARSIYVGNV 407
Query: 417 SFSVQRSDVENFFKDCGEVVDVRLATDEDGRFKGFGHVEFATAAAAQSALEYNGSELLQR 476
++ +V+ F+ CG V V + TD+ G+ KGF +VEF A A Q+AL N +EL R
Sbjct: 408 DYACTPEEVQQHFQSCGTVNRVTILTDKFGQPKGFAYVEFVEADAVQNALILNETELHGR 587
Query: 477 PVRLDLAR 484
+++ R
Sbjct: 588 QLKVSAKR 611
Score = 54.7 bits (130), Expect = 1e-07
Identities = 38/137 (27%), Positives = 72/137 (51%)
Frame = +3
Query: 476 RPVRLDLARERGAYTPNSGAANNSFQKGGRGQSQTVFVRGFDKFQGEDDIKSSLQEHFGS 535
R ++ + +E GA +G++ + +K +++++V D +++ Q+HF S
Sbjct: 288 REMQAKVEKEMGAVQDPAGSSVSQAEKE-EVDARSIYVGNVDYACTPEEV----QQHFQS 452
Query: 536 CGEITRISVPKDFESGNVKGFAYLDFSDTNSVNKALELHDSDLGGFTLAVDEAKPRDNQG 595
CG + R+++ D + G KGFAY++F + ++V AL L++++L G L V +
Sbjct: 453 CGTVNRVTILTD-KFGQPKGFAYVEFVEADAVQNALILNETELHGRQLKVSAKR---TNV 620
Query: 596 SGGRSGGGRSGGGRGGR 612
G + G R G RG R
Sbjct: 621 PGLKQYGRRPAGFRGRR 671
>TC86712 similar to GP|17528972|gb|AAL38696.1 putative RNA-binding protein
{Arabidopsis thaliana}, partial (51%)
Length = 2144
Score = 70.1 bits (170), Expect = 3e-12
Identities = 68/244 (27%), Positives = 101/244 (40%), Gaps = 8/244 (3%)
Frame = +3
Query: 411 LFVGNLSFSVQRSDVENFFKDCGEVVDVRLATDED-GRFKGFGHVEFATAAAAQSALEYN 469
LF+G +S+ + +F++ G+VV+ + D GR +GFG V FA + A+ +
Sbjct: 189 LFIGGISWDTNEDRLRQYFQNFGDVVEAVIMKDRTTGRARGFGFVVFADPSVAERVV-ME 365
Query: 470 GSELLQRPVRLDLARERGAYTPNSGAANNSFQKGGRG----QSQTVFVRGFDKFQGEDDI 525
+ R V A R N +NS G +++ +FV G E D
Sbjct: 366 KHVIDGRTVEAKKAVPRD--DQNVFTRSNSSSHGSPAPTPIRTKKIFVGGLASTVTESDF 539
Query: 526 KSSLQEHFGSCGEITRISVPKDFESGNVKGFAYLDFSDTNSVNKALELHDSDLGGFTLAV 585
K+ +F G IT + V D + +GF ++ + +V K L +L G + V
Sbjct: 540 KN----YFDQFGTITDVVVMYDHNTQRPRGFGFITYDSEEAVEKVLHKTFHELNGKMVEV 707
Query: 586 DEAKPRDNQGSGGRSG-GGRSGG--GRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGRGGG 642
A P+D S R GG S G R G F +G G GG G GR
Sbjct: 708 KRAVPKDLSPSPSRGQLGGFSYGTMSRVGSFSNGFAQGYNPSLIGGHGLRLDGRLSPANV 887
Query: 643 GRGG 646
GR G
Sbjct: 888 GRSG 899
Score = 42.0 bits (97), Expect = 8e-04
Identities = 30/115 (26%), Positives = 55/115 (47%), Gaps = 1/115 (0%)
Frame = +3
Query: 370 DEKSTKTPQKKVKDVEMVDASSGKNAPKTPAAQTENEGSKTLFVGNLSFSVQRSDVENFF 429
D ++ + + +D + V S ++ +PA +K +FVG L+ +V SD +N+F
Sbjct: 378 DGRTVEAKKAVPRDDQNVFTRSNSSSHGSPAPTPIR--TKKIFVGGLASTVTESDFKNYF 551
Query: 430 KDCGEVVDVRLATDED-GRFKGFGHVEFATAAAAQSALEYNGSELLQRPVRLDLA 483
G + DV + D + R +GFG + + + A + L EL + V + A
Sbjct: 552 DQFGTITDVVVMYDHNTQRPRGFGFITYDSEEAVEKVLHKTFHELNGKMVEVKRA 716
>TC76948 similar to SP|Q08935|ROC1_NICSY 29 kDa ribonucleoprotein A
chloroplast precursor (CP29A). [Wood tobacco] {Nicotiana
sylvestris}, partial (61%)
Length = 1374
Score = 69.3 bits (168), Expect = 4e-12
Identities = 58/195 (29%), Positives = 95/195 (47%), Gaps = 11/195 (5%)
Frame = +1
Query: 408 SKTLFVGNLSFSVQRSDVENFFKDCGEVVDVRLATDED-GRFKGFGHVEFATAAAAQSAL 466
++ LFVGNL FSV + + F++ G+V V + D+ GR +GFG V ++AA ++A
Sbjct: 337 NQRLFVGNLPFSVDSAQLAEIFENAGDVEMVEVIYDKSTGRSRGFGFVTMSSAAEVEAAA 516
Query: 467 EY-NGSELLQRPVRLDLARERGAYTPNSGAANNSFQKGGR--GQSQTVFVRGFDKFQ--- 520
+ NG + R +R++ + NS N G R G + G ++
Sbjct: 517 QQLNGYVVDGRELRVNAGPPPPPRSENSRFGENPRFGGDRPRGPPRGGSSDGDNRVHVGN 696
Query: 521 ---GEDDIKSSLQEHFGSCGEITRISVPKDFESGNVKGFAYLDFSDTNSVNKALE-LHDS 576
G D++ +L+ FG G++ V D ESG +GF ++ FS + V+ A+ L +
Sbjct: 697 LAWGVDNL--ALESLFGEQGQVLEAKVIYDRESGRSRGFGFVTFSSADEVDSAIRTLDGA 870
Query: 577 DLGGFTLAVDEAKPR 591
DL G + V A R
Sbjct: 871 DLNGRAIRVSPADSR 915
>TC84888 similar to GP|21322752|dbj|BAB78536. cold shock protein-1 {Triticum
aestivum}, partial (58%)
Length = 612
Score = 68.6 bits (166), Expect = 8e-12
Identities = 41/82 (50%), Positives = 47/82 (57%), Gaps = 10/82 (12%)
Frame = +2
Query: 577 DLGGFTLAVDEAKPRDNQGSGGRSGGGRSGGGRGGRFDSGRGGGRGRFG---SGGRGGDR 633
D+ G A + PR G GG GGGR GGG GG +D G GGG G +G GGRGG R
Sbjct: 245 DVTGPNGAPPQGAPRQEMGYGG--GGGRGGGGYGGGYDQGYGGGGGSYGGGFGGGRGGGR 418
Query: 634 GG----RGGRGGGG---RGGRG 648
GG +GG GGGG +GG G
Sbjct: 419 GGGGYXQGGYGGGGGYXQGGYG 484
Score = 50.4 bits (119), Expect = 2e-06
Identities = 30/56 (53%), Positives = 35/56 (61%), Gaps = 3/56 (5%)
Frame = +2
Query: 593 NQGSGGRSGGGRSGGGRGGRFDSGRGGG---RGRFGSGGRGGDRGGRGGRGGGGRG 645
+QG GG GGG GGG GG GRGGG +G +G GG G +GG GG+GG G G
Sbjct: 350 DQGYGG--GGGSYGGGFGGGRGGGRGGGGYXQGGYGGGG-GYXQGGYGGQGGYGGG 508
Score = 46.6 bits (109), Expect = 3e-05
Identities = 26/64 (40%), Positives = 31/64 (47%)
Frame = +2
Query: 579 GGFTLAVDEAKPRDNQGSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGG 638
GG+ D+ GG GGGR GG GG + G GG G + GG GG+GG
Sbjct: 329 GGYGGGYDQGYGGGGGSYGGGFGGGRGGGRGGGGYXQGGYGGGGGYXQGG----YGGQGG 496
Query: 639 RGGG 642
GGG
Sbjct: 497 YGGG 508
>TC78976 similar to GP|10178188|dbj|BAB11662. poly(A)-binding protein
II-like {Arabidopsis thaliana}, partial (67%)
Length = 1676
Score = 68.2 bits (165), Expect = 1e-11
Identities = 45/148 (30%), Positives = 77/148 (51%)
Frame = +2
Query: 337 DSDDEDSSDSDSEVKKDKMDVDESDSSEDSDEEDEKSTKTPQKKVKDVEMVDASSGKNAP 396
++DD D +D+ ++ D+D+ E++ + K Q KV+ EM NA
Sbjct: 86 ENDDVDMIGADN----NEADLDDMKKRLKEMEDEAAALKEMQAKVEK-EMGSVQDPANAS 250
Query: 397 KTPAAQTENEGSKTLFVGNLSFSVQRSDVENFFKDCGEVVDVRLATDEDGRFKGFGHVEF 456
+ + E + ++++FVGN+ ++ DV+ F+ CG V V + TD+ G+ KG+ +VEF
Sbjct: 251 ASQINREEVD-ARSIFVGNVDYACTPEDVQQHFQSCGTVNRVTIRTDKFGQPKGYAYVEF 427
Query: 457 ATAAAAQSALEYNGSELLQRPVRLDLAR 484
A Q AL N SEL R +++ R
Sbjct: 428 VEVEAVQEALLLNESELHGRQLKVTAKR 511
Score = 50.8 bits (120), Expect = 2e-06
Identities = 27/78 (34%), Positives = 48/78 (60%)
Frame = +2
Query: 508 SQTVFVRGFDKFQGEDDIKSSLQEHFGSCGEITRISVPKDFESGNVKGFAYLDFSDTNSV 567
++++FV D +D+ Q+HF SCG + R+++ D + G KG+AY++F + +V
Sbjct: 281 ARSIFVGNVDYACTPEDV----QQHFQSCGTVNRVTIRTD-KFGQPKGYAYVEFVEVEAV 445
Query: 568 NKALELHDSDLGGFTLAV 585
+AL L++S+L G L V
Sbjct: 446 QEALLLNESELHGRQLKV 499
>BQ140761 weakly similar to GP|18076245|emb phosphophoryn {Rattus
norvegicus}, partial (9%)
Length = 620
Score = 68.2 bits (165), Expect = 1e-11
Identities = 55/157 (35%), Positives = 73/157 (46%), Gaps = 4/157 (2%)
Frame = +1
Query: 214 AVKPAVQKKK---EESSSDDSSSESEDEKPAVKVSAPSKVVPAAVKNGAADSDSSDSDSS 270
A K A Q +E SSD S S+S D S+ + + + DS S SD S
Sbjct: 196 ATKQAAQSDAMDVDEKSSDSSDSDSSD-------SSDGSDSDSDASDDSDDSSDSSSDDS 354
Query: 271 SDEDDDKKKPATKVAKKLPAAAPIKKVEKSESSDDESSESSDEENDSKPATTTVAKP-SA 329
S +++ P + AKK A KS SS +SS+ S E++S+P V P S
Sbjct: 355 SSDEEPVPAPKKEKAKKAKKA-------KSVSSSSDSSDDSSSESESEPEPXNVPLPESD 513
Query: 330 VKKSASSDSDDEDSSDSDSEVKKDKMDVDESDSSEDS 366
SASSDSD SSDS S+ D + DSS +S
Sbjct: 514 DSSSASSDSD--SSSDSSSDSSSDSSSXSDXDSSSES 618
Score = 65.9 bits (159), Expect = 5e-11
Identities = 64/202 (31%), Positives = 86/202 (41%), Gaps = 2/202 (0%)
Frame = +1
Query: 1 MPKSSKKSATKVEAAAPVAAAPKSAKKAKRAAEDEVEKQVSAKKQKIAEVAAKQKKETKL 60
M KS KKSAT E +S K+ D+ ++ A A KQ ++
Sbjct: 64 MAKSDKKSATAPEDVQTDLDLVQSF--LKQTGHDKAASTLTG----FAAWATKQAAQSDA 225
Query: 61 QKVKKESSSDDDSSSEDEKPAPKVAPLKTSVKNGTTPAKKAKPAPSSSSSDEDSSDEEEE 120
V ++SS DS S D + + A S SSSD+ SSD EE
Sbjct: 226 MDVDEKSSDSSDSDSSDSSDG----------SDSDSDASDDSDDSSDSSSDDSSSD-EEP 372
Query: 121 VIAKKPTKVVVPKKEESSSDEDSSSDEDSSSESDDEKPAAKVAVPAK--KAPAKNGSVTA 178
V A K K KK +S S SSD DSSSES+ E V +P + A + S ++
Sbjct: 373 VPAPKKEKAKKAKKAKSVSSSSDSSD-DSSSESESEPEPXNVPLPESDDSSSASSDSDSS 549
Query: 179 PAKKAQPAPSSSSESEDDDSSE 200
+ + SSS S+ D SSE
Sbjct: 550 SDSSSDSSSDSSSXSDXDSSSE 615
Score = 64.7 bits (156), Expect = 1e-10
Identities = 60/182 (32%), Positives = 69/182 (36%), Gaps = 2/182 (1%)
Frame = +1
Query: 131 VPKKEESSSDEDSSSDEDSSSESDDEKPAAKVAVPAKKAPAKNGSVTAPAKKAQPAPSSS 190
V +K SSD DSS D S D A S
Sbjct: 232 VDEKSSDSSDSDSSDSSDGSDSDSD---------------------------ASDDSDDS 330
Query: 191 SESEDDDSSEEDEPSAKQPKKEVAVKPAVQKKKEES--SSDDSSSESEDEKPAVKVSAPS 248
S+S DDSS ++EP PKKE A K K S SSDDSSSESE E V P
Sbjct: 331 SDSSSDDSSSDEEP-VPAPKKEKAKKAKKAKSVSSSSDSSDDSSSESESEPEPXNVPLP- 504
Query: 249 KVVPAAVKNGAADSDSSDSDSSSDEDDDKKKPATKVAKKLPAAAPIKKVEKSESSDDESS 308
++ + S SSDSDSSSD D ++ S S D SS
Sbjct: 505 -------ESDDSSSASSDSDSSSDSSSDSSSDSS-----------------SXSDXDSSS 612
Query: 309 ES 310
ES
Sbjct: 613 ES 618
Score = 63.5 bits (153), Expect = 2e-10
Identities = 54/188 (28%), Positives = 84/188 (43%), Gaps = 3/188 (1%)
Frame = +1
Query: 199 SEEDEPSAKQPKKEVAVKPAVQKKKEESSSDDSSSESEDEKP-AVKVSAPSKVVPAAVKN 257
++ D+ SA P+ VQ +++ D ++S A K +A S + K+
Sbjct: 67 AKSDKKSATAPEDVQTDLDLVQSFLKQTGHDKAASTLTGFAAWATKQAAQSDAMDVDEKS 246
Query: 258 G-AADSDSSDSDSSSDEDDDKKKPATKVAKKLPAAAPIKKVEKSESSDDESSESSDEEND 316
++DSDSSDS SD D D + + S+SS D+S SSDEE
Sbjct: 247 SDSSDSDSSDSSDGSDSDSDASDDSD---------------DSSDSSSDDS--SSDEEPV 375
Query: 317 SKPATTTVAKPSAVKK-SASSDSDDEDSSDSDSEVKKDKMDVDESDSSEDSDEEDEKSTK 375
P K K S+SSDS D+ SS+S+SE + + + ESD S + + + S+
Sbjct: 376 PAPKKEKAKKAKKAKSVSSSSDSSDDSSSESESEPEPXNVPLPESDDSSSASSDSDSSSD 555
Query: 376 TPQKKVKD 383
+ D
Sbjct: 556 SSSDSSSD 579
Score = 43.9 bits (102), Expect = 2e-04
Identities = 40/130 (30%), Positives = 55/130 (41%), Gaps = 4/130 (3%)
Frame = +1
Query: 281 ATKVAKKLPAAAPIKKVEKSESSDDESSESSDEENDSKPATTTVAKPSAVKKSASSDSDD 340
ATK A + A +K S+SSD +SS+SSD SDSD
Sbjct: 196 ATKQAAQSDAMDVDEK--SSDSSDSDSSDSSD----------------------GSDSDS 303
Query: 341 EDSSDSDSEVKKDKMDVDESDSSEDSDEEDEKSTKTPQ----KKVKDVEMVDASSGKNAP 396
+ S DSD D SDSS D DE+ P+ KK K + V +SS +
Sbjct: 304 DASDDSD----------DSSDSSSDDSSSDEEPVPAPKKEKAKKAKKAKSVSSSSDSSDD 453
Query: 397 KTPAAQTENE 406
+ +++E E
Sbjct: 454 SSSESESEPE 483
>TC91772 weakly similar to GP|17380998|gb|AAL36311.1 unknown protein
{Arabidopsis thaliana}, partial (17%)
Length = 1237
Score = 68.2 bits (165), Expect = 1e-11
Identities = 82/300 (27%), Positives = 122/300 (40%), Gaps = 17/300 (5%)
Frame = +2
Query: 108 SSSDEDSSDEEEEVIAKKPTKVVVPKKEESSSDEDSSSDEDSSS-ESDDEKPAAKVAVPA 166
S+ +D E + + T V K + S D D D+ +S + KP V
Sbjct: 2 SARGKDPKLERSTIKKQDGTDSPVLSKGNNLSGNDERDDMDTEKIDSKEPKPERSVRRKG 181
Query: 167 KKA-------PAKNGSVTAPAKKAQPAPSSSSESEDDDSSEEDE----PSAKQPKKEVAV 215
KKA P+K +V + + + A S SS+ E S ED + KE+
Sbjct: 182 KKASSSKSTKPSKKSNVVSEKEAEKTADSKSSKKEVPISLNEDSVVEATGTSENDKEIKA 361
Query: 216 K---PAVQKKKEESSSDDSSSESEDEKPAVKVSAPSKVVPAAVKNGAADSDSSDSDSSSD 272
K P + +++ S SES ++ K +K ++ K AA+ S S
Sbjct: 362 KISSPKAGGLESDAAGSPSPSESNHDENRSKKRVRTKKNDSSAKEVAAEDISKKV--SEG 535
Query: 273 EDDDKKKPATKVAKKLP-AAAPIKKVEKSESSDDESSESSDEENDSKPATTTVAKPSAVK 331
D K KPA AKK P ++ +K V + +D SS E+ K T K S+ K
Sbjct: 536 TSDSKVKPARPSAKKGPIRSSDVKTVVHAVMADVGSSSLKPEDKKKK----THVKGSSEK 703
Query: 332 KSASSDSDDEDSSDSDSEVKKDKMDVDESDSSEDSDEEDEKSTKTPQK-KVKDVEMVDAS 390
A S ++DED S K DE SE++ + K +TP K K D + D S
Sbjct: 704 GLAKSSAEDEDKVTVSSLKSATKTTKDE--HSEETPKTTLKRKRTPGKEKGSDTKKNDQS 877
Score = 55.1 bits (131), Expect = 9e-08
Identities = 82/359 (22%), Positives = 136/359 (37%), Gaps = 25/359 (6%)
Frame = +2
Query: 73 SSSEDEKPAPK--VAPLKTSVKNGTTPAKKAKPAPSSSSSDE-----DSSDEEEEVIAKK 125
SSS+ KP+ K V K + K + + K K P S + D +S+ ++E+ A
Sbjct: 191 SSSKSTKPSKKSNVVSEKEAEKTADSKSSK-KEVPISLNEDSVVEATGTSENDKEIKA-- 361
Query: 126 PTKVVVPKKEESSSDEDSSSDEDSSSESDDEKPAAKVAVPAKKAPAKNGSVTAPAKKAQP 185
K+ PK SD ++ S SES+ ++ +K K+ K +A A+
Sbjct: 362 --KISSPKAGGLESD---AAGSPSPSESNHDENRSK-----KRVRTKKNDSSAKEVAAED 511
Query: 186 APSSSSESEDDDSSEEDEPSAKQ-PKKEVAVKPAVQKKKEESSSDDSSSESEDEKPAVKV 244
SE D + PSAK+ P + VK V + S E + +K VK
Sbjct: 512 ISKKVSEGTSDSKVKPARPSAKKGPIRSSDVKTVVHAVMADVGSSSLKPEDKKKKTHVKG 691
Query: 245 SAPSKVVPAAVKNGAADSDSSDSDSSSDEDDDKKKPATKVAKKLPAAAPIKKVEKSESSD 304
S+ + G A S + D D + K ATK K + K K + +
Sbjct: 692 SS---------EKGLAKSSAEDEDKVT---VSSLKSATKTTKDEHSEETPKTTLKRKRTP 835
Query: 305 DESSESSDEENDSK----------PATTTVAKPSAVKKSASSDSDDEDSSDSDSEV---K 351
+ S ++ND P K +S+ D D E+ K
Sbjct: 836 GKEKGSDTKKNDQSLVGKRVKVWWPDDNMFYKGVVDSFDSSTKKHKVLYDDGDEEILNFK 1015
Query: 352 KDKMDVDESDSSEDSDEEDEKSTKTPQKK----VKDVEMVDASSGKNAPKTPAAQTENE 406
++K ++ E D+ D D E+ +P+ +K +A K K +++ E +
Sbjct: 1016EEKYEIVEVDADADPDVEEGSHRASPEPSADMPLKKKGKTNAGESKKEVKKESSKVEEQ 1192
Score = 37.0 bits (84), Expect = 0.024
Identities = 52/246 (21%), Positives = 92/246 (37%)
Frame = +2
Query: 4 SSKKSATKVEAAAPVAAAPKSAKKAKRAAEDEVEKQVSAKKQKIAEVAAKQKKETKLQKV 63
S S +KV+ A P SAKK + D V+ V A + + K + + K V
Sbjct: 527 SEGTSDSKVKPARP------SAKKGPIRSSD-VKTVVHAVMADVGSSSLKPEDKKKKTHV 685
Query: 64 KKESSSDDDSSSEDEKPAPKVAPLKTSVKNGTTPAKKAKPAPSSSSSDEDSSDEEEEVIA 123
K S SS +++ V+ LK++ K TT + ++ P ++ + + +E+
Sbjct: 686 KGSSEKGLAKSSAEDEDKVTVSSLKSATK--TTKDEHSEETPKTTLKRKRTPGKEKGSDT 859
Query: 124 KKPTKVVVPKKEESSSDEDSSSDEDSSSESDDEKPAAKVAVPAKKAPAKNGSVTAPAKKA 183
KK + +V K+ + +D+ + D KV N +
Sbjct: 860 KKNDQSLVGKRVKVWWPDDNMFYKGVVDSFDSSTKKHKVLYDDGDEEILNFK-EEKYEIV 1036
Query: 184 QPAPSSSSESEDDDSSEEDEPSAKQPKKEVAVKPAVQKKKEESSSDDSSSESEDEKPAVK 243
+ + + E+ EPSA P K+ A + KKE E
Sbjct: 1037EVDADADPDVEEGSHRASPEPSADMPLKKKGKTNAGESKKEVKKESSKVEEQHPVNQRHH 1216
Query: 244 VSAPSK 249
+ +P+K
Sbjct: 1217LQSPTK 1234
>TC76331 similar to GP|7673355|gb|AAF66823.1| poly(A)-binding protein
{Nicotiana tabacum}, partial (97%)
Length = 2581
Score = 67.8 bits (164), Expect = 1e-11
Identities = 49/203 (24%), Positives = 92/203 (45%), Gaps = 7/203 (3%)
Frame = +3
Query: 377 PQKKVKDVEMVDASSGKNAP-------KTPAAQTENEGSKTLFVGNLSFSVQRSDVENFF 429
PQ + +++++ + N P + P+ + +G+ +F+ NL ++ + + F
Sbjct: 597 PQDAARALDVLNFTPLNNRPIRIMYSHRDPSIRKSGQGN--IFIKNLDKAIDHKALHDTF 770
Query: 430 KDCGEVVDVRLATDEDGRFKGFGHVEFATAAAAQSALEYNGSELLQRPVRLDLARERGAY 489
G ++ ++A D G+ KG+G V+F T AAQ A+E LL ++ Y
Sbjct: 771 SSFGNILSCKVAVDGSGQSKGYGFVQFDTEEAAQKAIEKLNGMLL---------NDKQVY 923
Query: 490 TPNSGAANNSFQKGGRGQSQTVFVRGFDKFQGEDDIKSSLQEHFGSCGEITRISVPKDFE 549
G R + VFV+ + +D++K + FG G IT V +D +
Sbjct: 924 VGPFLRKQERESTGDRAKFNNVFVKNLSESTTDDELKKT----FGEFGTITSAVVMRDGD 1091
Query: 550 SGNVKGFAYLDFSDTNSVNKALE 572
G K F +++F T+ +A+E
Sbjct: 1092-GKSKCFGFVNFESTDDAARAVE 1157
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.298 0.120 0.311
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,326,429
Number of Sequences: 36976
Number of extensions: 199582
Number of successful extensions: 10050
Number of sequences better than 10.0: 868
Number of HSP's better than 10.0 without gapping: 2904
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6100
length of query: 664
length of database: 9,014,727
effective HSP length: 102
effective length of query: 562
effective length of database: 5,243,175
effective search space: 2946664350
effective search space used: 2946664350
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 44 (22.0 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0135.11


