

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0133.10
(382 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
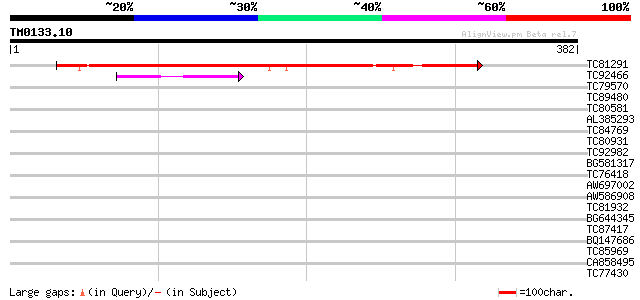
Score E
Sequences producing significant alignments: (bits) Value
TC81291 similar to GP|13374857|emb|CAC34491. putative protein {A... 358 2e-99
TC92466 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis... 42 3e-04
TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (AR... 40 0.001
TC89480 similar to GP|20260684|gb|AAM13240.1 unknown protein {Ar... 38 0.006
TC80581 similar to GP|19683021|gb|AAL92644.1 hypothetical protei... 38 0.006
AL385293 similar to GP|5737842|gb| adenylyl cyclase {Dictyosteli... 38 0.007
TC84769 similar to PIR|E96665|E96665 protein F22C12.16 [imported... 37 0.010
TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem tra... 37 0.017
TC92982 similar to GP|23332794|gb|AAN27343.1 Sequence 13 from pa... 35 0.048
BG581317 homologue to OMNI|NTL01EC003 IS2 hypothetical protein {... 35 0.063
TC76418 similar to GP|16226228|gb|AAL16109.1 At1g20110/T20H2_10 ... 35 0.063
AW697002 similar to GP|10728183|gb CG11122 gene product {Drosoph... 33 0.14
AW586908 weakly similar to GP|2104814|emb| pleiotropic effects o... 33 0.14
TC81932 similar to GP|13430778|gb|AAK26011.1 unknown protein {Ar... 33 0.14
BG644345 weakly similar to GP|19697333|gb putative protein poten... 33 0.18
TC87417 homologue to SP|O22607|MSI4_ARATH WD-40 repeat protein M... 33 0.24
BQ147686 similar to PIR|T02909|T02 hypothetical protein T13J8.19... 33 0.24
TC85969 similar to GP|21553908|gb|AAM62991.1 unknown {Arabidopsi... 32 0.31
CA858495 32 0.41
TC77430 similar to GP|13676413|dbj|BAB41197. hypothetical protei... 32 0.41
>TC81291 similar to GP|13374857|emb|CAC34491. putative protein {Arabidopsis
thaliana}, partial (14%)
Length = 1051
Score = 358 bits (919), Expect = 2e-99
Identities = 201/305 (65%), Positives = 232/305 (75%), Gaps = 18/305 (5%)
Frame = +3
Query: 32 GAYMRSLVKQLRTN---------DQGCVVNSDASSHGQNLTKHGKIGKACQSQQSTQQPQ 82
G + SLVKQL T+ +Q CVVN D S GQNL+K GK+ A +Q+ + QPQ
Sbjct: 156 GMELDSLVKQLNTSRAKDLMNPKNQHCVVN-DGVSVGQNLSKIGKVVDAQHAQKRSTQPQ 332
Query: 83 QHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQ-QQQQLQQQQQQ 141
++KKQVRRRLHT +PYQERLLNMAEARREIVTALKFHRASMKEASEQQ QQQQ Q+QQQ
Sbjct: 333 EYKKQVRRRLHTVKPYQERLLNMAEARREIVTALKFHRASMKEASEQQKQQQQEAQEQQQ 512
Query: 142 RAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSC--NTNFSSYMGDFS----SYPPAFVPNS 195
R SPQ SQ+ SFDQDGRYKSRRNPRIYPSC TN SSY+ DFS S+ P VPNS
Sbjct: 513 RQSPSPQPSQHQSFDQDGRYKSRRNPRIYPSCTTKTNSSSYLNDFSYPYLSHIPPSVPNS 692
Query: 196 YPVPVASPIAPPPLMAENPNFILPNQPLGLNLNFHDFNNLDATIHLSNTSLSSNSFSSAT 255
+ P ASPI PPPL+AENPNFILPNQ LGLNLNFHDFNNL+ T+HL+N+S S++S+SS T
Sbjct: 693 HTWPAASPINPPPLLAENPNFILPNQTLGLNLNFHDFNNLEFTVHLNNSS-STSSYSSGT 869
Query: 256 SS--SQEVPSVELSQGDGISSLVNSTESNAASQVNAGLHAAMDEEAIAEIRSLGEQYQME 313
SS SQ+VPSV Q +G S +S+AA+ V GLH MD+EA+AEIRSLG QYQME
Sbjct: 870 SSSASQDVPSVGTLQAEGF-----SIDSHAATHVTGGLHTVMDDEAMAEIRSLGNQYQME 1034
Query: 314 WNDTM 318
WNDTM
Sbjct: 1035WNDTM 1049
>TC92466 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis thaliana},
partial (14%)
Length = 632
Score = 42.4 bits (98), Expect = 3e-04
Identities = 25/85 (29%), Positives = 39/85 (45%)
Frame = +1
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
+ QQ QQ QQ + Q ++ H + Q+ + + + M+ ++QQQQ
Sbjct: 367 REQQQQQQQQQQQPQPQQSQHAQQQQQQHM--------------QMQQLLMQRHAQQQQQ 504
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQ 157
QQ QQQ Q P PQ Q + D+
Sbjct: 505 QQQHQQQPQSQPQQPQPQQQQNRDR 579
Score = 30.0 bits (66), Expect = 1.6
Identities = 18/33 (54%), Positives = 20/33 (60%)
Frame = +1
Query: 125 EASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQ 157
+A EQQQQQQ QQQQ Q PQ SQ+ Q
Sbjct: 361 KAREQQQQQQQQQQQPQ-----PQQSQHAQQQQ 444
Score = 28.5 bits (62), Expect = 4.5
Identities = 18/60 (30%), Positives = 28/60 (46%), Gaps = 8/60 (13%)
Frame = +1
Query: 120 RASMKEASEQQQQQQLQQQQQQRAPDSPQ--------LSQNPSFDQDGRYKSRRNPRIYP 171
+A ++ +QQQQQQ Q QQ Q A Q L Q + Q + + ++ P+ P
Sbjct: 361 KAREQQQQQQQQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQPQSQP 540
>TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (15%)
Length = 1143
Score = 40.4 bits (93), Expect = 0.001
Identities = 48/193 (24%), Positives = 77/193 (39%), Gaps = 1/193 (0%)
Frame = +2
Query: 52 NSDASSHGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARRE 111
N +A+ + + + +A Q +Q P Q + Q ++ +Q+ L + +
Sbjct: 404 NGNAALMQNQMLQQSQPHQAFPKNQESQHPSQSQAQTQQ-------FQQLLQH-----QH 547
Query: 112 IVTALKFHRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYP 171
T +H ++ +QQQQQQ QQ QQQ+ S Q Q Q
Sbjct: 548 SFTNQNYHMQQQQQQQQQQQQQQQQQHQQQQQQQSQQQQQVVDHQQ-------------- 685
Query: 172 SCNTNFSSYMGDFSSYPPAFVPNSYPVPVASPIAPPPLMAE-NPNFILPNQPLGLNLNFH 230
+N S M F S PP+ ++ P+ S I ++ N N + P L+
Sbjct: 686 --MSNAVSTMSQFVSAPPS--QSTQPMQAISSIGHQQNFSDSNGNPVSP-----LHNMLG 838
Query: 231 DFNNLDATIHLSN 243
F+N D T HL N
Sbjct: 839 SFSN-DETSHLLN 874
>TC89480 similar to GP|20260684|gb|AAM13240.1 unknown protein {Arabidopsis
thaliana}, partial (43%)
Length = 1068
Score = 38.1 bits (87), Expect = 0.006
Identities = 28/83 (33%), Positives = 43/83 (51%), Gaps = 3/83 (3%)
Frame = +1
Query: 73 QSQ-QSTQQPQQHKKQVRRRLHTSR--PYQERLLNMAEARREIVTALKFHRASMKEASEQ 129
QSQ QS Q P + +Q ++ LH + P+Q+ T +F + ++ +Q
Sbjct: 124 QSQSQSNQDPNLYLQQQQQFLHQQQQQPFQQSQ----------TTQSQFQQQQQQQLYQQ 273
Query: 130 QQQQQLQQQQQQRAPDSPQLSQN 152
QQQQ++ QQQQQ+ PQ QN
Sbjct: 274 QQQQRILQQQQQQL-QQPQQQQN 339
>TC80581 similar to GP|19683021|gb|AAL92644.1 hypothetical protein
{Dictyostelium discoideum}, partial (5%)
Length = 850
Score = 38.1 bits (87), Expect = 0.006
Identities = 32/100 (32%), Positives = 48/100 (48%), Gaps = 20/100 (20%)
Frame = +1
Query: 75 QQSTQQPQQHKKQVRRRLHTSRPYQ---ERLLNMAEARREIVTA-------LKFHRASMK 124
Q QQ QH++ ++++L + YQ ++L + A+ ++ +T L SM
Sbjct: 298 QPQLQQQLQHQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMG 477
Query: 125 EASEQQQQQQLQQQQ----------QQRAPDSPQLSQNPS 154
+ QQQQQQL QQQ QQ+ S QL QN S
Sbjct: 478 QPLLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSS 597
Score = 32.7 bits (73), Expect = 0.24
Identities = 26/88 (29%), Positives = 44/88 (49%), Gaps = 9/88 (10%)
Frame = +1
Query: 73 QSQQSTQQP---QQHKKQVRRRLHTSRPYQE------RLLNMAEARREIVTALKFHRASM 123
Q QQ QQ QQ ++++++L +S Q+ +L + + ++ + L +
Sbjct: 496 QQQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQ 675
Query: 124 KEASEQQQQQQLQQQQQQRAPDSPQLSQ 151
++ QQQQ LQQQQQQ+ QL Q
Sbjct: 676 QQLLLQQQQLVLQQQQQQQQQQRTQLLQ 759
>AL385293 similar to GP|5737842|gb| adenylyl cyclase {Dictyostelium
discoideum}, partial (1%)
Length = 467
Score = 37.7 bits (86), Expect = 0.007
Identities = 29/92 (31%), Positives = 42/92 (45%)
Frame = +1
Query: 128 EQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYMGDFSSY 187
+QQ QQQ QQQ QQ + D Q Q PS D D + + P TN + G S
Sbjct: 64 QQQHQQQQQQQNQQNSQDQSQQIQLPSKDDDSTFIAPTPPTQLNQIRTNVVN-TGSASPS 240
Query: 188 PPAFVPNSYPVPVASPIAPPPLMAENPNFILP 219
+ P +SP+A P +++P+ + P
Sbjct: 241 AALGRKSIRSNPASSPLALGP--SKSPHSLKP 330
>TC84769 similar to PIR|E96665|E96665 protein F22C12.16 [imported] -
Arabidopsis thaliana, partial (4%)
Length = 723
Score = 37.4 bits (85), Expect = 0.010
Identities = 31/89 (34%), Positives = 38/89 (41%), Gaps = 8/89 (8%)
Frame = +2
Query: 194 NSYPVPVASPIAPPPLMAENPNFILPNQPL--------GLNLNFHDFNNLDATIHLSNTS 245
N P V SP APP NPNFI N P G + + IH S S
Sbjct: 263 NHSPTSVPSP-APPSAETNNPNFIRKNLPSPWAQVVRGGSGWDTESQSQSPTGIHQSLPS 439
Query: 246 LSSNSFSSATSSSQEVPSVELSQGDGISS 274
SS+S SS T+ Q PS + + +SS
Sbjct: 440 SSSSSSSSLTTVDQPPPSDDSPKAAVVSS 526
>TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem transcription
factor M1 {Apium graveolens}, partial (67%)
Length = 1193
Score = 36.6 bits (83), Expect = 0.017
Identities = 23/98 (23%), Positives = 49/98 (49%)
Frame = +2
Query: 52 NSDASSHGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARRE 111
NS+ ++ ++ I Q Q QQ QQ Q+++ S+ Q+++ + + +++
Sbjct: 506 NSNKCNNTSRCNRNSSIFPQLQQQMQQQQQQQQLPQLQQNQQLSQ-IQQQIPQLQQQQQQ 682
Query: 112 IVTALKFHRASMKEASEQQQQQQLQQQQQQRAPDSPQL 149
+ + + +++ +QQQ QLQQ Q Q+ P Q+
Sbjct: 683 LPQLQQQQLSQLQQ--QQQQLPQLQQLQHQQLPQQQQM 790
Score = 28.9 bits (63), Expect = 3.5
Identities = 16/29 (55%), Positives = 17/29 (58%)
Frame = +2
Query: 129 QQQQQQLQQQQQQRAPDSPQLSQNPSFDQ 157
Q QQQ QQQQQQ+ PQL QN Q
Sbjct: 563 QLQQQMQQQQQQQQL---PQLQQNQQLSQ 640
Score = 28.1 bits (61), Expect = 5.9
Identities = 12/18 (66%), Positives = 15/18 (82%)
Frame = +1
Query: 125 EASEQQQQQQLQQQQQQR 142
+ S QQQQQQ+QQQQ Q+
Sbjct: 472 QLSGQQQQQQMQQQQMQQ 525
>TC92982 similar to GP|23332794|gb|AAN27343.1 Sequence 13 from patent US
6440698, partial (1%)
Length = 795
Score = 35.0 bits (79), Expect = 0.048
Identities = 27/93 (29%), Positives = 44/93 (47%)
Frame = -1
Query: 99 QERLLNMAEARREIVTALKFHRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQD 158
Q++ ++ + + R+S + S+QQ QQ QQQQQQ+ + Q Q S+
Sbjct: 699 QQQTASLQSFNMRVSASASQSRSSSTKYSQQQAQQAQQQQQQQQPQQTQQTQQTQSY--- 529
Query: 159 GRYKSRRNPRIYPSCNTNFSSYMGDFSSYPPAF 191
G Y P+ PS + S+ G S+ P +F
Sbjct: 528 GSYP----PQAQPS-HPEQHSWYGFGSNAPSSF 445
>BG581317 homologue to OMNI|NTL01EC003 IS2 hypothetical protein {Escherichia
coli K12-MG1655}, partial (55%)
Length = 808
Score = 34.7 bits (78), Expect = 0.063
Identities = 35/133 (26%), Positives = 58/133 (43%), Gaps = 5/133 (3%)
Frame = +2
Query: 147 PQLSQNPS-FDQDGRYKSRRNPRIYPSCN--TNFSSYMGDFSSYPPAFVPNSYPVPVASP 203
P + QNP+ +S P Y S T Y +S YPP N VP ++P
Sbjct: 113 PHMQQNPNPIPNPTPPQSDPIPNHYASAPPFTPNYDYSSTYSPYPPH---NPDHVPSSNP 283
Query: 204 -IAPPPLMAENPNFILPNQPLGLNLNFHDFNNLDAT-IHLSNTSLSSNSFSSATSSSQEV 261
PPP + NP + P+QP + ++ T ++ + + + N ++S+ S +
Sbjct: 284 SFNPPPFESSNPLYQQPSQPY-----YPPYDQQHQTPLNYAPPNPNPNLYNSSPYSVPPI 448
Query: 262 PSVELSQGDGISS 274
PS E S + + S
Sbjct: 449 PSYETSYENPVKS 487
>TC76418 similar to GP|16226228|gb|AAL16109.1 At1g20110/T20H2_10
{Arabidopsis thaliana}, partial (54%)
Length = 2020
Score = 34.7 bits (78), Expect = 0.063
Identities = 35/133 (26%), Positives = 58/133 (43%), Gaps = 5/133 (3%)
Frame = +3
Query: 147 PQLSQNPS-FDQDGRYKSRRNPRIYPSCN--TNFSSYMGDFSSYPPAFVPNSYPVPVASP 203
P + QNP+ +S P Y S T Y +S YPP N VP ++P
Sbjct: 69 PHMQQNPNPIPNPTPPQSDPIPNHYASAPPFTPNYDYSSTYSPYPPH---NPDHVPSSNP 239
Query: 204 -IAPPPLMAENPNFILPNQPLGLNLNFHDFNNLDAT-IHLSNTSLSSNSFSSATSSSQEV 261
PPP + NP + P+QP + ++ T ++ + + + N ++S+ S +
Sbjct: 240 SFNPPPFESSNPLYQQPSQPY-----YPPYDQQHQTPLNYAPPNPNPNLYNSSPYSVPPI 404
Query: 262 PSVELSQGDGISS 274
PS E S + + S
Sbjct: 405 PSYETSYENPVKS 443
>AW697002 similar to GP|10728183|gb CG11122 gene product {Drosophila
melanogaster}, partial (0%)
Length = 857
Score = 33.5 bits (75), Expect = 0.14
Identities = 22/50 (44%), Positives = 29/50 (58%)
Frame = +3
Query: 107 EARREIVTALKFHRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFD 156
E +RE + +K R S KE +QQQQL QQQQQ+A + S+ S D
Sbjct: 123 EDKREEPSQVKSPRESKKE----KQQQQLHQQQQQQASVDEKYSEWKSLD 260
>AW586908 weakly similar to GP|2104814|emb| pleiotropic effects on cellular
differentiation and slug behaviour {Dictyostelium
discoideum}, partial (8%)
Length = 668
Score = 33.5 bits (75), Expect = 0.14
Identities = 32/109 (29%), Positives = 48/109 (43%), Gaps = 17/109 (15%)
Frame = +2
Query: 60 QNLT-KHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKF 118
QN T +G IG Q QQP + Q++++L + ++ L A ++T+L
Sbjct: 218 QNTTMNNGTIGSNQIPNQCVQQPVTYS-QLQQQLLSGSMQSQQNLQSAGKNGLMMTSLPQ 394
Query: 119 HRASMKEASEQQ------QQQQ----------LQQQQQQRAPDSPQLSQ 151
++ +QQ QQQQ LQQ QR P PQ+SQ
Sbjct: 395 DSQFQQQIDQQQAGLLQRQQQQNQLQQSSLQLLQQSMLQRGPQQPQMSQ 541
Score = 29.3 bits (64), Expect = 2.6
Identities = 28/98 (28%), Positives = 41/98 (41%), Gaps = 10/98 (10%)
Frame = +2
Query: 60 QNLTKHGKIGKAC----QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEAR--REIV 113
QNL GK G Q Q QQ Q + + +R Q+ L + + +
Sbjct: 341 QNLQSAGKNGLMMTSLPQDSQFQQQIDQQQAGLLQRQQQQNQLQQSSLQLLQQSMLQRGP 520
Query: 114 TALKFHRASMKEASEQQQQ----QQLQQQQQQRAPDSP 147
+ + + S+QQQQ Q+LQQQQQQ+ P
Sbjct: 521 QQPQMSQTVPQNISDQQQQLQLLQKLQQQQQQQQQQQP 634
>TC81932 similar to GP|13430778|gb|AAK26011.1 unknown protein {Arabidopsis
thaliana}, partial (18%)
Length = 1139
Score = 33.5 bits (75), Expect = 0.14
Identities = 28/89 (31%), Positives = 43/89 (47%), Gaps = 14/89 (15%)
Frame = -2
Query: 184 FSSYPPAFVPNSYPVPVA------SPIAP----PPLMAENPNFILPNQPLG----LNLNF 229
FS + P FV ++ P+ ++ SP+ P PPL+ P + N PLG L L F
Sbjct: 622 FSFFSPQFV-SAKPLSLSDFGAGFSPLTPSCNSPPLLFLEPPAPISNPPLGIIGFLGLKF 446
Query: 230 HDFNNLDATIHLSNTSLSSNSFSSATSSS 258
H F + + ++ S S++ F S SS
Sbjct: 445 HGFCTISSAPPPASASSSASRFLSPALSS 359
>BG644345 weakly similar to GP|19697333|gb putative protein potential
transcriptional repressor Not4hp - Mus musculus, partial
(7%)
Length = 635
Score = 33.1 bits (74), Expect = 0.18
Identities = 15/25 (60%), Positives = 18/25 (72%)
Frame = +1
Query: 125 EASEQQQQQQLQQQQQQRAPDSPQL 149
+ +QQQQQQLQQQQQQ+ QL
Sbjct: 340 QQQQQQQQQQLQQQQQQQLQQQQQL 414
Score = 30.8 bits (68), Expect = 0.91
Identities = 18/36 (50%), Positives = 21/36 (58%), Gaps = 2/36 (5%)
Frame = +1
Query: 128 EQQQQQQLQQQQQQRAPDSPQ--LSQNPSFDQDGRY 161
+QQQQQQLQQQQQ + Q L Q+ S Q Y
Sbjct: 373 QQQQQQQLQQQQQLQLQQQQQQLLQQHQSSQQQQLY 480
Score = 28.9 bits (63), Expect = 3.5
Identities = 14/25 (56%), Positives = 16/25 (64%)
Frame = +1
Query: 127 SEQQQQQQLQQQQQQRAPDSPQLSQ 151
S+QQQQ Q QQQQQQ+ Q Q
Sbjct: 313 SQQQQQLQFQQQQQQQQQQLQQQQQ 387
>TC87417 homologue to SP|O22607|MSI4_ARATH WD-40 repeat protein MSI4.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (36%)
Length = 743
Score = 32.7 bits (73), Expect = 0.24
Identities = 21/45 (46%), Positives = 27/45 (59%)
Frame = +2
Query: 107 EARREIVTALKFHRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQ 151
E +RE + +K R S KE +QQQQL QQQQQ+A + SQ
Sbjct: 131 EDKREEPSQVKSPRESKKE----KQQQQLHQQQQQQASVDEKYSQ 253
>BQ147686 similar to PIR|T02909|T02 hypothetical protein T13J8.190 -
Arabidopsis thaliana, partial (2%)
Length = 669
Score = 32.7 bits (73), Expect = 0.24
Identities = 45/213 (21%), Positives = 80/213 (37%), Gaps = 12/213 (5%)
Frame = +3
Query: 132 QQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYMGDFSSYPPAF 191
++ L Q R P P++S + R RN +PS + N +S + + F
Sbjct: 60 RRSLHQSATSRVPYGPRISGGYNRHYGNRVP--RNKNAFPSSDGNPNSPPTIMNPHATEF 233
Query: 192 VPNSYPVPVASPIAPPPLMAENPNFILPNQPLGLNLNFHDFNNLDATIHLSNTSLSSNSF 251
VP VP +A P + +PN P P N+ L++T ++ N F
Sbjct: 234 VPGQTWVPPNEYMASPNSILVSPNSSPPVSP----------NDEIPPNSLNDTPVNQNEF 383
Query: 252 SSATSSSQEVPSVELSQGDGI----SSLVNSTESNAASQVNAGLHAAMDEEAIAEIRSLG 307
+++ S+ + Q + + + S N+ Q+ H E +
Sbjct: 384 ATSPCSNDSAQVEVILQNEKVVDEQVTEAFSIRVNSEKQLVPQQHPPASNENCCPRQEEK 563
Query: 308 EQY-------QMEWNDTMNLVK-SACWFKFLRN 332
+ Y ++ NDT++ VK CW + N
Sbjct: 564 DIYLSHPIEDEVVNNDTVDKVKPDKCWGDYSDN 662
>TC85969 similar to GP|21553908|gb|AAM62991.1 unknown {Arabidopsis thaliana},
partial (31%)
Length = 2756
Score = 32.3 bits (72), Expect = 0.31
Identities = 39/166 (23%), Positives = 71/166 (42%), Gaps = 8/166 (4%)
Frame = +2
Query: 41 QLRTNDQGCVVNSDASSHGQNLTKHGKIGK--ACQSQQSTQQPQQHKKQVRRRLHTSR-- 96
Q +T Q +S G +L + + + S Q+P +++Q++ + T+R
Sbjct: 1355 QAQTQSQAPQFQQKSSGGGSDLPSPDSVSSDSSLSNAMSRQKPVVYQEQIQIQPGTTRVS 1534
Query: 97 -PYQERL-LNMAEARREIVTALKFHRASMKEASEQQQQQQ--LQQQQQQRAPDSPQLSQN 152
P +L L+ +R ++ ++ +++ E QQQQQ QQQQQ+ PQ SQ
Sbjct: 1535 NPVDPKLNLSDPHSRIQVQQHVQDPGYLLQQQFELQQQQQQFELQQQQQQPQHQPQQSQP 1714
Query: 153 PSFDQDGRYKSRRNPRIYPSCNTNFSSYMGDFSSYPPAFVPNSYPV 198
Q + + + + G + + PA +P YPV
Sbjct: 1715 QHQPQQQQQLQHHQQQQF--------IHGGHYIHHNPA-IPTYYPV 1825
>CA858495
Length = 763
Score = 32.0 bits (71), Expect = 0.41
Identities = 22/80 (27%), Positives = 43/80 (53%), Gaps = 4/80 (5%)
Frame = +3
Query: 210 MAENPNFILPNQPLGL----NLNFHDFNNLDATIHLSNTSLSSNSFSSATSSSQEVPSVE 265
+AE N +L N+ GL N+ ++ L+ +++ ++LSS FS + S + ++
Sbjct: 57 LAEKTNLLL-NEKAGLVEKVNMLLNEKEGLEQKVNILESNLSS--FSEKEAGSVDTTNLL 227
Query: 266 LSQGDGISSLVNSTESNAAS 285
L + +G+ +N ESN +S
Sbjct: 228 LKEKEGLEQKLNILESNLSS 287
>TC77430 similar to GP|13676413|dbj|BAB41197. hypothetical protein {Glycine
max}, partial (71%)
Length = 2529
Score = 32.0 bits (71), Expect = 0.41
Identities = 37/175 (21%), Positives = 69/175 (39%), Gaps = 31/175 (17%)
Frame = +2
Query: 121 ASMKEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSY 180
+S+++ QQ + QQ P S Q + + + P ++ +
Sbjct: 746 SSLQQPLPSAMQQPMPSSMQQPMPSSMQQPMPSLYAAADAFIYAAAIAVSPVLVPHYQAG 925
Query: 181 MGDFSSYPPAFVP-----------NSYPVPVASPIAPPPLMA-------------ENPNF 216
+F +P AF+P ++ P+ +++ I PP A + P
Sbjct: 926 PSNFPEFPSAFLPVGTSSPNITSTSAPPLNLSTTILPPAPSATLAPETFQASVSNKAPTV 1105
Query: 217 ILPNQPLGLNL-NFHDF----NNLDATIHLSN--TSLSSNSFSSATSSSQEVPSV 264
LP LG NL + F ++++A + L+N ++S +S S T Q PS+
Sbjct: 1106SLPAATLGANLPSLAPFTSGGSDINAAVPLANKPNAISGSSLSYQTVPQQMTPSI 1270
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.312 0.126 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,921,512
Number of Sequences: 36976
Number of extensions: 190899
Number of successful extensions: 2409
Number of sequences better than 10.0: 129
Number of HSP's better than 10.0 without gapping: 1637
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2000
length of query: 382
length of database: 9,014,727
effective HSP length: 98
effective length of query: 284
effective length of database: 5,391,079
effective search space: 1531066436
effective search space used: 1531066436
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0133.10


