

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
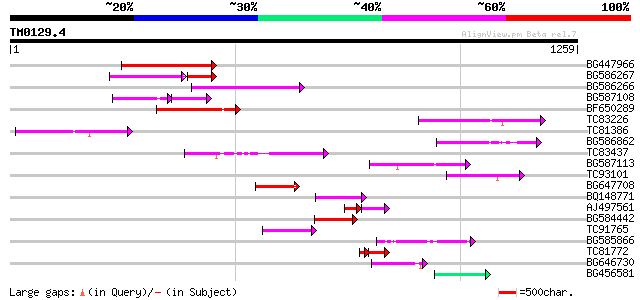
Query= TM0129.4
(1259 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [impo... 230 2e-60
BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At... 113 4e-39
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 153 4e-37
BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - ... 89 1e-33
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 137 3e-32
TC83226 weakly similar to PIR|G86419|G86419 probable reverse tra... 130 4e-30
TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR re... 112 7e-25
BG586862 102 9e-22
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 97 5e-20
BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At... 90 5e-18
TC93101 89 1e-17
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 84 4e-16
BQ148771 74 4e-13
AJ497561 weakly similar to GP|10140689|gb putative non-LTR retro... 56 2e-11
BG584442 68 2e-11
TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative ret... 67 6e-11
BG585866 62 1e-09
TC81772 53 2e-07
BG646730 54 4e-07
BG456581 54 4e-07
>BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 687
Score = 230 bits (587), Expect = 2e-60
Identities = 116/211 (54%), Positives = 144/211 (67%)
Frame = +2
Query: 248 WLKHGDKNTRFFHQKANQRRKRNLIEVLKDDNGREVEDDADIARVLGDYFGGLFTSSNPE 307
WLK GDKNT+FFH KA+QRRK N I+ LKD+ G + + ++ R+L YF LFTSSNP
Sbjct: 5 WLKDGDKNTKFFHSKASQRRKVNEIKKLKDETGNWCKGEENVERLLITYFNNLFTSSNPT 184
Query: 308 GIEETTNLVAGRVSETHLTVLGEPFTREEVEEALFQMHPTKAPGLDGFPTLFYQKYWDII 367
IEET +V G++S H+ + FT EEV EA+ QMHP KAPG DG P LF+QKYW I+
Sbjct: 185 AIEETCEVVKGKLSHEHIVWCEKEFTEEEVLEAINQMHPVKAPGPDGLPALFFQKYWHIV 364
Query: 368 GDDVSAFCLQVLQGVIPPGMINQTLLVLIPKIMKPEHATQFRPISLCNVLFKIITKTIAN 427
G +V LQVL + +N+T +VLIPK P +RPISLCNV+ KIITK IAN
Sbjct: 365 GKEVQQMVLQVLNNSMETEELNKTFIVLIPKGKNPNTPKDYRPISLCNVVMKIITKVIAN 544
Query: 428 RLKIILPDIICQTQSAFVPGRLIIDNALVVY 458
R+K LPD+I QSAFV GRLI DNAL+ +
Sbjct: 545 RVKQTLPDVIDVEQSAFVQGRLITDNALIAW 637
>BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (16%)
Length = 794
Score = 113 bits (283), Expect(2) = 4e-39
Identities = 60/171 (35%), Positives = 99/171 (57%), Gaps = 1/171 (0%)
Frame = +3
Query: 222 INVAERDLDSLLEQEEVWWKQRSRASWLKHGDKNTRFFHQKANQRRKRNLIEVLKDDNGR 281
+++ +L+ EE++W Q+SR +WL+ GD+NT+FFH RR +N I L DD+ +
Sbjct: 81 LSLLRSELNEEYHNEEIFWMQKSRLNWLRSGDRNTKFFHAVTKNRRAQNRILSLIDDDDK 260
Query: 282 EVEDDADIARVLGDYFGGLFTSSNPEGIEETTNLVAGRVSETHLTVLGEPFTREEVEEAL 341
E + D+ R+ +F L++S + E N + V+E L +REEV EA+
Sbjct: 261 EWFVEEDLGRLADSHFKLLYSSEDVGITLEDWNSIPAIVTEEQNAQLMAQISREEVREAV 440
Query: 342 FQMHPTKAPGLDGFPTLFYQKYWDIIGDDVSAFCLQVLQ-GVIPPGMINQT 391
F ++P K PG DG F+Q++WD +GDD+++ + L+ G + G IN+T
Sbjct: 441 FDINPHKCPGPDGMNVFFFQQFWDTMGDDLTSMAQEFLRTGKLEEG-INKT 590
Score = 67.8 bits (164), Expect(2) = 4e-39
Identities = 31/64 (48%), Positives = 47/64 (73%)
Frame = +1
Query: 395 LIPKIMKPEHATQFRPISLCNVLFKIITKTIANRLKIILPDIICQTQSAFVPGRLIIDNA 454
L+PK ++ + +FRPISLCNV +KI++K ++ RLK +LP II +TQ+AF +LI DN
Sbjct: 601 LVPKKLEAKRLVEFRPISLCNVAYKIVSKVLSKRLKSVLPWIITETQAAFGRRQLISDNI 780
Query: 455 LVVY 458
L+ +
Sbjct: 781 LIAH 792
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 153 bits (387), Expect = 4e-37
Identities = 88/254 (34%), Positives = 141/254 (54%), Gaps = 1/254 (0%)
Frame = -3
Query: 403 EHATQFRPISLCNVLFKIITKTIANRLKIILPDIICQTQSAFVPGRLIIDNALVVYECFH 462
+ +++R I+ CN +KII K ++ R++ +L II +QSAFVPGR I DN L+ ++ H
Sbjct: 784 KRVSEYRTIAPCNTQYKIIAKILSKRMQPLLRSIISPSQSAFVPGRAISDNVLITHKILH 605
Query: 463 FMKKRISGRNGMMTLKLDMSKAYDRVEWSFLQGVLQKMGFPPSWVSLIMSVSPLLGFPLC 522
++++ + ++ M +K DM+KAYDR+ W+FL+ VL ++GF W+S IM + +
Sbjct: 604 YLRQSGAKKHVSMAVKTDMTKAYDRIAWNFLREVLTRLGFHGIWISWIMECVSTVSYSFL 425
Query: 523 LTEIP-SRVLLLLEAYVRVILFRLIFLSYVERCSQLLLRSVWLLMLHGIKVANRAPVISH 581
+ P RVL L +F+ E S L +++ L G+KVA P I+H
Sbjct: 424 INGGPQGRVLPSRGLRQGDPLSPYLFILCTEVLSGLCQQALRKGTLPGVKVARNCPPINH 245
Query: 582 LLFADDSIIFARANTQEAECVLNILSTYERASGQVINLDKSQLSVSRNVPQNGFNELKQL 641
LLFADD++ F ++N +L+I+ Y ASG+ IN KS ++ S Q + +K
Sbjct: 244 LLFADDTMFFGKSNASSCAILLSIMDKYRAASGRCIN*TKSAITFSSKTSQAIIDRVKGE 65
Query: 642 LNVNAVESFDKYLG 655
L + KYLG
Sbjct: 64 LKIAKEGGTGKYLG 23
>BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - Arabidopsis
thaliana, partial (17%)
Length = 677
Score = 88.6 bits (218), Expect(2) = 1e-33
Identities = 50/136 (36%), Positives = 73/136 (52%), Gaps = 4/136 (2%)
Frame = +2
Query: 229 LDSLLEQEEVWWKQRSRASWLKHGDKNTRFFHQKANQRRKRNLIEVLKDDNGREVEDDAD 288
L + EE +W+Q+SR W GD N +F+H QR RN I L D +G + ++
Sbjct: 20 LQEAYKDEEDYWQQKSRNMWHISGDLNKKFYHALTKQRHARNRIVGLYDYDGNWITEEQG 199
Query: 289 IARVLGDYFGGLFTSSNPEG----IEETTNLVAGRVSETHLTVLGEPFTREEVEEALFQM 344
+ +V DYF LF + P G ++E T+ + ++++ L + T EEV ALF M
Sbjct: 200 VEKVAVDYFEDLFQRTTPTGFDGFLDEITSSITPQMNQRLLRLA----TEEEVRLALFIM 367
Query: 345 HPTKAPGLDGFPTLFY 360
HP KAPG DG TL +
Sbjct: 368 HPEKAPGPDGMTTLLF 415
Score = 74.7 bits (182), Expect(2) = 1e-33
Identities = 40/89 (44%), Positives = 52/89 (57%)
Frame = +3
Query: 359 FYQKYWDIIGDDVSAFCLQVLQGVIPPGMINQTLLVLIPKIMKPEHATQFRPISLCNVLF 418
F+Q W II D+ L +N T + LIPK +P T+ RPISLCNV +
Sbjct: 411 FFQHSWHIIKMDLLKMVNSFLASGKLDTRLNTTNICLIPKKKRPTRMTELRPISLCNVGY 590
Query: 419 KIITKTIANRLKIILPDIICQTQSAFVPG 447
KII+K + RLK+ LP +I +TQSAFV G
Sbjct: 591 KIISKVLCQRLKVCLPSLISETQSAFVHG 677
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 137 bits (344), Expect = 3e-32
Identities = 71/187 (37%), Positives = 116/187 (61%), Gaps = 1/187 (0%)
Frame = +3
Query: 327 VLGEPFTREEVEEALFQMHPTKAPGLDGFPTLFYQKYWDIIGDDVSAFCLQVLQGVIPPG 386
+L FT EV+ ALF M +KAPG+DG+ F++ W+IIGD V L + P
Sbjct: 9 LLCSEFTAVEVKNALFSMDSSKAPGIDGYNVHFFKCSWNIIGDSVIDAILDFFKTGFMPK 188
Query: 387 MINQTLLVLIPKIMKPEHATQFRPISLCNVLFKIITKTIANRLKIILPDIICQTQSAFVP 446
+IN T + L+PK + FRPI+ C+V++KII+K + +R++ +L ++ + QSAFV
Sbjct: 189 IINCTYVTLLPKEVNVTSVKNFRPIACCSVIYKIISKILTSRMQGVLNSVVSENQSAFVK 368
Query: 447 GRLIIDNALVVYECF-HFMKKRISGRNGMMTLKLDMSKAYDRVEWSFLQGVLQKMGFPPS 505
GR+I DN ++ +E + +K IS R +K+D+ KAYD EW F++ ++ ++GFP
Sbjct: 369 GRVIFDNIILSHELVKSYSRKGISPR---CMVKIDLXKAYDSXEWPFIKHLMLELGFPYK 539
Query: 506 WVSLIMS 512
+V+ +M+
Sbjct: 540 FVNWVMA 560
>TC83226 weakly similar to PIR|G86419|G86419 probable reverse transcriptase
100033-105622 [imported] - Arabidopsis thaliana, partial
(2%)
Length = 885
Score = 130 bits (326), Expect = 4e-30
Identities = 91/298 (30%), Positives = 140/298 (46%), Gaps = 15/298 (5%)
Frame = +3
Query: 907 MWPETVDGNYCSKSGYIFVRKNFLGACSSTYSQPSLPAPLWKKFWRTSAQPRCKEVAWRV 966
MW G Y KSGY +R ++T S S +WKK W PR K + WR+
Sbjct: 3 MWMHNPTGIYSVKSGYNTLRTWQTQQINNT-STSSDETLIWKKIWSLHTIPRHKVLLWRI 179
Query: 967 VSSLLPIRAALHRRGMDVDPLCPVCANEEETVFHLFVSCPVAQNFWFGSPLSLRVHGFSS 1026
++ LP+R++L +RG+ PLCP C ++ ET+ HLF+SCP+++ WFGS L + +
Sbjct: 180 LNDSLPVRSSLRKRGIQCYPLCPRCHSKTETITHLFMSCPLSKRVWFGSNLCINFDNLPN 359
Query: 1027 MEDFLADFFRAA--DDDALALWQAG-VYALWEMRNRVVFRDGEIPVPVAAMIQRCSMLAA 1083
+F+ + A D+ + + A +Y LW RN V D I +IQR S +
Sbjct: 360 -PNFIN*LYEAIL*KDECITI*IAAIIYNLWHARNLSVLEDQTI--LEMDIIQRASNCIS 530
Query: 1084 APVSEMTVVP------------RPAPLLQATWARPPYGIYKLNFDAAVATTGEVGFGLIV 1131
T P + P W RP G+ K+N DA + G+ G G+I+
Sbjct: 531 DYKQANTQAPPSMARTGYDPRSQHRPAKNTKWKRPNLGLVKVNTDANLQNHGKWGLGIII 710
Query: 1132 RNMLGEVLASAAQYLLHAASAILGEALVFRWSMQLTIQMEFRRVLFETDCLQLFQLWK 1189
R+ +G V+A++ A+ EA M+ F +V FE D +L ++ K
Sbjct: 711 RDEVGLVMAASTWETDGNDRALEAEAYALLTGMRFAKDCGFXKVXFEGDNEKLMKMVK 884
>TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR retroelement
reverse transcriptase {Oryza sativa (japonica
cultivar-group)}, partial (1%)
Length = 798
Score = 112 bits (281), Expect = 7e-25
Identities = 76/269 (28%), Positives = 122/269 (45%), Gaps = 9/269 (3%)
Frame = -1
Query: 13 KHRTWGLIRRFRPGDDIPWLCIGDFNDILSPADKLGGDLPDMGRMQVATCACSDCALHVV 72
+ + W I + + W CIGDFN IL + G P M L +
Sbjct: 795 RRQLWSAISNIQTQHKLSWCCIGDFNTILGSHEHQGSHTPARLPMLDFQQWSDVNNLFHL 616
Query: 73 DFTGRRFTWTNNRPKPGTVEERLDYALVNDAWESLWPSNKVCHVHRHCSDHSPILLMCAD 132
G FTWTN R +RLD ++VN S C + + SDH PIL
Sbjct: 615 PTRGSAFTWTNGRRGRNNTRKRLDRSIVNQLMIDNCESLSACTLTKLRSDHFPILFELQT 436
Query: 133 RRKRKQSKREYMFRFEELWLQKNEECKEVIAETWGRSLTEVP-----DRLSDVSGLLGRW 187
+ + S F+F ++W + +C +I + W + P +L ++ +L W
Sbjct: 435 QNIQFSSS----FKFMKMW-SAHPDCINIIKQCWANQVVGCPMFVLNQKLKNLKEVLKVW 271
Query: 188 GKEQFGDLPRKISDGQALLQELQRKDQT---KEVLM-AINVAERDLDSLLEQEEVWWKQR 243
K FG++ ++ + L ++Q K + +VLM A+ +L+S L EEV+W ++
Sbjct: 270 NKNTFGNVHSQVDNAYKELDDIQVKIDSIGYSDVLMDQEKAAQLNLESALNIEEVFWHEK 91
Query: 244 SRASWLKHGDKNTRFFHQKANQRRKRNLI 272
S+ +W GD+NT FFH+ A +R +LI
Sbjct: 90 SKVNWHCEGDRNTAFFHRVAKIKRTSSLI 4
>BG586862
Length = 804
Score = 102 bits (254), Expect = 9e-22
Identities = 73/239 (30%), Positives = 110/239 (45%), Gaps = 6/239 (2%)
Frame = -1
Query: 948 KKFWRTSAQPRCKEVAWRVVSSLLPIRAALHRRGMDVDPLCPVCANEEETVFHLFVSCPV 1007
+K W PR K WR++ + LP++ LH+RG+ LCP C ++ ETV HLF++C V
Sbjct: 660 EKVWGIKTIPRHKSFLWRLLHNALPVKDELHKRGIRCSLLCPRCESKIETVQHLFLNCEV 481
Query: 1008 AQNFWFGSPLSLRVH--GFSSMEDFLADFFRAADDDALALWQAGVYALWEMRNRVVFRDG 1065
Q WFGS L + H G D++ +F D++ + A +Y++W RN+ VF +
Sbjct: 480 TQKEWFGSQLGINFHSSGVLHFHDWITNFILKNDEETIIALTALLYSIWHARNQKVFEN- 304
Query: 1066 EIPVPVAAMIQRCS----MLAAAPVSEMTVVPRPAPLLQATWARPPYGIYKLNFDAAVAT 1121
I VP +IQR S A VS+ +V+P + A P Y ++
Sbjct: 303 -IDVPGDVVIQRASSSLHSFKMAQVSD-SVLP--------SNAIPSYSLW---------- 184
Query: 1122 TGEVGFGLIVRNMLGEVLASAAQYLLHAASAILGEALVFRWSMQLTIQMEFRRVLFETD 1180
G G++ RN G +AS A EA +M F + FE+D
Sbjct: 183 ----GIGVVARNCEGLAMASGTWLRHGIPCATTAEAWGIYQAMVFAGDCGFSKFEFESD 19
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 96.7 bits (239), Expect = 5e-20
Identities = 87/335 (25%), Positives = 146/335 (42%), Gaps = 14/335 (4%)
Frame = +2
Query: 388 INQTLLVLIPKIMKPEHATQFRPISLCNVLFKIITKTIANRLKIILPDIICQTQSAFVPG 447
IN T + LIPK+ P+ FRPISL L+KI+ K +ANRL++++ +I QSAFV
Sbjct: 50 INSTFIALIPKVDNPQRLNDFRPISLVGSLYKILGKLLANRLRVVIGSVISDAQSAFVKN 229
Query: 448 RLIIDNALV--------------VYECFHFMKKRISGRNGMMTLKLDMSKAYDRVEWSFL 493
R I++ + ++ C ++ KR ++TL + + V SFL
Sbjct: 230 RQILEMVFL*QMRLWMRLRN*RKIFCCLRWILKR------LITLSIGLIWILF*VGMSFL 391
Query: 494 QGVLQKMGFPPSWVSLIMSVSPLLGFPLCLTEIPSRVLLLLEAYVRVILFRLIFLSYVER 553
VL + W+ +S + + + P+ V L+++ V+ LF V
Sbjct: 392 --VLWR-----KWIKECVSTATT---SVLVNGSPTNV--LMKSLVQTQLFTRYSFGVV-- 529
Query: 554 CSQLLLRSVWLLMLHGIKVANRAPVISHLLFADDSIIFARANTQEAECVLNILSTYERAS 613
V+SHL FA+D+++ N + L + S
Sbjct: 530 ---------------------NPVVVSHLQFANDTLLLETKNWANIRALRAALVIF*AMS 646
Query: 614 GQVINLDKSQLSVSRNVPQNGFNELKQLLNVNAVESFDKYLGLPTMIGKFKTRIFNFVKD 673
G +N KS L V N+ + +E +L+ + YLG+P + + + +
Sbjct: 647 GLKVNFHKSGL-VCVNIAPSWLSEAASVLSWKVGKVPFLYLGMPIEGNSRRLSFWEPIVN 823
Query: 674 RVWKKLKGWKESTLSRAGREVLIKSVVQAIPSYVM 708
R+ +L GW LS GR VL+KSV+ ++ Y +
Sbjct: 824 RIKARLTGWNSRFLSFGGRLVLLKSVLTSLSVYAL 928
>BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (10%)
Length = 767
Score = 90.1 bits (222), Expect = 5e-18
Identities = 66/235 (28%), Positives = 103/235 (43%), Gaps = 10/235 (4%)
Frame = -3
Query: 799 LGAKRGYRPSYAWSSILQSSWIFQEGGRWKIGDGSQVDILHDQWLPQGVPVIGSQDLMAE 858
L A G SYAW SI + + ++G + IG+G +I + + V + +
Sbjct: 762 LNAPLGSWASYAWRSIHSAQHLIKQGAKVIIGNGENTNIWEREMAWKLTCVTNHPNKHSS 583
Query: 859 --------FGVSKVSHLIDHAAKSWKYALVDFIFHPATVSHILKIHLPLNGSVDTLMWPE 910
+G D + L++ IF T IL IH D+ W
Sbjct: 582 RAY*APTLYGYEGCRS-DDPMRRERNANLINSIFPEGTRRKILSIHPQGPIGEDSYSWEY 406
Query: 911 TVDGNYCSKSGYIFVRKNFLGACSS--TYSQPSLPAPLWKKFWRTSAQPRCKEVAWRVVS 968
+ G+Y KSGY +V+ N + A + T QPSL L+++ W+ + P+ + WR +S
Sbjct: 405 SKSGHYSVKSGY-YVQTNIIAAANQRGTVDQPSLD-DLYQRVWKYNTSPKVRHFLWRCIS 232
Query: 969 SLLPIRAALHRRGMDVDPLCPVCANEEETVFHLFVSCPVAQNFWFGSPLSLRVHG 1023
+ LP A + R + D C C E ETV H+ CP A+ W SP+ +G
Sbjct: 231 NSLPTAANMRSRHISKDGSCSRCGMESETVNHILFQCPYARLIWATSPIHAPPYG 67
>TC93101
Length = 675
Score = 88.6 bits (218), Expect = 1e-17
Identities = 56/190 (29%), Positives = 87/190 (45%), Gaps = 18/190 (9%)
Frame = -1
Query: 971 LPIRAALHRRGMDVDPLCPVCANEEETVFHLFVSCPVAQNFWFGSPLSLRVHGFSSM--E 1028
LP+R L++RG++ PLCP C ET H+F+SC Q WFGS LS+R S++
Sbjct: 663 LPVRXELNKRGVNCPPLCPRCYFNLETTNHIFMSCERTQRVWFGSQLSIRFPDNSTINFS 484
Query: 1029 DFLADFFRAADDDALALWQAGVYALWEMRNRVVFRDGEIPVPVAAMIQRCSMLA------ 1082
D+L D ++ + A Y++W RN+ +F + + + S+LA
Sbjct: 483 DWLFDAISNQTEEIIIKISAITYSIWHARNKAIFENQFVSEDTIIQ*AQNSILAYEQATI 304
Query: 1083 ----------AAPVSEMTVVPRPAPLLQATWARPPYGIYKLNFDAAVATTGEVGFGLIVR 1132
+ + T R +++ W +P I K N DA + G G G I+R
Sbjct: 303 KPQNPNIVLSSLSATSNTNTTRRRSNVRSRWQKPLNNILKANCDANLQVQGRWGLGCIIR 124
Query: 1133 NMLGEVLASA 1142
N GE +A
Sbjct: 123 NADGEAKVTA 94
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 83.6 bits (205), Expect = 4e-16
Identities = 44/97 (45%), Positives = 65/97 (66%)
Frame = +1
Query: 546 IFLSYVERCSQLLLRSVWLLMLHGIKVANRAPVISHLLFADDSIIFARANTQEAECVLNI 605
+F+ S LL R LHGI+VA P I+HLLFADDS++FARAN EA ++ +
Sbjct: 46 LFILCANVLSGLLKREGNKQNLHGIQVARSDPKITHLLFADDSLLFARANLTEAATIMQV 225
Query: 606 LSTYERASGQVINLDKSQLSVSRNVPQNGFNELKQLL 642
L +Y+ ASGQ++N +KS++S S+NVP N+ K+++
Sbjct: 226 LHSYQSASGQLVNFEKSEVSYSQNVP----NQEKEMI 324
Score = 35.4 bits (80), Expect(2) = 0.008
Identities = 19/63 (30%), Positives = 28/63 (44%)
Frame = +3
Query: 860 GVSKVSHLIDHAAKSWKYALVDFIFHPATVSHILKIHLPLNGSVDTLMWPETVDGNYCSK 919
G V LID+ K W L+ F+ I+KI L + D ++W DG Y +
Sbjct: 390 GSLSVDELIDYDTKQWNRDLIFHSFNNYAAHQIIKIPLSMRQPEDKIIWHWEKDGIYSVR 569
Query: 920 SGY 922
S +
Sbjct: 570 SAH 578
Score = 23.1 bits (48), Expect(2) = 0.008
Identities = 7/21 (33%), Positives = 10/21 (47%)
Frame = +2
Query: 946 LWKKFWRTSAQPRCKEVAWRV 966
+WK W T A + WR+
Sbjct: 641 IWKAIWNTPASRSVQNFLWRL 703
>BQ148771
Length = 680
Score = 73.9 bits (180), Expect = 4e-13
Identities = 38/114 (33%), Positives = 58/114 (50%)
Frame = -3
Query: 679 LKGWKESTLSRAGREVLIKSVVQAIPSYVMSCFILPDSLCADIERMVSRFYWGGDVDKRG 738
L WK + LS A R L KSV++A+P Y M I+P + +I+++ +F WG R
Sbjct: 573 LANWKANHLSLARRVTLAKSVIEAVPLYPMMTTIIPKACIEEIQKLQRKFVWGDTEVSRR 394
Query: 739 LQWASWKKLTRSKFDGGLGFRDFKSFNIALVAKNWWRIYTQPETLLAQVFKGVY 792
W+ +++ K GLG R N A + K W IY+ +L +V +G Y
Sbjct: 393 YHAVGWETMSKPKTIYGLGLRRLDVMNKACIMKLGWSIYSGSNSLCTEVMRGKY 232
>AJ497561 weakly similar to GP|10140689|gb putative non-LTR retroelement
reverse transcriptase {Oryza sativa (japonica
cultivar-group)}, partial (2%)
Length = 621
Score = 56.2 bits (134), Expect(2) = 2e-11
Identities = 24/62 (38%), Positives = 37/62 (58%)
Frame = +2
Query: 782 TLLAQVFKGVYFAQGDLLGAKRGYRPSYAWSSILQSSWIFQEGGRWKIGDGSQVDILHDQ 841
+LL+++ K YF Q D A G+ PS+ W S+L + + G RW IGDGSQ+++
Sbjct: 410 SLLSKILKFKYFPQWDFSYANLGHNPSFTWRSLLSTQSLLTLGHRWMIGDGSQINVSSMS 589
Query: 842 WL 843
W+
Sbjct: 590 WI 595
Score = 32.0 bits (71), Expect(2) = 2e-11
Identities = 14/37 (37%), Positives = 24/37 (64%)
Frame = +3
Query: 744 WKKLTRSKFDGGLGFRDFKSFNIALVAKNWWRIYTQP 780
+ L R K GGLG ++ + FNI+++ K W++ +QP
Sbjct: 303 FNNLARHKSVGGLGLQN-QEFNISMLGKQRWKLLSQP 410
>BG584442
Length = 775
Score = 68.2 bits (165), Expect = 2e-11
Identities = 38/96 (39%), Positives = 60/96 (61%), Gaps = 1/96 (1%)
Frame = +1
Query: 677 KKLKGWKESTLSRAGREVLIKSVVQAIPSYVMSCFILPDSLCADIERMVSRFYW-GGDVD 735
KK+ + LS+ EV+IK +Q+I SYVMS F+L +S +IE++++ F W +
Sbjct: 379 KKINF*RNKCLSKVM*EVMIKYALQSISSYVMSIFLLLNSQVDEIEKIMNTFSWVHVGEN 558
Query: 736 KRGLQWASWKKLTRSKFDGGLGFRDFKSFNIALVAK 771
++G+ W S +KL K GG+GF DF +FNI ++ K
Sbjct: 559 RKGMHWMS*EKLFVHKNYGGMGFTDFTTFNIPMLGK 666
>TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative retroelement
{Oryza sativa (japonica cultivar-group)}, partial (1%)
Length = 625
Score = 66.6 bits (161), Expect = 6e-11
Identities = 42/119 (35%), Positives = 58/119 (48%)
Frame = +3
Query: 562 VWLLMLHGIKVANRAPVISHLLFADDSIIFARANTQEAECVLNILSTYERASGQVINLDK 621
VW R + LLF + R A+ + NIL YE SG+ I+L K
Sbjct: 219 VWRACPS*FPTPRRGAIPMALLFEGAPLRSLRVEEHHAQIMKNILILYEEDSGKAISLRK 398
Query: 622 SQLSVSRNVPQNGFNELKQLLNVNAVESFDKYLGLPTMIGKFKTRIFNFVKDRVWKKLK 680
S + SRNVP + +L V + KYLGLP+MIG+ +T F+ +K VW+K K
Sbjct: 399 S*IYCSRNVPDILKTSITYILGVQFMLGTCKYLGLPSMIGRDRTTTFSSIKGGVWQKNK 575
Score = 29.6 bits (65), Expect = 7.7
Identities = 12/29 (41%), Positives = 20/29 (68%)
Frame = +1
Query: 479 LDMSKAYDRVEWSFLQGVLQKMGFPPSWV 507
L +SK Y+RV+ +L+ ++ KMGF W+
Sbjct: 1 LYISKVYNRVD*DYLKEIMIKMGFNNRWI 87
>BG585866
Length = 828
Score = 62.4 bits (150), Expect = 1e-09
Identities = 55/228 (24%), Positives = 92/228 (40%), Gaps = 8/228 (3%)
Frame = +3
Query: 814 ILQSSWIFQEGGRWKIGDGSQVDILHDQWLPQGVPVIGSQDLMAEFGVSKVSHLI--DHA 871
I++ + + G W+ G G+ + W G+ +G+Q A F HL D
Sbjct: 84 IIRDKNVLKSGYTWRAGSGNS-SFWYTNWSSLGL--LGTQ---APFVDIHDLHLTVKDVF 245
Query: 872 AKSWKYALVDFIFHPATVSHILK-IHLPLNGSV-DTLMWPETVDGNYCSKSGYIFVRKNF 929
++ + P ++ ++ HL N S+ D +WP +G Y +KSGY ++
Sbjct: 246 TTGGQHTQSLYTILPTDIAEVINNTHLNFNASIGDAYIWPHNSNGVYTAKSGYSWILSQ- 422
Query: 930 LGACSSTYSQPSLPAPLWKKFWRTSAQPRCKEVAWRVVSSLLPIRAALHRRGMDVDPLCP 989
+ Y+ S W WR + K W + +P + L+ R M +C
Sbjct: 423 --TETVNYNNSS-----WSWIWRLKIPEKYKFFLWLACHNAVPTLSLLNHRNMVNSAICS 581
Query: 990 VCANEEETVFHLFVSCPVAQNFW----FGSPLSLRVHGFSSMEDFLAD 1033
C EE+ FH C ++ W F SP SS++D+L D
Sbjct: 582 RCGEHEESFFHCVRDCRFSKIIWHKIGFSSP---DFFSSSSVQDWLKD 716
>TC81772
Length = 982
Score = 52.8 bits (125), Expect(2) = 2e-07
Identities = 22/47 (46%), Positives = 29/47 (60%)
Frame = -2
Query: 797 DLLGAKRGYRPSYAWSSILQSSWIFQEGGRWKIGDGSQVDILHDQWL 843
+LLG G+ PSY W S+ + +EG WKIGDGS ++I D WL
Sbjct: 735 ELLGVVIGHNPSYIWCSV*TLRMVLEEGHHWKIGDGSSINIWTDHWL 595
Score = 21.9 bits (45), Expect(2) = 2e-07
Identities = 8/24 (33%), Positives = 17/24 (70%)
Frame = -3
Query: 776 IYTQPETLLAQVFKGVYFAQGDLL 799
I QP++++++VF+ YF+ + L
Sbjct: 797 IVNQPKSIVSRVFEVKYFSHWNFL 726
>BG646730
Length = 799
Score = 53.9 bits (128), Expect = 4e-07
Identities = 38/129 (29%), Positives = 53/129 (40%), Gaps = 6/129 (4%)
Frame = +1
Query: 804 GYRPSYAWSSILQSSWIFQEGGRWKIGDGSQVDILHDQWLPQGVPVIGSQDLMAEFGVSK 863
G +PSYAW S+ + G RW I +G V I D WLP ++ +
Sbjct: 388 GNQPSYAWRSMFNIKDVIDLGSRWSISNGQNVRIWKDDWLPNQTGFKVCSPMVDFEEATC 567
Query: 864 VSHLIDHAAKSWKYALVDFIFHPATVSHILKIHLPLNGSVDTLMWPE------TVDGNYC 917
+S LID KS K LV +F IL I L +WP+ D NY
Sbjct: 568 ISELIDTNTKSSKRDLVRNVFSAFEAKQILNIPLSWR------LWPDRRILYWERDRNYS 729
Query: 918 SKSGYIFVR 926
+S + ++
Sbjct: 730 VRSAHHLIK 756
>BG456581
Length = 683
Score = 53.9 bits (128), Expect = 4e-07
Identities = 36/129 (27%), Positives = 51/129 (38%), Gaps = 5/129 (3%)
Frame = +2
Query: 944 APLWKKFWRTSAQPRCKEVAWRVVSSLLPIRAALHRRGMDVDPLCPVCANEEETVFHLFV 1003
AP W + P + WR+ LP L RG + +C C + ET HLF+
Sbjct: 62 APWASTIWNSCIPPSHSFICWRLAHDRLPTDDNLSSRGCALVSMCSFCLEQVETSDHLFL 241
Query: 1004 SCPVAQNFWFGSPLSLRVH-GFSSMEDFLADFFRAADDDALALWQAG----VYALWEMRN 1058
C W LRV FSS + L+ R L+ A V+++W RN
Sbjct: 242 RCKFVVTLWSWLCSQLRVGLDFSSFKALLSSLPRHCSSQVRDLYVAAVVHMVHSIWWARN 421
Query: 1059 RVVFRDGEI 1067
V F ++
Sbjct: 422 NVRFSSAKV 448
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.139 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 44,472,971
Number of Sequences: 36976
Number of extensions: 703125
Number of successful extensions: 4511
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 3317
Number of HSP's successfully gapped in prelim test: 172
Number of HSP's that attempted gapping in prelim test: 1095
Number of HSP's gapped (non-prelim): 3618
length of query: 1259
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1152
effective length of database: 5,058,295
effective search space: 5827155840
effective search space used: 5827155840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0129.4


