

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0100a.10
(700 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
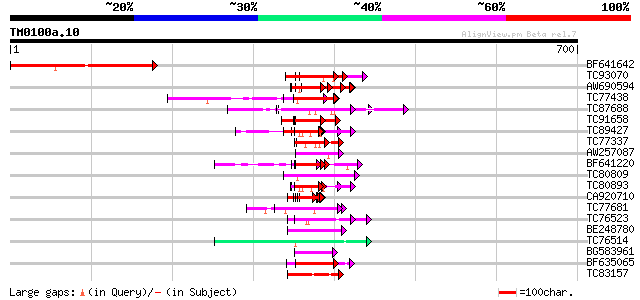
Score E
Sequences producing significant alignments: (bits) Value
BF641642 weakly similar to GP|16604595|gb| unknown protein {Arab... 226 2e-59
TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown prot... 62 6e-10
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 62 1e-09
TC77438 homologue to PIR|T06807|T06807 nucleosome assembly prote... 61 2e-09
TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~un... 60 3e-09
TC91658 similar to GP|14329812|emb|CAC40753. putative nucleosome... 57 2e-08
TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed pr... 57 3e-08
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 56 4e-08
AW257087 homologue to GP|6683126|dbj KIAA0339 protein {Homo sapi... 56 4e-08
BF641220 weakly similar to PIR|G86203|G86 probable N-arginine di... 56 4e-08
TC80809 similar to GP|22136852|gb|AAM91770.1 unknown protein {Ar... 55 9e-08
TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {... 55 1e-07
CA920710 similar to PIR|S64314|S643 probable membrane protein YG... 52 8e-07
TC77681 similar to GP|22597156|gb|AAN03465.1 nucleolar histone d... 51 1e-06
TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 50 2e-06
BE248780 similar to GP|5929884|gb|A nucleolin-related protein NR... 50 2e-06
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 50 3e-06
BG583961 similar to GP|14599408|em probable major surface glycop... 50 3e-06
BF635065 weakly similar to GP|11037008|gb latent nuclear antigen... 50 3e-06
TC83157 similar to GP|21595368|gb|AAM66095.1 unknown {Arabidopsi... 49 5e-06
>BF641642 weakly similar to GP|16604595|gb| unknown protein {Arabidopsis
thaliana}, partial (12%)
Length = 563
Score = 226 bits (577), Expect = 2e-59
Identities = 117/189 (61%), Positives = 144/189 (75%), Gaps = 8/189 (4%)
Frame = +2
Query: 2 ASMSGPTAPNLTFPNKNLSIP-FTLKRPSLLPFPSHPPCRCLSSASDDKNDSSDN----- 55
+S+S + P+LT P +NLS F LKRPSLL FPS P C CL+S SDD ++S++N
Sbjct: 5 SSISSLSTPHLTXPKQNLSTTTFPLKRPSLLHFPSQPLCLCLNSISDDNHNSTNNNDADG 184
Query: 56 --RRWDSLLQDFLTGAAKQFDSFMNALRDGRSAEDRAATAGGGSDEDWDWDRWRQHFDEV 113
RRWDS+LQ+F+TGA KQFDS++N LR G +A + D+DWDW+RWR +FD+V
Sbjct: 185 NNRRWDSVLQEFVTGAIKQFDSYLNLLRGGSAAAKEGSDV---HDDDWDWNRWRHYFDQV 355
Query: 114 DDQERLLSVLKSQLGRAVYVEDYEDAARLKVAIAAASNNDSVGRVMSHLNRAIKEERYGD 173
DDQERLL +LKSQL AVYVEDYE+AA+LKVAIAAA+NNDSVG+V + L RAIKEERY D
Sbjct: 356 DDQERLLIILKSQLRHAVYVEDYEEAAKLKVAIAAAANNDSVGKVKTLLKRAIKEERYND 535
Query: 174 AAFLRDKAG 182
AAFL DKAG
Sbjct: 536 AAFLXDKAG 562
>TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown protein
{Arabidopsis thaliana}, partial (19%)
Length = 679
Score = 62.4 bits (150), Expect = 6e-10
Identities = 30/72 (41%), Positives = 45/72 (61%), Gaps = 7/72 (9%)
Frame = +1
Query: 353 EEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVD-------MKSETDQ 405
+ DDE E+ D ++DEDED+D+DED+D+D+D+D+E D D G E D +KS+T
Sbjct: 220 DSDDEIEEMDFEDDEDEDEDEDEDDDDDDDDDDEDDDDDGFAEPTDLNAKDKRLKSKTVP 399
Query: 406 EADDEVEINTGL 417
+ E E G+
Sbjct: 400 DRQQEKEKEKGV 435
Score = 55.8 bits (133), Expect = 5e-08
Identities = 26/69 (37%), Positives = 44/69 (63%), Gaps = 3/69 (4%)
Frame = +1
Query: 341 DLISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEE---KDSDTGSLELV 397
D ++ E ++++DEDEDEDED+D+D+DDDD++D+D+ E + KD S +
Sbjct: 220 DSDDEIEEMDFEDDEDEDEDEDEDDDDDDDDDDEDDDDDGFAEPTDLNAKDKRLKSKTVP 399
Query: 398 DMKSETDQE 406
D + E ++E
Sbjct: 400 DRQQEKEKE 426
Score = 50.1 bits (118), Expect = 3e-06
Identities = 27/83 (32%), Positives = 45/83 (53%)
Frame = +1
Query: 359 EDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEVEINTGLG 418
+ +DE E+ D +DD+DEDEDEDED+D++ D D + TD A D+ + +
Sbjct: 220 DSDDEIEEMDFEDDEDEDEDEDEDDDDDDDDDDEDDDDDGFAEPTDLNAKDKRLKSKTVP 399
Query: 419 TSDREDQNEIAVKVVIGGIVQKL 441
+E + E V+ + G ++L
Sbjct: 400 DRQQEKEKEKGVRSLNNGQSKRL 468
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 61.6 bits (148), Expect = 1e-09
Identities = 24/51 (47%), Positives = 41/51 (80%)
Frame = +3
Query: 348 EQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVD 398
E+ DE D+E+E+E+E+E+EDE DD++E+E+E+E+E+EE D + G + +D
Sbjct: 105 EEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEEDDDEEGEEDEID 257
Score = 59.3 bits (142), Expect = 5e-09
Identities = 28/72 (38%), Positives = 48/72 (65%), Gaps = 4/72 (5%)
Frame = +3
Query: 347 IEQIIDEEDD----EDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSE 402
+E+ +EED+ E+EDE ++E+EDE D+++E+E+E+E+EDE D + E + + E
Sbjct: 42 VEEHNEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEE 221
Query: 403 TDQEADDEVEIN 414
D E +E EI+
Sbjct: 222 DDDEEGEEDEID 257
Score = 54.7 bits (130), Expect = 1e-07
Identities = 28/73 (38%), Positives = 47/73 (64%)
Frame = +3
Query: 353 EEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEVE 412
EE +E+EDE E E+E ED+ D+E+EDE ++E+EE++ + E D + E ++E ++E E
Sbjct: 45 EEHNEEEDEGEVEEE-EDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEE 221
Query: 413 INTGLGTSDREDQ 425
+ G D D+
Sbjct: 222 DDDEEGEEDEIDR 260
Score = 45.1 bits (105), Expect = 9e-05
Identities = 15/42 (35%), Positives = 36/42 (85%)
Frame = +3
Query: 348 EQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDS 389
E+ +EE+DE +DE+E+E+E+E++++++D++E E+++ ++ S
Sbjct: 141 EEEEEEEEDEHDDEEEEEEEEEEEEEEDDDEEGEEDEIDRIS 266
Score = 43.1 bits (100), Expect = 4e-04
Identities = 21/67 (31%), Positives = 39/67 (57%)
Frame = +3
Query: 361 EDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEVEINTGLGTS 420
E+ +E+EDE + ++E+++ DE+E++E D + E + + E + E DDE E
Sbjct: 45 EEHNEEEDEGEVEEEEDEHDEEEEDEHDEE----EEEEEEEEEEDEHDDEEEEEEEEEEE 212
Query: 421 DREDQNE 427
+ ED +E
Sbjct: 213 EEEDDDE 233
Score = 39.3 bits (90), Expect = 0.005
Identities = 18/64 (28%), Positives = 34/64 (53%)
Frame = +3
Query: 364 DEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEVEINTGLGTSDRE 423
+E +E+D+ + +E+EDE ++EE+D E + + E D+ D+E E + +
Sbjct: 45 EEHNEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEED 224
Query: 424 DQNE 427
D E
Sbjct: 225 DDEE 236
>TC77438 homologue to PIR|T06807|T06807 nucleosome assembly protein 1 -
garden pea, partial (93%)
Length = 1607
Score = 60.8 bits (146), Expect = 2e-09
Identities = 56/204 (27%), Positives = 89/204 (43%), Gaps = 6/204 (2%)
Frame = +2
Query: 196 VNNPHGLIIRITPEHGRYVARSYSPRQLAMSAAGVPLFEFFLKMDKK---GEFKSQAVYL 252
+++P G + + Y + S + M P+ E + + + G+ +Q V
Sbjct: 602 IDDPKGFKLEFFFDTNPYFSNSILTKTYHMVDEDEPILEKAIGTEIQWLPGKCLTQKVLK 781
Query: 253 KR--KGAFHGPPTTSSKALDAAERLSSVESTEDRSELFVVSTEDPESGDDRNDGSDPAER 310
K+ KGA + P T ++ ++ F E PE DD + D AE
Sbjct: 782 KKPKKGAKNAKPITKTETCESFFN-------------FFNPPEVPE--DDEDIDEDMAEE 916
Query: 311 MPGFQNGLKDMIPGVKVKIFKVITPEKVDKDLISKVIEQIIDEEDDEDEDEDED-EDEDE 369
+ D+ ++ KI P V + + DDED+DED+D ED+DE
Sbjct: 917 LQNQMEQDYDIGSTIRDKII----PHAVSWFTGEAAQGEEFGDLDDEDDDEDDDAEDDDE 1084
Query: 370 DDDDDEDEDEDEDEDEEKDSDTGS 393
D+D+DEDED+DEDE+E K S
Sbjct: 1085DEDEDEDEDDDEDEEETKTKKKSS 1156
Score = 59.7 bits (143), Expect = 4e-09
Identities = 24/55 (43%), Positives = 38/55 (68%)
Frame = +2
Query: 351 IDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQ 405
+D+EDD+++D+ ED+DEDED+D+DED+DEDE+E + K + + E Q
Sbjct: 1034 LDDEDDDEDDDAEDDDEDEDEDEDEDDDEDEEETKTKKKSSAKKSGIAQLGEGQQ 1198
Score = 48.9 bits (115), Expect = 7e-06
Identities = 28/77 (36%), Positives = 45/77 (58%), Gaps = 8/77 (10%)
Frame = +2
Query: 339 DKDLISKVIEQIIDE-------EDDEDEDEDEDEDEDEDDDDD-EDEDEDEDEDEEKDSD 390
D D+ S + ++II E + E+ + +DED+D+DDD ED+DEDEDEDE++D D
Sbjct: 938 DYDIGSTIRDKIIPHAVSWFTGEAAQGEEFGDLDDEDDDEDDDAEDDDEDEDEDEDEDDD 1117
Query: 391 TGSLELVDMKSETDQEA 407
E K + +++
Sbjct: 1118 EDEEETKTKKKSSAKKS 1168
Score = 28.9 bits (63), Expect = 7.0
Identities = 22/88 (25%), Positives = 36/88 (40%), Gaps = 26/88 (29%)
Frame = +2
Query: 354 EDDEDEDEDEDEDEDEDDDDDED----------------------EDED----EDEDEEK 387
EDDED DED E+ + D D + E+ +DED+++
Sbjct: 878 EDDEDIDEDMAEELQNQMEQDYDIGSTIRDKIIPHAVSWFTGEAAQGEEFGDLDDEDDDE 1057
Query: 388 DSDTGSLELVDMKSETDQEADDEVEINT 415
D D + + + E + + +DE E T
Sbjct: 1058DDDAEDDDEDEDEDEDEDDDEDEEETKT 1141
>TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~unknown
protein {Arabidopsis thaliana}, partial (46%)
Length = 2077
Score = 60.1 bits (144), Expect = 3e-09
Identities = 48/183 (26%), Positives = 86/183 (46%), Gaps = 3/183 (1%)
Frame = +2
Query: 270 DAAERLSSVESTEDRSELFVVSTEDPESGDDRNDGSDPAERMPGFQNGLKDMIPGVKVKI 329
+ E+ + +E + ED E +D +D D E++ G G++D +KI
Sbjct: 431 EGVEKKKAKGGSEGEDDFEEEDDEDEEGSEDEDDEEDEKEKVKG--GGIEDKF----LKI 592
Query: 330 FKVITPEKVDKDLISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDS 389
++ + ++D K E+ DE D++ E++DE E E + DDED+D+D++EDEE +
Sbjct: 593 DELTEYLEKEEDNYEKGEER--DEADEDSEEDDELEKAGEFEMDDEDDDDDDEEDEEAE- 763
Query: 390 DTGSLELVDM---KSETDQEADDEVEINTGLGTSDREDQNEIAVKVVIGGIVQKLSSNIS 446
D G++ D K+E + D++ +G D +D K +K S
Sbjct: 764 DMGNVRYEDFFGGKNEKGSKRKDQLLEVSG----DEDDMESTKQKKRTASTYEKQRGKNS 931
Query: 447 TRD 449
+D
Sbjct: 932 IKD 940
Score = 44.3 bits (103), Expect = 2e-04
Identities = 33/161 (20%), Positives = 77/161 (47%)
Frame = +2
Query: 332 VITPEKVDKDLISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDT 391
V PE++ + + + + ++++ + +E+E +D DE+ DDD+D+ E ++ + K
Sbjct: 290 VKNPEEIAQFKVPLDVGKKLEKKKRVELEEEESDDFDEELDDDDDDFEGVEKKKAKGGSE 469
Query: 392 GSLELVDMKSETDQEADDEVEINTGLGTSDREDQNEIAVKVVIGGIVQKLSSNISTRDLL 451
G + + ++E D++ E G+ D +D+ + KV GGI K L
Sbjct: 470 G-------EDDFEEEDDEDEE-----GSEDEDDEEDEKEKVKGGGIEDK---------FL 586
Query: 452 RVPAKLEIKKRGSFSFTVEKEVNQQDGQAKGKSSSDKSTKF 492
++ E ++ ++ +E ++ D ++ +K+ +F
Sbjct: 587 KIDELTEYLEKEEDNYEKGEERDEADEDSEEDDELEKAGEF 709
Score = 42.7 bits (99), Expect = 5e-04
Identities = 34/124 (27%), Positives = 59/124 (47%), Gaps = 26/124 (20%)
Frame = +2
Query: 330 FKVITPEKVDKDLISKVIEQIIDEEDDEDEDEDEDEDED---------------EDDDDD 374
FKV P V K L K ++ +EE D+ ++E +D+D+D EDD ++
Sbjct: 317 FKV--PLDVGKKLEKKKRVELEEEESDDFDEELDDDDDDFEGVEKKKAKGGSEGEDDFEE 490
Query: 375 ED-------EDEDEDEDEEKDSDTGSLE----LVDMKSETDQEADDEVEINTGLGTSDRE 423
ED EDED++EDE++ G +E +D +E ++ +D E +D +
Sbjct: 491 EDDEDEEGSEDEDDEEDEKEKVKGGGIEDKFLKIDELTEYLEKEEDNYEKGEERDEADED 670
Query: 424 DQNE 427
+ +
Sbjct: 671 SEED 682
>TC91658 similar to GP|14329812|emb|CAC40753. putative nucleosome assembly
protein 1 {Atropa belladonna}, partial (46%)
Length = 583
Score = 57.0 bits (136), Expect = 2e-08
Identities = 24/56 (42%), Positives = 38/56 (67%)
Frame = +2
Query: 353 EEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEAD 408
E D++DED DE++D+D+DDDDD++EDED++E++E + S K DQ +
Sbjct: 80 EVDEDDEDGDEEDDDDDDDDDDDEEDEDDEEEDEGKGKSKSKRGSKAKPGKDQSTE 247
Score = 55.8 bits (133), Expect = 5e-08
Identities = 23/59 (38%), Positives = 42/59 (70%), Gaps = 1/59 (1%)
Frame = +2
Query: 351 IDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKD-SDTGSLELVDMKSETDQEAD 408
+DE+D++ ++ED+D+D+D+DDD+++++DE+EDE + K S GS T++ AD
Sbjct: 83 VDEDDEDGDEEDDDDDDDDDDDEEDEDDEEEDEGKGKSKSKRGSKAKPGKDQSTERPAD 259
Score = 54.7 bits (130), Expect = 1e-07
Identities = 20/39 (51%), Positives = 34/39 (86%)
Frame = +2
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSD 390
D +D E +++DED DE++DDDDD+D+D++EDED+E++ +
Sbjct: 65 DFDDIEVDEDDEDGDEEDDDDDDDDDDDEEDEDDEEEDE 181
Score = 49.3 bits (116), Expect = 5e-06
Identities = 19/55 (34%), Positives = 38/55 (68%)
Frame = +2
Query: 336 EKVDKDLISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSD 390
+K+ +S + + + D+ E +++DED DE+DDDD+D+D+D++EDE+ + +
Sbjct: 11 DKIIPHAVSWFTGEAGESDFDDIEVDEDDEDGDEEDDDDDDDDDDDEEDEDDEEE 175
Score = 34.7 bits (78), Expect = 0.13
Identities = 18/52 (34%), Positives = 29/52 (55%)
Frame = +2
Query: 369 EDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEVEINTGLGTS 420
E D DD + DED+++ +E+D D D + D+E +D+ E + G G S
Sbjct: 59 ESDFDDIEVDEDDEDGDEEDDDD------DDDDDDDEEDEDDEEEDEGKGKS 196
>TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed protein
{Arabidopsis thaliana}, partial (50%)
Length = 753
Score = 56.6 bits (135), Expect = 3e-08
Identities = 26/55 (47%), Positives = 34/55 (61%), Gaps = 12/55 (21%)
Frame = +1
Query: 348 EQIIDEEDDEDED------------EDEDEDEDEDDDDDEDEDEDEDEDEEKDSD 390
E +EDDED D D+D+++D+DDDDD D+ EDEDEDEE+D D
Sbjct: 370 EDFSGDEDDEDADPEDDPVPNGAGGSDDDDEDDDDDDDDNDDGEDEDEDEEEDDD 534
Score = 55.8 bits (133), Expect = 5e-08
Identities = 31/76 (40%), Positives = 46/76 (59%)
Frame = +1
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEV 411
+ EDD+DED+D+D + DEDDD+DED DED DE+ D + + S+ D E DD+
Sbjct: 304 ETEDDDDEDDDDDVN-DEDDDNDEDFSGDED-DEDADPEDDPVPNGAGGSDDDDEDDDDD 477
Query: 412 EINTGLGTSDREDQNE 427
+ + G + ED+ E
Sbjct: 478 DDDNDDGEDEDEDEEE 525
Score = 55.5 bits (132), Expect = 7e-08
Identities = 24/58 (41%), Positives = 39/58 (66%), Gaps = 8/58 (13%)
Frame = +1
Query: 339 DKDLISKVIEQIIDEEDD--------EDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKD 388
D+D ++ D EDD D+D+++D+D+D+D+DD EDEDEDE+ED+++D
Sbjct: 367 DEDFSGDEDDEDADPEDDPVPNGAGGSDDDDEDDDDDDDDNDDGEDEDEDEEEDDDED 540
Score = 53.5 bits (127), Expect = 3e-07
Identities = 28/73 (38%), Positives = 42/73 (57%), Gaps = 14/73 (19%)
Frame = +1
Query: 352 DEEDDEDEDEDEDEDEDED---DDDDEDEDEDED-----------EDEEKDSDTGSLELV 397
D+EDD+D+ DED+D DED D+DDED D ++D +DE+ D D +
Sbjct: 319 DDEDDDDDVNDEDDDNDEDFSGDEDDEDADPEDDPVPNGAGGSDDDDEDDDDDDDDNDDG 498
Query: 398 DMKSETDQEADDE 410
+ + E ++E DDE
Sbjct: 499 EDEDEDEEEDDDE 537
Score = 53.5 bits (127), Expect = 3e-07
Identities = 19/36 (52%), Positives = 31/36 (85%)
Frame = +1
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEK 387
D+EDD+D+D+D D+ EDED+D++ED+DED+ ++K
Sbjct: 454 DDEDDDDDDDDNDDGEDEDEDEEEDDDEDQPPSKKK 561
Score = 53.1 bits (126), Expect = 3e-07
Identities = 18/36 (50%), Positives = 31/36 (86%)
Frame = +1
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEK 387
D++D++D+D+D+D D+ ED+D+DE+ED+DED+ K
Sbjct: 448 DDDDEDDDDDDDDNDDGEDEDEDEEEDDDEDQPPSK 555
Score = 53.1 bits (126), Expect = 3e-07
Identities = 23/76 (30%), Positives = 43/76 (56%)
Frame = +1
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEV 411
+++DDED+D+D ++++D++D+D +++DED D E D D E D + DD+
Sbjct: 310 EDDDDEDDDDDVNDEDDDNDEDFSGDEDDEDADPEDDPVPNGAGGSDDDDEDDDDDDDDN 489
Query: 412 EINTGLGTSDREDQNE 427
+ + ED +E
Sbjct: 490 DDGEDEDEDEEEDDDE 537
Score = 52.8 bits (125), Expect = 5e-07
Identities = 36/109 (33%), Positives = 53/109 (48%)
Frame = +1
Query: 279 ESTEDRSELFVVSTEDPESGDDRNDGSDPAERMPGFQNGLKDMIPGVKVKIFKVITPEKV 338
E +D SE TED + DD +D +D + +G +D
Sbjct: 283 EENKDGSE-----TEDDDDEDDDDDVNDEDDDNDEDFSGDED----------------DE 399
Query: 339 DKDLISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEK 387
D D + DD+DED D+D+D+D DD +DEDEDE+ED+DE++
Sbjct: 400 DADPEDDPVPNGAGGSDDDDED-DDDDDDDNDDGEDEDEDEEEDDDEDQ 543
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 56.2 bits (134), Expect = 4e-08
Identities = 29/54 (53%), Positives = 41/54 (75%), Gaps = 10/54 (18%)
Frame = +3
Query: 352 DEEDDED-EDEDEDEDEDEDDDDDED---EDEDE------DEDEEKDSDTGSLE 395
+EEDDED +D+DED+DEDEDDDD+E+ EDE+E +E+EE+D D +L+
Sbjct: 594 EEEDDEDGDDQDEDDDEDEDDDDEEEGGEEDEEEGVDEEDNEEEEEDEDEEALQ 755
Score = 53.1 bits (126), Expect = 3e-07
Identities = 24/59 (40%), Positives = 41/59 (68%), Gaps = 1/59 (1%)
Frame = +3
Query: 355 DDEDEDEDEDEDEDEDDDDDEDE-DEDEDEDEEKDSDTGSLELVDMKSETDQEADDEVE 412
D+ E+ED+++ +D+D+DDDEDE D+DE+E E+D + G VD + ++E D++ E
Sbjct: 582 DENGEEEDDEDGDDQDEDDDEDEDDDDEEEGGEEDEEEG----VDEEDNEEEEEDEDEE 746
Score = 44.3 bits (103), Expect = 2e-04
Identities = 27/105 (25%), Positives = 43/105 (40%), Gaps = 44/105 (41%)
Frame = +3
Query: 352 DEEDDEDEDEDE--------------------------------------------DEDE 367
D DDEDE++D+ E+E
Sbjct: 423 DGNDDEDEEDDDGDGAFGEGEDELSSEDGGGYGNNSNNKSNSKKAPEGGAGGADENGEEE 602
Query: 368 DEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEVE 412
D++D DD+DED+DEDED++ + + G + + E D E ++E E
Sbjct: 603 DDEDGDDQDEDDDEDEDDDDEEEGGEEDEEEGVDEEDNEEEEEDE 737
Score = 36.6 bits (83), Expect = 0.034
Identities = 27/108 (25%), Positives = 46/108 (42%), Gaps = 28/108 (25%)
Frame = +3
Query: 352 DEEDDEDEDED----EDEDEDEDDDD------------------------DEDEDEDEDE 383
D+ED+ED+D D E EDE +D DE+ +E++DE
Sbjct: 432 DDEDEEDDDGDGAFGEGEDELSSEDGGGYGNNSNNKSNSKKAPEGGAGGADENGEEEDDE 611
Query: 384 DEEKDSDTGSLELVDMKSETDQEADDEVEINTGLGTSDREDQNEIAVK 431
D + + + D E E D+E ++ + ED++E A++
Sbjct: 612 DGDDQDEDDDEDEDDDDEEEGGEEDEEEGVDEEDNEEEEEDEDEEALQ 755
Score = 35.4 bits (80), Expect = 0.075
Identities = 28/111 (25%), Positives = 46/111 (41%), Gaps = 36/111 (32%)
Frame = +3
Query: 353 EEDDEDEDEDEDEDEDEDDDDD------EDE----------------------------- 377
+ D E ED ++DEDE EDDD D EDE
Sbjct: 402 QHDAEGEDGNDDEDE-EDDDGDGAFGEGEDELSSEDGGGYGNNSNNKSNSKKAPEGGAGG 578
Query: 378 -DEDEDEDEEKDSDTGSLELVDMKSETDQEADDEVEINTGLGTSDREDQNE 427
DE+ +E++++D D + + + + D+E E + G+ D E++ E
Sbjct: 579 ADENGEEEDDEDGDDQDEDDDEDEDDDDEEEGGEEDEEEGVDEEDNEEEEE 731
Score = 32.3 bits (72), Expect = 0.63
Identities = 15/76 (19%), Positives = 37/76 (47%)
Frame = +3
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEV 411
D + + + + E ED +DD+DE++D+ + E + + S + + ++ +++ +
Sbjct: 375 DSKTQSQDKQHDAEGEDGNDDEDEEDDDGDGAFGEGEDELSSEDGGGYGNNSNNKSNSKK 554
Query: 412 EINTGLGTSDREDQNE 427
G G +D + E
Sbjct: 555 APEGGAGGADENGEEE 602
>AW257087 homologue to GP|6683126|dbj KIAA0339 protein {Homo sapiens},
partial (2%)
Length = 466
Score = 56.2 bits (134), Expect = 4e-08
Identities = 29/72 (40%), Positives = 42/72 (58%), Gaps = 12/72 (16%)
Frame = -3
Query: 353 EEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTG------------SLELVDMK 400
+++DEDEDEDEDEDEDED+DD+E +E E E E S +LE ++
Sbjct: 380 DDEDEDEDEDEDEDEDEDEDDEELLEESESESESDSSFLAGVAFAAETAGFFTLETEELV 201
Query: 401 SETDQEADDEVE 412
E ++E++ E E
Sbjct: 200 EELEEESESESE 165
>BF641220 weakly similar to PIR|G86203|G86 probable N-arginine dibasic
convertase [imported] - Arabidopsis thaliana, partial
(5%)
Length = 634
Score = 56.2 bits (134), Expect = 4e-08
Identities = 44/134 (32%), Positives = 66/134 (48%), Gaps = 1/134 (0%)
Frame = +2
Query: 253 KRKGAFHGPPTTSSKALDAAERLSSVESTEDRSELFVVSTEDPESGDDRNDGSDPAERMP 312
K G P ++ A AA LSS + ++ V S D + R+
Sbjct: 149 KESMGLKGAPAAATTAATAAVALSSSD------DVIVKSPND-----------NRLYRLV 277
Query: 313 GFQNGLKDMIPGVKVKIFKVITPEKVDKDLISKVIEQIIDEEDDEDEDEDEDEDEDEDDD 372
+NGL+ +I I PE KD IDE+D+E++DEDE++DE++DD+
Sbjct: 278 HLKNGLQALIVHDPE-----IYPEGAPKD-------GSIDEDDEEEDDEDEEDDEEDDDE 421
Query: 373 DDEDEDED-EDEDE 385
++DEDE+ EDEDE
Sbjct: 422 GEDDEDEEXEDEDE 463
Score = 50.8 bits (120), Expect = 2e-06
Identities = 21/43 (48%), Positives = 33/43 (75%)
Frame = +2
Query: 353 EEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLE 395
+EDDE+ED DEDE++DE+DDD+ ++DEDE+ ++E + E
Sbjct: 359 DEDDEEED-DEDEEDDEEDDDEGEDDEDEEXEDEDEXXVXGRE 484
Score = 49.3 bits (116), Expect = 5e-06
Identities = 17/39 (43%), Positives = 32/39 (81%)
Frame = +2
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSD 390
D DED++E++DEDE++D++DD++ ++DEDE+ E + +
Sbjct: 347 DGSIDEDDEEEDDEDEEDDEEDDDEGEDDEDEEXEDEDE 463
Score = 46.2 bits (108), Expect = 4e-05
Identities = 15/37 (40%), Positives = 31/37 (83%)
Frame = +2
Query: 354 EDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSD 390
+D +++DE+ED+++++DD+ED+DE ED+++E+ D
Sbjct: 344 KDGSIDEDDEEEDDEDEEDDEEDDDEGEDDEDEEXED 454
Score = 43.5 bits (101), Expect = 3e-04
Identities = 29/92 (31%), Positives = 46/92 (49%), Gaps = 5/92 (5%)
Frame = +2
Query: 349 QIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEAD 408
Q + D E E +D D+DD+E++DEDE++DEE D D G + + D+E +
Sbjct: 296 QALIVHDPEIYPEGAPKDGSIDEDDEEEDDEDEEDDEE-DDDEG-------EDDEDEEXE 451
Query: 409 DEVEINT-----GLGTSDREDQNEIAVKVVIG 435
DE E G G +++ + + V IG
Sbjct: 452 DEDEXXVXGREGGKGAANQSKKAAXXMCVXIG 547
>TC80809 similar to GP|22136852|gb|AAM91770.1 unknown protein {Arabidopsis
thaliana}, partial (11%)
Length = 681
Score = 55.1 bits (131), Expect = 9e-08
Identities = 34/99 (34%), Positives = 48/99 (48%), Gaps = 6/99 (6%)
Frame = +3
Query: 339 DKDLISKVIEQIIDE----EDDEDEDED-EDEDEDEDDDDDEDEDEDEDEDEEKDSDTGS 393
D DL S +Q+ D+ DDED D D +DE D + DEDED D + ++
Sbjct: 195 DSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARK 374
Query: 394 LELVDMKSETD-QEADDEVEINTGLGTSDREDQNEIAVK 431
+L+D K E D Q A DE++ N D D+ + K
Sbjct: 375 SKLIDRKRERDSQAAADELQTNIQTNIQDESDEFTLPTK 491
>TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {Arabidopsis
thaliana}, partial (39%)
Length = 883
Score = 54.7 bits (130), Expect = 1e-07
Identities = 29/79 (36%), Positives = 43/79 (53%), Gaps = 3/79 (3%)
Frame = +3
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDE---EKDSDTGSLELVDMKSETDQEAD 408
D EDDED+D+D+D +++DD ++ED DE E+E E D + D + D + D
Sbjct: 318 DTEDDEDDDDDDDVQDEDDDGEEEDYSGDEGEEEGDPEDDPEANGAGGSDDGEDDDDDGD 497
Query: 409 DEVEINTGLGTSDREDQNE 427
+E E G D ED+ E
Sbjct: 498 EEDE-EDGEDEEDEEDEEE 551
Score = 50.8 bits (120), Expect = 2e-06
Identities = 29/77 (37%), Positives = 43/77 (55%), Gaps = 14/77 (18%)
Frame = +3
Query: 348 EQIIDEEDDEDE-----DEDEDEDEDEDD---------DDDEDEDEDEDEDEEKDSDTGS 393
+ + DE+DD +E DE E+E + EDD DD ED+D+D DE++E+D +
Sbjct: 351 DDVQDEDDDGEEEDYSGDEGEEEGDPEDDPEANGAGGSDDGEDDDDDGDEEDEEDGE--- 521
Query: 394 LELVDMKSETDQEADDE 410
D + E D+E DDE
Sbjct: 522 ----DEEDEEDEEEDDE 560
Score = 50.4 bits (119), Expect = 2e-06
Identities = 25/82 (30%), Positives = 47/82 (56%), Gaps = 1/82 (1%)
Frame = +3
Query: 347 IEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDED-EDEEKDSDTGSLELVDMKSETDQ 405
+E ++ + E++ + ED+++DDDDD+ +DED+D E+E+ D G E +
Sbjct: 270 LEHLLGRKPAYKENKSDTEDDEDDDDDDDVQDEDDDGEEEDYSGDEG---------EEEG 422
Query: 406 EADDEVEINTGLGTSDREDQNE 427
+ +D+ E N G+ D ED ++
Sbjct: 423 DPEDDPEANGAGGSDDGEDDDD 488
Score = 46.2 bits (108), Expect = 4e-05
Identities = 22/48 (45%), Positives = 33/48 (67%), Gaps = 8/48 (16%)
Frame = +3
Query: 352 DEEDD--------EDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDT 391
D EDD D+ ED+D+D DE+D++D EDE+++EDEE+D +T
Sbjct: 423 DPEDDPEANGAGGSDDGEDDDDDGDEEDEED-GEDEEDEEDEEEDDET 563
Score = 43.1 bits (100), Expect = 4e-04
Identities = 14/36 (38%), Positives = 29/36 (79%)
Frame = +3
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEK 387
+++DD+ ++EDE++ EDE+D++DE+ED++ + K
Sbjct: 474 EDDDDDGDEEDEEDGEDEEDEEDEEEDDETPQPPAK 581
Score = 41.6 bits (96), Expect = 0.001
Identities = 14/36 (38%), Positives = 28/36 (76%)
Frame = +3
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEK 387
D++DD DE+++ED +++ED++D+E++DE +K
Sbjct: 477 DDDDDGDEEDEEDGEDEEDEEDEEEDDETPQPPAKK 584
>CA920710 similar to PIR|S64314|S643 probable membrane protein YGR023w -
yeast (Saccharomyces cerevisiae), partial (7%)
Length = 774
Score = 52.0 bits (123), Expect = 8e-07
Identities = 20/34 (58%), Positives = 31/34 (90%)
Frame = -2
Query: 351 IDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDED 384
I+ E + +E+E+E++DEDED+++DEDEDEDEDE+
Sbjct: 386 IEAEAEAEEEEEEEKDEDEDENEDEDEDEDEDEE 285
Score = 51.6 bits (122), Expect = 1e-06
Identities = 20/33 (60%), Positives = 30/33 (90%)
Frame = -2
Query: 354 EDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEE 386
E + + +E+E+E++DED+D++EDEDEDEDEDEE
Sbjct: 383 EAEAEAEEEEEEEKDEDEDENEDEDEDEDEDEE 285
Score = 50.8 bits (120), Expect = 2e-06
Identities = 21/37 (56%), Positives = 31/37 (83%)
Frame = -2
Query: 344 SKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDED 380
S IE + E++E+E++DEDEDE+ED+D+DEDEDE+
Sbjct: 395 SGYIEAEAEAEEEEEEEKDEDEDENEDEDEDEDEDEE 285
Score = 46.6 bits (109), Expect = 3e-05
Identities = 16/33 (48%), Positives = 30/33 (90%)
Frame = -2
Query: 356 DEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKD 388
+ + + +E+E+E++D+D+DE+EDEDEDEDE+++
Sbjct: 383 EAEAEAEEEEEEEKDEDEDENEDEDEDEDEDEE 285
Score = 45.1 bits (105), Expect = 9e-05
Identities = 16/33 (48%), Positives = 28/33 (84%)
Frame = -2
Query: 358 DEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSD 390
+ + + +E+E+E+ D+DEDE+EDEDEDE++D +
Sbjct: 383 EAEAEAEEEEEEEKDEDEDENEDEDEDEDEDEE 285
>TC77681 similar to GP|22597156|gb|AAN03465.1 nucleolar histone deacetylase
HD2-P39 {Glycine max}, partial (47%)
Length = 1233
Score = 51.2 bits (121), Expect = 1e-06
Identities = 34/137 (24%), Positives = 65/137 (46%), Gaps = 14/137 (10%)
Frame = +2
Query: 293 EDPESGDDRNDGSDPAERMPGF---QNGLKDMIPGVKVKIFKVITPEKVD-------KDL 342
ED + ++ + E +P QNG P K + KV P+K D K+
Sbjct: 365 EDNDDSEEEGETDSEDEDIPLITTEQNGK----PEPKAEELKVSEPKKADAKNAAPAKNA 532
Query: 343 ISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDD----EDEDEDEDEDEEKDSDTGSLELVD 398
+ +++D +D++ +D+DE +DE DD+ + + EDED+ +E D + ++ VD
Sbjct: 533 ATAKHVKVVDPKDEDSDDDDESDDEIGSSDDEMENADSDSEDEDDSDEDDEEETPVKKVD 712
Query: 399 MKSETDQEADDEVEINT 415
+ E+ + I++
Sbjct: 713 QGKKRPNESASKTPISS 763
Score = 43.9 bits (102), Expect = 2e-04
Identities = 23/83 (27%), Positives = 41/83 (48%)
Frame = +2
Query: 328 KIFKVITPEKVDKDLISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEK 387
K KV+ P+ D D + ++I +D+ + + + EDED+ D+DDE+E + D+ K
Sbjct: 542 KHVKVVDPKDEDSDDDDESDDEIGSSDDEMENADSDSEDEDDSDEDDEEETPVKKVDQGK 721
Query: 388 DSDTGSLELVDMKSETDQEADDE 410
S + S+ + A E
Sbjct: 722 KRPNESASKTPISSKKSKNATPE 790
>TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (69%)
Length = 1615
Score = 50.4 bits (119), Expect = 2e-06
Identities = 28/87 (32%), Positives = 44/87 (50%), Gaps = 11/87 (12%)
Frame = +3
Query: 352 DEEDDEDEDEDED-----------EDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMK 400
DEE DE+ DEDED + +D D +DED+D DE+K E+ + +
Sbjct: 171 DEESDEESDEDEDAKPVSKPAAVAKKSKKDSSDSDDEDDDSSSDEDKKPVAAKKEVSESE 350
Query: 401 SETDQEADDEVEINTGLGTSDREDQNE 427
S++ +DDE ++N SD ++ E
Sbjct: 351 SDS---SDDEDKMNVDKDGSDSDESEE 422
Score = 43.5 bits (101), Expect = 3e-04
Identities = 30/114 (26%), Positives = 51/114 (44%), Gaps = 10/114 (8%)
Frame = +3
Query: 343 ISKVIEQIIDEEDDEDEDEDEDEDE----------DEDDDDDEDEDEDEDEDEEKDSDTG 392
++K ++ + DDED+D DED+ + + D +DED+ + + DSD
Sbjct: 237 VAKKSKKDSSDSDDEDDDSSSDEDKKPVAAKKEVSESESDSSDDEDKMNVDKDGSDSDES 416
Query: 393 SLELVDMKSETDQEADDEVEINTGLGTSDREDQNEIAVKVVIGGIVQKLSSNIS 446
E D S+T Q+ +VE+ G S ++ N A + G N+S
Sbjct: 417 EEESEDEPSKTPQKKIKDVEM-VDAGKSGKKAPNTPATPIENSGSKTLFVGNLS 575
Score = 32.7 bits (73), Expect = 0.49
Identities = 16/61 (26%), Positives = 32/61 (52%)
Frame = +3
Query: 339 DKDLISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVD 398
D+D ++ + E + + D+++ + D+D D DE E+E EDE + ++ V+
Sbjct: 300 DEDKKPVAAKKEVSESESDSSDDEDKMNVDKDGSD-SDESEEESEDEPSKTPQKKIKDVE 476
Query: 399 M 399
M
Sbjct: 477 M 479
>BE248780 similar to GP|5929884|gb|A nucleolin-related protein NRP {Rattus
norvegicus}, partial (4%)
Length = 561
Score = 50.4 bits (119), Expect = 2e-06
Identities = 26/72 (36%), Positives = 41/72 (56%)
Frame = +2
Query: 344 SKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSET 403
SK + I + E EDED+D D D+DDDDD+ ++E+ D ++E D + +E
Sbjct: 287 SKQDDHIKEGGTHEIEDEDDDSDGDDDDDDDDYDEEEGDYEDEDDDTVAVSSRKKVYTEE 466
Query: 404 DQEADDEVEINT 415
++EA+ E NT
Sbjct: 467 EKEAEAEAIWNT 502
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 50.1 bits (118), Expect = 3e-06
Identities = 46/202 (22%), Positives = 80/202 (38%), Gaps = 9/202 (4%)
Frame = +3
Query: 254 RKGAFHGPPTTSSKALDAAERLSSVESTEDRSELFVVSTEDPESGDDRNDGSDPAERMPG 313
+K A T S D + + ++ ++ D ES D+ ++ SD ++ P
Sbjct: 615 KKAASDEEDTDESSDEDEEDEKPAAKAVPSKNGSVPAKKADTESSDEDSESSDEEDKKPA 794
Query: 314 FQNGLKDMIPGVKVKIFK-VITPEKVDKDLISKVIEQII--------DEEDDEDEDEDED 364
+ P K + E+ D+D +K + + D D +DED+D
Sbjct: 795 AKASKNVSAPTKKAASSSDEESDEESDEDEDAKPVSKPAAVAKKSKKDSSDSDDEDDDSS 974
Query: 365 EDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEVEINTGLGTSDRED 424
DED+ + ED+ + + DSD E D S+T Q+ +VE+ G S ++
Sbjct: 975 SDEDKKPVAAKKEDKMNVDKDGSDSDQSEEESEDEPSKTPQKKIKDVEM-VDAGKSGKKA 1151
Query: 425 QNEIAVKVVIGGIVQKLSSNIS 446
N A + G N+S
Sbjct: 1152PNTPATPIENSGSKTLFVGNLS 1217
Score = 33.5 bits (75), Expect = 0.28
Identities = 61/299 (20%), Positives = 107/299 (35%), Gaps = 36/299 (12%)
Frame = +3
Query: 219 SPRQLAMSAAGVPLFEFFLKMDKKGEFKSQ----AVYLKRKGAFHGPPTTSSKALDAAER 274
S ++ A P+ +K KKG+ +++ AV K++ + + E
Sbjct: 111 SSKKSATKVDAAPVVVSPVKSGKKGKRQAEEEVKAVSAKKQKVEEVAAKQKALKVVKKEE 290
Query: 275 LSSVESTEDRSELFVVSTEDPESGDDRNDGSDPAERMPGFQNGLKDMIPGVKVKIFKVIT 334
SS ES+E E VV P +++ P + +K P + +
Sbjct: 291 SSSEESSESEDEQPVVKAPAP------------SKKTPAKKGNVKKAQPETTSEESDSDS 434
Query: 335 PEKVDKDLISKVIEQIIDEEDDEDEDE--DEDEDEDEDDDDDED---------------- 376
D++ + K + + + ++ + D E+ED ++ DED
Sbjct: 435 SSS-DEEEVKKPVSKAVPSKNGSAPAKKVDTSEEEDSEESSDEDKKPAAKAVPSKNGSAP 611
Query: 377 --------EDEDEDEDEEKDSDTGSLELVDMKS------ETDQEADDEVEINTGLGTSDR 422
ED DE DE+++ + + + V K+ + D E+ DE +SD
Sbjct: 612 AKKAASDEEDTDESSDEDEEDEKPAAKAVPSKNGSVPAKKADTESSDE-----DSESSDE 776
Query: 423 EDQNEIAVKVVIGGIVQKLSSNISTRDLLRVPAKLEIKKRGSFSFTVEKEVNQQDGQAK 481
ED+ A K S N+S KK S S E + +D AK
Sbjct: 777 EDKKPAA----------KASKNVSA----------PTKKAASSSDEESDEESDEDEDAK 893
>BG583961 similar to GP|14599408|em probable major surface glycoprotein
{Pneumocystis carinii}, partial (4%)
Length = 803
Score = 50.1 bits (118), Expect = 3e-06
Identities = 20/53 (37%), Positives = 29/53 (53%)
Frame = +1
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETD 404
D ++D D D D D D D D+D+D D D D D D + D+D + + D + D
Sbjct: 598 DNDNDNDHDNDNDNDNDNDNDNDHDNDNDNDNDHDNDNDHDHVNVNDNVNVND 756
Score = 37.0 bits (84), Expect = 0.026
Identities = 18/57 (31%), Positives = 29/57 (50%), Gaps = 3/57 (5%)
Frame = +1
Query: 368 DEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEAD---DEVEINTGLGTSD 421
D D+D+D D D D D D + D+D + D ++ D + D D V +N + +D
Sbjct: 598 DNDNDNDHDNDNDNDNDNDNDNDHDN----DNDNDNDHDNDNDHDHVNVNDNVNVND 756
>BF635065 weakly similar to GP|11037008|gb latent nuclear antigen {Human
herpesvirus 8}, partial (3%)
Length = 681
Score = 50.1 bits (118), Expect = 3e-06
Identities = 22/54 (40%), Positives = 34/54 (62%)
Frame = +2
Query: 353 EEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQE 406
E DD+D+D ++DE+ DE+DD++E E E++ EEK L +D E D+E
Sbjct: 362 EHDDDDDDVEDDEETDEEDDNEEAAVEQEEQKEEKQVTMNELVQMDNVDEVDKE 523
Score = 43.5 bits (101), Expect = 3e-04
Identities = 23/84 (27%), Positives = 45/84 (53%)
Frame = +2
Query: 342 LISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKS 401
L +K +E ++ + + E E DDDD+D ++DE+ DEE D++ ++E + K
Sbjct: 281 LYAKAMEHLMAFKSQHIVNNPVIPQEQEHDDDDDDVEDDEETDEEDDNEEAAVEQEEQKE 460
Query: 402 ETDQEADDEVEINTGLGTSDREDQ 425
E ++ V+++ + D+E Q
Sbjct: 461 EKQVTMNELVQMD-NVDEVDKEKQ 529
>TC83157 similar to GP|21595368|gb|AAM66095.1 unknown {Arabidopsis
thaliana}, partial (57%)
Length = 1108
Score = 49.3 bits (116), Expect = 5e-06
Identities = 29/69 (42%), Positives = 42/69 (60%)
Frame = +3
Query: 344 SKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSET 403
SK + I + E EDED+D D D+DDDDD D DE+E + E++D DT ++ K T
Sbjct: 294 SKQDDHIKEGGTHEIEDEDDDSDGDDDDDDD-DYDEEEGDYEDEDDDTVAVS-SRKKVYT 467
Query: 404 DQEADDEVE 412
++E + E E
Sbjct: 468 EEEKEAEAE 494
Score = 31.2 bits (69), Expect = 1.4
Identities = 19/80 (23%), Positives = 40/80 (49%), Gaps = 2/80 (2%)
Frame = +3
Query: 363 EDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEVEINT--GLGTS 420
+ +D ++ E EDED+D D + D D + + + + + E DD V +++ + T
Sbjct: 297 KQDDHIKEGGTHEIEDEDDDSDGDDDDDDDDYD--EEEGDYEDEDDDTVAVSSRKKVYTE 470
Query: 421 DREDQNEIAVKVVIGGIVQK 440
+ ++ A+ + G +QK
Sbjct: 471 EEKEAEAEAIGYKVVGPLQK 530
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.135 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,399,724
Number of Sequences: 36976
Number of extensions: 262660
Number of successful extensions: 10827
Number of sequences better than 10.0: 402
Number of HSP's better than 10.0 without gapping: 2124
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5466
length of query: 700
length of database: 9,014,727
effective HSP length: 103
effective length of query: 597
effective length of database: 5,206,199
effective search space: 3108100803
effective search space used: 3108100803
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0100a.10


