

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
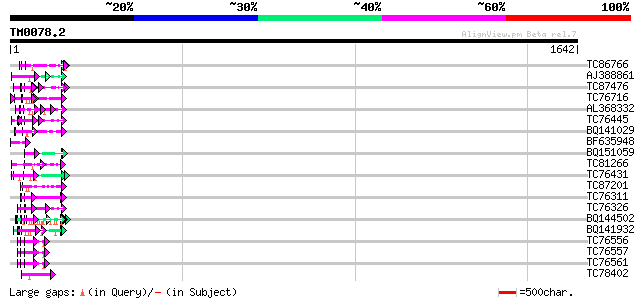
Query= TM0078.2
(1642 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E... 72 1e-12
AJ388861 weakly similar to GP|1754989|gb| proline-rich protein P... 65 3e-10
TC87476 homologue to PIR|T11622|T11622 extensin class 1 precurso... 64 5e-10
TC76716 similar to PIR|T10863|T10863 extensin precursor - kidney... 64 5e-10
AL368332 weakly similar to GP|21322711|e pherophorin-dz1 protein... 64 5e-10
TC76445 similar to PIR|T07623|T07623 extensin homolog HRGP2 - so... 63 1e-09
BQ141029 similar to GP|8132441|gb| extensin {Pisum sativum}, par... 60 5e-09
BF635948 similar to GP|21622312|emb putative protein {Neurospora... 60 5e-09
BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melano... 60 7e-09
TC81266 weakly similar to GP|16415994|emb|CAB88653. hypothetical... 59 2e-08
TC76431 similar to GP|8132441|gb|AAF73291.1| extensin {Pisum sat... 59 2e-08
TC87201 similar to PIR|F86388|F86388 hypothetical protein AAG292... 59 2e-08
TC76311 homologue to GP|3204132|emb|CAA07235.1 extensin {Cicer a... 58 3e-08
TC76326 homologue to PIR|C29356|C29356 hydroxyproline-rich glyco... 58 3e-08
BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 58 3e-08
BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 57 5e-08
TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 57 5e-08
TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 57 5e-08
TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 57 5e-08
TC78402 weakly similar to PIR|B84853|B84853 hypothetical protein... 57 6e-08
>TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E8.80 -
Arabidopsis thaliana, partial (18%)
Length = 1260
Score = 72.4 bits (176), Expect = 1e-12
Identities = 51/161 (31%), Positives = 68/161 (41%), Gaps = 18/161 (11%)
Frame = +2
Query: 29 YAPLGPQSHHHLSQPPPPL----PPPHAPPPPPH-PPPLSYRNIHTPPPPPLYNTPPVHP 83
++P P +++ S PPPP PPP +PPPPPH P P Y + PPPPP+++ PP
Sbjct: 83 FSPPPPVPYYYSSPPPPPAHSPPPPPXSPPPPPHSPTPPVYPYLSPPPPPPVHSPPPPVY 262
Query: 84 SRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPPLD 143
S P P P P P P + +P P +P + PP
Sbjct: 263 SPPPPSPPPCVEPPPPPPPPCVEPPPPSSPAPHQTPYHPPPSPSPPPSPVYAYPSPPPPV 442
Query: 144 YDRE-----FNHHRPQQP----PPPPPSY----PPVDSLRY 171
Y + + P P PPPPP Y PPV + Y
Sbjct: 443 YTSPPPSPVYAYPSPPPPVYSSPPPPPVYEGPIPPVFGISY 565
Score = 59.7 bits (143), Expect = 9e-09
Identities = 42/126 (33%), Positives = 55/126 (43%), Gaps = 2/126 (1%)
Frame = +2
Query: 45 PPLPPPHAPPPPPHPPPLSYRNIHTPPPPP--LYNTPPVHPSRQLQFYPHHRSAPEERPV 102
PP PPP++PPPPP P P + +PPPP Y++PP P+ P P P
Sbjct: 29 PPPPPPNSPPPPPPPAP-----VFSPPPPVPYYYSSPPPPPAHSPPPPPXSPPPPPHSPT 193
Query: 103 PTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPPLDYDREFNHHRPQQPPPPPPS 162
P +P+ L P P + P P ++P PP + P PPPPP
Sbjct: 194 PPVYPY---LSPPPPPPVHS--PPPPVYSPPPPS---PPPCVE-------PPPPPPPPCV 328
Query: 163 YPPVDS 168
PP S
Sbjct: 329 EPPPPS 346
Score = 57.4 bits (137), Expect = 5e-08
Identities = 47/150 (31%), Positives = 57/150 (37%), Gaps = 18/150 (12%)
Frame = +2
Query: 34 PQSHHHLSQPPPPLPPPHAPPPPP-----HPPPLSY-------RNIHTPPPPPLYNTPPV 81
P + + PPPP P PPPPP PPP+ Y H+PPPPP PP
Sbjct: 2 PTPPYCVRSPPPPPPNSPPPPPPPAPVFSPPPPVPYYYSSPPPPPAHSPPPPPXSPPPPP 181
Query: 82 HPSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPP 141
H S YP+ P P P + P P P P P P ++ P
Sbjct: 182 H-SPTPPVYPYLSPPP---PPPVHSPPPPVYSPPPPSPPPCVEPPPPPPPPCVEP-PPPS 346
Query: 142 LDYDREFNHHRPQQPPPPP------PSYPP 165
+ +H P P PPP PS PP
Sbjct: 347 SPAPHQTPYHPPPSPSPPPSPVYAYPSPPP 436
Score = 37.7 bits (86), Expect = 0.037
Identities = 27/81 (33%), Positives = 34/81 (41%), Gaps = 26/81 (32%)
Frame = +2
Query: 22 PQQHRTRYAPLG-----PQSHHHLSQPPPPL---PPPH------APPPPPH---PPPLSY 64
P H+T Y P P + PPPP+ PPP +PPPP + PPP Y
Sbjct: 350 PAPHQTPYHPPPSPSPPPSPVYAYPSPPPPVYTSPPPSPVYAYPSPPPPVYSSPPPPPVY 529
Query: 65 R---------NIHTPPPPPLY 76
+ +PPPPP Y
Sbjct: 530 EGPIPPVFGISYASPPPPPFY 592
Score = 37.0 bits (84), Expect = 0.063
Identities = 34/122 (27%), Positives = 43/122 (34%), Gaps = 20/122 (16%)
Frame = +3
Query: 9 RHYHH----------------HHQHQQQHPQQHRTRYAPLGPQSHHHLSQPPPPLPPPHA 52
RH HH HH H+Q+H + T + P HHH P P +
Sbjct: 273 RHLHHLV*NLHHLHRHLV*NLHHHHRQRHIKHLITHHHP--HHHHHHQYMPIPHRHHRYT 446
Query: 53 PPPPPHP----PPLSYRNIHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPEERPVPTNHPF 108
P P H P L +R IH LY H S + + HH +HPF
Sbjct: 447 PHPHHHQYMHIPHLHHRYIHR-RHHHLYMKGLYHLSLESHMHHHH-----------HHPF 590
Query: 109 SD 110
D
Sbjct: 591 ID 596
>AJ388861 weakly similar to GP|1754989|gb| proline-rich protein PRP2
precursor {Lupinus luteus}, partial (24%)
Length = 491
Score = 64.7 bits (156), Expect = 3e-10
Identities = 47/160 (29%), Positives = 61/160 (37%)
Frame = -3
Query: 6 PRDRHYHHHHQHQQQHPQQHRTRYAPLGPQSHHHLSQPPPPLPPPHAPPPPPHPPPLSYR 65
P++ H Q + P +++ P H H PPP PPH PPP H PP +
Sbjct: 402 PQEYQPLHEKPPQMKPPSEYQP---PHEKPPHEH---PPPEYQPPHVHPPPEHQPP--HE 247
Query: 66 NIHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRN 125
N PPP Y P P PH+ P E P HP P+Y +
Sbjct: 246 NPPHENPPPEYQPPHEKP-------PHYEHPPPENQPPHVHP----------PPEYQPPH 118
Query: 126 PEPAWNPSLDDLNYPPLDYDREFNHHRPQQPPPPPPSYPP 165
+P +PP +Y H +P Q PPP PP
Sbjct: 117 EKPPH-------EHPPPEYQPP--HEKPPQENPPPEYKPP 25
Score = 52.0 bits (123), Expect = 2e-06
Identities = 28/85 (32%), Positives = 36/85 (41%), Gaps = 4/85 (4%)
Frame = -3
Query: 6 PRDRHYHHHHQHQQQHPQQHRTRYAPLGPQSHHHLSQPPPPLPPPHAPPPPPHPPPLSYR 65
P + H + + Q P + Y P++ PPP PPH PP HPPP Y+
Sbjct: 255 PHENPPHENPPPEYQPPHEKPPHYEHPPPENQPPHVHPPPEYQPPHEKPPHEHPPP-EYQ 79
Query: 66 NIHTPP----PPPLYNTPPVHPSRQ 86
H P PPP Y P P R+
Sbjct: 78 PPHEKPPQENPPPEYKPPHEKPPRE 4
Score = 48.9 bits (115), Expect = 2e-05
Identities = 37/124 (29%), Positives = 48/124 (37%), Gaps = 12/124 (9%)
Frame = -3
Query: 6 PRDRHYHHHHQHQQQHPQQHRT-RYAPLGPQSHHHLSQPPPPLPPPHAPPP--------- 55
P ++ H H + Q P H + P P + PPP PPH PP
Sbjct: 336 PHEKPPHEHPPPEYQPPHVHPPPEHQP--PHENPPHENPPPEYQPPHEKPPHYEHPPPEN 163
Query: 56 -PPHP-PPLSYRNIHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPEERPVPTNHPFSDDLL 113
PPH PP Y+ H PP + PP ++ P H P+E P P P +
Sbjct: 162 QPPHVHPPPEYQPPHEKPP---HEHPPP------EYQPPHEKPPQENPPPEYKP-PHEKP 13
Query: 114 PRRN 117
PR N
Sbjct: 12 PREN 1
Score = 31.6 bits (70), Expect = 2.7
Identities = 15/58 (25%), Positives = 22/58 (37%)
Frame = -3
Query: 3 QHYPRDRHYHHHHQHQQQHPQQHRTRYAPLGPQSHHHLSQPPPPLPPPHAPPPPPHPP 60
+H P + H H + P + + P+ +PP PPP PP PP
Sbjct: 183 EHPPPENQPPHVHPPPEYQPPHEKPPHEHPPPEYQPPHEKPPQENPPPEYKPPHEKPP 10
>TC87476 homologue to PIR|T11622|T11622 extensin class 1 precursor - cowpea,
partial (48%)
Length = 939
Score = 63.9 bits (154), Expect = 5e-10
Identities = 53/172 (30%), Positives = 72/172 (41%), Gaps = 10/172 (5%)
Frame = +3
Query: 12 HHHHQHQQQHPQQHRTRYAPLGPQSHHHLSQPPPPLPPP---HAPPPPPH---PPPLSYR 65
H HH H + H + L + HH +PP P PPP + PPPP PPP Y+
Sbjct: 282 HPHHLHTFINLLLHHLLHHRLHISTSHH--RPPSPSPPPPYVYKSPPPPSPSPPPPYVYK 455
Query: 66 N----IHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRNIPDY 121
+ +PPPP +Y +PP PS +++S P P P P Y
Sbjct: 456 SPPPPSPSPPPPYVYKSPP-PPSASPPPPYYYKSPPPPSPSPP--------------PPY 590
Query: 122 DRRNPEPAWNPSLDDLNYPPLDYDREFNHHRPQQPPPPPPSYPPVDSLRYDA 173
++P P +PS PP Y + PPPP PS PP Y++
Sbjct: 591 GYKSPPPP-SPSPP----PPYIY---------KSPPPPSPSPPPYHPYLYNS 704
Score = 52.8 bits (125), Expect = 1e-06
Identities = 47/144 (32%), Positives = 62/144 (42%), Gaps = 12/144 (8%)
Frame = +2
Query: 34 PQSHHHLSQPPP-PLPPP---HAPPPPPH---PPPLSYRN----IHTPPPPPLYNTPPVH 82
P + + S PPP P PPP + PPPP PPP Y++ +PPPP +Y +PP
Sbjct: 2 PPPYIYKSPPPPSPSPPPPYVYKSPPPPSPSPPPPYVYKSPPPPSPSPPPPYVYKSPP-- 175
Query: 83 PSRQLQFYPH-HRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPP 141
P P+ ++S P P P P Y ++P P +PS PP
Sbjct: 176 PPTPSPPPPYIYKSPPPPSPSPP--------------PPYVYKSPPPP-SPSPP----PP 298
Query: 142 LDYDREFNHHRPQQPPPPPPSYPP 165
Y + PPPP PS PP
Sbjct: 299 YVY---------KSPPPPSPSPPP 343
Score = 46.2 bits (108), Expect = 1e-04
Identities = 25/76 (32%), Positives = 36/76 (46%), Gaps = 21/76 (27%)
Frame = +3
Query: 43 PPPPL-----PPPHAPPPPPH------------PPPLSYRN----IHTPPPPPLYNTPPV 81
PPPP PPP A PPPP+ PPP Y++ +PPPP +Y +PP
Sbjct: 480 PPPPYVYKSPPPPSASPPPPYYYKSPPPPSPSPPPPYGYKSPPPPSPSPPPPYIYKSPPP 659
Query: 82 HPSRQLQFYPHHRSAP 97
++P+ ++P
Sbjct: 660 PSPSPPPYHPYLYNSP 707
Score = 44.3 bits (103), Expect = 4e-04
Identities = 21/52 (40%), Positives = 29/52 (55%), Gaps = 5/52 (9%)
Frame = +3
Query: 30 APLGPQSHHHLSQPPP-PLPPP----HAPPPPPHPPPLSYRNIHTPPPPPLY 76
+P P + + S PPP P PPP +PPPP PP + ++ PPPP+Y
Sbjct: 567 SPSPPPPYGYKSPPPPSPSPPPPYIYKSPPPPSPSPPPYHPYLYNSPPPPIY 722
Score = 44.3 bits (103), Expect = 4e-04
Identities = 25/61 (40%), Positives = 33/61 (53%), Gaps = 11/61 (18%)
Frame = +2
Query: 31 PLGPQSHHHLSQPPP-PLPPP---HAPPPPPH---PPPLSYRN----IHTPPPPPLYNTP 79
P P + + S PPP P PPP + PPPP PPP Y++ +PPPP +Y +P
Sbjct: 185 PSPPPPYIYKSPPPPSPSPPPPYVYKSPPPPSPSPPPPYVYKSPPPPSPSPPPPYIYKSP 364
Query: 80 P 80
P
Sbjct: 365 P 367
Score = 34.3 bits (77), Expect = 0.41
Identities = 25/72 (34%), Positives = 33/72 (45%), Gaps = 10/72 (13%)
Frame = +2
Query: 30 APLGPQSHHHLSQPPP-PLPPP---HAPPPPP---HPPPLSYRNIHTPPPPPLYNTPPV- 81
+P P + + S PPP P PPP + PPPP PPP Y+ +PP + T +
Sbjct: 230 SPSPPPPYVYKSPPPPSPSPPPPYVYKSPPPPSPSPPPPYIYK---SPPTTIPFTTTSIC 400
Query: 82 --HPSRQLQFYP 91
PS F P
Sbjct: 401 L*VPSSSFSFTP 436
>TC76716 similar to PIR|T10863|T10863 extensin precursor - kidney bean,
partial (48%)
Length = 1275
Score = 63.9 bits (154), Expect = 5e-10
Identities = 46/167 (27%), Positives = 71/167 (41%), Gaps = 16/167 (9%)
Frame = +3
Query: 15 HQHQQQHPQQHRTRYAPLGPQSHHHLSQPPPPLPPPHAPPPP---PHPPPLSYRN----I 67
++++ P ++ + P P+ + PPPP+ +PPPP P PP Y++ +
Sbjct: 351 YKYKSPPPPVYKYKSPPPPPKKPYKYPSPPPPVYKYKSPPPPVYSPPPPVYKYKSPPPPV 530
Query: 68 HTPPPPPLYNTPP----VHPSRQLQFYPHHRSAP-----EERPVPTNHPFSDDLLPRRNI 118
H+PPPP Y +PP +PS Y + P + P P P+ P
Sbjct: 531 HSPPPPYKYKSPPPPPYKYPSPPPPVYKYKSPPPPVYKYKSPPPPPKKPYK---YPSPPP 701
Query: 119 PDYDRRNPEPAWNPSLDDLNYPPLDYDREFNHHRPQQPPPPPPSYPP 165
P Y ++P P P + PP + H+ PPPP S PP
Sbjct: 702 PVYKYKSPPP---PVYKYKSPPPPVHSPPPPHYVYSSPPPPVYSPPP 833
Score = 63.5 bits (153), Expect = 6e-10
Identities = 44/168 (26%), Positives = 72/168 (42%), Gaps = 18/168 (10%)
Frame = +3
Query: 15 HQHQQQHPQQHRTRYAPLGPQSHHHLSQPPPPLPPPHAPPPPPHPPPLSYR------NIH 68
++++ P ++ + P P+ + PPPP+ +PPPP + PP Y+ ++
Sbjct: 96 YKYKSPPPPVYKYKSPPPPPKKPYKYPSPPPPVYKYKSPPPPVYSPPPVYKYKSPPPPVY 275
Query: 69 TPPPPPLYNTPP----VHPSRQLQFYPHHRSAP-----EERPVPTNHPFSDDLLPRRNIP 119
+PPPP Y +PP +PS Y + P + P P P+ P P
Sbjct: 276 SPPPPYKYKSPPPPPYKYPSPPPPVYKYKSPPPPVYKYKSPPPPPKKPYK---YPSPPPP 446
Query: 120 DYDRRN-PEPAWNPSLDDLNY--PPLDYDREFNHHRPQQPPPPPPSYP 164
Y ++ P P ++P Y PP ++ + PPPPP YP
Sbjct: 447 VYKYKSPPPPVYSPPPPVYKYKSPPPPVHSPPPPYKYKSPPPPPYKYP 590
Score = 55.8 bits (133), Expect = 1e-07
Identities = 40/144 (27%), Positives = 58/144 (39%), Gaps = 12/144 (8%)
Frame = +3
Query: 34 PQSHHHLSQPPPPLPPPHAPPPP-------PHPPPLSYRNIHTPPPPPLYNTPPVHPSRQ 86
P + PPPP+ +PPPP P PPP Y+ + PPPP+Y+ PPV+ +
Sbjct: 84 PPPVYKYKSPPPPVYKYKSPPPPPKKPYKYPSPPPPVYK--YKSPPPPVYSPPPVYKYKS 257
Query: 87 LQFYPHHRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPPLDYDR 146
+ P + P P+ P P Y ++P P + PP +
Sbjct: 258 PPPPVYSPPPPYKYKSPPPPPYK---YPSPPPPVYKYKSPP----PPVYKYKSPPPPPKK 416
Query: 147 EFNHHRPQQP-----PPPPPSYPP 165
+ + P P PPPP Y P
Sbjct: 417 PYKYPSPPPPVYKYKSPPPPVYSP 488
Score = 53.5 bits (127), Expect = 7e-07
Identities = 27/79 (34%), Positives = 43/79 (54%), Gaps = 11/79 (13%)
Frame = +3
Query: 15 HQHQQQHPQQHRTRYAPLGPQSHHHLSQPPPPL------PPP----HAPPPPPH-PPPLS 63
++++ P ++ + P P+ + PPPP+ PPP +PPPP H PPP
Sbjct: 609 YKYKSPPPPVYKYKSPPPPPKKPYKYPSPPPPVYKYKSPPPPVYKYKSPPPPVHSPPPPH 788
Query: 64 YRNIHTPPPPPLYNTPPVH 82
Y +++ PPPP+Y+ PP H
Sbjct: 789 Y--VYSSPPPPVYSPPPPH 839
Score = 52.4 bits (124), Expect = 1e-06
Identities = 39/134 (29%), Positives = 54/134 (40%), Gaps = 6/134 (4%)
Frame = +3
Query: 37 HHHLSQPPPPLPPPHAPPPPPH----PPPLSYRNIHTPPPPPLYNTPPVHPSRQLQFYPH 92
H + PPPP+ +PPPP + PPP Y+ PPP Y +PP P + + YP
Sbjct: 3 HINTPSPPPPVYKYKSPPPPVYKYKSPPPPVYKYKSPPPPVYKYKSPPPPPKKPYK-YP- 176
Query: 93 HRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRN--PEPAWNPSLDDLNYPPLDYDREFNH 150
P P + + P + P + P P ++P PP Y
Sbjct: 177 -------SPPPPVYKYKSPPPPVYSPPPVYKYKSPPPPVYSPP------PPYKY------ 299
Query: 151 HRPQQPPPPPPSYP 164
+ PPPPP YP
Sbjct: 300 ---KSPPPPPYKYP 332
Score = 42.7 bits (99), Expect = 0.001
Identities = 28/91 (30%), Positives = 39/91 (42%), Gaps = 12/91 (13%)
Frame = +3
Query: 31 PLGPQSHHHLSQPPPPLPPPHAPPPPPH--------PPPLSYRNIHTPPPPPLYN----T 78
P P + PPPP+ +PPPPP PPP+ Y+ + PPPP+Y
Sbjct: 588 PSPPPPVYKYKSPPPPVYKYKSPPPPPKKPYKYPSPPPPV-YK--YKSPPPPVYKYKSPP 758
Query: 79 PPVHPSRQLQFYPHHRSAPEERPVPTNHPFS 109
PPVH + P P P ++ +S
Sbjct: 759 PPVHSPPPPHYVYSSPPPPVYSPPPPHYIYS 851
Score = 41.2 bits (95), Expect(2) = 1e-04
Identities = 22/52 (42%), Positives = 30/52 (57%), Gaps = 8/52 (15%)
Frame = +3
Query: 34 PQSHHHLSQPPPPL---PPPH----APPPPPH-PPPLSYRNIHTPPPPPLYN 77
P + PPPP+ PPPH +PPPP + PPP Y I++ PPPP ++
Sbjct: 726 PPPVYKYKSPPPPVHSPPPPHYVYSSPPPPVYSPPPPHY--IYSSPPPPYHS 875
Score = 35.0 bits (79), Expect(2) = 0.012
Identities = 18/41 (43%), Positives = 22/41 (52%), Gaps = 7/41 (17%)
Frame = +3
Query: 34 PQSHHHLSQPPPPL---PPPH----APPPPPHPPPLSYRNI 67
P H+ S PPPP+ PPPH +PPPP H Y +I
Sbjct: 777 PPPHYVYSSPPPPVYSPPPPHYIYSSPPPPYHS*ITVYHSI 899
Score = 23.9 bits (50), Expect(2) = 1e-04
Identities = 7/17 (41%), Positives = 10/17 (58%)
Frame = +1
Query: 1 MDQHYPRDRHYHHHHQH 17
+ H+PR+ HHH H
Sbjct: 652 LHHHHPRNHTSTHHHHH 702
Score = 23.1 bits (48), Expect(2) = 0.012
Identities = 8/30 (26%), Positives = 10/30 (32%)
Frame = +1
Query: 12 HHHHQHQQQHPQQHRTRYAPLGPQSHHHLS 41
HHHH H + + P H S
Sbjct: 655 HHHHPRNHTSTHHHHHPFTNISPLLHQFTS 744
>AL368332 weakly similar to GP|21322711|e pherophorin-dz1 protein {Volvox
carteri f. nagariensis}, partial (10%)
Length = 478
Score = 63.9 bits (154), Expect = 5e-10
Identities = 47/135 (34%), Positives = 56/135 (40%), Gaps = 10/135 (7%)
Frame = +1
Query: 41 SQPPPPLPPPHAPPPPPH---PPPLSYRNIHTPPPP-PLYNTPPVHPSRQLQFYPHHRSA 96
+QP P PH PPPP H PPP + I+ PP P +N PP H + P H S
Sbjct: 31 NQPNFYFPFPHLPPPPSHPLNPPPSPFHPINPPPSPFHPFNPPPPHSIKSPPPAPSHSSP 210
Query: 97 P------EERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPPLDYDREFNH 150
P + P P +HPFS P +P P +P PP H
Sbjct: 211 PPPPPPRKXPPPPPSHPFS---------PPPPHNHPPP--SPHHPIAPSPP--------H 333
Query: 151 HRPQQPPPPPPSYPP 165
RP PPP PPS P
Sbjct: 334 VRPSPPPPLPPSPSP 378
Score = 51.6 bits (122), Expect = 2e-06
Identities = 29/78 (37%), Positives = 33/78 (42%), Gaps = 3/78 (3%)
Frame = +1
Query: 29 YAPLGPQSHHHLSQPPPPLPPPHAPPPPPHPPPLSYRNIHTPPPPPLYNTPPVHPSRQLQ 88
+ P P H + PPP P H+ PPPP PP PPPPP + P P
Sbjct: 139 FHPFNPPPPHSIKSPPP--APSHSSPPPPPPPR------KXPPPPPSHPFSPPPPHNHPP 294
Query: 89 FYPHH---RSAPEERPVP 103
PHH S P RP P
Sbjct: 295 PSPHHPIAPSPPHVRPSP 348
Score = 51.2 bits (121), Expect = 3e-06
Identities = 39/114 (34%), Positives = 46/114 (40%), Gaps = 12/114 (10%)
Frame = +1
Query: 32 LGPQSHHHLSQPPPPLPPPHAPP--------PPPH----PPPLSYRNIHTPPPPPLYNTP 79
L P H L+ PP P P + PP PPPH PPP + +PPPPP
Sbjct: 64 LPPPPSHPLNPPPSPFHPINPPPSPFHPFNPPPPHSIKSPPPAPSHS--SPPPPPPPRKX 237
Query: 80 PVHPSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPS 133
P P P H +P P P NHP P P + R +P P PS
Sbjct: 238 PPPP-------PSHPFSP---PPPHNHPPPSPHHPIAPSPPHVRPSPPPPLPPS 369
Score = 47.0 bits (110), Expect = 6e-05
Identities = 31/87 (35%), Positives = 37/87 (41%), Gaps = 19/87 (21%)
Frame = +1
Query: 17 HQQQHPQQHRTRYAPLGPQSHHHLSQPPPPLPPPHAPPPP-----------PHPPPLSYR 65
H P H + P P SH S PPPP PP PPPP HPPP +
Sbjct: 142 HPFNPPPPHSIKSPPPAP-SH---SSPPPPPPPRKXPPPPPSHPFSPPPPHNHPPPSPHH 309
Query: 66 -------NIHTPPPPPLYNTP-PVHPS 84
++ PPPPL +P P HP+
Sbjct: 310 PIAPSPPHVRPSPPPPLPPSPSPYHPT 390
>TC76445 similar to PIR|T07623|T07623 extensin homolog HRGP2 - soybean
(fragment), partial (76%)
Length = 821
Score = 62.8 bits (151), Expect = 1e-09
Identities = 47/143 (32%), Positives = 59/143 (40%), Gaps = 11/143 (7%)
Frame = +2
Query: 34 PQSHHHLSQPPPPLPPPHAP-----PPPPH---PPPLSYRNIHTP---PPPPLYNTPPVH 82
P ++ PPPP P P +P PPPP PPP Y++ P PPPP Y P
Sbjct: 8 PPPPYYYKSPPPPSPSPPSPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYKSPPP 187
Query: 83 PSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPPL 142
PS H++S P P P +HP P+Y + P P+ +P PP
Sbjct: 188 PSPSPPPPYHYQSPPP--PSPISHP-----------PNYYKSPPPPSPSPP------PPY 310
Query: 143 DYDREFNHHRPQQPPPPPPSYPP 165
Y PPPP S PP
Sbjct: 311 HY---------VSPPPPVKSPPP 352
Score = 54.7 bits (130), Expect = 3e-07
Identities = 44/131 (33%), Positives = 57/131 (42%), Gaps = 8/131 (6%)
Frame = +2
Query: 43 PPPPLPPPHAPPPPPHPPPLS---YRNIHTP---PPPPLY--NTPPVHPSRQLQFYPHHR 94
P PP P + PPPP P P S Y++ P PPPP Y + PP PS +Y ++
Sbjct: 2 PSPPPPYYYKSPPPPSPSPPSPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPYY--YK 175
Query: 95 SAPEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPPLDYDREFNHHRPQ 154
S P P P P Y ++P P PS +++PP Y +
Sbjct: 176 SPPPPSPSPP--------------PPYHYQSPPP---PS--PISHPPNYY---------K 271
Query: 155 QPPPPPPSYPP 165
PPPP PS PP
Sbjct: 272 SPPPPSPSPPP 304
Score = 50.4 bits (119), Expect = 6e-06
Identities = 29/67 (43%), Positives = 34/67 (50%), Gaps = 9/67 (13%)
Frame = +2
Query: 22 PQQHRTRYAPLGPQSH--HHLSQPPPPLPPPHAP-----PPPP--HPPPLSYRNIHTPPP 72
P H P P SH ++ PPPP P P P PPPP PPP +Y I+ PP
Sbjct: 206 PPYHYQSPPPPSPISHPPNYYKSPPPPSPSPPPPYHYVSPPPPVKSPPPPAY--IYASPP 379
Query: 73 PPLYNTP 79
PP+YN P
Sbjct: 380 PPIYN*P 400
Score = 47.4 bits (111), Expect(2) = 2e-07
Identities = 28/75 (37%), Positives = 36/75 (47%), Gaps = 11/75 (14%)
Frame = +2
Query: 34 PQSHHHLSQPPPPLPPPHAP-----PPPPHP-PPLSYRNIHTPPPPPLYNTPP---VHPS 84
P +H PPPP P H P PPPP P PP Y + PPPP+ + PP ++ S
Sbjct: 200 PPPPYHYQSPPPPSPISHPPNYYKSPPPPSPSPPPPYH--YVSPPPPVKSPPPPAYIYAS 373
Query: 85 RQLQFY--PHHRSAP 97
Y PH+ +P
Sbjct: 374 PPPPIYN*PHNTKSP 418
Score = 27.3 bits (59), Expect(2) = 2e-07
Identities = 14/37 (37%), Positives = 17/37 (45%)
Frame = +3
Query: 4 HYPRDRHYHHHHQHQQQHPQQHRTRYAPLGPQSHHHL 40
H + H+HHHH HP T + L P H HL
Sbjct: 66 HITTNLHHHHHH----PHPLPTITNHLHL-PHHHRHL 161
>BQ141029 similar to GP|8132441|gb| extensin {Pisum sativum}, partial (54%)
Length = 613
Score = 60.5 bits (145), Expect = 5e-09
Identities = 44/147 (29%), Positives = 62/147 (41%), Gaps = 17/147 (11%)
Frame = +2
Query: 36 SHHHLSQPPPPLP--------PPHAPPPPPH----PPPLSYRNIHTPPPPPLYNTPPVHP 83
S+ + S PPPP P P H+PPPP H PPP Y+ + PPPP+Y+ PPV+
Sbjct: 98 SYIYSSPPPPPKPYYYHSPPPPVHSPPPPYHYSSPPPPPVYK--YKSPPPPVYSPPPVYK 271
Query: 84 SRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPPLD 143
+ + P + P P+ P P Y ++P P + PP
Sbjct: 272 YKSPPPPVYSPPPPYKYKSPPPPPYK---YPSPPPPVYKYKSPP----PPVYKYKSPPPP 430
Query: 144 YDREFNHHRPQQP-----PPPPPSYPP 165
+ + + P P PPPP Y P
Sbjct: 431 PKKPYKYPSPPPPVYKYKSPPPPVYSP 511
Score = 45.4 bits (106), Expect = 2e-04
Identities = 24/70 (34%), Positives = 34/70 (48%), Gaps = 6/70 (8%)
Frame = +2
Query: 17 HQQQHPQQHRTRYAPLGPQSHHHLSQPPP------PLPPPHAPPPPPHPPPLSYRNIHTP 70
++ + P +Y P + + S PPP P PPP P P PPP Y+ +
Sbjct: 314 YKYKSPPPPPYKYPSPPPPVYKYKSPPPPVYKYKSPPPPPKKPYKYPSPPPPVYK--YKS 487
Query: 71 PPPPLYNTPP 80
PPPP+Y+ PP
Sbjct: 488 PPPPVYSPPP 517
Score = 44.3 bits (103), Expect = 4e-04
Identities = 20/66 (30%), Positives = 37/66 (55%), Gaps = 1/66 (1%)
Frame = +2
Query: 15 HQHQQQHPQQHRTRYAPLGPQSHHHLSQPPPPLPPPHAPPPPPH-PPPLSYRNIHTPPPP 73
++++ P ++ + P P+ + PPPP+ +PPPP + PPP Y+ + PPP
Sbjct: 374 YKYKSPPPPVYKYKSPPPPPKKPYKYPSPPPPVYKYKSPPPPVYSPPPPVYK--YKSPPP 547
Query: 74 PLYNTP 79
P+++ P
Sbjct: 548 PVHSPP 565
Score = 38.5 bits (88), Expect = 0.022
Identities = 23/70 (32%), Positives = 32/70 (44%), Gaps = 8/70 (11%)
Frame = +2
Query: 15 HQHQQQHPQQHRTRYAPLGPQSHHHLSQPPPPL---PPP----HAPPPPPHPPPLSY-RN 66
++++ P + P P + PPPP+ PPP +PPPP H PP +
Sbjct: 404 YKYKSPPPPPKKPYKYPSPPPPVYKYKSPPPPVYSPPPPVYKYKSPPPPVHSPPSTI*VQ 583
Query: 67 IHTPPPPPLY 76
I PPPP Y
Sbjct: 584 ILPPPPPYKY 613
Score = 34.3 bits (77), Expect = 0.41
Identities = 23/68 (33%), Positives = 28/68 (40%)
Frame = +3
Query: 1 MDQHYPRDRHYHHHHQHQQQHPQQHRTRYAPLGPQSHHHLSQPPPPLPPPHAPPPPPHPP 60
+ H+PR+ HHH HQ T +PL HHL Q H P HPP
Sbjct: 417 LHHHHPRNHTSIHHHHHQ-------FTNISPLLHLYTHHLHQFTSISLLLH--PSIHHPP 569
Query: 61 PLSYRNIH 68
P Y+ H
Sbjct: 570 PYKYKFSH 593
>BF635948 similar to GP|21622312|emb putative protein {Neurospora crassa},
partial (6%)
Length = 538
Score = 60.5 bits (145), Expect = 5e-09
Identities = 28/59 (47%), Positives = 35/59 (58%), Gaps = 1/59 (1%)
Frame = +1
Query: 2 DQHYPRDRHYHH-HHQHQQQHPQQHRTRYAPLGPQSHHHLSQPPPPLPPPHAPPPPPHP 59
+ +YP D H+H HQHQQQH QQ++T + P H +S P PPL PP P P P P
Sbjct: 277 NNNYPYDHHHHQLQHQHQQQHHQQNQTNF----PIQHRSISFPTPPLQPPQQPQPIPPP 441
Score = 36.6 bits (83), Expect = 0.083
Identities = 38/133 (28%), Positives = 48/133 (35%), Gaps = 12/133 (9%)
Frame = +1
Query: 44 PPPLPPP---HAPPPPPHPPPLSYRNIHT--------PPPPPLYNTPPVHPSRQLQFYPH 92
PPP P P H PP+P P NI + PPPP ++ P + Y H
Sbjct: 121 PPPSPTPFDMHTFFNPPNPNPNPNPNISSQFPSSAPSPPPPSSSSSYPFPHNNNNYPYDH 300
Query: 93 HRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPPLDYDREFNHHR 152
H + + +H + + N P R P PPL
Sbjct: 301 HHHQLQHQHQQQHHQQN-----QTNFPIQHRSISFPT----------PPL--------QP 411
Query: 153 PQQPPP-PPPSYP 164
PQQP P PPPS P
Sbjct: 412 PQQPQPIPPPSNP 450
>BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melanogaster},
partial (26%)
Length = 308
Score = 60.1 bits (144), Expect = 7e-09
Identities = 44/123 (35%), Positives = 44/123 (35%)
Frame = -1
Query: 43 PPPPLPPPHAPPPPPHPPPLSYRNIHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPEERPV 102
PPPP PPP PPPPP PPP PPPPP PP P P P P
Sbjct: 305 PPPPPPPPPPPPPPPPPPP--------PPPPPPPPPPPPPP-------PPPPPPPPPPPP 171
Query: 103 PTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPPLDYDREFNHHRPQQPPPPPPS 162
P P P P P P P PP P PPPPPP
Sbjct: 170 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPP-------PP-----------PPPPPPPPPP 45
Query: 163 YPP 165
PP
Sbjct: 44 PPP 36
Score = 60.1 bits (144), Expect = 7e-09
Identities = 44/123 (35%), Positives = 44/123 (35%)
Frame = -3
Query: 43 PPPPLPPPHAPPPPPHPPPLSYRNIHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPEERPV 102
PPPP PPP PPPPP PPP PPPPP PP P P P P
Sbjct: 306 PPPPPPPPPPPPPPPPPPP--------PPPPPPPPPPPPPP-------PPPPPPPPPPPP 172
Query: 103 PTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPPLDYDREFNHHRPQQPPPPPPS 162
P P P P P P P PP P PPPPPP
Sbjct: 171 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPP-------PP-----------PPPPPPPPPP 46
Query: 163 YPP 165
PP
Sbjct: 45 PPP 37
Score = 59.7 bits (143), Expect = 9e-09
Identities = 43/123 (34%), Positives = 44/123 (34%)
Frame = -3
Query: 43 PPPPLPPPHAPPPPPHPPPLSYRNIHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPEERPV 102
PPPP PPP PPPPP PPP PPPPP PP P P P P
Sbjct: 300 PPPPPPPPPPPPPPPPPPP-------PPPPPPPPPPPPPPPP------PPPPPPPPPPPP 160
Query: 103 PTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPPLDYDREFNHHRPQQPPPPPPS 162
P P P P P P P PP P PPPPPP
Sbjct: 159 PPPPP------PPPPPPPPPPPPPPPPPPPPPPPPPPPP-----------PPPPPPPPPG 31
Query: 163 YPP 165
+ P
Sbjct: 30 FSP 22
Score = 58.2 bits (139), Expect = 3e-08
Identities = 44/124 (35%), Positives = 44/124 (35%)
Frame = -2
Query: 43 PPPPLPPPHAPPPPPHPPPLSYRNIHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPEERPV 102
PPPP PPP PPPPP PPP PPPPP PP P P P P
Sbjct: 301 PPPPPPPPPPPPPPPPPPP--------PPPPPPPPPPPPPP-------PPPPPPPPPPPP 167
Query: 103 PTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPPLDYDREFNHHRPQQPPPPPPS 162
P P P P P P P PP P PPPPPP
Sbjct: 166 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPP-------PP-----------PPPPPPPPPP 41
Query: 163 YPPV 166
P V
Sbjct: 40 PPGV 29
Score = 52.0 bits (123), Expect = 2e-06
Identities = 23/43 (53%), Positives = 24/43 (55%)
Frame = -2
Query: 43 PPPPLPPPHAPPPPPHPPPLSYRNIHTPPPPPLYNTPPVHPSR 85
PPPP PPP PPPPP PPP PPPPP P V P +
Sbjct: 145 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPGVFPGK 17
>TC81266 weakly similar to GP|16415994|emb|CAB88653. hypothetical protein
{Neurospora crassa}, partial (2%)
Length = 644
Score = 58.9 bits (141), Expect = 2e-08
Identities = 36/102 (35%), Positives = 45/102 (43%), Gaps = 4/102 (3%)
Frame = +3
Query: 6 PRDRHYHHHHQHQQQHPQQHRTRYAPLGPQSHHHLSQPPPPLPP----PHAPPPPPHPPP 61
P HHQHQ QH HH+ PPP PP P+ PPPPP PP
Sbjct: 405 PPQNSNFQHHQHQVQH----------------HHVPHAPPPPPPSNYYPYQPPPPPPPPD 536
Query: 62 LSYRNIHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPEERPVP 103
SY+ PPPPP +PS Q+ +H+ ++ P P
Sbjct: 537 NSYQ---PPPPPP---GSMYYPSNNNQY--NHQQQQQQPPPP 638
Score = 49.7 bits (117), Expect = 9e-06
Identities = 40/126 (31%), Positives = 52/126 (40%), Gaps = 6/126 (4%)
Frame = +3
Query: 43 PPPPLPPPHAPPPPPHPPPLS------YRNIHTPPPPPLYNTPPVHPSRQLQFYPHHRSA 96
PPP P APPPPP S Y + PP N+ H Q+Q + H A
Sbjct: 303 PPPASQNPIAPPPPPPQQRPSNWGGYGYSGVVEIPP---QNSNFQHHQHQVQHH-HVPHA 470
Query: 97 PEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPPLDYDREFNHHRPQQP 156
P P +P+ P P + P P P + YP + ++NH + QQ
Sbjct: 471 PPPPPPSNYYPYQP---PPPPPPPDNSYQPPP---PPPGSMYYP--SNNNQYNHQQQQQQ 626
Query: 157 PPPPPS 162
PPPPPS
Sbjct: 627 PPPPPS 644
Score = 35.0 bits (79), Expect = 0.24
Identities = 33/123 (26%), Positives = 46/123 (36%), Gaps = 4/123 (3%)
Frame = +3
Query: 57 PHPPPLSYRNIHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRR 116
P PPP S I PPPPP PS + + E P +N +
Sbjct: 297 PFPPPASQNPIAPPPPPPQQR-----PSNWGGY--GYSGVVEIPPQNSNFQHHQHQVQHH 455
Query: 117 NIPDYDRRNPEPAWNPSLDDLNYPPLDYDREFNHHRPQQPPPPPPSY----PPVDSLRYD 172
++P P P PP +Y + + P PPPP SY PP S+ Y
Sbjct: 456 HVP----HAPPPP----------PPSNY---YPYQPPPPPPPPDNSYQPPPPPPGSMYYP 584
Query: 173 ASD 175
+++
Sbjct: 585 SNN 593
>TC76431 similar to GP|8132441|gb|AAF73291.1| extensin {Pisum sativum},
partial (30%)
Length = 890
Score = 58.5 bits (140), Expect = 2e-08
Identities = 44/153 (28%), Positives = 57/153 (36%)
Frame = +1
Query: 11 YHHHHQHQQQHPQQHRTRYAPLGPQSHHHLSQPPPPLPPPHAPPPPPHPPPLSYRNIHTP 70
Y H H H HP + ++P P S + P P PP H P PHP H+P
Sbjct: 43 YPHPHPHPHPHPHPYPVHHSP--PPSPKKPYKYPSPPPPTHHHYPHPHPV------YHSP 198
Query: 71 PPPPLYNTPPVHPSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAW 130
PPP + P P + P++ S P P P+ P P Y P+P +
Sbjct: 199 PPPKYKYSSPPPPVHHVSPKPYYHSPP-----PPKKPYKYSSPPPPVHPPY---TPKPVY 354
Query: 131 NPSLDDLNYPPLDYDREFNHHRPQQPPPPPPSY 163
+ Y P H PPPP SY
Sbjct: 355 HSPPVHPPYTP--------HPVYHSPPPPVHSY 429
Score = 57.4 bits (137), Expect = 5e-08
Identities = 35/114 (30%), Positives = 47/114 (40%), Gaps = 34/114 (29%)
Frame = +1
Query: 4 HYPRDRHYHHHHQHQQQHPQQHRT--------RYAPLGPQSHHHLSQPPP----PLPPPH 51
++P + H H H +P H +Y P +HHH P P P PP +
Sbjct: 34 YHPYPHPHPHPHPHPHPYPVHHSPPPSPKKPYKYPSPPPPTHHHYPHPHPVYHSPPPPKY 213
Query: 52 ---APPPPPH-----------PPPLSYRNIHTPPPP--------PLYNTPPVHP 83
+PPPP H PPP +PPPP P+Y++PPVHP
Sbjct: 214 KYSSPPPPVHHVSPKPYYHSPPPPKKPYKYSSPPPPVHPPYTPKPVYHSPPVHP 375
Score = 55.1 bits (131), Expect = 2e-07
Identities = 47/142 (33%), Positives = 56/142 (39%), Gaps = 11/142 (7%)
Frame = +1
Query: 41 SQPPPPLPPPHAPPPPPHPPPLSYRNIHTPPPPPL--YNTPPVHPSRQLQFYPHHRSAPE 98
S PPPP P P P PHP P Y H+PPP P Y P P YPH
Sbjct: 16 SSPPPPYHPYPHPHPHPHPHPHPYPVHHSPPPSPKKPYKYPSPPPPTH-HHYPHPHPV-Y 189
Query: 99 ERPVPTNHPFSDDLLPRRNI-PDYDRRNPEPAWNPSLDDLNYPPL--DYDREFNHHRPQQ 155
P P + +S P ++ P +P P P PP+ Y + +H P
Sbjct: 190 HSPPPPKYKYSSPPPPVHHVSPKPYYHSPPPPKKPYKYSSPPPPVHPPYTPKPVYHSPPV 369
Query: 156 PPP--PPPSY----PPVDSLRY 171
PP P P Y PPV S Y
Sbjct: 370 HPPYTPHPVYHSPPPPVHSYIY 435
>TC87201 similar to PIR|F86388|F86388 hypothetical protein AAG29221.1
[imported] - Arabidopsis thaliana, partial (41%)
Length = 991
Score = 58.5 bits (140), Expect = 2e-08
Identities = 48/147 (32%), Positives = 63/147 (42%), Gaps = 12/147 (8%)
Frame = +1
Query: 31 PLGPQSHHHLSQPPPPL------PPPH---APPPPPHPPPLSYRNIHTPPPPPLYNTPPV 81
P P + + S PPPP PPP+ +PPPP PPP Y++ PPPP +Y PP
Sbjct: 136 PSPPPPYIYKSPPPPPYVYKSPPPPPYVYPSPPPPSPPPPYIYKS--PPPPPYVYTPPPY 309
Query: 82 HPSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPP 141
++S P P P P+ P P Y ++P P P + PP
Sbjct: 310 ----------VYKSPPPPSPSPP-PPYVYKSPPP---PPYVYKSPPP---PPYVYESPPP 438
Query: 142 LDYDREFNHHRP---QQPPPPPPSYPP 165
Y + P + PPPP PS PP
Sbjct: 439 PQYVYKSPPPPPYVYESPPPPSPSPPP 519
Score = 56.2 bits (134), Expect = 1e-07
Identities = 48/153 (31%), Positives = 58/153 (37%), Gaps = 23/153 (15%)
Frame = +1
Query: 36 SHHHLS-----QPPPPLPPPH---------------APPPPPHPPPLSYRNIHTPPPPPL 75
SHH L PPPP PPP +PPPP PPP Y++ PPPP +
Sbjct: 16 SHHLLRPYVYPSPPPPSPPPPIRIQITTRAPPYVYPSPPPPSPPPPYIYKS--PPPPPYV 189
Query: 76 YNTPPVHPSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRNPEP---AWNP 132
Y +PP P YP P P + P P Y ++P P + P
Sbjct: 190 YKSPPPPP----YVYP--------SPPPPSPP-----------PPYIYKSPPPPPYVYTP 300
Query: 133 SLDDLNYPPLDYDREFNHHRPQQPPPPPPSYPP 165
PP Y + PPPP PS PP
Sbjct: 301 -------PPYVY---------KSPPPPSPSPPP 351
Score = 55.5 bits (132), Expect = 2e-07
Identities = 48/157 (30%), Positives = 63/157 (39%), Gaps = 24/157 (15%)
Frame = +1
Query: 31 PLGPQSHHHLSQPPPPL------------PPPHAPPPPPH----PPPLSYRNIHTPPPPP 74
P P + + S PPPP PPP PPPP+ PPP Y PPPP
Sbjct: 238 PSPPPPYIYKSPPPPPYVYTPPPYVYKSPPPPSPSPPPPYVYKSPPPPPYVYKSPPPPPY 417
Query: 75 LYNTPPVHPSRQLQFYPH-----HRSAPEERPVPTNHPFSDDLLPR--RNIPDYDRRNPE 127
+Y +PP P + + P + S P P P + + P P Y ++P
Sbjct: 418 VYESPP--PPQYVYKSPPPPPYVYESPPPPSPSPPPPYYYKPIPPPYVYESPPYIYKSPP 591
Query: 128 PAWNPSLDDLNYPPLDYDREFNHHRP-QQPPPPPPSY 163
PA P + PP Y ++P Q PPPP Y
Sbjct: 592 PA--PYV--YKSPPYVYKSPPTPYKPYQYSSPPPPVY 690
Score = 42.7 bits (99), Expect = 0.001
Identities = 27/87 (31%), Positives = 42/87 (48%), Gaps = 6/87 (6%)
Frame = +1
Query: 17 HQQQHPQQHRTRYAPLGPQSHHHLSQPPPPLPPP--HAPPPPPH---PPPLSYRNIHTPP 71
++ P Q+ + P P + P P PPP + P PPP+ PP Y++ PP
Sbjct: 421 YESPPPPQYVYKSPPPPPYVYESPPPPSPSPPPPYYYKPIPPPYVYESPPYIYKS--PPP 594
Query: 72 PPPLYNTPP-VHPSRQLQFYPHHRSAP 97
P +Y +PP V+ S + P+ S+P
Sbjct: 595 APYVYKSPPYVYKSPPTPYKPYQYSSP 675
>TC76311 homologue to GP|3204132|emb|CAA07235.1 extensin {Cicer arietinum},
partial (77%)
Length = 973
Score = 58.2 bits (139), Expect = 3e-08
Identities = 46/137 (33%), Positives = 58/137 (41%), Gaps = 14/137 (10%)
Frame = +2
Query: 43 PPPPLPPP----HAPPPP-PHPPPLSYRNIHTP----PPPPLY--NTPPVHPSRQLQFYP 91
PP P PPP H+PPPP P PPP Y P PPPP Y + PP PS +Y
Sbjct: 2 PPSPSPPPPYYYHSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPYY- 178
Query: 92 HHRSAPEERPVP-TNHPFSDDLLPRRNIPD--YDRRNPEPAWNPSLDDLNYPPLDYDREF 148
++S P P P T + + P + P + P P +P PP
Sbjct: 179 -YQSPPPPSPTPHTPYHYKSPPPPTASPPPPYHYVSPPPPTSSPPPYHYTSPPPPSPAPA 355
Query: 149 NHHRPQQPPPPPPSYPP 165
+ + PPPP S PP
Sbjct: 356 PTYIYKSPPPPMKSPPP 406
Score = 57.8 bits (138), Expect = 3e-08
Identities = 44/141 (31%), Positives = 55/141 (38%), Gaps = 14/141 (9%)
Frame = +2
Query: 34 PQSHHHLSQPPPPLPPP-----HAPPPPPHPPPLSYRNIHTP----PPPPLY--NTPPVH 82
P ++H PP P PPP PPP P PPP Y P PPPP Y + PP
Sbjct: 23 PPYYYHSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYQSPPPPS 202
Query: 83 PSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRN---PEPAWNPSLDDLNY 139
P+ ++ P P P H S P + P Y + P PA P+ +
Sbjct: 203 PTPHTPYHYKSPPPPTASPPPPYHYVSPP-PPTSSPPPYHYTSPPPPSPAPAPTYIYKSP 379
Query: 140 PPLDYDREFNHHRPQQPPPPP 160
PP P + PPPP
Sbjct: 380 PP-----------PMKSPPPP 409
Score = 43.1 bits (100), Expect = 9e-04
Identities = 22/53 (41%), Positives = 27/53 (50%), Gaps = 7/53 (13%)
Frame = +2
Query: 31 PLGPQSHHHLSQPPPPLPPP-----HAPPPPP--HPPPLSYRNIHTPPPPPLY 76
P +H + PPPP P P + PPPP PPP Y I+ PPPP+Y
Sbjct: 293 PTSSPPPYHYTSPPPPSPAPAPTYIYKSPPPPMKSPPPPVY--IYASPPPPIY 445
>TC76326 homologue to PIR|C29356|C29356 hydroxyproline-rich glycoprotein
(clone Hyp2.13) - kidney bean (fragment), partial (32%)
Length = 777
Score = 58.2 bits (139), Expect = 3e-08
Identities = 45/132 (34%), Positives = 51/132 (38%), Gaps = 9/132 (6%)
Frame = +1
Query: 43 PPPPLPPP-----HAPPPPPHPPPLSYRNIHTP----PPPPLYNTPPVHPSRQLQFYPHH 93
PP P PPP PPP P PPP Y P PPPP Y P PS H+
Sbjct: 16 PPSPSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPYHY 195
Query: 94 RSAPEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPPLDYDREFNHHRP 153
+S P P P +HP P Y + P P+ +P PP Y
Sbjct: 196 QSPPP--PSPISHP-----------PYYYKSPPPPSPSPP------PPYHY--------- 291
Query: 154 QQPPPPPPSYPP 165
PPPP S PP
Sbjct: 292 VSPPPPVKSPPP 327
Score = 54.7 bits (130), Expect = 3e-07
Identities = 43/126 (34%), Positives = 55/126 (43%), Gaps = 5/126 (3%)
Frame = +1
Query: 45 PPLPPPHAPPPPPH----PPPLSYRNIHTPPPPPLYNT-PPVHPSRQLQFYPHHRSAPEE 99
P PPP PPPP+ PPP S +PPPP Y + PP PS +Y ++S P
Sbjct: 4 PGPPPPSPSPPPPYYYKSPPPPS----PSPPPPYYYKSPPPPSPSPPPPYY--YKSPPPP 165
Query: 100 RPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPPLDYDREFNHHRPQQPPPP 159
P P P Y ++P P PS +++PP Y + PPPP
Sbjct: 166 SPSPP--------------PPYHYQSPPP---PS--PISHPPYYY---------KSPPPP 261
Query: 160 PPSYPP 165
PS PP
Sbjct: 262 SPSPPP 279
Score = 48.1 bits (113), Expect = 3e-05
Identities = 33/100 (33%), Positives = 45/100 (45%), Gaps = 12/100 (12%)
Frame = +1
Query: 30 APLGPQSHHHLSQPPP-PLPPP---HAPPPPPHP-PPLSYRNIHTPPP-----PPLY--N 77
+P P +++ S PPP P PPP + PPPP P PP Y PPP PP Y +
Sbjct: 70 SPSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPYHYQSPPPPSPISHPPYYYKS 249
Query: 78 TPPVHPSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRN 117
PP PS ++ P + P P + ++ P N
Sbjct: 250 PPPPSPSPPPPYHYVSPPPPVKSPPPPTYIYASPPPPHYN 369
Score = 45.8 bits (107), Expect = 1e-04
Identities = 28/65 (43%), Positives = 34/65 (52%), Gaps = 9/65 (13%)
Frame = +1
Query: 22 PQQHRTRYAPLGPQSH--HHLSQPPPPLP---PPH---APPPP-PHPPPLSYRNIHTPPP 72
P H P P SH ++ PPPP P PP+ +PPPP PPP +Y I+ PP
Sbjct: 181 PPYHYQSPPPPSPISHPPYYYKSPPPPSPSPPPPYHYVSPPPPVKSPPPPTY--IYASPP 354
Query: 73 PPLYN 77
PP YN
Sbjct: 355 PPHYN 369
>BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (39%)
Length = 1358
Score = 57.8 bits (138), Expect = 3e-08
Identities = 54/166 (32%), Positives = 56/166 (33%), Gaps = 17/166 (10%)
Frame = -2
Query: 17 HQQQHPQQHRTRYAPLGPQSHHHLSQP-PPPLPPPHAPP-PPPHPPPLSYRNIHTPPPPP 74
H H R P P H +P PPP PPP AP PP PP PP PP
Sbjct: 577 HTPAHLAPPRQSPRPRPPPEPLHNPRPRPPPRPPPAAPARAPPSGPP-------APPTPP 419
Query: 75 LYNTPPVHPS----RQLQFYPHHRSAPEERPVP---TNHPFSDDLLPRRNIPDYDRRNPE 127
PPV S Q Q P R + PVP HP P DR P
Sbjct: 418 ATRAPPVRASPWCPGQGQETPQGRHTGVQSPVPCPALPHPPGPP-------PPLDRAPPP 260
Query: 128 P--------AWNPSLDDLNYPPLDYDREFNHHRPQQPPPPPPSYPP 165
P P PP RP PPPPPP PP
Sbjct: 259 PPAPLPLASRLRPRASPPPPPPPPGPAASAPPRPPPPPPPPPQRPP 122
Score = 52.4 bits (124), Expect = 1e-06
Identities = 49/155 (31%), Positives = 56/155 (35%), Gaps = 31/155 (20%)
Frame = -1
Query: 42 QPPPPLPPPHAPP---------PPPHPPPLSYRNIHTPPPPPLYNTPPVHPSRQLQ---F 89
+PPPP+PPP +PP PPP PPP R P PP P HP Q
Sbjct: 560 RPPPPVPPPPSPPRAPPQSEAAPPPPPPPRGARP-RAPLRPPSAPDPAGHPRAPRQGVAV 384
Query: 90 YPHHRS---------APEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPS------- 133
P RS PE RP+P P P R P P PA P+
Sbjct: 383 VPGPRSGNSPRAAHRGPEPRPLPRPPP------PTRP-PAAPGPGPPPAPGPTPLSKPPA 225
Query: 134 ---LDDLNYPPLDYDREFNHHRPQQPPPPPPSYPP 165
+ PP R P PP PP + PP
Sbjct: 224 AEGIPPATAPPPRARRLRPPSAPAPPPTPPAAAPP 120
Score = 51.2 bits (121), Expect = 3e-06
Identities = 44/137 (32%), Positives = 47/137 (34%), Gaps = 13/137 (9%)
Frame = -1
Query: 43 PPPPLPPPHAPPP--PPHPPPLSYRNIHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPEER 100
P PP PPP PPP PP PP S PPPPP P P R P
Sbjct: 572 PRPPRPPPPVPPPPSPPRAPPQS--EAAPPPPPPPRGARPRAPLR-----------PPSA 432
Query: 101 PVPTNHPFSDDLLPRR-----------NIPDYDRRNPEPAWNPSLDDLNYPPLDYDREFN 149
P P HP + PR+ N P R PEP P PP
Sbjct: 431 PDPAGHPRA----PRQGVAVVPGPRSGNSPRAAHRGPEPRPLPRPPPPTRPPA------- 285
Query: 150 HHRPQQPPPPPPSYPPV 166
P PPP P P+
Sbjct: 284 --APGPGPPPAPGPTPL 240
Score = 50.8 bits (120), Expect = 4e-06
Identities = 24/59 (40%), Positives = 28/59 (46%), Gaps = 8/59 (13%)
Frame = -3
Query: 33 GPQSHHHLSQPPPPLPPPHAPPPPPHPP--------PLSYRNIHTPPPPPLYNTPPVHP 83
G H P PP PPP P PPPHPP P + ++ PPPPP + PP P
Sbjct: 219 GHPPRHRPPPPGPPPPPPLGPRPPPHPPRSGPPRPHPPTRTSLSPPPPPPAASPPPPAP 43
Score = 50.4 bits (119), Expect = 6e-06
Identities = 48/147 (32%), Positives = 59/147 (39%), Gaps = 3/147 (2%)
Frame = -2
Query: 19 QQHPQ-QHRTRYAPLGPQSHHHLSQPPPPLPPPHAPPPPPHPPPLS--YRNIHTPPPPPL 75
Q+ PQ +H +P+ + H PPPPL APPPPP P PL+ R +PPPPP
Sbjct: 367 QETPQGRHTGVQSPVPCPALPHPPGPPPPLD--RAPPPPPAPLPLASRLRPRASPPPPP- 197
Query: 76 YNTPPVHPSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLD 135
PP P+ SAP P P P P+R P P P +
Sbjct: 196 ---PPPGPA---------ASAPPRPPPPPPPP------PQRPPP------PPPPHTHQPE 89
Query: 136 DLNYPPLDYDREFNHHRPQQPPPPPPS 162
PP +P P PPPS
Sbjct: 88 PPTSPP--------RRQPPAPRAPPPS 32
Score = 50.1 bits (118), Expect = 7e-06
Identities = 41/144 (28%), Positives = 53/144 (36%)
Frame = -3
Query: 19 QQHPQQHRTRYAPLGPQSHHHLSQPPPPLPPPHAPPPPPHPPPLSYRNIHTPPPPPLYNT 78
Q+HP + R + S PPPP PPP + PP P N P
Sbjct: 768 QRHPPRRTLRASAAQHPSRAPRRHPPPPPPPPPSSPPGMGTRPSCQENWWAAP------- 610
Query: 79 PPVHPSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLN 138
P PSR ++ P P P P + P +P ++ R P PA P
Sbjct: 609 PGARPSRPSRYIP-----PPTSPPPASPPAP---VPPQSPSTIRGRAPPPAPPPRRPPAR 454
Query: 139 YPPLDYDREFNHHRPQQPPPPPPS 162
PP RP++PP PPS
Sbjct: 453 PPPAP-----QRPRPRRPPARPPS 397
Score = 49.7 bits (117), Expect = 9e-06
Identities = 31/89 (34%), Positives = 35/89 (38%)
Frame = -2
Query: 31 PLGPQSHHHLSQPPPPLPPPHAPPPPPHPPPLSYRNIHTPPPPPLYNTPPVHPSRQLQFY 90
PL + S PPPP PP A PP PPP PPPPP PP P Q
Sbjct: 244 PLASRLRPRASPPPPPPPPGPAASAPPRPPP--------PPPPPPQRPPPPPPPHTHQPE 89
Query: 91 PHHRSAPEERPVPTNHPFSDDLLPRRNIP 119
P + P P P S L ++P
Sbjct: 88 PPTSPPRRQPPAPRAPPPSGI*LIAPSVP 2
Score = 47.4 bits (111), Expect = 5e-05
Identities = 42/133 (31%), Positives = 49/133 (36%), Gaps = 16/133 (12%)
Frame = -2
Query: 48 PPPHAPPPPPHPPPLSYRNI----HTPPPPPLYNTPPVHP------------SRQLQFYP 91
P PH P PPP P R TPPPPP ++PP P R+L P
Sbjct: 784 PSPHRPAPPPQAHPARERRTASLARTPPPPP--SSPPPAPLLPPRNGNASLLPRKLVGSP 611
Query: 92 HHRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPPLDYDREFNHH 151
RS+ + T L P R P R PEP NP
Sbjct: 610 ARRSSLSSESLHT----PAHLAPPRQSP-RPRPPPEPLHNP------------------- 503
Query: 152 RPQQPPPPPPSYP 164
RP+ PP PPP+ P
Sbjct: 502 RPRPPPRPPPAAP 464
Score = 47.0 bits (110), Expect = 6e-05
Identities = 59/205 (28%), Positives = 72/205 (34%), Gaps = 52/205 (25%)
Frame = -2
Query: 22 PQQHRTRYAPLGPQSHH-------HLSQPPPPLPPPHAPPPPPHPPP------------- 61
P HR AP PQ+H L++ PPP PP +PPP P PP
Sbjct: 784 PSPHRP--AP-PPQAHPARERRTASLARTPPP--PPSSPPPAPLLPPRNGNASLLPRKLV 620
Query: 62 --------LSYRNIHT--------------PPPPPLYNTPPVHPSRQLQFYPHHRSAPEE 99
LS ++HT PPP PL+N P P R P R+ P
Sbjct: 619 GSPARRSSLSSESLHTPAHLAPPRQSPRPRPPPEPLHNPRPRPPPRPPPAAP-ARAPPSG 443
Query: 100 RPVPTNHPFSDDLLPRRN--IPDYDRRNPE--------PAWNPSLDDLNYPPLDYDREFN 149
P P P + R + P + P+ P P+L PP DR
Sbjct: 442 PPAPPTPPATRAPPVRASPWCPGQGQETPQGRHTGVQSPVPCPALPHPPGPPPPLDR--- 272
Query: 150 HHRPQQPPPPPPSYPPVDSLRYDAS 174
PPPPP P LR AS
Sbjct: 271 -----APPPPPAPLPLASRLRPRAS 212
Score = 45.4 bits (106), Expect = 2e-04
Identities = 49/179 (27%), Positives = 59/179 (32%), Gaps = 42/179 (23%)
Frame = -3
Query: 30 APLGPQSHHHL-SQPPPPLPPPHAPPPPPHPPPLSYRNIHTPPPPPLYN----------- 77
AP+ PQS + + PPP PPP PP P P P R P PP
Sbjct: 537 APVPPQSPSTIRGRAPPPAPPPRRPPARPPPAPQRPRPRRPPARPPSGRRRGARAKVRKL 358
Query: 78 ----TP-----------PVHPSRQLQFY-------PH--------HRSAPEERPVPTNHP 107
TP P HP+ + + PH P RP P P
Sbjct: 357 PKGGTPGSRAPSPAPPSPTHPAPRRPWTGPPPRPRPHSP*QAACGRGHPPRHRPPPPGPP 178
Query: 108 FSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPPLDYDREFNHHRPQQPPPPPPSYPPV 166
L PR P R P P+ L+ PP P PPPP+ PP+
Sbjct: 177 PPPPLGPRPP-PHPPRSGPPRPHPPTRTSLSPPP----------PPPAASPPPPAPPPL 34
Score = 45.1 bits (105), Expect = 2e-04
Identities = 47/156 (30%), Positives = 56/156 (35%), Gaps = 19/156 (12%)
Frame = -3
Query: 30 APLGPQSHHHLSQPPPPLPPPHAPPPPPHPP--PLSYRNIHTPPPPPLYNTPPVHPSRQL 87
AP G + PPP PP A PP P PP P + R PPP P PP P
Sbjct: 615 APPGARPSRPSRYIPPPTSPPPASPPAPVPPQSPSTIRG-RAPPPAPPPRRPPARPPPAP 439
Query: 88 QFYPHHRSAPEERPVPTNHPFSDDL--LPRRNIPDYDRRNPEPAWNPSLDDLNYP---PL 142
Q P R P P + LP+ P +P P +P+ P P
Sbjct: 438 Q-RPRPRRPPARPPSGRRRGARAKVRKLPKGGTPGSRAPSPAPP-SPTHPAPRRPWTGPP 265
Query: 143 DYDREFN------------HHRPQQPPPPPPSYPPV 166
R + HRP PPP PP PP+
Sbjct: 264 PRPRPHSP*QAACGRGHPPRHRP--PPPGPPPPPPL 163
Score = 41.6 bits (96), Expect = 0.003
Identities = 23/50 (46%), Positives = 23/50 (46%)
Frame = -3
Query: 22 PQQHRTRYAPLGPQSHHHLSQPPPPLPPPHAPPPPPHPPPLSYRNIHTPP 71
P H R P P S PPP PP A PPPP PPPL N PP
Sbjct: 153 PPPHPPRSGPPRPHPPTRTSLSPPPPPPA-ASPPPPAPPPLLVYN**LPP 7
Score = 33.5 bits (75), Expect = 0.70
Identities = 50/189 (26%), Positives = 58/189 (30%), Gaps = 42/189 (22%)
Frame = -2
Query: 44 PPPLPPPHAPPPPPHPPPLSYRNIHTP--PPPPLYNTPPVHPSRQLQFYPHHRSAPEER- 100
P P PP HA P S R P P P +Y R+AP ER
Sbjct: 1135 PAPAPPAHAALRTRRSAPASRRRAPAPRRPAPSIYGG-----------LCAVRAAPSERA 989
Query: 101 ---PVPT-------------NHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPPLDY 144
P P+ P L+PR + + DR P P D PLD
Sbjct: 988 FSAPAPSGVCPVVSRSSTGARLPPRYSLVPRAHAREIDRA-PTFEHFPGPPDPASTPLDR 812
Query: 145 DREFNHHRPQQP-----------------------PPPPPSYPPVDSLRYDASDGANCRL 181
R RP P PPPPPS PP L N L
Sbjct: 811 ARACCARRPPSPHRPAPPPQAHPARERRTASLARTPPPPPSSPPPAPLL--PPRNGNASL 638
Query: 182 RTERLDGNP 190
+L G+P
Sbjct: 637 LPRKLVGSP 611
Score = 32.3 bits (72), Expect = 1.6
Identities = 34/124 (27%), Positives = 46/124 (36%)
Frame = -2
Query: 1361 PRTWHRTGNNPTTSLPRIKPSPGTAPPKRPILPRKANFQNTSTSYIRKGNNSLVRQPSPV 1420
P H T SL R P P ++PP P+LP R GN SL
Sbjct: 760 PPQAHPARERRTASLARTPPPPPSSPPPAPLLPP------------RNGNASL------- 638
Query: 1421 SALPQISSANQSPARSESRADVTDQSIYMKTGETYAPQHRQRTPSLPIDNKPKENISSPL 1480
LP+ SPAR R+ ++ +S+ + P H P P E + +P
Sbjct: 637 --LPR--KLVGSPAR---RSSLSSESL-------HTPAHLAPPRQSPRPRPPPEPLHNPR 500
Query: 1481 VDPP 1484
PP
Sbjct: 499 PRPP 488
Score = 32.0 bits (71), Expect = 2.0
Identities = 28/102 (27%), Positives = 34/102 (32%), Gaps = 11/102 (10%)
Frame = -1
Query: 17 HQQQHPQQHRTRYAPLGPQSHHHLSQPPPPLPPPHAPPP-----PP--HPPPLSYR---- 65
H+ P+ P P + PP P P P + PP PP PPP + R
Sbjct: 344 HRGPEPRPLPRPPPPTRPPAAPGPGPPPAPGPTPLSKPPAAEGIPPATAPPPRARRLRPP 165
Query: 66 NIHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPEERPVPTNHP 107
+ PPP P PP H P P P P
Sbjct: 164 SAPAPPPTPPAAAPPAPTPPHAPA*APHLPPPPPAPRPPRPP 39
Score = 31.2 bits (69), Expect = 3.5
Identities = 40/168 (23%), Positives = 55/168 (31%), Gaps = 46/168 (27%)
Frame = -1
Query: 44 PPPLPPPHAPPPP-------PHPPPLSYRNI-------HTPPPPPLYN------------ 77
P LP P + PP PH P ++ PPPPP +
Sbjct: 791 PSALPAPPSATPPGAPCARAPHSIPRAHPAATPLLPPPRPPPPPPEWERVPLAKKIGGQP 612
Query: 78 TPPVHPSRQLQFYPHHRSAPEERPVPTNHPFS---DDLLP---------RRNIPDYDRRN 125
P + P ++ YP P P P + P + + P R P
Sbjct: 611 RPALVPLVRVATYPRPPRPPPPVPPPPSPPRAPPQSEAAPPPPPPPRGARPRAPLRPPSA 432
Query: 126 PEPAWNPSLDDLNYPPLDYDREFN-----HHRPQQ---PPPPPPSYPP 165
P+PA +P + R N H P+ P PPPP+ PP
Sbjct: 431 PDPAGHPRAPRQGVAVVPGPRSGNSPRAAHRGPEPRPLPRPPPPTRPP 288
>BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (30%)
Length = 1338
Score = 57.4 bits (137), Expect = 5e-08
Identities = 46/143 (32%), Positives = 54/143 (37%), Gaps = 11/143 (7%)
Frame = -1
Query: 34 PQSHHH----LSQPPPPL--PPPHAP--PPPPHPPPLSYRNIHTPPPPPLYNTPPVHPSR 85
P HHH SQ P L P H P PPP PPP +PPPPP T P P R
Sbjct: 579 PAIHHHPNPPASQSPATLNRPTSHTPACPPPSRPPPHPRAPSLSPPPPPHPPTSPPRPGR 400
Query: 86 QLQFY---PHHRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYPPL 142
P HR + T+ +P +P P P+ P+L P
Sbjct: 399 PRSSTLAPPPHRQFHSPSALCTHCG-----IPPPAVPPPPPPTPPPSSPPALVSPPPRPF 235
Query: 143 DYDREFNHHRPQQPPPPPPSYPP 165
+ N P PPPPP PP
Sbjct: 234 LFPPPPNQPSPSPSPPPPPPLPP 166
Score = 55.8 bits (133), Expect = 1e-07
Identities = 47/162 (29%), Positives = 62/162 (38%), Gaps = 7/162 (4%)
Frame = -1
Query: 11 YHHHHQHQQQHPQQHRTRYAPLGPQSHHHLSQPPPPLPPPH------APPPPPHPPPLSY 64
+HH + Q P A L + H + PPP PPPH +PPPPPHPP
Sbjct: 570 HHHPNPPASQSP-------ATLNRPTSHTPACPPPSRPPPHPRAPSLSPPPPPHPPT--- 421
Query: 65 RNIHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRNIPDYDRR 124
+PP P + + P P HR + T+ +P +P
Sbjct: 420 ----SPPRPGRPRSSTLAP-------PPHRQFHSPSALCTHCG-----IPPPAVPPPPPP 289
Query: 125 NPEPAWNPSLDDLNYPPLDYDREFNHHRPQ-QPPPPPPSYPP 165
P P+ P+L P + N P PPPPPP PP
Sbjct: 288 TPPPSSPPALVSPPPRPFLFPPPPNQPSPSPSPPPPPPLPPP 163
Score = 50.4 bits (119), Expect = 6e-06
Identities = 47/148 (31%), Positives = 52/148 (34%), Gaps = 7/148 (4%)
Frame = -2
Query: 25 HRTRYAPLGPQSHHHLSQPPPPLPPPHAP-PPPPHP--PPLSYRNIHTPPPPPLYNTPPV 81
H L PQ H + PPP PP P P PPHP PP R P PP PP
Sbjct: 581 HPPSTTTLTPQ---HPNPPPP*TDPPATPLPAPPHPVRPPTHARLRSPPLRPPTLRPPPP 411
Query: 82 HPSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAW----NPSLDDL 137
P HR +P R + P +P P P P P L L
Sbjct: 410 APVAH-----GHRPSPPRRIGSSTPP--PPFVPIAGFPPPPSPPPPPPLPLHPPPQLSSL 252
Query: 138 NYPPLDYDREFNHHRPQQPPPPPPSYPP 165
P F PPPPPP +PP
Sbjct: 251 ---PPHAPSSFLLPLTSPPPPPPPPHPP 177
Score = 49.3 bits (116), Expect = 1e-05
Identities = 34/96 (35%), Positives = 40/96 (41%), Gaps = 10/96 (10%)
Frame = -1
Query: 22 PQQHRTRYAPLGPQSHHHLSQP---------PPPLPPPHAPPPPPHPPPLSYRNIHTPPP 72
P+ R R + L P H P PPP PP PPPP PPP S + +PPP
Sbjct: 414 PRPGRPRSSTLAPPPHRQFHSPSALCTHCGIPPPAVPP---PPPPTPPPSSPPALVSPPP 244
Query: 73 PP-LYNTPPVHPSRQLQFYPHHRSAPEERPVPTNHP 107
P L+ PP PS S P P+P P
Sbjct: 243 RPFLFPPPPNQPSPS-------PSPPPPPPLPPPPP 157
Score = 48.9 bits (115), Expect = 2e-05
Identities = 35/95 (36%), Positives = 39/95 (40%), Gaps = 15/95 (15%)
Frame = -2
Query: 28 RYAPLGPQSHHHLSQPPPPL-----PPPHAP----PPPPHPPPLSYRNIHTPPPPPLYNT 78
R P P +H H PP + PPP P PPPP PPP +H PPP L +
Sbjct: 425 RPPPPAPVAHGHRPSPPRRIGSSTPPPPFVPIAGFPPPPSPPPPPPLPLH--PPPQLSSL 252
Query: 79 PPVHPSRQLQFY------PHHRSAPEERPVPTNHP 107
PP PS L P P P P NHP
Sbjct: 251 PPHAPSSFLLPLTSPPPPPPPPHPPPSPPRPHNHP 147
Score = 46.2 bits (108), Expect = 1e-04
Identities = 29/82 (35%), Positives = 36/82 (43%), Gaps = 15/82 (18%)
Frame = -1
Query: 22 PQQHRTRYAPLGPQSHHHLSQP-----PPPLPPPHAPP----PPPHP------PPLSYRN 66
P HR ++P +H + P PPP PPP +PP PPP P P +
Sbjct: 378 PPPHRQFHSPSALCTHCGIPPPAVPPPPPPTPPPSSPPALVSPPPRPFLFPPPPNQPSPS 199
Query: 67 IHTPPPPPLYNTPPVHPSRQLQ 88
PPPPPL PP P + Q
Sbjct: 198 PSPPPPPPLPPPPPQPPQSRHQ 133
Score = 43.5 bits (101), Expect = 7e-04
Identities = 42/149 (28%), Positives = 55/149 (36%), Gaps = 14/149 (9%)
Frame = -1
Query: 28 RYAPLGPQSHHHLSQPPPPLPPPH--APPPPPHPPPLSYRNIHTPPPPPLYNTPPVHPSR 85
R+ P P + H ++ P L H AP P P PP + +H P P +T H
Sbjct: 1098 RHRPYPPLTSPHTARYSPSLSLSHYPAPRPVPRPPRAAVSILHLPLPTATTHTRHTHTPL 919
Query: 86 QLQFYPHHRSAP----EERPVPTNHPFSDDLLPR-RNIPDYDRRNPEPAWN-------PS 133
+ P H AP P+ P S D+ P R P R P+P P
Sbjct: 918 SPRSAPLHLLAPTFTTNTPPLIATRPRSTDITPTLRGQPPTPRPYPQPLRTRAHVGAYPR 739
Query: 134 LDDLNYPPLDYDREFNHHRPQQPPPPPPS 162
L D + L HR +PP PS
Sbjct: 738 LRDAAHAVL-----VKAHRSPRPPHVGPS 667
Score = 42.4 bits (98), Expect = 0.002
Identities = 51/170 (30%), Positives = 62/170 (36%), Gaps = 17/170 (10%)
Frame = -3
Query: 14 HHQHQQQHPQQHRTRYAPLGPQSHHHLSQPPPPLPPP--HAPPP-----------PPH-- 58
+ Q +HP P+ P+ QP P LPPP APPP PP
Sbjct: 595 NRQRPTRHPPPP*PPSIPI-PRHPEPTHQPHPCLPPPIPSAPPPTRAFALPPSAPPPSDL 419
Query: 59 -PPPLSYRNIHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRN 117
PPP S I PP P+ P L+ P R P P+ P S L P
Sbjct: 418 PPPPRSPTVIDPRPPAASAVPLPLRPLYPLRDSPPRRPPPPPPHSPSILPPSSRLSPPTP 239
Query: 118 IPDYDRRNPEPAWNPSLDDLNYPPLDYDREFNHHRPQQPPPPP-PSYPPV 166
+P +P PA L PP P PPPPP P+ P+
Sbjct: 238 LP--LSSSP*PAL-----PLPLPP-----------PTPPPPPPAPTTTPI 143
Score = 37.4 bits (85), Expect = 0.048
Identities = 38/110 (34%), Positives = 45/110 (40%), Gaps = 27/110 (24%)
Frame = -2
Query: 24 QHRTRYAPLGPQ------SHHHLSQPPP------PLPPPHA----PPPPPHPPPLSYRNI 67
Q RTR P P+ + H L P P PL P H P P P PLS R
Sbjct: 1148 QPRTRSLPYHPRPPPPLATDHTLLSPRPTPRATHPLSPSHTIPRLAPSPGLPAPLS-RYY 972
Query: 68 HTPPPPPLYN----TPPVHP-------SRQLQFYPHHRSAPEERPVPTNH 106
+ PPPL+ TPP P SR+L H S+P R +P H
Sbjct: 971 TSLSPPPLHTHDTLTPPSAPAPLPSISSRRLLRPTLHLSSPPARALPILH 822
Score = 36.2 bits (82), Expect = 0.11
Identities = 15/36 (41%), Positives = 19/36 (52%)
Frame = -1
Query: 32 LGPQSHHHLSQPPPPLPPPHAPPPPPHPPPLSYRNI 67
L P + S P P PPP PPPPP PP ++ +
Sbjct: 234 LFPPPPNQPSPSPSPPPPPPLPPPPPQPPQSRHQRL 127
Score = 35.8 bits (81), Expect = 0.14
Identities = 19/49 (38%), Positives = 23/49 (46%)
Frame = -2
Query: 21 HPQQHRTRYAPLGPQSHHHLSQPPPPLPPPHAPPPPPHPPPLSYRNIHT 69
HP + P P S PPP PPP PPP P P P ++ N+ T
Sbjct: 278 HPPPQLSSLPPHAPSSFLLPLTSPPPPPPPPHPPPSP-PRPHNHPNLGT 135
Score = 35.4 bits (80), Expect = 0.18
Identities = 35/139 (25%), Positives = 46/139 (32%)
Frame = -3
Query: 21 HPQQHRTRYAPLGPQSHHHLSQPPPPLPPPHAPPPPPHPPPLSYRNIHTPPPPPLYNTPP 80
H HR P + S PP P PP P H+ PP P +P
Sbjct: 1075 HLAPHRALLTLSLPLTLSRASPRPPASPRRCLDTTPPSPHRHYTHTTHSHPPQPPLRSP- 899
Query: 81 VHPSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRNIPDYDRRNPEPAWNPSLDDLNYP 140
PS + FY H ++ R P + + D P R+ P P S ++
Sbjct: 898 --PSPRADFYDQHSTS--HRHPPALYRYYTD--PPRSTPHSATLPPASTHTRSCRRVSAI 737
Query: 141 PLDYDREFNHHRPQQPPPP 159
R P PPPP
Sbjct: 736 ARRCTRRTREGSPFTPPPP 680
Score = 32.0 bits (71), Expect = 2.0
Identities = 32/147 (21%), Positives = 49/147 (32%), Gaps = 23/147 (15%)
Frame = -3
Query: 42 QPPPPLPPPHAPPPP------PHPPPLSYRNIHTPPPPPLYNTP---------------- 79
+PP P+ P + P PH P R + +TP
Sbjct: 1333 RPPAPVTPIDSSPDQTTLSARPHAPATYVRLVPPATTSSARSTPSSIYSSMLRRLVASRR 1154
Query: 80 PVHPSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRNIP-DYDRRNPEPAWNPSLDDLN 138
P++P+ P +P++H L ++P R +P P +P
Sbjct: 1153 PLNPAHARSHIIRVPPLPSPPTIPSSHLAPHRALLTLSLPLTLSRASPRPPASPRRCLDT 974
Query: 139 YPPLDYDREFNHHRPQQPPPPPPSYPP 165
PP + R + H PP PP PP
Sbjct: 973 TPPSPH-RHYTHTTHSHPPQPPLRSPP 896
Score = 30.0 bits (66), Expect = 7.7
Identities = 23/73 (31%), Positives = 33/73 (44%)
Frame = -2
Query: 1356 APISKPRTWHRTGNNPTTSLPRIKPSPGTAPPKRPILPRKANFQNTSTSYIRKGNNSLVR 1415
+P PR H +P+ ++PR+ PSPG LP + TS S + +
Sbjct: 1076 SPRPTPRATHPL--SPSHTIPRLAPSPG--------LPAPLSRYYTSLSPPPLHTHDTLT 927
Query: 1416 QPSPVSALPQISS 1428
PS + LP ISS
Sbjct: 926 PPSAPAPLPSISS 888
>TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 754
Score = 57.4 bits (137), Expect = 5e-08
Identities = 34/77 (44%), Positives = 38/77 (49%), Gaps = 5/77 (6%)
Frame = -2
Query: 44 PPPL---PPPHAPPPPPH--PPPLSYRNIHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPE 98
PPPL PPP+ PPPPP PPP R+ + PPPPP Y PP P R P + P
Sbjct: 555 PPPLLYPPPPYPPPPPPLL*PPPPPRRSPYPPPPPPPYPPPPP*PRRS----PPYPPPPP 388
Query: 99 ERPVPTNHPFSDDLLPR 115
P P P LPR
Sbjct: 387 P*PPPPRPPPPPPPLPR 337
Score = 48.5 bits (114), Expect = 2e-05
Identities = 37/106 (34%), Positives = 45/106 (41%), Gaps = 14/106 (13%)
Frame = -2
Query: 22 PQQHRTRYAPLGPQSHHHLSQPPPPLP---PPHAPPPPPHPPPLSYRNIHTPPPPPL--- 75
P R+ Y P P + PPPP P PP+ PPPPP PPP PPPPPL
Sbjct: 489 PPPRRSPYPPPPPPPY----PPPPP*PRRSPPYPPPPPP*PPPPR----PPPPPPPLPRL 334
Query: 76 --YNTPPVHPSRQLQFYPHHRS------APEERPVPTNHPFSDDLL 113
+ T + PS+ F S A P P + P S L+
Sbjct: 333 *AWFTVMLRPSKS*PFIASMASFMDFSLANVTNPKPLDRPVSRSLM 196
Score = 45.1 bits (105), Expect = 2e-04
Identities = 25/56 (44%), Positives = 27/56 (47%), Gaps = 3/56 (5%)
Frame = -2
Query: 31 PLGPQSHHHLSQPPPPLPPPHAPPPP---PHPPPLSYRNIHTPPPPPLYNTPPVHP 83
P P L PPPP P+ PPPP P PPP R+ PPPPP PP P
Sbjct: 528 PYPPPPPPLL*PPPPPRRSPYPPPPPPPYPPPPP*PRRSPPYPPPPPP*PPPPRPP 361
Score = 42.0 bits (97), Expect = 0.002
Identities = 35/109 (32%), Positives = 37/109 (33%)
Frame = -2
Query: 56 PPHPPPLSYRNIHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPR 115
PP PPPL Y PPPPP PP P R P+ P P P P
Sbjct: 564 PPSPPPLLYPPPPYPPPPPPLL*PPPPPRRS----PYPPPPPPPYPPPPP*P-------- 421
Query: 116 RNIPDYDRRNPEPAWNPSLDDLNYPPLDYDREFNHHRPQQPPPPPPSYP 164
R P Y P P P P +PPPPPP P
Sbjct: 420 RRSPPYP--PPPPP*PP--------------------PPRPPPPPPPLP 340
>TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 912
Score = 57.4 bits (137), Expect = 5e-08
Identities = 34/77 (44%), Positives = 38/77 (49%), Gaps = 5/77 (6%)
Frame = -2
Query: 44 PPPL---PPPHAPPPPPH--PPPLSYRNIHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPE 98
PPPL PPP+ PPPPP PPP R+ + PPPPP Y PP P R P + P
Sbjct: 713 PPPLLYPPPPYPPPPPPLL*PPPPPRRSPYPPPPPPPYPPPPP*PRRS----PPYPPPPP 546
Query: 99 ERPVPTNHPFSDDLLPR 115
P P P LPR
Sbjct: 545 P*PPPPRPPPPPPPLPR 495
Score = 48.5 bits (114), Expect = 2e-05
Identities = 37/106 (34%), Positives = 45/106 (41%), Gaps = 14/106 (13%)
Frame = -2
Query: 22 PQQHRTRYAPLGPQSHHHLSQPPPPLP---PPHAPPPPPHPPPLSYRNIHTPPPPPL--- 75
P R+ Y P P + PPPP P PP+ PPPPP PPP PPPPPL
Sbjct: 647 PPPRRSPYPPPPPPPY----PPPPP*PRRSPPYPPPPPP*PPPPR----PPPPPPPLPRL 492
Query: 76 --YNTPPVHPSRQLQFYPHHRS------APEERPVPTNHPFSDDLL 113
+ T + PS+ F S A P P + P S L+
Sbjct: 491 *AWFTVMLRPSKS*PFIASMASFMDFSLANVTNPKPLDRPVSRSLM 354
Score = 45.1 bits (105), Expect = 2e-04
Identities = 25/56 (44%), Positives = 27/56 (47%), Gaps = 3/56 (5%)
Frame = -2
Query: 31 PLGPQSHHHLSQPPPPLPPPHAPPPP---PHPPPLSYRNIHTPPPPPLYNTPPVHP 83
P P L PPPP P+ PPPP P PPP R+ PPPPP PP P
Sbjct: 686 PYPPPPPPLL*PPPPPRRSPYPPPPPPPYPPPPP*PRRSPPYPPPPPP*PPPPRPP 519
Score = 42.0 bits (97), Expect = 0.002
Identities = 35/109 (32%), Positives = 37/109 (33%)
Frame = -2
Query: 56 PPHPPPLSYRNIHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPR 115
PP PPPL Y PPPPP PP P R P+ P P P P
Sbjct: 722 PPSPPPLLYPPPPYPPPPPPLL*PPPPPRRS----PYPPPPPPPYPPPPP*P-------- 579
Query: 116 RNIPDYDRRNPEPAWNPSLDDLNYPPLDYDREFNHHRPQQPPPPPPSYP 164
R P Y P P P P +PPPPPP P
Sbjct: 578 RRSPPYP--PPPPP*PP--------------------PPRPPPPPPPLP 498
>TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 1054
Score = 57.4 bits (137), Expect = 5e-08
Identities = 34/77 (44%), Positives = 38/77 (49%), Gaps = 5/77 (6%)
Frame = -3
Query: 44 PPPL---PPPHAPPPPPH--PPPLSYRNIHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPE 98
PPPL PPP+ PPPPP PPP R+ + PPPPP Y PP P R P + P
Sbjct: 809 PPPLLYPPPPYPPPPPPLL*PPPPPRRSPYPPPPPPPYPPPPP*PRRS----PPYPPPPP 642
Query: 99 ERPVPTNHPFSDDLLPR 115
P P P LPR
Sbjct: 641 P*PPPPRPPPPPPPLPR 591
Score = 48.5 bits (114), Expect = 2e-05
Identities = 37/106 (34%), Positives = 45/106 (41%), Gaps = 14/106 (13%)
Frame = -3
Query: 22 PQQHRTRYAPLGPQSHHHLSQPPPPLP---PPHAPPPPPHPPPLSYRNIHTPPPPPL--- 75
P R+ Y P P + PPPP P PP+ PPPPP PPP PPPPPL
Sbjct: 743 PPPRRSPYPPPPPPPY----PPPPP*PRRSPPYPPPPPP*PPPPR----PPPPPPPLPRL 588
Query: 76 --YNTPPVHPSRQLQFYPHHRS------APEERPVPTNHPFSDDLL 113
+ T + PS+ F S A P P + P S L+
Sbjct: 587 *AWFTVMLRPSKS*PFIASMASFMDFSLANVTNPKPLDRPVSRSLM 450
Score = 45.1 bits (105), Expect = 2e-04
Identities = 25/56 (44%), Positives = 27/56 (47%), Gaps = 3/56 (5%)
Frame = -3
Query: 31 PLGPQSHHHLSQPPPPLPPPHAPPPP---PHPPPLSYRNIHTPPPPPLYNTPPVHP 83
P P L PPPP P+ PPPP P PPP R+ PPPPP PP P
Sbjct: 782 PYPPPPPPLL*PPPPPRRSPYPPPPPPPYPPPPP*PRRSPPYPPPPPP*PPPPRPP 615
Score = 42.0 bits (97), Expect = 0.002
Identities = 35/109 (32%), Positives = 37/109 (33%)
Frame = -3
Query: 56 PPHPPPLSYRNIHTPPPPPLYNTPPVHPSRQLQFYPHHRSAPEERPVPTNHPFSDDLLPR 115
PP PPPL Y PPPPP PP P R P+ P P P P
Sbjct: 818 PPSPPPLLYPPPPYPPPPPPLL*PPPPPRRS----PYPPPPPPPYPPPPP*P-------- 675
Query: 116 RNIPDYDRRNPEPAWNPSLDDLNYPPLDYDREFNHHRPQQPPPPPPSYP 164
R P Y P P P P +PPPPPP P
Sbjct: 674 RRSPPYP--PPPPP*PP--------------------PPRPPPPPPPLP 594
>TC78402 weakly similar to PIR|B84853|B84853 hypothetical protein At2g42370
[imported] - Arabidopsis thaliana, partial (14%)
Length = 1982
Score = 57.0 bits (136), Expect = 6e-08
Identities = 39/111 (35%), Positives = 50/111 (44%), Gaps = 11/111 (9%)
Frame = -1
Query: 34 PQSHHHLSQPPPPLPPPHAPPPP--------PHPPPLSYRNIHTPPPPPLY--NTPPVHP 83
P SHHH S PP P PPH P PP PHP P +H PPPPP++ TP
Sbjct: 560 PFSHHH-SHPPHPSRPPHLPLPPHLSPHPLPPHPSPHPLL-LHPPPPPPVFPRTTPCAAT 387
Query: 84 SRQLQFYPHHRSAPEERPVPTNHPFSDDLLPRRNIPDYDR-RNPEPAWNPS 133
H + P+E P + S + P + P++ NP A +PS
Sbjct: 386 *SNAANVMHSNTPPDEWPSIPSPACSTSIQPNPSSPNFPP*SNPTAA*HPS 234
Score = 40.0 bits (92), Expect = 0.007
Identities = 52/185 (28%), Positives = 60/185 (32%), Gaps = 39/185 (21%)
Frame = -1
Query: 33 GPQSHHHLSQPPPPLPP-PHAPPPPPHPPPLSYRNIHTPPPPPLYNT-----PPVHPSRQ 86
G Q H HL Q PPLPP HA PHP L + P P Y+T PP S Q
Sbjct: 896 GTQHHAHLLQHRPPLPPLAHALLLLPHPLSLVHSRY---PSYPCYDTASKRAPPHSSSDQ 726
Query: 87 LQFYPHHRSAPEERP----------------------------VPTNHPFSDDLLPR--- 115
P S + P +PT P D L P
Sbjct: 725 ETTSPFLLS*TAQNPSFVPLPPALFPPVKSQLYLAPDSIQYCVLPTPEP--DSLQPHPFS 552
Query: 116 --RNIPDYDRRNPEPAWNPSLDDLNYPPLDYDREFNHHRPQQPPPPPPSYPPVDSLRYDA 173
+ P + R P P L PP H PPPPPP +P
Sbjct: 551 HHHSHPPHPSRPPHLPLPPHLSPHPLPPHPSPHPLLLH----PPPPPPVFPRTTPCAAT* 384
Query: 174 SDGAN 178
S+ AN
Sbjct: 383 SNAAN 369
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.309 0.127 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 52,272,713
Number of Sequences: 36976
Number of extensions: 1077695
Number of successful extensions: 37306
Number of sequences better than 10.0: 1045
Number of HSP's better than 10.0 without gapping: 9708
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 19915
length of query: 1642
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1533
effective length of database: 4,984,343
effective search space: 7640997819
effective search space used: 7640997819
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0078.2


