

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0058a.3
(579 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
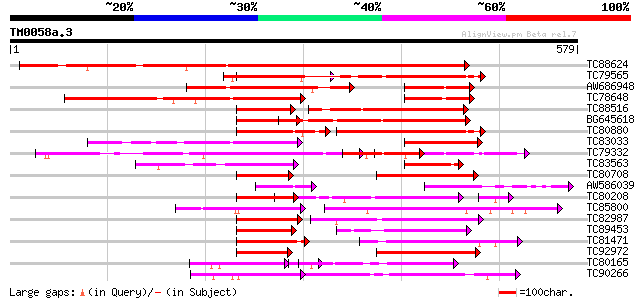
Score E
Sequences producing significant alignments: (bits) Value
TC88624 similar to PIR|S51529|S51529 SPF1 protein - sweet potato... 679 0.0
TC79565 similar to SP|Q9ZQ70|WRK3_ARATH Probable WRKY transcript... 235 4e-62
AW686948 homologue to PIR|S51529|S515 SPF1 protein - sweet potat... 232 3e-61
TC78648 similar to PIR|S51529|S51529 SPF1 protein - sweet potato... 231 8e-61
TC88516 similar to GP|14530681|dbj|BAB61053. WRKY DNA-binding pr... 218 7e-57
BG645618 similar to GP|18158619|gb WRKY-like drought-induced pro... 206 2e-53
TC80880 similar to SP|Q9ZQ70|WRK3_ARATH Probable WRKY transcript... 165 5e-41
TC83033 similar to PIR|S51529|S51529 SPF1 protein - sweet potato... 156 2e-38
TC79332 similar to PIR|JC6203|JC6203 SP8 binding protein homolog... 150 1e-36
TC83563 similar to PIR|T52092|T52092 DNA-binding protein WRKY2 [... 130 2e-30
TC80708 weakly similar to GP|14530685|dbj|BAB61055. WRKY DNA-bin... 123 2e-28
AW586039 similar to GP|14530681|dbj WRKY DNA-binding protein {Ni... 122 4e-28
TC80208 similar to GP|17064158|gb|AAL35286.1 WRKY transcription ... 113 2e-27
TC85800 similar to GP|11320830|dbj|BAB18313. putative WRKY DNA b... 117 9e-27
TC82987 similar to GP|15991740|gb|AAL13047.1 WRKY transcription ... 116 3e-26
TC89453 similar to GP|9187622|emb|CAB97004.1 WRKY DNA binding pr... 115 4e-26
TC81471 similar to PIR|A84913|A84913 probable WRKY-type DNA bind... 112 5e-25
TC92972 similar to GP|9187622|emb|CAB97004.1 WRKY DNA binding pr... 104 8e-23
TC80165 similar to PIR|A84913|A84913 probable WRKY-type DNA bind... 103 2e-22
TC90266 similar to SP|Q9C519|WRK6_ARATH WRKY transcription facto... 101 9e-22
>TC88624 similar to PIR|S51529|S51529 SPF1 protein - sweet potato, partial
(50%)
Length = 1496
Score = 679 bits (1752), Expect = 0.0
Identities = 358/479 (74%), Positives = 390/479 (80%), Gaps = 20/479 (4%)
Frame = +2
Query: 11 SASSFNNFTFSTHPFMTTSFSDLLASPISEDKPRGGLSDRIAERTGSGVPKFKSTPPPSL 70
S + FNNFTFSTHPFM+T+FSDLLASP S + R GVPKFKSTPPPSL
Sbjct: 101 SLNGFNNFTFSTHPFMSTTFSDLLASPASASASDDNNNSR-------GVPKFKSTPPPSL 259
Query: 71 PLSPPTIF------IPPGLSPAELLDSPVLLNSSNILPSPTTGAFAAQS-FNWKSSSGGN 123
PLSPP + IPPGLSPAE LDSPV+LNSSNILPSPTTGAFAAQS +NW ++S GN
Sbjct: 260 PLSPPPVSPSSYFSIPPGLSPAEFLDSPVMLNSSNILPSPTTGAFAAQSNYNWMNNSEGN 439
Query: 124 QQVVKEEDKSFSNFSFQTQQAPPPATTSTATYQSSNVASQTQQQPWSYQEATKQDS---- 179
QQ+ KE NFSFQTQQ P + ST T+QSS V QQQ WSY E T Q++
Sbjct: 440 QQMRKE------NFSFQTQQQGPVVSASTTTFQSSTVGGVQQQQQWSYIENTNQNAFSSE 601
Query: 180 --MMMKTENS-SSMQSFSPEIASVQTNH-NNGFQSDYNNYQPQQQQLQPQTLNRRSDDGY 235
M+ TEN+ SSMQSFSPEIASVQTN+ NNGFQSDY+NYQ QQQ QTL+RRSDDGY
Sbjct: 602 KNMIQTTENNNSSMQSFSPEIASVQTNNTNNGFQSDYSNYQQPQQQ-PTQTLSRRSDDGY 778
Query: 236 NWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDGQITEIVYKGTHNHPKPQATRR 295
NWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERS++GQ+TEIVYKGTHNHPKPQ TRR
Sbjct: 779 NWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSIEGQVTEIVYKGTHNHPKPQCTRR 958
Query: 296 NSSSSSSLPIPHSNHISNDIQDQSYATHGSGQMDSVATPENSSISI-GDDDFEQSS-QKC 353
NSSSSS+ + + N+I DQSYA+HG+GQMDS ATPENSSISI GDDDFEQSS Q+
Sbjct: 959 NSSSSSNALVVVPVNPINEIHDQSYASHGNGQMDSAATPENSSISIGGDDDFEQSSHQRS 1138
Query: 354 RSG--GDELDEDEPDSKRWKVEGENEGISA-PGSRTVREPRVVVQTTSDIDILDDGYRWR 410
RSG GDE DE+EP++KRWK EGENEGISA P SRTVREPRVVVQTTSDIDILDDGYRWR
Sbjct: 1139RSGGAGDEFDEEEPEAKRWKNEGENEGISAQPASRTVREPRVVVQTTSDIDILDDGYRWR 1318
Query: 411 KYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSG 469
KYGQKVVKGNPNPRSYYKCTHP CPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSG
Sbjct: 1319KYGQKVVKGNPNPRSYYKCTHPNCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSG 1495
>TC79565 similar to SP|Q9ZQ70|WRK3_ARATH Probable WRKY transcription factor
3 (WRKY DNA-binding protein 3). [Mouse-ear cress],
partial (35%)
Length = 1602
Score = 235 bits (599), Expect = 4e-62
Identities = 130/275 (47%), Positives = 171/275 (61%), Gaps = 7/275 (2%)
Frame = +2
Query: 219 QQQLQPQT---LNRRSDDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERS-LDG 274
+Q+LQ + +++ +DDGYNWRKYGQKQVKG E PRSYYKCT+P+C KKVER +DG
Sbjct: 626 EQRLQKSSFVNVDKANDDGYNWRKYGQKQVKGCEFPRSYYKCTHPSCLVTKKVERDPVDG 805
Query: 275 QITEIVYKGTHNHPKPQATRRNSSSSSSLPIPHSNHISNDIQDQSYATHGSGQMDSVATP 334
+T I+YKG H H +P+ ++ + +SS
Sbjct: 806 HVTAIIYKGEHIHQRPRPSKLTNDNSSV-------------------------------- 889
Query: 335 ENSSISIGDDDFEQSSQKCRSGGDELDED-EPDSKRWKVEGE--NEGISAPGSRTVREPR 391
+ G D E+ G E + D EP KR K E + N +S RTV +P+
Sbjct: 890 --QQVLSGTSDSEEE------GDHETEVDYEPGLKRRKTEAKLLNPALS---HRTVSKPK 1036
Query: 392 VVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVI 451
++VQTTSD+D+L+DGYRWRKYGQKVVKGNP PRSYYKCT PGC VRKHVER S D +AV+
Sbjct: 1037IIVQTTSDVDLLEDGYRWRKYGQKVVKGNPYPRSYYKCTTPGCNVRKHVERVSTDPKAVL 1216
Query: 452 TTYEGKHNHDVPAARGSGNHSVTRPLPNNTANAIR 486
TTYEGKHNHDVPAA+ + +H++ NN+A+ ++
Sbjct: 1217TTYEGKHNHDVPAAK-TNSHNLA---SNNSASQLK 1309
Score = 80.9 bits (198), Expect = 1e-15
Identities = 45/105 (42%), Positives = 60/105 (56%), Gaps = 4/105 (3%)
Frame = +2
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVER-SLDGQITEIVYKGTHNHPKP 290
+DGY WRKYGQK VKG+ PRSYYKCT P C +K VER S D + Y+G HNH P
Sbjct: 1073 EDGYRWRKYGQKVVKGNPYPRSYYKCTTPGCNVRKHVERVSTDPKAVLTTYEGKHNHDVP 1252
Query: 291 QATRRN---SSSSSSLPIPHSNHISNDIQDQSYATHGSGQMDSVA 332
A + +S++S+ + N I + Q++ G Q +VA
Sbjct: 1253 AAKTNSHNLASNNSASQLKSQNAIP---EMQNFNRRGQHQPSAVA 1378
>AW686948 homologue to PIR|S51529|S515 SPF1 protein - sweet potato, partial
(16%)
Length = 565
Score = 232 bits (591), Expect = 3e-61
Identities = 123/180 (68%), Positives = 137/180 (75%), Gaps = 8/180 (4%)
Frame = +1
Query: 181 MMKTENSSSMQSFSPEIASV--QTNHN-NGFQSDYNNYQPQQQQLQPQTLNRRSDDGYNW 237
M+K ENSSSMQSF+PE S Q N+N +G QS+YNNYQ Q Q Q L+RRSDDGYNW
Sbjct: 22 MVKAENSSSMQSFTPESNSTSEQNNYNKSGSQSNYNNYQSQPQV---QILSRRSDDGYNW 192
Query: 238 RKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDGQITEIVYKGTHNHPKPQATRRNS 297
RKYGQKQVKGSENPRSYYKCTYPNCPTKKKVER LDGQITEIVYKG+HNHPKP A +RN+
Sbjct: 193 RKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERGLDGQITEIVYKGSHNHPKPVANKRNT 372
Query: 298 SSSSSLPIPH-----SNHISNDIQDQSYATHGSGQMDSVATPENSSISIGDDDFEQSSQK 352
+S SS + H SNH N+I QMD VATPENSSISIGDD+FEQ+S K
Sbjct: 373 NSMSSSSLSHANPPPSNHFGNEI-----------QMDLVATPENSSISIGDDEFEQTSHK 519
Score = 87.0 bits (214), Expect = 2e-17
Identities = 39/71 (54%), Positives = 47/71 (65%)
Frame = +1
Query: 404 DDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVP 463
DDGY WRKYGQK VKG+ NPRSYYKCT+P CP +K VER D + Y+G HNH P
Sbjct: 175 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERGL-DGQITEIVYKGSHNHPKP 351
Query: 464 AARGSGNHSVT 474
A +S++
Sbjct: 352 VANKRNTNSMS 384
>TC78648 similar to PIR|S51529|S51529 SPF1 protein - sweet potato, partial
(18%)
Length = 990
Score = 231 bits (588), Expect = 8e-61
Identities = 136/260 (52%), Positives = 163/260 (62%), Gaps = 14/260 (5%)
Frame = +1
Query: 57 SGVPKFKSTPPPSLPLSP-PTIFIP--PGLSPAELLDSPVLLNSSNILPSPTTGAFAAQS 113
+G PKFKS PPSLPLS P+ +P SP + L SP L+S N+ SPTT AFA QS
Sbjct: 223 NGAPKFKSVQPPSLPLSSSPSFHLPFSSTFSPTDFLISPFFLSSPNVFASPTTEAFANQS 402
Query: 114 FNWKSSSGGN--QQVVKEEDKSFSNFSFQTQQAPPPATTSTATYQSSNVASQTQ---QQP 168
FNW +S G QQ K+++K+ S+FSF TQ P S S+N+ Q Q Q
Sbjct: 403 FNWNKNSLGEEEQQGDKKDEKNLSDFSFPTQTKPESVFQS-----STNMFQQEQTKKQDI 567
Query: 169 WSYQEATKQ-DSMMMKTENSS---SMQSFSPEIASVQTN-HNNGFQ-SDYNNYQPQQQQL 222
W + E KQ D +T + S S QS+S EI ++ H+N S Y NY Q
Sbjct: 568 WKFNEPKKQTDFSSERTASKSEFQSTQSYSSEIVPIKPEIHSNSVTGSGYYNYNNNASQF 747
Query: 223 QPQTLNRRSDDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDGQITEIVYK 282
+ +RS+DGYNWRKYGQKQVKGSENPRSYYKCT PNC KKKVER LDGQITEIVYK
Sbjct: 748 VRE--QKRSEDGYNWRKYGQKQVKGSENPRSYYKCTNPNCSMKKKVERDLDGQITEIVYK 921
Query: 283 GTHNHPKPQATRRNSSSSSS 302
GTHNHPKPQ+ RRN+S +S
Sbjct: 922 GTHNHPKPQSNRRNNSQPTS 981
Score = 82.4 bits (202), Expect = 4e-16
Identities = 40/73 (54%), Positives = 47/73 (63%), Gaps = 2/73 (2%)
Frame = +1
Query: 404 DDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVIT--TYEGKHNHD 461
+DGY WRKYGQK VKG+ NPRSYYKCT+P C ++K VER DL IT Y+G HNH
Sbjct: 769 EDGYNWRKYGQKQVKGSENPRSYYKCTNPNCSMKKKVER---DLDGQITEIVYKGTHNHP 939
Query: 462 VPAARGSGNHSVT 474
P + N T
Sbjct: 940 KPQSNRRNNSQPT 978
>TC88516 similar to GP|14530681|dbj|BAB61053. WRKY DNA-binding protein
{Nicotiana tabacum}, partial (17%)
Length = 1085
Score = 218 bits (554), Expect = 7e-57
Identities = 108/163 (66%), Positives = 130/163 (79%)
Frame = +2
Query: 306 PHSNHISNDIQDQSYATHGSGQMDSVATPENSSISIGDDDFEQSSQKCRSGGDELDEDEP 365
P S+ ++ I DQS MD V+ E+SS S+G+++FEQ+SQ SGG++ + P
Sbjct: 14 PTSSCTNSGISDQS-------AMDHVSIQEDSSASVGEEEFEQTSQTSYSGGND-NALVP 169
Query: 366 DSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRS 425
++KRWK + ENEG A SRTV+EPRVVVQTTS+IDILDDG+RWRKYGQKVVKGNPN RS
Sbjct: 170 EAKRWKGDNENEGYCASASRTVKEPRVVVQTTSEIDILDDGFRWRKYGQKVVKGNPNARS 349
Query: 426 YYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGS 468
YYKCT PGC VRKHVERA+HD++AVITTYEGKHNHDVPAARGS
Sbjct: 350 YYKCTAPGCNVRKHVERAAHDIKAVITTYEGKHNHDVPAARGS 478
Score = 75.9 bits (185), Expect = 4e-14
Identities = 35/62 (56%), Positives = 41/62 (65%), Gaps = 1/62 (1%)
Frame = +2
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSL-DGQITEIVYKGTHNHPKP 290
DDG+ WRKYGQK VKG+ N RSYYKCT P C +K VER+ D + Y+G HNH P
Sbjct: 284 DDGFRWRKYGQKVVKGNPNARSYYKCTAPGCNVRKHVERAAHDIKAVITTYEGKHNHDVP 463
Query: 291 QA 292
A
Sbjct: 464 AA 469
>BG645618 similar to GP|18158619|gb WRKY-like drought-induced protein {Retama
raetam}, partial (37%)
Length = 747
Score = 206 bits (525), Expect = 2e-53
Identities = 110/196 (56%), Positives = 134/196 (68%)
Frame = -1
Query: 275 QITEIVYKGTHNHPKPQATRRNSSSSSSLPIPHSNHISNDIQDQSYATHGSGQMDSVATP 334
QI EI+YKGTH+HPKPQ + R S+ S + + +D + S A +S P
Sbjct: 747 QIIEIIYKGTHDHPKPQPSNRYSAGS----VMSTQGERSDNRASSLAVRDDKASNS---P 589
Query: 335 ENSSISIGDDDFEQSSQKCRSGGDELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVV 394
E S ++ D E + D +D+D+P SK+ K+E N I P + +REPRVVV
Sbjct: 588 EQSVVATNDLSPEGAGFVSTRTNDGVDDDDPFSKQRKMELGNADI-IPVVKPIREPRVVV 412
Query: 395 QTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTY 454
QT S+IDILDDGYRWRKYGQKVV+GNPNPRSYYKCT+ GCPVRKHVERASHD +AVITTY
Sbjct: 411 QTMSEIDILDDGYRWRKYGQKVVRGNPNPRSYYKCTNAGCPVRKHVERASHDPKAVITTY 232
Query: 455 EGKHNHDVPAARGSGN 470
EGKHNHDVPAAR S +
Sbjct: 231 EGKHNHDVPAARSSSH 184
Score = 81.3 bits (199), Expect = 1e-15
Identities = 40/68 (58%), Positives = 46/68 (66%), Gaps = 1/68 (1%)
Frame = -1
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVER-SLDGQITEIVYKGTHNHPKP 290
DDGY WRKYGQK V+G+ NPRSYYKCT CP +K VER S D + Y+G HNH P
Sbjct: 384 DDGYRWRKYGQKVVRGNPNPRSYYKCTNAGCPVRKHVERASHDPKAVITTYEGKHNHDVP 205
Query: 291 QATRRNSS 298
A R+SS
Sbjct: 204 AA--RSSS 187
>TC80880 similar to SP|Q9ZQ70|WRK3_ARATH Probable WRKY transcription factor
3 (WRKY DNA-binding protein 3). [Mouse-ear cress],
partial (24%)
Length = 907
Score = 165 bits (417), Expect = 5e-41
Identities = 85/155 (54%), Positives = 106/155 (67%), Gaps = 2/155 (1%)
Frame = +2
Query: 334 PENSSISIGDDDFEQSSQKCRSGGDELDED--EPDSKRWKVEGENEGISAPGSRTVREPR 391
PE+S ++ S+ E+ E EPDSKR E + RTV EP+
Sbjct: 20 PESSQATVEHLSGTSDSEDVGDRETEVHEKRIEPDSKRRNTEVTVSNPTTSSHRTVTEPK 199
Query: 392 VVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVI 451
++VQTTS++D+LDDGYRWRKYGQKVVKGNP PRSYYKCT PGC VRKHVERAS D +AVI
Sbjct: 200 IIVQTTSEVDLLDDGYRWRKYGQKVVKGNPYPRSYYKCTTPGCNVRKHVERASTDPKAVI 379
Query: 452 TTYEGKHNHDVPAARGSGNHSVTRPLPNNTANAIR 486
TTYEGKHNHDVPAA+ + +H++ NN A+ ++
Sbjct: 380 TTYEGKHNHDVPAAK-TNSHTIA----NNNASQLK 469
Score = 80.5 bits (197), Expect = 2e-15
Identities = 45/100 (45%), Positives = 60/100 (60%), Gaps = 4/100 (4%)
Frame = +2
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVER-SLDGQITEIVYKGTHNHPKP 290
DDGY WRKYGQK VKG+ PRSYYKCT P C +K VER S D + Y+G HNH P
Sbjct: 236 DDGYRWRKYGQKVVKGNPYPRSYYKCTTPGCNVRKHVERASTDPKAVITTYEGKHNHDVP 415
Query: 291 QATRRNS---SSSSSLPIPHSNHISNDIQDQSYATHGSGQ 327
A + NS +++++ + N IS + S+ + G G+
Sbjct: 416 -AAKTNSHTIANNNASQLKSQNTIS---EKTSFGSIGIGE 523
>TC83033 similar to PIR|S51529|S51529 SPF1 protein - sweet potato, partial
(17%)
Length = 900
Score = 156 bits (395), Expect = 2e-38
Identities = 97/222 (43%), Positives = 126/222 (56%), Gaps = 2/222 (0%)
Frame = +3
Query: 80 PPGLSPAELLDSPVLLNSSNILPSPTTGAFAAQSFNWKSSSGGNQQVVKE-EDKSFSNFS 138
PPG SP+ELL+SP L+SSNI S T QS N QQ KE E+++FS S
Sbjct: 186 PPGFSPSELLNSPSFLSSSNIPSSYTNEPSTVQSMN--------QQHSKELEERNFSEPS 341
Query: 139 FQTQQAPPPATTSTATYQSSNVASQTQQQPWSYQEATKQDSMMMKTENSSSMQSFSPEIA 198
FQTQ+ T +QSS Q ++ ++ D+++ + SF
Sbjct: 342 FQTQKQ-----TMHDLFQSSTSMFQVEEPLKTH------DTLIFNESTKQTNFSFEGTTK 488
Query: 199 SVQTNHNNGFQSDYNNYQPQQQQLQPQTLNRRSDDGYNWRKYGQKQVKGSENPRSYYKCT 258
+++ S Y N ++ Q + +DGY WRKYG+KQVKGSENPRSYYKCT
Sbjct: 489 HEAQTNSSVPNSSYFNNTSASMSIREQ--RKSEEDGYKWRKYGEKQVKGSENPRSYYKCT 662
Query: 259 YPNCPTKKKVERSL-DGQITEIVYKGTHNHPKPQATRRNSSS 299
P C KKVERSL DGQIT+IVYKG+HNHPKPQ T+R ++S
Sbjct: 663 NPICSMXKKVERSLDDGQITQIVYKGSHNHPKPQCTKRRANS 788
Score = 81.3 bits (199), Expect = 1e-15
Identities = 37/80 (46%), Positives = 50/80 (62%)
Frame = +3
Query: 404 DDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVP 463
+DGY+WRKYG+K VKG+ NPRSYYKCT+P C + K VER+ D + Y+G HNH P
Sbjct: 582 EDGYKWRKYGEKQVKGSENPRSYYKCTNPICSMXKKVERSLDDGQITQIVYKGSHNHPKP 761
Query: 464 AARGSGNHSVTRPLPNNTAN 483
+S P+++ N
Sbjct: 762 QCTKRRANSQYIHQPSSSCN 821
>TC79332 similar to PIR|JC6203|JC6203 SP8 binding protein homolog -
cucumber, partial (49%)
Length = 1486
Score = 150 bits (379), Expect = 1e-36
Identities = 117/348 (33%), Positives = 161/348 (45%), Gaps = 12/348 (3%)
Frame = +2
Query: 27 TTSFSDLLA----SPI---SEDKPRGGLSDRIAERTGSGVPKFKSTPPPSLPLSPPTIF- 78
+ +FS LLA SP+ S G S + GS FK + P +L ++ +F
Sbjct: 200 SATFSQLLAGAMASPLAFSSSSSLAGEYSFGKEDDGGSLNGGFKQSRPMNLVIARSPVFT 379
Query: 79 IPPGLSPAELLDSPVLLNSSNILPSPTTGAFAAQSFNWKSSSGGNQQVVKEEDKSFSNFS 138
+PPGLSP+ L+SP G F+ QS F
Sbjct: 380 VPPGLSPSGFLNSP--------------GFFSPQS----------------------PFG 451
Query: 139 FQTQQAPPPATTSTATYQSSNVASQTQQQPWSYQEATKQDSMMMKTENSSSMQSFSPE-- 196
QQA T QS N+ Q + Q SY+ T++ E S ++ +PE
Sbjct: 452 MSHQQALAQVTAQAVLAQSQNMHMQPEYQLVSYEAPTER-----LAEQPSYTRNEAPEQQ 616
Query: 197 -IASVQTNHNNGFQSDYNNYQPQQQQLQPQTLNRRSDDGYNWRKYGQKQVKGSENPRSYY 255
A V N ++ + ++ Q +++ +DDGYNWRKYGQKQVKGSE PRSYY
Sbjct: 617 VTAPVSEPRNAQMETSEITHSDKKYQPSSLPIDKPADDGYNWRKYGQKQVKGSEYPRSYY 796
Query: 256 KCTYPNCPTKKKVERSLDGQITEIVYKGTHNHPKPQATRRNSSSSSSLPIPHSNHISNDI 315
KCT+ NCP KKKVER+ DG ITEI+YKG HNH KPQ RR ++S L + +D
Sbjct: 797 KCTHLNCPVKKKVERAPDGHITEIIYKGQHNHEKPQPNRRVKENNSDLNGNANVQPKSDS 976
Query: 316 QDQSYATHGSGQMDSVATPENSSISIGDD-DFEQSSQKCRSGGDELDE 362
Q + G+ S P++S D Q + + R G E +E
Sbjct: 977 NSQGW--FGNSNKISEIVPDSSPPEPESDLTSNQGAIRPRPGSSESEE 1114
Score = 93.2 bits (230), Expect = 2e-19
Identities = 65/175 (37%), Positives = 85/175 (48%), Gaps = 17/175 (9%)
Frame = +2
Query: 373 EGENEGISAPGSRTVREPRVVVQTTSDIDI---------------LDDGYRWRKYGQKVV 417
E + ++AP V EPR TS+I DDGY WRKYGQK V
Sbjct: 599 EAPEQQVTAP----VSEPRNAQMETSEITHSDKKYQPSSLPIDKPADDGYNWRKYGQKQV 766
Query: 418 KGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPA--ARGSGNHSVTR 475
KG+ PRSYYKCTH CPV+K VERA D Y+G+HNH+ P R N+S
Sbjct: 767 KGSEYPRSYYKCTHLNCPVKKKVERAP-DGHITEIIYKGQHNHEKPQPNRRVKENNSDL- 940
Query: 476 PLPNNTANAIRPSSVTHQHHHNTNNNSLQSLRPQQAPDQVQSSPFTLEMLQQNQG 530
N AN ++P S ++ N+N + + P +P + +S L NQG
Sbjct: 941 ---NGNAN-VQPKSDSNSQGWFGNSNKISEIVPDSSPPEPESD------LTSNQG 1075
Score = 80.5 bits (197), Expect = 2e-15
Identities = 39/86 (45%), Positives = 55/86 (63%), Gaps = 3/86 (3%)
Frame = +3
Query: 341 IGDDDFEQSSQKCRSGGDELDED---EPDSKRWKVEGENEGISAPGSRTVREPRVVVQTT 397
+ D D +Q + + +E EP+ KR +E + P +TV EP+++VQT
Sbjct: 1077 LSDHDLDQVRARKVGNAENKEEGVDCEPNPKRRSIEPAVPEVP-PSQKTVTEPKIIVQTR 1253
Query: 398 SDIDILDDGYRWRKYGQKVVKGNPNP 423
S++D+LDDGYRWRKYGQKVVKGNP+P
Sbjct: 1254 SEVDLLDDGYRWRKYGQKVVKGNPHP 1331
Score = 39.3 bits (90), Expect = 0.004
Identities = 15/20 (75%), Positives = 17/20 (85%)
Frame = +3
Query: 232 DDGYNWRKYGQKQVKGSENP 251
DDGY WRKYGQK VKG+ +P
Sbjct: 1272 DDGYRWRKYGQKVVKGNPHP 1331
>TC83563 similar to PIR|T52092|T52092 DNA-binding protein WRKY2 [imported] -
common tobacco, partial (21%)
Length = 838
Score = 130 bits (326), Expect = 2e-30
Identities = 76/174 (43%), Positives = 103/174 (58%), Gaps = 7/174 (4%)
Frame = +1
Query: 129 EEDKSFSNFSFQTQQAPPPATT----STATYQSSNVA-SQTQQQPWSYQEATKQDSMMMK 183
++ KSFS + +P P T S+ + V+ TQQQP + A + S
Sbjct: 361 DDGKSFSQLLAGSMVSPAPLTAGGLDSSGFFSHPQVSFGMTQQQPLAQVSAHEGPS---- 528
Query: 184 TENSSSMQSFSPEIASVQTNHNNGFQSDYNNYQPQQQQLQPQTLN--RRSDDGYNWRKYG 241
+++M + + ++ V T+ ++ S+ Q+LQP +LN + +DDGYNWRKYG
Sbjct: 529 ---NTNMHNQAEHLSYVPTSTDHAPLSE--------QRLQPSSLNVDKPADDGYNWRKYG 675
Query: 242 QKQVKGSENPRSYYKCTYPNCPTKKKVERSLDGQITEIVYKGTHNHPKPQATRR 295
QKQVKGSE PRSYYKCT+PNCP KKKVERSL G IT I+YKG HNH P +R
Sbjct: 676 QKQVKGSEFPRSYYKCTHPNCPVKKKVERSLAGHITAIIYKGEHNHLLPNPNKR 837
Score = 85.5 bits (210), Expect = 5e-17
Identities = 39/61 (63%), Positives = 48/61 (77%), Gaps = 1/61 (1%)
Frame = +1
Query: 404 DDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERA-SHDLRAVITTYEGKHNHDV 462
DDGY WRKYGQK VKG+ PRSYYKCTHP CPV+K VER+ + + A+I Y+G+HNH +
Sbjct: 646 DDGYNWRKYGQKQVKGSEFPRSYYKCTHPNCPVKKKVERSLAGHITAII--YKGEHNHLL 819
Query: 463 P 463
P
Sbjct: 820 P 822
>TC80708 weakly similar to GP|14530685|dbj|BAB61055. WRKY DNA-binding
protein {Nicotiana tabacum}, partial (20%)
Length = 864
Score = 123 bits (309), Expect = 2e-28
Identities = 55/105 (52%), Positives = 77/105 (72%), Gaps = 1/105 (0%)
Frame = +2
Query: 375 ENEGISAPGSRTVR-EPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPG 433
+N A G+ + E RV+V+TTS+ I++DGYRWRKYGQK+VKGN NPR+YY+C+ PG
Sbjct: 101 DNSNTDATGADVLTGESRVIVRTTSESGIVNDGYRWRKYGQKMVKGNTNPRNYYRCSSPG 280
Query: 434 CPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSGNHSVTRPLP 478
CPV+KHVE++S + VITTYEG+H+H P RG +++ + P
Sbjct: 281 CPVKKHVEKSSQNTTTVITTYEGQHDHAPPTGRGVLDNTAVKLTP 415
Score = 79.7 bits (195), Expect = 3e-15
Identities = 34/60 (56%), Positives = 42/60 (69%), Gaps = 1/60 (1%)
Frame = +2
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDGQITEI-VYKGTHNHPKP 290
+DGY WRKYGQK VKG+ NPR+YY+C+ P CP KK VE+S T I Y+G H+H P
Sbjct: 191 NDGYRWRKYGQKMVKGNTNPRNYYRCSSPGCPVKKHVEKSSQNTTTVITTYEGQHDHAPP 370
>AW586039 similar to GP|14530681|dbj WRKY DNA-binding protein {Nicotiana
tabacum}, partial (9%)
Length = 413
Score = 122 bits (306), Expect = 4e-28
Identities = 75/153 (49%), Positives = 88/153 (57%), Gaps = 1/153 (0%)
Frame = -2
Query: 424 RSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSGNHSVTRPLPNNTAN 483
RSYYKCT+PGCPVRKHVERAS DLRAVITTYEGKH HDVPA RGSGNHS+ +PLP T
Sbjct: 397 RSYYKCTNPGCPVRKHVERASQDLRAVITTYEGKHTHDVPAPRGSGNHSINKPLPIQTT- 221
Query: 484 AIRPSSVTHQHHHNTNNNSLQSLRPQQAPDQVQSSPFTLEMLQQNQGGFGFSSGFGNSMG 543
N NNS+ + V + F EM+Q N GF+ F
Sbjct: 220 ------------MNNPNNSINN-------TSVNNGFFNHEMMQNN----GFTVDF----- 125
Query: 544 SYNMNNQQQQQLPDNVFSSRAKEEP-RDDTFFD 575
NMN QQ +N+ RAK+E +D FF+
Sbjct: 124 YMNMN----QQRVNNIIRGRAKQETVPNDPFFE 38
Score = 50.4 bits (119), Expect = 2e-06
Identities = 27/63 (42%), Positives = 36/63 (56%), Gaps = 1/63 (1%)
Frame = -2
Query: 252 RSYYKCTYPNCPTKKKVER-SLDGQITEIVYKGTHNHPKPQATRRNSSSSSSLPIPHSNH 310
RSYYKCT P CP +K VER S D + Y+G H H P A R + + S + P+P
Sbjct: 397 RSYYKCTNPGCPVRKHVERASQDLRAVITTYEGKHTHDVP-APRGSGNHSINKPLPIQTT 221
Query: 311 ISN 313
++N
Sbjct: 220 MNN 212
>TC80208 similar to GP|17064158|gb|AAL35286.1 WRKY transcription factor 28
{Arabidopsis thaliana}, partial (31%)
Length = 1294
Score = 113 bits (283), Expect(2) = 2e-27
Identities = 77/200 (38%), Positives = 100/200 (49%), Gaps = 7/200 (3%)
Frame = +1
Query: 271 SLDGQITEIVYKGTHNH-PKPQATRRNSSSSSSLPIPHSNHISNDIQDQSYATHGSGQMD 329
S + TE + G +H P AT S SS + S N Q S G G D
Sbjct: 208 SYNMSFTESLQGGNMDHYNSPLATSFGVLSPSSSEVFSSVDQGN--QKPSAGELGGG--D 375
Query: 330 SVATPENSSIS------IGDDDFEQSSQKCRSGGDELDEDEPDSKRWKVEGENEGISAPG 383
NSSIS G + E S ++ ++ G +E +S + G N+ G
Sbjct: 376 QTLATLNSSISNSSSSEAGGVEEEDSGKRKKNIGQVKEEPGQESSK---NGNNKVDKKKG 546
Query: 384 SRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERA 443
+ +EPR T S++D L+DGYRWRKYGQK VK +P PRSYY+CT C V+K VER+
Sbjct: 547 EKKQKEPRFAFMTKSEVDQLEDGYRWRKYGQKAVKNSPYPRSYYRCTTQKCTVKKRVERS 726
Query: 444 SHDLRAVITTYEGKHNHDVP 463
D VITTYEG+HN P
Sbjct: 727 YEDPTTVITTYEGQHNASCP 786
Score = 73.9 bits (180), Expect = 2e-13
Identities = 35/65 (53%), Positives = 41/65 (62%), Gaps = 1/65 (1%)
Frame = +1
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDGQITEI-VYKGTHNHPKP 290
+DGY WRKYGQK VK S PRSYY+CT C KK+VERS + T I Y+G HN P
Sbjct: 607 EDGYRWRKYGQKAVKNSPYPRSYYRCTTQKCTVKKRVERSYEDPTTVITTYEGQHNASCP 786
Query: 291 QATRR 295
+R
Sbjct: 787 HFPKR 801
Score = 27.7 bits (60), Expect(2) = 2e-27
Identities = 15/43 (34%), Positives = 23/43 (52%), Gaps = 7/43 (16%)
Frame = +3
Query: 479 NNTANAIRPSSVTHQH-------HHNTNNNSLQSLRPQQAPDQ 514
+NT N +P++ T + H+N NN+ LQ + Q PDQ
Sbjct: 906 HNTFNITQPTTHTTNNVAGSVYSHNNINNSFLQQYQQQLPPDQ 1034
>TC85800 similar to GP|11320830|dbj|BAB18313. putative WRKY DNA binding
protein {Oryza sativa (japonica cultivar-group)}, partial
(37%)
Length = 2028
Score = 117 bits (294), Expect = 9e-27
Identities = 91/292 (31%), Positives = 133/292 (45%), Gaps = 49/292 (16%)
Frame = +1
Query: 322 THGSGQMDSVATPENSSISIGDDDFEQSSQKCRSGGDELDEDE-----PDSKRWKVEGEN 376
T+G+ +++ + +S + + + ++G ++ E E P+ + + N
Sbjct: 760 TNGTTEVEDQVSNSSSDERTRSNTPQMRNSNGKTGREDSPESETQGWGPNKSQKILNSSN 939
Query: 377 EGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHP-GCP 435
A T+R+ RV V+ S+ ++ DG +WRKYGQK+ KGNP PR+YY+CT GCP
Sbjct: 940 VADQANTEATMRKARVSVRARSEASMISDGCQWRKYGQKMAKGNPCPRAYYRCTMAVGCP 1119
Query: 436 VRKHVERASHDLRAVITTYEGKHNHDVPAA----------------RGSGNHSVTRPLPN 479
VRK V+R + D ++TTYEG HNH +P A GS + + PN
Sbjct: 1120 VRKQVQRCAEDKTILVTTYEGTHNHPLPPAAMAMASTTSAAASMLLSGSMSSADGIMTPN 1299
Query: 480 NTANAIRPSS--------------VTHQHHHNTNNNSLQSLRPQQAP-------DQVQSS 518
A AI P S VT N+N N LQ RPQ AP Q Q+
Sbjct: 1300 LLARAILPCSTSMATLSASAPFPTVTLDLTQNSNPNPLQFQRPQHAPFHQVPSFFQGQNQ 1479
Query: 519 PFTLEML----QQNQGGFGFSSGFGNSMGSYNMNNQQQQQ--LPDNVFSSRA 564
F Q G S G+S + + QQQQQ L D+V ++ A
Sbjct: 1480 NFAQAAASLYNQSKFSGLQLSQEVGSSHLTTQASTQQQQQPSLADSVSAATA 1635
Score = 77.8 bits (190), Expect = 1e-14
Identities = 51/147 (34%), Positives = 74/147 (49%), Gaps = 14/147 (9%)
Frame = +1
Query: 170 SYQEATKQDSMMMKTENSSSMQSFSPEIASVQTNHNNGFQSDYNNYQPQQQQLQPQTLNR 229
S E T+ ++ M+ N + + SPE + Q N Q N+ Q T+ +
Sbjct: 802 SSDERTRSNTPQMRNSNGKTGREDSPE-SETQGWGPNKSQKILNSSNVADQANTEATMRK 978
Query: 230 -------RSD-----DGYNWRKYGQKQVKGSENPRSYYKCTYP-NCPTKKKVER-SLDGQ 275
RS+ DG WRKYGQK KG+ PR+YY+CT CP +K+V+R + D
Sbjct: 979 ARVSVRARSEASMISDGCQWRKYGQKMAKGNPCPRAYYRCTMAVGCPVRKQVQRCAEDKT 1158
Query: 276 ITEIVYKGTHNHPKPQATRRNSSSSSS 302
I Y+GTHNHP P A +S++S+
Sbjct: 1159ILVTTYEGTHNHPLPPAAMAMASTTSA 1239
>TC82987 similar to GP|15991740|gb|AAL13047.1 WRKY transcription factor 71
{Arabidopsis thaliana}, partial (33%)
Length = 975
Score = 116 bits (290), Expect = 3e-26
Identities = 67/183 (36%), Positives = 103/183 (55%), Gaps = 6/183 (3%)
Frame = +1
Query: 308 SNHISNDIQDQSYATHGSGQMDSVA----TPENSSISIGDDD-FEQSSQKCRSGGDELDE 362
S+ + + I D + +G + S+ TP++S S ++ E+ S + +L
Sbjct: 388 SSEVISSIDDNNTKKSSAGDLSSLVGISETPKSSESSSSNEAVIEEDSTNKKDKQPKLGC 567
Query: 363 DEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPN 422
++ D ++ K E + + + V+EPR T S+ID L+DGYRWRKYGQK VK +P
Sbjct: 568 EDGDDEKSKKENK----AKKKEKKVKEPRFAFLTKSEIDHLEDGYRWRKYGQKAVKNSPY 735
Query: 423 PRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAA-RGSGNHSVTRPLPNNT 481
PRSYY+CT C V+K VER+ D V+TTYEG+HNH PA RG+ + + P+
Sbjct: 736 PRSYYRCTSQKCIVKKRVERSYQDPSIVMTTYEGQHNHHCPATLRGNASSMLLSSSPSLF 915
Query: 482 ANA 484
A++
Sbjct: 916 ASS 924
Score = 80.9 bits (198), Expect = 1e-15
Identities = 39/69 (56%), Positives = 44/69 (63%), Gaps = 1/69 (1%)
Frame = +1
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSL-DGQITEIVYKGTHNHPKP 290
+DGY WRKYGQK VK S PRSYY+CT C KK+VERS D I Y+G HNH P
Sbjct: 679 EDGYRWRKYGQKAVKNSPYPRSYYRCTSQKCIVKKRVERSYQDPSIVMTTYEGQHNHHCP 858
Query: 291 QATRRNSSS 299
R N+SS
Sbjct: 859 ATLRGNASS 885
>TC89453 similar to GP|9187622|emb|CAB97004.1 WRKY DNA binding protein
{Solanum tuberosum}, partial (56%)
Length = 701
Score = 115 bits (289), Expect = 4e-26
Identities = 59/138 (42%), Positives = 76/138 (54%)
Frame = +1
Query: 334 PENSSISIGDDDFEQSSQKCRSGGDELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVV 393
P N+ IS+ DD E K G + D+ D K+ G + ++P+
Sbjct: 295 PTNNEISVTHDDHEDV--KGEEGSVNVIIDQQDVKK------------KGEKKAKKPKYA 432
Query: 394 VQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITT 453
QT S +DILDDGYRWRKYGQK VK N PRSYY+CTH GC V+K V+R + D V+TT
Sbjct: 433 FQTRSQVDILDDGYRWRKYGQKAVKNNKFPRSYYRCTHQGCNVKKQVQRLTKDEGVVVTT 612
Query: 454 YEGKHNHDVPAARGSGNH 471
YEG H H + + H
Sbjct: 613 YEGVHTHPIEKTTDNFEH 666
Score = 73.9 bits (180), Expect = 2e-13
Identities = 32/63 (50%), Positives = 42/63 (65%), Gaps = 1/63 (1%)
Frame = +1
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVER-SLDGQITEIVYKGTHNHPKP 290
DDGY WRKYGQK VK ++ PRSYY+CT+ C KK+V+R + D + Y+G H HP
Sbjct: 463 DDGYRWRKYGQKAVKNNKFPRSYYRCTHQGCNVKKQVQRLTKDEGVVVTTYEGVHTHPIE 642
Query: 291 QAT 293
+ T
Sbjct: 643 KTT 651
>TC81471 similar to PIR|A84913|A84913 probable WRKY-type DNA binding protein
[imported] - Arabidopsis thaliana, partial (43%)
Length = 1274
Score = 112 bits (279), Expect = 5e-25
Identities = 67/182 (36%), Positives = 92/182 (49%), Gaps = 16/182 (8%)
Frame = +2
Query: 358 DELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVV 417
DE +E + +K+ K + N + REPR T S++D L+DGYRWRKYGQK V
Sbjct: 440 DEEEEKQKTNKQLKTKKTN-------LKRQREPRFAFMTKSEVDHLEDGYRWRKYGQKAV 598
Query: 418 KGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSGNHSVTRPL 477
K +P PRSYY+CT C V+K VER+ D V+TTYEG+H H P S V P
Sbjct: 599 KNSPFPRSYYRCTTASCNVKKRVERSYTDPSIVVTTYEGQHTHPSPTMSRSAFAGVQIPQ 778
Query: 478 P----------NNTANAIRPSSVTHQH-----HHNTNNNSLQSL-RPQQAPDQVQSSPFT 521
P N + ++ + ++ H H N+L SL P ++SPFT
Sbjct: 779 PAGVVSGGFSTTNFGSVLQGNYLSQYHQQPYQHQQLLVNTLSSLSHPYNNSSSFKNSPFT 958
Query: 522 LE 523
+
Sbjct: 959 TQ 964
Score = 79.3 bits (194), Expect = 4e-15
Identities = 39/76 (51%), Positives = 48/76 (62%), Gaps = 1/76 (1%)
Frame = +2
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERS-LDGQITEIVYKGTHNHPKP 290
+DGY WRKYGQK VK S PRSYY+CT +C KK+VERS D I Y+G H HP P
Sbjct: 557 EDGYRWRKYGQKAVKNSPFPRSYYRCTTASCNVKKRVERSYTDPSIVVTTYEGQHTHPSP 736
Query: 291 QATRRNSSSSSSLPIP 306
+R S+ + + IP
Sbjct: 737 TMSR---SAFAGVQIP 775
>TC92972 similar to GP|9187622|emb|CAB97004.1 WRKY DNA binding protein
{Solanum tuberosum}, partial (46%)
Length = 672
Score = 104 bits (260), Expect = 8e-23
Identities = 50/106 (47%), Positives = 68/106 (63%)
Frame = +2
Query: 375 ENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGC 434
+ E S G + + + + V QT S IDILDDG+RWRKYG+K+VK N PRSYYKCT+ GC
Sbjct: 143 KKETSSQKGGKEIMKHKYVFQTRSQIDILDDGFRWRKYGEKMVKDNKFPRSYYKCTYQGC 322
Query: 435 PVRKHVERASHDLRAVITTYEGKHNHDVPAARGSGNHSVTRPLPNN 480
V+K ++R S D + V T+YEG H H V + S + + + NN
Sbjct: 323 KVKKQIQRHSKDEQVVETSYEGMHIHPVEKSAESFDQILRNFITNN 460
Score = 77.8 bits (190), Expect = 1e-14
Identities = 33/58 (56%), Positives = 42/58 (71%), Gaps = 1/58 (1%)
Frame = +2
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVER-SLDGQITEIVYKGTHNHP 288
DDG+ WRKYG+K VK ++ PRSYYKCTY C KK+++R S D Q+ E Y+G H HP
Sbjct: 230 DDGFRWRKYGEKMVKDNKFPRSYYKCTYQGCKVKKQIQRHSKDEQVVETSYEGMHIHP 403
>TC80165 similar to PIR|A84913|A84913 probable WRKY-type DNA binding protein
[imported] - Arabidopsis thaliana, partial (32%)
Length = 1013
Score = 103 bits (256), Expect = 2e-22
Identities = 63/168 (37%), Positives = 92/168 (54%), Gaps = 5/168 (2%)
Frame = +1
Query: 296 NSSSSSSLPIP---HSNHISNDIQDQSYATHGSGQMDSVATPENSSISIGDDDF--EQSS 350
N+ S S L +P + +D + Y+ + Q ATP +SSIS + ++ +
Sbjct: 337 NNYSDSLLDLPVVVKEPPLESDGNGKEYSEVLNSQQQP-ATPNSSSISSASSEAINDEHN 513
Query: 351 QKCRSGGDELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWR 410
+ ++L+ K+ K + N+ + RE R+ T S++D L+DGYRWR
Sbjct: 514 KTVDQTNNQLN------KQLKAKKTNQ-------KKPREARIAFMTKSEVDHLEDGYRWR 654
Query: 411 KYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKH 458
KYGQK VK +P PRSYY+CT C V+KHVER+ D V+TTYEGKH
Sbjct: 655 KYGQKAVKNSPFPRSYYRCTSVSCNVKKHVERSLSDPTIVVTTYEGKH 798
Score = 73.6 bits (179), Expect(2) = 2e-14
Identities = 47/115 (40%), Positives = 60/115 (51%), Gaps = 13/115 (11%)
Frame = +1
Query: 184 TENSSSMQSFSPEIASVQTNH-----NNGFQSDY-----NNYQPQQQQLQPQTLNR--RS 231
T NSSS+ S S E + + N NN N +P++ ++ T +
Sbjct: 454 TPNSSSISSASSEAINDEHNKTVDQTNNQLNKQLKAKKTNQKKPREARIAFMTKSEVDHL 633
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSL-DGQITEIVYKGTH 285
+DGY WRKYGQK VK S PRSYY+CT +C KK VERSL D I Y+G H
Sbjct: 634 EDGYRWRKYGQKAVKNSPFPRSYYRCTSVSCNVKKHVERSLSDPTIVVTTYEGKH 798
Score = 23.9 bits (50), Expect(2) = 2e-14
Identities = 11/36 (30%), Positives = 19/36 (52%)
Frame = +2
Query: 285 HNHPKPQATRRNSSSSSSLPIPHSNHISNDIQDQSY 320
+ HP P +R ++ + SL P + +N DQ+Y
Sbjct: 794 NTHPNPIMSRSSAVRAGSLLPPPAECTTNFASDQNY 901
>TC90266 similar to SP|Q9C519|WRK6_ARATH WRKY transcription factor 6 (WRKY
DNA-binding protein 6) (AtWRKY6). [Mouse-ear cress],
partial (15%)
Length = 1376
Score = 101 bits (251), Expect = 9e-22
Identities = 72/230 (31%), Positives = 107/230 (46%), Gaps = 7/230 (3%)
Frame = +2
Query: 299 SSSSLPIPHSNHISNDIQDQSYATHGSGQMDSVATPENSSISIGDDDFEQSSQKC--RSG 356
SSS H+ + + QD + + Q +P N S + G E +
Sbjct: 152 SSSLSQNNHNLLLKENTQDAGKSVLPTRQFFDEPSPSNCSKNNGFAIVENNENNMGRNLA 331
Query: 357 GDELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKV 416
+ ++E E +SK +E ++ + R RV ++ SD + DG +WRKYGQK
Sbjct: 332 CEYINEGEINSK---IEDQSSEVGC------RRARVSIRARSDFAFMVDGCQWRKYGQKT 484
Query: 417 VKGNPNPRSYYKCT-HPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSGNHSVTR 475
KGNP PR+YY+C+ CPVRK V+R D ITTYEG HNH +P A +
Sbjct: 485 AKGNPCPRAYYRCSMGTSCPVRKQVQRCFKDESVFITTYEGNHNHQLPPA--------AK 640
Query: 476 PLPNNTANAIR----PSSVTHQHHHNTNNNSLQSLRPQQAPDQVQSSPFT 521
P+ N T++A+ SS T Q + N N+ P P+ + F+
Sbjct: 641 PIANLTSSALNTFLPTSSTTLQQYGNNLTNTFLFSSPLSPPNSNAIATFS 790
Score = 67.8 bits (164), Expect = 1e-11
Identities = 46/139 (33%), Positives = 68/139 (48%), Gaps = 21/139 (15%)
Frame = +2
Query: 185 ENSSSMQSFSPEIASVQTNHNN---GFQSDYNNYQPQQQQLQPQT-----------LNRR 230
E S S S + A V+ N NN +Y N +++ Q+ + R
Sbjct: 248 EPSPSNCSKNNGFAIVENNENNMGRNLACEYINEGEINSKIEDQSSEVGCRRARVSIRAR 427
Query: 231 SD-----DGYNWRKYGQKQVKGSENPRSYYKCTY-PNCPTKKKVERSL-DGQITEIVYKG 283
SD DG WRKYGQK KG+ PR+YY+C+ +CP +K+V+R D + Y+G
Sbjct: 428 SDFAFMVDGCQWRKYGQKTAKGNPCPRAYYRCSMGTSCPVRKQVQRCFKDESVFITTYEG 607
Query: 284 THNHPKPQATRRNSSSSSS 302
HNH P A + ++ +SS
Sbjct: 608 NHNHQLPPAAKPIANLTSS 664
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.309 0.125 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,830,453
Number of Sequences: 36976
Number of extensions: 334915
Number of successful extensions: 3581
Number of sequences better than 10.0: 193
Number of HSP's better than 10.0 without gapping: 2957
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3405
length of query: 579
length of database: 9,014,727
effective HSP length: 101
effective length of query: 478
effective length of database: 5,280,151
effective search space: 2523912178
effective search space used: 2523912178
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0058a.3


