

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
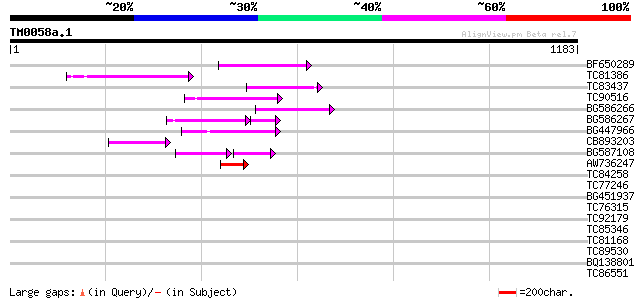
Query= TM0058a.1
(1183 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 127 2e-29
TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR re... 107 3e-23
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 103 5e-22
TC90516 100 4e-21
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 98 2e-20
BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At... 75 4e-20
BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [impo... 97 5e-20
CB893203 95 2e-19
BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - ... 59 6e-19
AW736247 weakly similar to PIR|T14619|T146 reverse transcriptase... 57 5e-08
TC84258 32 1.1
TC77246 similar to PIR|C71408|C71408 probable protein kinase - A... 32 1.5
BG451937 similar to GP|9758872|dbj| contains similarity to salt-... 32 1.9
TC76315 similar to GP|10121845|gb|AAG13395.1 cold acclimation pr... 30 5.5
TC92179 30 7.2
TC85346 similar to GP|14335160|gb|AAK59860.1 AT3g52120/F4F15_230... 30 7.2
TC81168 similar to GP|17979303|gb|AAL49877.1 unknown protein {Ar... 30 7.2
TC89530 29 9.4
BQ138801 similar to GP|12039384|gb putative urea active transpor... 29 9.4
TC86551 similar to SP|P48628|FD6C_SOYBN Omega-6 fatty acid desat... 29 9.4
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 127 bits (320), Expect = 2e-29
Identities = 61/196 (31%), Positives = 111/196 (56%), Gaps = 1/196 (0%)
Frame = +3
Query: 436 LVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRG 495
L F VE+K A++ +K+PG DG+N F + W+++ V+ A+ +F+ G P+
Sbjct: 12 LCSEFTAVEVKNALFSMDSSKAPGIDGYNVHFFKCSWNIIGDSVIDAILDFFKTGFMPKI 191
Query: 496 SNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGN 555
N +++ L+PK + +FRPI+ IYK++ K+ R++ V+ V+ E Q FV
Sbjct: 192 INCTYVTLLPKEVNVTSVKNFRPIACCSVIYKIISKILTSRMQGVLNSVVSENQSAFVKG 371
Query: 556 RNMLDSVVVLNEVMHEAKVKK-RPSIFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGW 614
R + D++++ +E++ K P K+D KAY+S EW F++++M + F K++ W
Sbjct: 372 RVIFDNIILSHELVKSYSRKGISPRCMVKIDLXKAYDSXEWPFIKHLMLELGFPYKFVNW 551
Query: 615 IMGCLNSATVSVLVNG 630
+M L +A+ + NG
Sbjct: 552 VMAXLTTASYTFNXNG 599
>TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR retroelement
reverse transcriptase {Oryza sativa (japonica
cultivar-group)}, partial (1%)
Length = 798
Score = 107 bits (267), Expect = 3e-23
Identities = 72/269 (26%), Positives = 125/269 (45%), Gaps = 4/269 (1%)
Frame = -1
Query: 118 KRAMWDALCAWRGTCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRREMREFNVFIANMD 177
+R +W A+ + T WC GDFN + + E +GS + R M +F + +
Sbjct: 795 RRQLWSAISNIQ-TQHKLSWCCIGDFNTILGSHEHQGSH---TPARLPMLDFQQWSDVNN 628
Query: 178 LVDIPLTGRRFTW---RRANDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLM 234
L +P G FTW RR + + R+DR +V+ D + S L + SDH P+L
Sbjct: 627 LFHLPTRGSAFTWTNGRRGRNNTRKRLDRSIVNQLMIDNCESLSACTLTKLRSDHFPILF 448
Query: 235 R-QVVADWGPKPFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI* 293
Q F+ + W P ++Q W ++ G +VL +KLK L+ L++
Sbjct: 447 ELQTQNIQFSSSFKFMKMWSAHPDCINIIKQCWAN-QVVGCPMFVLNQKLKNLKEVLKVW 271
Query: 294 NKEVFGDLKTRREKVVQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQK 353
NK FG++ ++ + ++++ + VK + +G + + + + L +E F +K
Sbjct: 270 NKNTFGNVHSQVDNAYKELDDIQVKIDSIGYSDVLMDQEKAAQLNLESALNIEEVFWHEK 91
Query: 354 ACMKWLVEGDLNSGFFHSTINWKRRADSI 382
+ + W EGD N+ FFH KR + I
Sbjct: 90 SKVNWHCEGDRNTAFFHRVAKIKRTSSLI 4
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 103 bits (256), Expect = 5e-22
Identities = 67/160 (41%), Positives = 97/160 (59%), Gaps = 2/160 (1%)
Frame = +2
Query: 494 RGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFV 553
+G N++FI LIPKV PQ LNDFRPISL+G +YK++ KL A RLR+VIG VI + Q FV
Sbjct: 44 KGINSTFIALIPKVDNPQRLNDFRPISLVGSLYKILGKLLANRLRVVIGSVISDAQSAFV 223
Query: 554 GNRNMLDSVVVLNEVMHEAKVKKRPSIFFKVDY-EKAYNSVEWGFLEYMM-GRMNFCAKW 611
NR +L+ +V L ++ +++ IF + + K ++ G + + M+F W
Sbjct: 224 KNRQILE-MVFL*QMRLWMRLRN*RKIFCCLRWILKRLITLSIGLIWILF*VGMSFLVLW 400
Query: 612 IGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQGILLRRF 651
WI C+++AT SVLVNGSP+ + K L Q L R+
Sbjct: 401 RKWIKECVSTATTSVLVNGSPTNV--LMKSLVQTQLFTRY 514
>TC90516
Length = 983
Score = 100 bits (248), Expect = 4e-21
Identities = 64/205 (31%), Positives = 105/205 (51%)
Frame = -2
Query: 365 NSGFFHSTINWKRRADSIVGLMVDGMWEDEPVRVKAAIHDFFQKKFKSVAWLRPKLDGVQ 424
N+ +F + + + +SI V+ W D+ + K AI +FF F RP + +
Sbjct: 742 NTTYFLACVKNRGMRNSISAPRVE-RWMDDVAKNKQAIVNFFTHHFSDP*TYRPTMGDID 566
Query: 425 FSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVR 484
F I++ DN L F +++ V G +SP PDGFN F R ++L D+
Sbjct: 565 FFHISNLDNVLLSAQFLVSKMELVVSSLDGNESPRPDGFNLNFFIRLRNMLKADIEIMFE 386
Query: 485 EFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKV 544
+F+ + + F+ LI KV+ P LL DF +S +G + K++ K+ A+RL ++ K+
Sbjct: 385 QFYTPANLLKIFS*YFLTLISKVEYPILLGDFSLMSFLGSL*KLMAKVLALRLAHIMEKI 206
Query: 545 IDERQFTFVGNRNMLDSVVVLNEVM 569
I Q TFV R +D VV +NE++
Sbjct: 205 IFVNQSTFVRGRQHVDGVVAINEII 131
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 98.2 bits (243), Expect = 2e-20
Identities = 53/171 (30%), Positives = 93/171 (53%), Gaps = 6/171 (3%)
Frame = -3
Query: 513 LNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMH-- 570
++++R I+ YK++ K+ + R++ ++ +I Q FV R + D+V++ ++++H
Sbjct: 778 VSEYRTIAPCNTQYKIIAKILSKRMQPLLRSIISPSQSAFVPGRAISDNVLITHKILHYL 599
Query: 571 -EAKVKKRPSIFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVN 629
++ KK S+ K D KAY+ + W FL ++ R+ F WI WIM C+++ + S L+N
Sbjct: 598 RQSGAKKHVSMAVKTDMTKAYDRIAWNFLREVLTRLGFHGIWISWIMECVSTVSYSFLIN 419
Query: 630 GSPSGEFHMEKGLRQGILLRRFFS*LLLKA*MGYSRMQLELGNL---KVSR 677
G P G +GLRQG L + L + G + L G L KV+R
Sbjct: 418 GGPQGRVLPSRGLRQGDPLSPYLFILCTEVLSGLCQQALRKGTLPGVKVAR 266
>BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (16%)
Length = 794
Score = 75.5 bits (184), Expect(2) = 4e-20
Identities = 52/178 (29%), Positives = 83/178 (46%), Gaps = 3/178 (1%)
Frame = +3
Query: 327 EELVEMRDLLIEFWRVLRYQESFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSIVGLM 386
EEL +R L E + +E F QK+ + WL GD N+ FFH+ +R + I+ L+
Sbjct: 75 EELSLLRSELNEEYH---NEEIFWMQKSRLNWLRSGDRNTKFFHAVTKNRRAQNRILSLI 245
Query: 387 VDGMWE---DEPVRVKAAIHDFFQKKFKSVAWLRPKLDGVQFSTITDRDNTELVGPFDEV 443
D E +E + A H + V + + + +T+ N +L+
Sbjct: 246 DDDDKEWFVEEDLGRLADSHFKLLYSSEDVGITLEDWNSIP-AIVTEEQNAQLMAQISRE 422
Query: 444 EIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFI 501
E+++AV++ K PGPDG N F Q+FWD + D+ +EF G G N + I
Sbjct: 423 EVREAVFDINPHKCPGPDGMNVFFFQQFWDTMGDDLTSMAQEFLRTGKLEEGINKTNI 596
Score = 42.0 bits (97), Expect(2) = 4e-20
Identities = 22/62 (35%), Positives = 37/62 (59%)
Frame = +1
Query: 503 LIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLDSV 562
L+PK + L +FRPISL YK+V K+ + RL+ V+ +I E Q F + + D++
Sbjct: 601 LVPKKLEAKRLVEFRPISLCNVAYKIVSKVLSKRLKSVLPWIITETQAAFGRRQLISDNI 780
Query: 563 VV 564
++
Sbjct: 781 LI 786
>BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 687
Score = 96.7 bits (239), Expect = 5e-20
Identities = 63/212 (29%), Positives = 102/212 (47%), Gaps = 5/212 (2%)
Frame = +2
Query: 358 WLVEGDLNSGFFHSTINWKRRADSIVGLMVD-GMWEDEPVRVKAAIHDFFQKKFKSVAWL 416
WL +GD N+ FFHS + +R+ + I L + G W V+ + +F F S
Sbjct: 5 WLKDGDKNTKFFHSKASQRRKVNEIKKLKDETGNWCKGEENVERLLITYFNNLFTSS--- 175
Query: 417 RPKLDGVQFSTITDRDNTELV----GPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFW 472
P + + + E + F E E+ +A+ + K+PGPDG F Q++W
Sbjct: 176 NPTAIEETCEVVKGKLSHEHIVWCEKEFTEEEVLEAINQMHPVKAPGPDGLPALFFQKYW 355
Query: 473 DVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKL 532
++ +V + V + N +FIVLIPK K P D+RPISL + K++ K+
Sbjct: 356 HIVGKEVQQMVLQVLNNSMETEELNKTFIVLIPKGKNPNTPKDYRPISLCNVVMKIITKV 535
Query: 533 FAIRLRLVIGKVIDERQFTFVGNRNMLDSVVV 564
A R++ + VID Q FV R + D+ ++
Sbjct: 536 IANRVKQTLPDVIDVEQSAFVQGRLITDNALI 631
>CB893203
Length = 800
Score = 94.7 bits (234), Expect = 2e-19
Identities = 47/130 (36%), Positives = 69/130 (52%)
Frame = -3
Query: 206 VSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQVVADWGPKPFRVLNCWLDDPRLFPFVEQS 265
VS SWC WPN L R +SD CP+++ +WGP+ +L CW D P FV+
Sbjct: 798 VSESWCIKWPNMIQRALFRGLSDRCPIMLTIDEGNWGPRLHHMLKCWADLPGYHLFVKDK 619
Query: 266 WKGMKIQGWGAYVLKEKLKGLRGSLRI*NKEVFGDLKTRREKVVQKINLLDVKEEEVGLQ 325
W ++ WG YVLKEK K +R +L+ + +L R + + I LD K E++ L
Sbjct: 618 WNSFQVSRWGGYVLKEK*KMVRNNLKDWHHNHTRNLDARINDIRESIAYLDSKGEDLTLD 439
Query: 326 PEELVEMRDL 335
EE+ ++ L
Sbjct: 438 TEEVDDLHTL 409
>BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - Arabidopsis
thaliana, partial (17%)
Length = 677
Score = 58.5 bits (140), Expect(2) = 6e-19
Identities = 31/87 (35%), Positives = 45/87 (51%)
Frame = +3
Query: 467 FIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIY 526
F Q W ++ D++K V F G N + I LIPK K P + + RPISL Y
Sbjct: 411 FFQHSWHIIKMDLLKMVNSFLASGKLDTRLNTTNICLIPKKKRPTRMTELRPISLCNVGY 590
Query: 527 KVVPKLFAIRLRLVIGKVIDERQFTFV 553
K++ K+ RL++ + +I E Q FV
Sbjct: 591 KIISKVLCQRLKVCLPSLISETQSAFV 671
Score = 55.1 bits (131), Expect(2) = 6e-19
Identities = 38/120 (31%), Positives = 60/120 (49%), Gaps = 3/120 (2%)
Frame = +2
Query: 346 QESFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSIVGLM-VDGMWEDEPVRVKAAIHD 404
+E + QK+ W + GDLN F+H+ + + IVGL DG W E V+ D
Sbjct: 41 EEDYWQQKSRNMWHISGDLNKKFYHALTKQRHARNRIVGLYDYDGNWITEEQGVEKVAVD 220
Query: 405 FFQKKFK--SVAWLRPKLDGVQFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDG 462
+F+ F+ + LD + S+IT + N L+ E E++ A++ K+PGPDG
Sbjct: 221 YFEDLFQRTTPTGFDGFLDEIT-SSITPQMNQRLLRLATEEEVRLALFIMHPEKAPGPDG 397
>AW736247 weakly similar to PIR|T14619|T146 reverse transcriptase - beet
retrotransposon (fragment), partial (4%)
Length = 305
Score = 56.6 bits (135), Expect = 5e-08
Identities = 23/60 (38%), Positives = 38/60 (63%), Gaps = 2/60 (3%)
Frame = +1
Query: 440 FDEVEIKQAVWECGGTKS--PGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSN 497
F E E+++AVW+C + S PGPDG NF F++ +W+++ D ++ V EF G +G +
Sbjct: 121 FSEEEVRKAVWDCDSSHS*NPGPDGVNFTFVKEYWELIKVDFLRVVMEFHTNGKLGKGDD 300
>TC84258
Length = 758
Score = 32.3 bits (72), Expect = 1.1
Identities = 17/44 (38%), Positives = 24/44 (53%), Gaps = 2/44 (4%)
Frame = -2
Query: 794 LLGGVFVWFRVCYL--LCLCFFYLFSEFLAVLSGSAIRSCVIFC 835
++G F+ F + +L C CFFY F F G +R CV+FC
Sbjct: 199 VIGFWFIGFLLVFLQHYCCCFFYQFCGF-----GWPVRFCVLFC 83
>TC77246 similar to PIR|C71408|C71408 probable protein kinase - Arabidopsis
thaliana, partial (65%)
Length = 1715
Score = 32.0 bits (71), Expect = 1.5
Identities = 13/38 (34%), Positives = 23/38 (60%)
Frame = +1
Query: 668 LELGNLKVSRLGQEAILWCLYCSLLMILCFWVRLHFRI 705
L LG + G+ +LW L C L+++C W+ L++R+
Sbjct: 1099 LRLGRMVQLDWGKGRLLWLLRCLRLLMICCWLLLNWRM 1212
>BG451937 similar to GP|9758872|dbj| contains similarity to salt-inducible
protein~gene_id:K24G6.6 {Arabidopsis thaliana}, partial
(18%)
Length = 670
Score = 31.6 bits (70), Expect = 1.9
Identities = 22/65 (33%), Positives = 34/65 (51%), Gaps = 3/65 (4%)
Frame = -1
Query: 801 WFRVCYLLCLCFFYLFSEFLAVLSGSAIRSCVIFCAVLLRGEG---KLRG*NGLMSANLR 857
+FR+ +LL FFY S L +++R +I+C + GEG ++ NGL S +
Sbjct: 331 YFRILHLLS--FFYFLSFHLLHFLRNSLRPFIIYC*LNCSGEG*RWRIGLGNGLCSGSET 158
Query: 858 MLVGW 862
VGW
Sbjct: 157 GGVGW 143
>TC76315 similar to GP|10121845|gb|AAG13395.1 cold acclimation protein
WCOR413-like protein {Oryza sativa} [Oryza sativa
(japonica cultivar-group), partial (69%)
Length = 908
Score = 30.0 bits (66), Expect = 5.5
Identities = 14/30 (46%), Positives = 16/30 (52%)
Frame = +1
Query: 790 ISWYLLGGVFVWFRVCYLLCLCFFYLFSEF 819
I WY LG F+W +LC C YLF F
Sbjct: 187 IRWYRLGRFFLW-----ILCCCCCYLFVGF 261
>TC92179
Length = 754
Score = 29.6 bits (65), Expect = 7.2
Identities = 13/31 (41%), Positives = 17/31 (53%), Gaps = 1/31 (3%)
Frame = -3
Query: 799 FVWFRVCYLLCLCFFYLF-SEFLAVLSGSAI 828
F WFR C C CFF+ F S L L +++
Sbjct: 233 FFWFR*CKF*CYCFFFFFWSMMLLFLKNTSV 141
>TC85346 similar to GP|14335160|gb|AAK59860.1 AT3g52120/F4F15_230
{Arabidopsis thaliana}, partial (12%)
Length = 1182
Score = 29.6 bits (65), Expect = 7.2
Identities = 16/41 (39%), Positives = 23/41 (56%)
Frame = -3
Query: 394 EPVRVKAAIHDFFQKKFKSVAWLRPKLDGVQFSTITDRDNT 434
EP+ K I D +++ V WLR K++ V FS D DN+
Sbjct: 562 EPLH-KRTIIDKEGRRWSLVHWLRSKINNVAFSNSLDEDNS 443
>TC81168 similar to GP|17979303|gb|AAL49877.1 unknown protein {Arabidopsis
thaliana}, partial (34%)
Length = 489
Score = 29.6 bits (65), Expect = 7.2
Identities = 13/35 (37%), Positives = 19/35 (54%)
Frame = +1
Query: 786 HLGSISWYLLGGVFVWFRVCYLLCLCFFYLFSEFL 820
H + +W+ V W CYLLC C++ L S F+
Sbjct: 217 HSFTRTWFDYDFVGSWCCWCYLLCCCYYQLCSFFV 321
>TC89530
Length = 1614
Score = 29.3 bits (64), Expect = 9.4
Identities = 15/24 (62%), Positives = 21/24 (87%), Gaps = 2/24 (8%)
Frame = -3
Query: 803 RVCYLL-CLCFFYLF-SEFLAVLS 824
+VCYLL CL +F+LF S+FL+V+S
Sbjct: 760 QVCYLLICLFYFFLFTSQFLSVVS 689
>BQ138801 similar to GP|12039384|gb putative urea active transport protein
{Oryza sativa}, partial (20%)
Length = 517
Score = 29.3 bits (64), Expect = 9.4
Identities = 25/81 (30%), Positives = 40/81 (48%), Gaps = 2/81 (2%)
Frame = +2
Query: 923 IGLMKLFLSRFGEVMQLHSGWMIGPGGVLLGSSFIACIICPL*SVLLLRA--VVTGIKTN 980
+GL+ + L++ G + GWM GVL+GS+ I P+ +LL R + I
Sbjct: 107 MGLLAVILNKAGVSL----GWMYLAMGVLIGSAVI-----PIAFMLLWRKANAIGAILGT 259
Query: 981 GGGIYLGDGRWVCVSKAGWGR 1001
G LG W+ V+K +G+
Sbjct: 260 VFGCVLGIITWLSVTKIEYGK 322
>TC86551 similar to SP|P48628|FD6C_SOYBN Omega-6 fatty acid desaturase
chloroplast precursor (EC 1.14.99.-). [Soybean] {Glycine
max}, partial (84%)
Length = 1629
Score = 29.3 bits (64), Expect = 9.4
Identities = 17/76 (22%), Positives = 31/76 (40%)
Frame = +3
Query: 178 LVDIPLTGRRFTWRRANDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQV 237
L +PL WR +D+ ++ + T+W VW +D + P L + +
Sbjct: 597 LAFLPLIYPYEPWRFKHDKHHAKTNMLKEDTAWQPVW---------QDEFESSPFLRKAI 749
Query: 238 VADWGPKPFRVLNCWL 253
+ +GP CW+
Sbjct: 750 IYGYGP-----FRCWM 782
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.339 0.151 0.528
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 45,758,174
Number of Sequences: 36976
Number of extensions: 805057
Number of successful extensions: 6847
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 3755
Number of HSP's successfully gapped in prelim test: 339
Number of HSP's that attempted gapping in prelim test: 2870
Number of HSP's gapped (non-prelim): 4457
length of query: 1183
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1076
effective length of database: 5,058,295
effective search space: 5442725420
effective search space used: 5442725420
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0058a.1


