

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
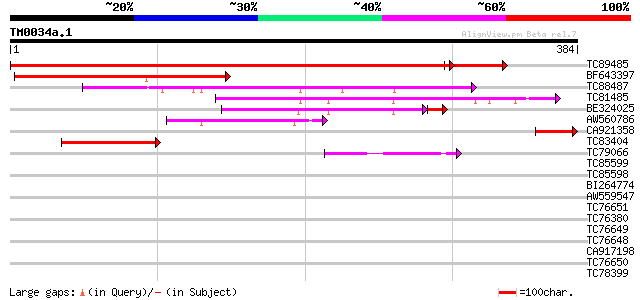
Query= TM0034a.1
(384 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89485 homologue to SP|O04278|GBA1_PEA Guanine nucleotide-bindi... 560 e-177
BF643397 homologue to GP|22770980|gb G protein alpha I subunit {... 179 2e-45
TC88487 similar to PIR|T01135|T01135 probable GTP-binding protei... 147 7e-36
TC81485 weakly similar to PIR|T01135|T01135 probable GTP-binding... 128 3e-30
BE324025 similar to PIR|E86443|E86 probable G-protein alpha subu... 84 1e-17
AW560786 similar to GP|7363278|dbj ESTs C74776(E51022) C26123(C1... 60 2e-09
CA921358 GP|22476942|gb G protein alpha II subunit {Pisum sativu... 57 1e-08
TC83404 similar to PIR|E86443|E86443 probable G-protein alpha su... 53 2e-07
TC79066 homologue to PIR|T51561|T51561 ADP-ribosylation factor-l... 42 4e-04
TC85599 homologue to SP|O04834|SARA_ARATH GTP-binding protein SA... 40 0.002
TC85598 homologue to SP|O04834|SARA_ARATH GTP-binding protein SA... 39 0.003
BI264774 homologue to PIR|D49993|D49 ADP-ribosylation factor - A... 39 0.004
AW559547 weakly similar to GP|12654451|gb| ADP-ribosylation fact... 39 0.004
TC76651 PIR|T52341|T52341 ADP-ribosylation factor [imported] - r... 38 0.007
TC76380 homologue to PIR|T52341|T52341 ADP-ribosylation factor [... 38 0.007
TC76649 homologue to GP|20161472|dbj|BAB90396. ADP-ribosylation ... 38 0.007
TC76648 homologue to PIR|T52341|T52341 ADP-ribosylation factor [... 38 0.007
CA917198 GP|20161472|db ADP-ribosylation factor {Oryza sativa (j... 38 0.007
TC76650 GP|965483|dbj|BAA08259.1| DcARF1 {Daucus carota}, complete 38 0.007
TC78399 homologue to SP|O04834|SARA_ARATH GTP-binding protein SA... 37 0.010
>TC89485 homologue to SP|O04278|GBA1_PEA Guanine nucleotide-binding protein
alpha-1 subunit (GP-alpha-1). [Garden pea] {Pisum
sativum}, partial (87%)
Length = 1164
Score = 560 bits (1444), Expect(2) = e-177
Identities = 281/301 (93%), Positives = 287/301 (94%)
Frame = +2
Query: 1 MGLLCSKNRRYNDADTEENAQTAEIERRIELETKAEKHIQKLLLLGAGESGKSTIFKQIK 60
MGLLCSK+RRYNDA+TEENAQTAEIERRIELETKAEKHIQKLLLLGAGESGKSTIFKQIK
Sbjct: 152 MGLLCSKSRRYNDANTEENAQTAEIERRIELETKAEKHIQKLLLLGAGESGKSTIFKQIK 331
Query: 61 LLFQTGFDEAELKSYQPVIHANVYQTIKLLHDGAKELAQNDVDFSKYVISDENKEIGEKL 120
LLFQTGFDEAELKSY PVIHANVYQTIKLLHDG+KE AQNDVDFSKYVIS ENK+IGEKL
Sbjct: 332 LLFQTGFDEAELKSYLPVIHANVYQTIKLLHDGSKEFAQNDVDFSKYVISGENKDIGEKL 511
Query: 121 SEIGGRLDYPCLTKELALEIENLWKDAAIQETYARGNELQVPDCTHYFMENLHRLSDANY 180
SEIGGRLDYP LTKELA EIE LWKD AIQETY+RGNELQVPDCTHYFMENL RLSDANY
Sbjct: 512 SEIGGRLDYPRLTKELAQEIECLWKDPAIQETYSRGNELQVPDCTHYFMENLQRLSDANY 691
Query: 181 VPTKDDVLYARVRTTGVVEIQFSPVGENKKSGEVYRLFDVGGQRNERRKWIHLFEGVSAV 240
VPTK+DVL ARVRTTGVVEIQFSPVGENKKSGEVYRLFDVGGQRNERRKWIHLFEGVSAV
Sbjct: 692 VPTKEDVLLARVRTTGVVEIQFSPVGENKKSGEVYRLFDVGGQRNERRKWIHLFEGVSAV 871
Query: 241 IFCAAISEYDQTLFEDENKNRMMETKELFEWVLKQPCFEKTSFMLFLNKFDIFEKKILKV 300
IFC AISEYDQTLFEDENKNRMMETKELFEWVLKQ CFEKTSFMLFLNKFDIFEKKI
Sbjct: 872 IFCVAISEYDQTLFEDENKNRMMETKELFEWVLKQQCFEKTSFMLFLNKFDIFEKKIPGC 1051
Query: 301 P 301
P
Sbjct: 1052P 1054
Score = 79.3 bits (194), Expect(2) = e-177
Identities = 35/43 (81%), Positives = 38/43 (87%)
Frame = +3
Query: 295 KKILKVPLNVCEWFKDYQPVSTGKQEIEHAYEFVKKKFEESYF 337
++ L VPLNVCEW KDY PVSTGKQEIEHAY+FV KKFEESYF
Sbjct: 1035 RRSLDVPLNVCEWXKDYXPVSTGKQEIEHAYQFVXKKFEESYF 1163
>BF643397 homologue to GP|22770980|gb G protein alpha I subunit {Pisum
sativum}, partial (38%)
Length = 531
Score = 179 bits (454), Expect = 2e-45
Identities = 100/153 (65%), Positives = 113/153 (73%), Gaps = 7/153 (4%)
Frame = +2
Query: 4 LCSKNRRYNDADTEENAQTAEIERRIELETKAEKHIQKLLLLGAGESGKSTIFKQIKLLF 63
LCSK+RRYNDA+TEENAQTAEIERRIELETKAEKHIQKLLLLGAGESGKSTIFKQIKLLF
Sbjct: 71 LCSKSRRYNDANTEENAQTAEIERRIELETKAEKHIQKLLLLGAGESGKSTIFKQIKLLF 250
Query: 64 QTGFDEAELKSYQPVIHANVYQTIKLLH-------DGAKELAQNDVDFSKYVISDENKEI 116
QTGFDEAELKSY PVIHANVYQTIK+ + K L + + F + + + +
Sbjct: 251 QTGFDEAELKSYLPVIHANVYQTIKVRNTWKGYFMTDRKSLPRMMLIFRSMLYPVKIRTL 430
Query: 117 GEKLSEIGGRLDYPCLTKELALEIENLWKDAAI 149
+ ++ L P KELA EIE LWKD AI
Sbjct: 431 AKSYQKLEAGLITPVSLKELAQEIECLWKDPAI 529
>TC88487 similar to PIR|T01135|T01135 probable GTP-binding protein (extra
large) [imported] - Arabidopsis thaliana, partial (41%)
Length = 1513
Score = 147 bits (371), Expect = 7e-36
Identities = 99/311 (31%), Positives = 163/311 (51%), Gaps = 44/311 (14%)
Frame = +3
Query: 50 SGKSTIFKQIKLLFQT-GFDEAELKSYQPVIHANVYQTIKLLHDGAKELAQNDV------ 102
SG STIFKQ K+L+++ F E E ++ I +NVY + +L +G +E ++++
Sbjct: 45 SGTSTIFKQAKILYKSIPFSEDEHENIILTIQSNVYTYLGILLEG-RERFEDEILADLTK 221
Query: 103 ----------------DFSKYVISDENKEIGEKLSEI--GGRLD--YPCLTKELALEIEN 142
D + Y I K + L + G+L+ +P T+E A IE
Sbjct: 222 RQSSMLDTTGTNPKPDDKTVYSIGPRLKAFSDWLLKTMASGKLEAIFPAATREYAPLIEE 401
Query: 143 LWKDAAIQETYARGNELQV-PDCTHYFMENLHRLSDANYVPTKDDVLYARVRTT--GVVE 199
LW D AI+ TY R +EL++ P YF+E ++ +Y P+ D+LYA T+ G+
Sbjct: 402 LWNDTAIEATYERRSELEMLPSVATYFLERAVKILRTDYEPSDLDILYAEGVTSSNGLAC 581
Query: 200 IQFS-PVGENKKSGEVYRLFDVGGQR-----------NERRKWIHLFEGVSAVIFCAAIS 247
++FS P +++ + +D R E KW+ +FE V VIFC ++S
Sbjct: 582 VEFSFPQSAPEETVDTTDQYDSLASRYQLIRVHARGLGENCKWLEMFEDVEMVIFCVSLS 761
Query: 248 EYDQTLFEDENK--NRMMETKELFEWVLKQPCFEKTSFMLFLNKFDIFEKKILKVPLNVC 305
+YDQ + N+M+ + + FE ++ P FE+ F+L LNKFD+FE+K+ +VPL C
Sbjct: 762 DYDQFSVDGNGSLTNKMILSMKFFETMVTHPTFEQMEFLLILNKFDLFEEKVEQVPLTKC 941
Query: 306 EWFKDYQPVST 316
+WF D+ P+++
Sbjct: 942 DWFSDFHPITS 974
>TC81485 weakly similar to PIR|T01135|T01135 probable GTP-binding protein
(extra large) [imported] - Arabidopsis thaliana, partial
(21%)
Length = 1181
Score = 128 bits (322), Expect = 3e-30
Identities = 86/275 (31%), Positives = 136/275 (49%), Gaps = 41/275 (14%)
Frame = +2
Query: 140 IENLWKDAAIQETYARGNELQV-PDCTHYFMENLHRLSDANYVPTKDDVLYARVRTT--G 196
+E LW D+AI+ TY R NE+++ P YF+E ++ +Y P+ D+LYA T+ G
Sbjct: 26 VEELWNDSAIKATYERRNEIEMLPSVASYFLERAVQILTTDYEPSDLDILYAEGVTSSNG 205
Query: 197 VVEIQFS-PVGENKKSGEV---------YRLFDVGGQR-NERRKWIHLFEGVSAVIFCAA 245
+ ++FS P +++G+ Y L + + + KW+ +FE V VIFC +
Sbjct: 206 LASVEFSFPQSTPEETGDTDYLHDSLARYELITIHARGLGDNCKWLEMFEDVGLVIFCVS 385
Query: 246 ISEYDQTLFEDEN--KNRMMETKELFEWVLKQPCFEKTSFMLFLNKFDIFEKKILKVPLN 303
+S+Y+Q + N+M+ +++LFE ++ P FEK F+L LNK D FE+KI +VPL
Sbjct: 386 LSDYNQFSTDGNGCVTNKMILSRKLFETIVTHPTFEKMDFLLILNKLDEFEQKIEQVPLT 565
Query: 304 VCEWFKDYQPV------STGKQEIEH-------AYEFVKKKFEESYFQNTA--------- 341
C+WF D+ PV + I + A ++ KF+ Y T
Sbjct: 566 QCDWFSDFHPVISHNRPGSNNNSINNNPSISQLASHYIAVKFKRLYSSLTGQNLYVSLVK 745
Query: 342 ---PDRVDRVFKIYRTTALDQKVVKKTFKLVDETL 373
PD VD K Y L K F L D+++
Sbjct: 746 GLEPDSVDASLK-YGKEILKWNEEKPNFSLSDDSM 847
>BE324025 similar to PIR|E86443|E86 probable G-protein alpha subunit
[imported] - Arabidopsis thaliana, partial (19%)
Length = 540
Score = 84.0 bits (206), Expect(2) = 1e-17
Identities = 57/164 (34%), Positives = 88/164 (52%), Gaps = 24/164 (14%)
Frame = +2
Query: 144 WKDAAIQETYARGNELQ-VPDCTHYFMENLHRLSDANYVPTKDDVLYARVRT--TGVVEI 200
WKD AIQET+ R +EL +PD YF+ +S +Y P++ D+LYA T G+ +
Sbjct: 8 WKDPAIQETFKRKDELHFLPDVAEYFLSRAVEISSNDYEPSERDILYAEGVTQGNGLAFM 187
Query: 201 QFSPVGENKKSGEV-------------YRLFDVGGQ-RNERRKWIHLFEGVSAVIFCAAI 246
+FS + K Y+L V + +E KW+ +FE V AV+FC ++
Sbjct: 188 EFSLDDRSPKFDAYTDNLDAQLQPMTKYQLIRVNAKGMSEGCKWVEMFEDVRAVVFCVSL 367
Query: 247 SEYDQ-TLFEDEN------KNRMMETKELFEWVLKQPCFEKTSF 283
S+YDQ +L D N +N+M+++KELF + P F++ SF
Sbjct: 368 SDYDQLSLAPDSNGSGTLLQNKMIQSKELF*NNGETPLFQRHSF 499
Score = 23.1 bits (48), Expect(2) = 1e-17
Identities = 8/13 (61%), Positives = 12/13 (91%)
Frame = +1
Query: 284 MLFLNKFDIFEKK 296
+L LNK+D+FE+K
Sbjct: 502 ILVLNKYDVFEEK 540
>AW560786 similar to GP|7363278|dbj ESTs C74776(E51022) C26123(C116681)
correspond to a region of the predicted gene.~Similar
to, partial (16%)
Length = 525
Score = 59.7 bits (143), Expect = 2e-09
Identities = 36/116 (31%), Positives = 58/116 (49%), Gaps = 7/116 (6%)
Frame = +3
Query: 107 YVISDENKEIGEKLSEIGGRLD----YPCLTKELALEIENLWKDAAIQETYARGNEL-QV 161
Y I+ K + L +I D +P T+E A ++ +WKD A+ ETY R EL +
Sbjct: 165 YSINQRFKHFSDWLLDIMATGDLEAFFPAATREYAPMVDEIWKDPAVHETYKRREELHNL 344
Query: 162 PDCTHYFMENLHRLSDANYVPTKDDVLYAR--VRTTGVVEIQFSPVGENKKSGEVY 215
PD YF++ +S Y P+ D+LYA ++ G+ ++FS + E+Y
Sbjct: 345 PDVAKYFLDRAIEISSNEYEPSDKDILYAERVTQSNGLAFMEFS-FDDRSPMSEIY 509
>CA921358 GP|22476942|gb G protein alpha II subunit {Pisum sativum}, partial
(7%)
Length = 410
Score = 57.0 bits (136), Expect = 1e-08
Identities = 28/28 (100%), Positives = 28/28 (100%)
Frame = -3
Query: 357 LDQKVVKKTFKLVDETLRRRNLFEAGLL 384
LDQKVVKKTFKLVDETLRRRNLFEAGLL
Sbjct: 408 LDQKVVKKTFKLVDETLRRRNLFEAGLL 325
>TC83404 similar to PIR|E86443|E86443 probable G-protein alpha subunit
[imported] - Arabidopsis thaliana, partial (38%)
Length = 1140
Score = 52.8 bits (125), Expect = 2e-07
Identities = 27/67 (40%), Positives = 41/67 (60%)
Frame = +2
Query: 36 EKHIQKLLLLGAGESGKSTIFKQIKLLFQTGFDEAELKSYQPVIHANVYQTIKLLHDGAK 95
+K QKLLLLG SG ST+FKQ K L+ F + EL+ + +I +N+Y+ + +L DG +
Sbjct: 764 QKKTQKLLLLGIQGSGTSTMFKQAKFLYGNKFTDQELEDVKLMIQSNMYKYLSILLDGRE 943
Query: 96 ELAQNDV 102
+ V
Sbjct: 944 RFEEEVV 964
>TC79066 homologue to PIR|T51561|T51561 ADP-ribosylation factor-like protein
- Arabidopsis thaliana, partial (95%)
Length = 839
Score = 42.0 bits (97), Expect = 4e-04
Identities = 22/93 (23%), Positives = 44/93 (46%)
Frame = +2
Query: 214 VYRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVL 273
V+ ++DVGGQ R W H F +I+ + + ++ R+ + K+ F+ ++
Sbjct: 263 VFTVWDVGGQEKLRPLWRHYFNNTDGLIY----------VVDSLDRERISQAKQEFQAII 412
Query: 274 KQPCFEKTSFMLFLNKFDIFEKKILKVPLNVCE 306
P + ++F NK D+ + P+ +CE
Sbjct: 413 NDPFMLNSVILVFANKQDL---RGAMTPMEICE 502
>TC85599 homologue to SP|O04834|SARA_ARATH GTP-binding protein SAR1A.
[Mouse-ear cress] {Arabidopsis thaliana}, complete
Length = 999
Score = 39.7 bits (91), Expect = 0.002
Identities = 25/90 (27%), Positives = 41/90 (44%), Gaps = 1/90 (1%)
Frame = +2
Query: 204 PVGENKKSGEV-YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRM 262
P E G++ ++ FD+GG + RR W + V AV++ + YD K R
Sbjct: 314 PTSEELSIGKIKFKAFDLGGHQIARRVWKDYYAKVDAVVY--LVDAYD--------KERF 463
Query: 263 METKELFEWVLKQPCFEKTSFMLFLNKFDI 292
E+K+ + +L F++ NK DI
Sbjct: 464 AESKKELDALLSDESLANVPFLILGNKIDI 553
>TC85598 homologue to SP|O04834|SARA_ARATH GTP-binding protein SAR1A.
[Mouse-ear cress] {Arabidopsis thaliana}, complete
Length = 900
Score = 39.3 bits (90), Expect = 0.003
Identities = 25/90 (27%), Positives = 41/90 (44%), Gaps = 1/90 (1%)
Frame = +3
Query: 204 PVGENKKSGEV-YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRM 262
P E G++ ++ FD+GG + RR W + V AV++ + YD K R
Sbjct: 333 PTSEELSIGKIKFKAFDLGGHQIARRVWKDYYAQVDAVVY--LVDAYD--------KERF 482
Query: 263 METKELFEWVLKQPCFEKTSFMLFLNKFDI 292
E+K+ + +L F++ NK DI
Sbjct: 483 AESKKELDALLSDESLANVPFLVLGNKIDI 572
>BI264774 homologue to PIR|D49993|D49 ADP-ribosylation factor - Ajellomyces
capsulata, partial (91%)
Length = 665
Score = 38.5 bits (88), Expect = 0.004
Identities = 22/78 (28%), Positives = 37/78 (47%)
Frame = +3
Query: 215 YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLK 274
+ ++DVGGQ R W H F+ +IF ++ D+ + E RM+ EL + +L
Sbjct: 153 FTVWDVGGQDKIRPLWRHYFQNTQGIIFVVDSNDRDRIVEAREELQRMLNEDELRDAIL- 329
Query: 275 QPCFEKTSFMLFLNKFDI 292
++F NK D+
Sbjct: 330 ---------LVFANKQDL 356
>AW559547 weakly similar to GP|12654451|gb| ADP-ribosylation factor-like 7
{Homo sapiens}, partial (22%)
Length = 394
Score = 38.5 bits (88), Expect = 0.004
Identities = 17/60 (28%), Positives = 30/60 (49%)
Frame = +2
Query: 209 KKSGEVYRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKEL 268
KK G + +D+GGQ R +W G +I+ + +DQ + +R++E +EL
Sbjct: 176 KKGGVQMKCWDIGGQAQYRSEWGRYTRGCDVIIYVVDANAFDQISLARKELHRLLEDREL 355
>TC76651 PIR|T52341|T52341 ADP-ribosylation factor [imported] - rice,
complete
Length = 1044
Score = 37.7 bits (86), Expect = 0.007
Identities = 22/78 (28%), Positives = 38/78 (48%)
Frame = +2
Query: 215 YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLK 274
+ ++DVGGQ R W H F+ +IF ++ D+ + + +RM+ EL + VL
Sbjct: 299 FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSNDRDRVVEARDELHRMLNEDELRDAVL- 475
Query: 275 QPCFEKTSFMLFLNKFDI 292
++F NK D+
Sbjct: 476 ---------LVFANKQDL 502
>TC76380 homologue to PIR|T52341|T52341 ADP-ribosylation factor [imported] -
rice, complete
Length = 990
Score = 37.7 bits (86), Expect = 0.007
Identities = 22/78 (28%), Positives = 38/78 (48%)
Frame = +1
Query: 215 YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLK 274
+ ++DVGGQ R W H F+ +IF ++ D+ + + +RM+ EL + VL
Sbjct: 268 FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSNDRDRVVEARDELHRMLNEDELRDAVL- 444
Query: 275 QPCFEKTSFMLFLNKFDI 292
++F NK D+
Sbjct: 445 ---------LVFANKQDL 471
>TC76649 homologue to GP|20161472|dbj|BAB90396. ADP-ribosylation factor
{Oryza sativa (japonica cultivar-group)}, partial (48%)
Length = 1133
Score = 37.7 bits (86), Expect = 0.007
Identities = 22/78 (28%), Positives = 38/78 (48%)
Frame = +1
Query: 215 YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLK 274
+ ++DVGGQ R W H F+ +IF ++ D+ + + +RM+ EL + VL
Sbjct: 307 FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSNDRDRVVEARDELHRMLNEDELRDAVL- 483
Query: 275 QPCFEKTSFMLFLNKFDI 292
++F NK D+
Sbjct: 484 ---------LVFANKQDL 510
>TC76648 homologue to PIR|T52341|T52341 ADP-ribosylation factor [imported] -
rice, complete
Length = 1357
Score = 37.7 bits (86), Expect = 0.007
Identities = 22/78 (28%), Positives = 38/78 (48%)
Frame = +3
Query: 215 YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLK 274
+ ++DVGGQ R W H F+ +IF ++ D+ + + +RM+ EL + VL
Sbjct: 696 FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSNDRDRVVEARDELHRMLNEDELRDAVL- 872
Query: 275 QPCFEKTSFMLFLNKFDI 292
++F NK D+
Sbjct: 873 ---------LVFANKQDL 899
>CA917198 GP|20161472|db ADP-ribosylation factor {Oryza sativa (japonica
cultivar-group)}, partial (43%)
Length = 780
Score = 37.7 bits (86), Expect = 0.007
Identities = 22/78 (28%), Positives = 38/78 (48%)
Frame = +3
Query: 215 YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLK 274
+ ++DVGGQ R W H F+ +IF ++ D+ + + +RM+ EL + VL
Sbjct: 483 FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSNDRDRVVEARDELHRMLNEDELRDAVL- 659
Query: 275 QPCFEKTSFMLFLNKFDI 292
++F NK D+
Sbjct: 660 ---------LVFANKQDL 686
>TC76650 GP|965483|dbj|BAA08259.1| DcARF1 {Daucus carota}, complete
Length = 839
Score = 37.7 bits (86), Expect = 0.007
Identities = 22/78 (28%), Positives = 38/78 (48%)
Frame = +2
Query: 215 YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLK 274
+ ++DVGGQ R W H F+ +IF ++ D+ + + +RM+ EL + VL
Sbjct: 275 FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSNDRDRVVEARDELHRMLNEDELRDAVL- 451
Query: 275 QPCFEKTSFMLFLNKFDI 292
++F NK D+
Sbjct: 452 ---------LVFANKQDL 478
>TC78399 homologue to SP|O04834|SARA_ARATH GTP-binding protein SAR1A.
[Mouse-ear cress] {Arabidopsis thaliana}, complete
Length = 1164
Score = 37.4 bits (85), Expect = 0.010
Identities = 24/90 (26%), Positives = 40/90 (43%), Gaps = 1/90 (1%)
Frame = +2
Query: 204 PVGENKKSGEV-YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRM 262
P E G + ++ FD+GG + RR W + V AV++ L + +K R
Sbjct: 233 PTSEELSIGRIKFKAFDLGGHQIARRVWRDYYAQVDAVVY----------LVDAFDKERF 382
Query: 263 METKELFEWVLKQPCFEKTSFMLFLNKFDI 292
E+K+ + +L F++ NK DI
Sbjct: 383 PESKKELDALLADESLGNVPFLILGNKIDI 472
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,361,064
Number of Sequences: 36976
Number of extensions: 127444
Number of successful extensions: 702
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 690
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 691
length of query: 384
length of database: 9,014,727
effective HSP length: 98
effective length of query: 286
effective length of database: 5,391,079
effective search space: 1541848594
effective search space used: 1541848594
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0034a.1


