

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
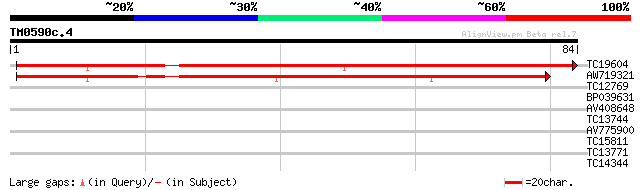
Query= TM0590c.4
(84 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC19604 similar to UP|Q04129 (Q04129) Wound induced protein (Fra... 82 2e-17
AW719321 44 5e-06
TC12769 similar to UP|Q8MRP6 (Q8MRP6) GH07623p, partial (3%) 29 0.13
BP039631 25 3.3
AV408648 24 4.3
TC13744 weakly similar to UP|Q39126 (Q39126) HYP1 protein (T4P13... 24 5.6
AV775900 23 7.3
TC15811 similar to UP|RL13_ARATH (P41127) 60S ribosomal protein ... 23 7.3
TC13771 23 7.3
TC14344 similar to UP|Q40310 (Q40310) Chalcone reductase, complete 23 9.5
>TC19604 similar to UP|Q04129 (Q04129) Wound induced protein (Fragment),
partial (63%)
Length = 500
Score = 81.6 bits (200), Expect = 2e-17
Identities = 48/90 (53%), Positives = 61/90 (67%), Gaps = 7/90 (7%)
Frame = +1
Query: 2 SAATRSLIV----GAVEALKDKLGVYNRRRNYALRSLQQRVVKNNNITPNG---NSSSAA 54
+AATR+ +V G VEALKD+LGV R NYALRSLQQ N +S+S+A
Sbjct: 73 TAATRAWVVASSVGVVEALKDQLGVC--RWNYALRSLQQHAKTNIRSYAQAKKLSSASSA 246
Query: 55 AIPSKVKRSNQEFMRKVMDLNCWGPSTTKF 84
A+ +KVKR+ E M++VMDLNCWGPST +F
Sbjct: 247 AVSNKVKRTKDESMKRVMDLNCWGPSTARF 336
>AW719321
Length = 356
Score = 43.9 bits (102), Expect = 5e-06
Identities = 36/90 (40%), Positives = 54/90 (60%), Gaps = 11/90 (12%)
Frame = +2
Query: 2 SAATRSLIV----GAVEALKDKLGVYNRRRNYALRSLQQRV---VKNNNITPNGNSSSAA 54
SAA+++ IV GAVEALKD+ G+ R NY L+ QQ + V + + T + SS+A
Sbjct: 44 SAASKAWIVAASVGAVEALKDQ-GIC--RWNYVLKHAQQHLKNRVGSLSQTRILSCSSSA 214
Query: 55 AIPSKVK----RSNQEFMRKVMDLNCWGPS 80
+K+K + +E +R VM L+CWGP+
Sbjct: 215 LFANKLKGEKAKQAEESLRTVMYLSCWGPN 304
>TC12769 similar to UP|Q8MRP6 (Q8MRP6) GH07623p, partial (3%)
Length = 469
Score = 29.3 bits (64), Expect = 0.13
Identities = 20/57 (35%), Positives = 29/57 (50%), Gaps = 3/57 (5%)
Frame = -1
Query: 25 RRRNYALRSLQQ-RVVKNNNITPNGNSSSAAAIPSKVKRSNQ--EFMRKVMDLNCWG 78
R+ A+RS+ Q R + +G + + A P R Q E +RKVM +NCWG
Sbjct: 343 RKCKTAVRSIHQNRSRLFAGVVGSGVTGAMAGNPDAEHRLRQTDESLRKVMYMNCWG 173
>BP039631
Length = 554
Score = 24.6 bits (52), Expect = 3.3
Identities = 13/36 (36%), Positives = 17/36 (47%)
Frame = +1
Query: 48 GNSSSAAAIPSKVKRSNQEFMRKVMDLNCWGPSTTK 83
GNS + A SNQ +R ++ LNC G K
Sbjct: 211 GNSVGSLACVIAASDSNQTLVRGIVLLNCAGGMNNK 318
>AV408648
Length = 430
Score = 24.3 bits (51), Expect = 4.3
Identities = 12/30 (40%), Positives = 19/30 (63%)
Frame = -1
Query: 25 RRRNYALRSLQQRVVKNNNITPNGNSSSAA 54
RRR R Q++ KN TP+G++++AA
Sbjct: 136 RRRREL*RLRQKKRSKNETRTPSGHAAAAA 47
>TC13744 weakly similar to UP|Q39126 (Q39126) HYP1 protein (T4P13.21
protein), partial (26%)
Length = 744
Score = 23.9 bits (50), Expect = 5.6
Identities = 13/34 (38%), Positives = 18/34 (52%)
Frame = -1
Query: 33 SLQQRVVKNNNITPNGNSSSAAAIPSKVKRSNQE 66
SLQ ++ NNI NG ++ I +KRS E
Sbjct: 609 SLQA*ILNPNNIQENGTQATPKKIK*NLKRS*ME 508
>AV775900
Length = 523
Score = 23.5 bits (49), Expect = 7.3
Identities = 12/32 (37%), Positives = 18/32 (55%)
Frame = +2
Query: 2 SAATRSLIVGAVEALKDKLGVYNRRRNYALRS 33
SA + + +VGA EA + G RRR L++
Sbjct: 314 SAGSEAGLVGATEAQTGRPGYRRRRRRRCLKT 409
>TC15811 similar to UP|RL13_ARATH (P41127) 60S ribosomal protein L13 (BBC1
protein homolog), complete
Length = 981
Score = 23.5 bits (49), Expect = 7.3
Identities = 9/17 (52%), Positives = 14/17 (81%)
Frame = +2
Query: 33 SLQQRVVKNNNITPNGN 49
SL+ +VK+NN+ PNG+
Sbjct: 20 SLR*AMVKHNNVIPNGH 70
>TC13771
Length = 474
Score = 23.5 bits (49), Expect = 7.3
Identities = 11/29 (37%), Positives = 17/29 (57%)
Frame = -1
Query: 23 YNRRRNYALRSLQQRVVKNNNITPNGNSS 51
+ + RNY L SL ++ +N+T NSS
Sbjct: 246 FTKIRNYFLSSLSHTILLVSNLTKLRNSS 160
>TC14344 similar to UP|Q40310 (Q40310) Chalcone reductase, complete
Length = 1180
Score = 23.1 bits (48), Expect = 9.5
Identities = 12/32 (37%), Positives = 17/32 (52%)
Frame = +1
Query: 49 NSSSAAAIPSKVKRSNQEFMRKVMDLNCWGPS 80
NSS+ P + Q + RKV+ NC+G S
Sbjct: 400 NSSTGVLGPLSDPLAPQFYTRKVLISNCYGGS 495
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.126 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,197,875
Number of Sequences: 28460
Number of extensions: 11912
Number of successful extensions: 69
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 63
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 65
length of query: 84
length of database: 4,897,600
effective HSP length: 60
effective length of query: 24
effective length of database: 3,190,000
effective search space: 76560000
effective search space used: 76560000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 48 (23.1 bits)
Lotus: description of TM0590c.4


