

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0340.18
(967 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
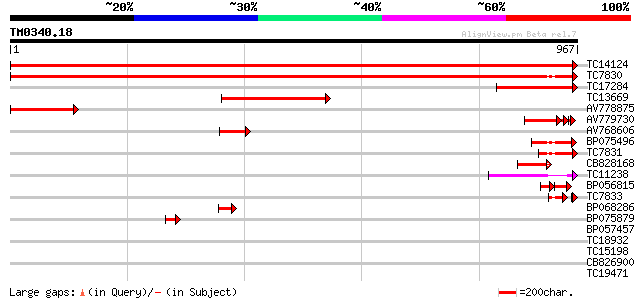
Score E
Sequences producing significant alignments: (bits) Value
TC14124 UP|Q8H945 (Q8H945) Phosphoenolpyruvate carboxylase , co... 1928 0.0
TC7830 UP|Q8H946 (Q8H946) Phosphoenolpyruvate carboxylase , com... 1751 0.0
TC17284 similar to UP|Q93WZ9 (Q93WZ9) Phosphoenolpyruvate carbox... 226 1e-59
TC13669 similar to UP|Q8GVE8 (Q8GVE8) Phosphoenolpyruvate carbox... 184 8e-47
AV778875 181 6e-46
AV779730 112 2e-28
AV768606 106 2e-23
BP075496 100 2e-21
TC7831 similar to UP|Q8H946 (Q8H946) Phosphoenolpyruvate carboxy... 93 2e-19
CB828168 91 1e-18
TC11238 similar to UP|Q8GVE8 (Q8GVE8) Phosphoenolpyruvate carbox... 81 9e-16
BP056815 49 3e-11
TC7833 homologue to UP|Q8H946 (Q8H946) Phosphoenolpyruvate carbo... 50 4e-07
BP068286 51 8e-07
BP075879 44 9e-05
BP057457 36 0.033
TC18932 31 0.82
TC15198 similar to UP|IF35_ARATH (O04202) Eukaryotic translation... 31 1.1
CB826900 29 3.1
TC19471 29 3.1
>TC14124 UP|Q8H945 (Q8H945) Phosphoenolpyruvate carboxylase , complete
Length = 3367
Score = 1928 bits (4995), Expect = 0.0
Identities = 965/967 (99%), Positives = 965/967 (99%)
Frame = +2
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV
Sbjct: 128 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 307
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK
Sbjct: 308 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 487
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVL AHPTQSVRRS
Sbjct: 488 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLAAHPTQSVRRS 667
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM
Sbjct: 668 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 847
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 848 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 1027
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN
Sbjct: 1028 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 1207
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC
Sbjct: 1208 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 1387
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDR TDVLDAITKHLGIGSYLEWSEE
Sbjct: 1388 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRLTDVLDAITKHLGIGSYLEWSEE 1567
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL
Sbjct: 1568 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 1747
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS
Sbjct: 1748 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 1927
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI
Sbjct: 1928 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 2107
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA
Sbjct: 2108 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 2287
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT
Sbjct: 2288 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 2467
Query: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY
Sbjct: 2468 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 2647
Query: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC
Sbjct: 2648 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 2827
Query: 901 QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 960
QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA
Sbjct: 2828 QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 3007
Query: 961 AGMQNTG 967
AGMQNTG
Sbjct: 3008 AGMQNTG 3028
>TC7830 UP|Q8H946 (Q8H946) Phosphoenolpyruvate carboxylase , complete
Length = 3376
Score = 1751 bits (4536), Expect = 0.0
Identities = 875/967 (90%), Positives = 918/967 (94%)
Frame = +2
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MANRNLEKMASIDAQLRQL PAKVSEDDKL+EYDALLLDRFLDILQDLHGEDLKETVQEV
Sbjct: 98 MANRNLEKMASIDAQLRQLAPAKVSEDDKLIEYDALLLDRFLDILQDLHGEDLKETVQEV 277
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYERKH+ ++LE LG +ITSLDAGDSIVVAKSF+HMLNLANLAEEVQI+ RRR K
Sbjct: 278 YELSAEYERKHDHEKLEELGKVITSLDAGDSIVVAKSFSHMLNLANLAEEVQISRRRRNK 457
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDFADE NATTESDIEET KKLV ++KKSPQEVFD LKNQTVDLVLTAHPTQSVRRS
Sbjct: 458 LKKGDFADENNATTESDIEETLKKLVFKLKKSPQEVFDALKNQTVDLVLTAHPTQSVRRS 637
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKH RIRN L+QLYAKDITPDDKQELDEALQREIQAAFRTDEI+RT PTPQDEMRAGM
Sbjct: 638 LLQKHARIRNSLSQLYAKDITPDDKQELDEALQREIQAAFRTDEIKRTPPTPQDEMRAGM 817
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 818 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 997
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMA+NLYYSQIEDLMFELSMWRCNDELRV A+EL +SNKD VAKHYIEFWK
Sbjct: 998 DVCLLARMMASNLYYSQIEDLMFELSMWRCNDELRVHADELHRNSNKDEVAKHYIEFWKK 1177
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
+PP+EPYRVVLGEVR+RLY+TRE SR LLA YSDI EE TFTNVEEFLEPLELCYRSLC
Sbjct: 1178 VPPNEPYRVVLGEVRDRLYKTREHSRILLAQGYSDIPEEATFTNVEEFLEPLELCYRSLC 1357
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
CGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDV+DAITKHL IGSY EWSEE
Sbjct: 1358 DCGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVMDAITKHLEIGSYQEWSEE 1537
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
KRQ+WLL ELSGKRPLFGPDLPQTEEI+DVLDTFHVIAELPSDNFGAYIISMATAPSDVL
Sbjct: 1538 KRQEWLLGELSGKRPLFGPDLPQTEEIRDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 1717
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQRE H+KNPLRVVPLFEKL DLE APAALARLFS+EWYRNRINGKQEVMIGYSDS
Sbjct: 1718 AVELLQRESHIKNPLRVVPLFEKLDDLEAAPAALARLFSIEWYRNRINGKQEVMIGYSDS 1897
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGRFSAAWQLYKAQE+LIKVAK++GVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI
Sbjct: 1898 GKDAGRFSAAWQLYKAQEDLIKVAKKFGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 2077
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HGSLRVTVQGEVIEQSFGEEHLCFRTLQR+TAATLEHGM+PP SPKPEWRALMD++AVIA
Sbjct: 2078 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRFTAATLEHGMNPPFSPKPEWRALMDQLAVIA 2257
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
TEEYRSIVF+EPRFVEYFRLATPELEYGRMNIGSRPAKR+P+GGIETLRAIPWIFAWTQT
Sbjct: 2258 TEEYRSIVFQEPRFVEYFRLATPELEYGRMNIGSRPAKRRPTGGIETLRAIPWIFAWTQT 2437
Query: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
RFHLPVWLGFGAAF+ +I KD+RNL+ LQEMYNQWPFFRVTIDLVEMVFAKGDPGIAAL
Sbjct: 2438 RFHLPVWLGFGAAFRQLIEKDVRNLHTLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALN 2617
Query: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
DRLLVSEDLW FGEQLR +EETK+LLLQVA HK++LEGDPYLKQRLRLRDSYITT+NV
Sbjct: 2618 DRLLVSEDLWPFGEQLRNKYEETKKLLLQVAGHKEVLEGDPYLKQRLRLRDSYITTMNVF 2797
Query: 901 QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 960
QAYTLKRIRDPNY+VK HISKE SKPADELV LNPTSEYAPGLEDTLILTMKGIA
Sbjct: 2798 QAYTLKRIRDPNYDVK---HISKEK---SKPADELVRLNPTSEYAPGLEDTLILTMKGIA 2959
Query: 961 AGMQNTG 967
AGMQNTG
Sbjct: 2960 AGMQNTG 2980
>TC17284 similar to UP|Q93WZ9 (Q93WZ9) Phosphoenolpyruvate carboxylase
housekeeping isozyme pepc2 (Fragment) , partial (35%)
Length = 577
Score = 226 bits (576), Expect = 1e-59
Identities = 112/137 (81%), Positives = 122/137 (88%)
Frame = +1
Query: 831 KGDPGIAALYDRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLR 890
KGDPGIAALYD+LLV+ +LW FGE+LR FEE K LLQVA H+DLLEGDPYLKQRLRLR
Sbjct: 1 KGDPGIAALYDKLLVTGELWPFGERLREKFEECKSFLLQVAGHRDLLEGDPYLKQRLRLR 180
Query: 891 DSYITTLNVCQAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLED 950
DSYITTLNV QAYTLKRIRDP+Y+VKLRPH+SK+ + SKPA ELV LNP SEYAPGLED
Sbjct: 181 DSYITTLNVLQAYTLKRIRDPDYHVKLRPHLSKDYTESSKPAAELVKLNPKSEYAPGLED 360
Query: 951 TLILTMKGIAAGMQNTG 967
TLILTMKGIAAGMQNTG
Sbjct: 361 TLILTMKGIAAGMQNTG 411
>TC13669 similar to UP|Q8GVE8 (Q8GVE8) Phosphoenolpyruvate carboxylase ,
partial (19%)
Length = 610
Score = 184 bits (466), Expect = 8e-47
Identities = 93/187 (49%), Positives = 128/187 (67%)
Frame = +3
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
+P PYRVVLG V+ +L ++R R LL + D + ++ LEPL +CY SL
Sbjct: 48 LPGIAPYRVVLGNVKAKLERSRRRLELLLEDVLCDYDPLDYYETSDQLLEPLLICYESLQ 227
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
+CG +ADG L D +R+V+TFG+ L++LD+RQES RH++ LDAIT +L +G+Y EW EE
Sbjct: 228 SCGSGVLADGRLADLIRRVATFGMVLMKLDLRQESGRHSETLDAITSYLDMGTYSEWDEE 407
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
K+ +L EL GKRPL + ++K+VLDTF + AEL SD+ GAY+ISMA+ SDVL
Sbjct: 408 KKLDFLTRELKGKRPLVPVSIEVPADVKEVLDTFRIAAELGSDSLGAYVISMASNASDVL 587
Query: 541 AVELLQR 547
AVELLQ+
Sbjct: 588 AVELLQK 608
>AV778875
Length = 464
Score = 181 bits (458), Expect = 6e-46
Identities = 89/116 (76%), Positives = 104/116 (88%)
Frame = +1
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MA R +EKMASIDAQLR L P KVS+DDK VEYDALLLDRFLDILQDLHGED+++TVQ+
Sbjct: 115 MATRKIEKMASIDAQLRLLAPRKVSDDDKWVEYDALLLDRFLDILQDLHGEDIRQTVQDC 294
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHR 116
YELSAEYE ++P++LE LGN++T LDAGDSIV+AKSF HML+LANLAEEVQIA+R
Sbjct: 295 YELSAEYEGNYKPEKLEELGNMLTGLDAGDSIVIAKSFTHMLSLANLAEEVQIAYR 462
>AV779730
Length = 481
Score = 112 bits (279), Expect(3) = 2e-28
Identities = 54/63 (85%), Positives = 59/63 (92%)
Frame = -3
Query: 879 GDPYLKQRLRLRDSYITTLNVCQAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTL 938
GDPYL+ RLRLRDSYITTL+VC AYTLKRIRDP+YNVKLRP +SKEAIDVS+PAD LVTL
Sbjct: 479 GDPYLEPRLRLRDSYITTLHVCPAYTLKRIRDPDYNVKLRPPLSKEAIDVSEPADGLVTL 300
Query: 939 NPT 941
NPT
Sbjct: 299 NPT 291
Score = 27.3 bits (59), Expect(3) = 2e-28
Identities = 12/19 (63%), Positives = 14/19 (73%)
Frame = -1
Query: 934 ELVTLNPTSEYAPGLEDTL 952
+L+ P S YAPGLEDTL
Sbjct: 313 DLLL*TPPSAYAPGLEDTL 257
Score = 24.6 bits (52), Expect(3) = 2e-28
Identities = 10/13 (76%), Positives = 12/13 (91%)
Frame = -2
Query: 953 ILTMKGIAAGMQN 965
+LT+KGIAAGM N
Sbjct: 255 LLTLKGIAAGMPN 217
>AV768606
Length = 566
Score = 106 bits (264), Expect = 2e-23
Identities = 51/52 (98%), Positives = 51/52 (98%)
Frame = -1
Query: 359 KNIPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLE 410
K IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLE
Sbjct: 158 KKIPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLE 3
>BP075496
Length = 432
Score = 99.8 bits (247), Expect = 2e-21
Identities = 55/77 (71%), Positives = 62/77 (80%)
Frame = -2
Query: 890 RDSYITTLNVCQAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLE 949
RDSYITT+N AYTLKRIRDPNY+V+ HISKE S+PAD LV L+PT+EYA GL
Sbjct: 431 RDSYITTMNGFLAYTLKRIRDPNYDVE---HISKEK---SQPADGLVRLDPTTEYASGLG 270
Query: 950 DTLILTMKGIAAGMQNT 966
D LILTMKGIAAGMQN+
Sbjct: 269 DNLILTMKGIAAGMQNS 219
>TC7831 similar to UP|Q8H946 (Q8H946) Phosphoenolpyruvate carboxylase ,
partial (6%)
Length = 489
Score = 93.2 bits (230), Expect = 2e-19
Identities = 50/66 (75%), Positives = 56/66 (84%)
Frame = +2
Query: 902 AYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIAA 961
AYTLKRIRDP+Y+VK H+SKE S PADELV L+PTS YAPGLEDTLILT+KGIAA
Sbjct: 2 AYTLKRIRDPHYDVK---HLSKEK---SPPADELVRLDPTSGYAPGLEDTLILTLKGIAA 163
Query: 962 GMQNTG 967
GMQ+TG
Sbjct: 164 GMQDTG 181
>CB828168
Length = 260
Score = 90.5 bits (223), Expect = 1e-18
Identities = 46/59 (77%), Positives = 51/59 (85%)
Frame = +3
Query: 866 LLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVCQAYTLKRIRDPNYNVKLRPHISKE 924
L +VA HK++LEG PYLKQRLRLRDSYITT+NV QAYTLKRIRDPNY+VK HISKE
Sbjct: 90 LFWKVAGHKEVLEGYPYLKQRLRLRDSYITTMNVFQAYTLKRIRDPNYDVK---HISKE 257
>TC11238 similar to UP|Q8GVE8 (Q8GVE8) Phosphoenolpyruvate carboxylase ,
partial (12%)
Length = 532
Score = 80.9 bits (198), Expect = 9e-16
Identities = 52/151 (34%), Positives = 75/151 (49%)
Frame = +1
Query: 817 FFRVTIDLVEMVFAKGDPGIAALYDRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDL 876
FF+ TIDL+EMV K D IA YD +LVSE+ G QLR+ E T + +L V + L
Sbjct: 1 FFQSTIDLIEMVLGKADSPIAKHYDDVLVSEERKELGRQLRSELETTGKFVLVVCGQEKL 180
Query: 877 LEGDPYLKQRLRLRDSYITTLNVCQAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELV 936
L+ + L++ + R ++ +N+ Q LKR+R + N K R
Sbjct: 181 LQNNRTLRRLIENRLPFLNPMNILQVEILKRLRCDDDNHKAR------------------ 306
Query: 937 TLNPTSEYAPGLEDTLILTMKGIAAGMQNTG 967
D L++T+ GIAAGM+NTG
Sbjct: 307 -------------DALLITINGIAAGMKNTG 360
>BP056815
Length = 411
Score = 48.5 bits (114), Expect(2) = 3e-11
Identities = 22/23 (95%), Positives = 22/23 (95%)
Frame = -1
Query: 906 KRIRDPNYNVKLRPHISKEAIDV 928
KRIRDPNYNVKLRPHI KEAIDV
Sbjct: 411 KRIRDPNYNVKLRPHIFKEAIDV 343
Score = 37.4 bits (85), Expect(2) = 3e-11
Identities = 21/31 (67%), Positives = 22/31 (70%), Gaps = 1/31 (3%)
Frame = -2
Query: 929 SKPADELVTLNPTSEYA-PGLEDTLILTMKG 958
SKPADEL P + PGLEDTLILTMKG
Sbjct: 341 SKPADELGYSGPHKGISHPGLEDTLILTMKG 249
>TC7833 homologue to UP|Q8H946 (Q8H946) Phosphoenolpyruvate carboxylase ,
partial (4%)
Length = 456
Score = 49.7 bits (117), Expect(2) = 4e-07
Identities = 25/32 (78%), Positives = 27/32 (84%)
Frame = +3
Query: 920 HISKEAIDVSKPADELVTLNPTSEYAPGLEDT 951
HISKE SKPADELV LNP+SEYAPGL+DT
Sbjct: 6 HISKEK---SKPADELVRLNPSSEYAPGLDDT 92
Score = 21.9 bits (45), Expect(2) = 4e-07
Identities = 9/10 (90%), Positives = 9/10 (90%)
Frame = +2
Query: 958 GIAAGMQNTG 967
GIAAGMQN G
Sbjct: 113 GIAAGMQNPG 142
>BP068286
Length = 187
Score = 51.2 bits (121), Expect = 8e-07
Identities = 22/31 (70%), Positives = 27/31 (86%)
Frame = -1
Query: 357 FWKNIPPSEPYRVVLGEVRNRLYQTRERSRH 387
F + PP+EPYRVVLGEVR+RLY+TRE SR+
Sbjct: 94 FGRRFPPNEPYRVVLGEVRDRLYKTREHSRY 2
>BP075879
Length = 114
Score = 44.3 bits (103), Expect = 9e-05
Identities = 19/24 (79%), Positives = 21/24 (87%)
Frame = -2
Query: 267 INERVPYNAPLIQFSSWMGGDRDG 290
I+ERVPY+APLIQF SWMGGD G
Sbjct: 74 IDERVPYHAPLIQFFSWMGGDLGG 3
>BP057457
Length = 547
Score = 35.8 bits (81), Expect = 0.033
Identities = 14/24 (58%), Positives = 20/24 (83%)
Frame = -2
Query: 140 ETFKKLVGEMKKSPQEVFDELKNQ 163
E+FK+LV E+KK+P E+ D +KNQ
Sbjct: 120 ESFKRLVAELKKTPLEILDAMKNQ 49
>TC18932
Length = 284
Score = 31.2 bits (69), Expect = 0.82
Identities = 14/16 (87%), Positives = 15/16 (93%)
Frame = +3
Query: 952 LILTMKGIAAGMQNTG 967
LILT+KGIAAGMQN G
Sbjct: 3 LILTLKGIAAGMQNPG 50
>TC15198 similar to UP|IF35_ARATH (O04202) Eukaryotic translation initiation
factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p32 subunit)
(eIF3f), partial (35%)
Length = 668
Score = 30.8 bits (68), Expect = 1.1
Identities = 17/54 (31%), Positives = 28/54 (51%)
Frame = +3
Query: 129 EGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRSLL 182
EG +++I + VG + KSP VFD+L N ++ L + S+ R+ L
Sbjct: 111 EGRIAPDNNIGKFISDAVGSLPKSPPAVFDKLVNDSLQDHLLSLYLSSITRTQL 272
>CB826900
Length = 599
Score = 29.3 bits (64), Expect = 3.1
Identities = 14/34 (41%), Positives = 18/34 (52%)
Frame = -2
Query: 465 ITKHLGIGSYLEWSEEKRQQWLLSELSGKRPLFG 498
ITKH G G W+E++ L L GK P +G
Sbjct: 256 ITKHTGPGRNRGWTEKRPNNEDLCSLRGKTPFWG 155
>TC19471
Length = 514
Score = 29.3 bits (64), Expect = 3.1
Identities = 16/71 (22%), Positives = 32/71 (44%)
Frame = +3
Query: 83 ITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIKLKKGDFADEGNATTESDIEETF 142
I+ L+ D++++ + F N+ EE+Q+ + ++ K A + + E
Sbjct: 186 ISELEKEDALIIEEEFKCEDPKRNIIEEIQVLSKEALEKGKETAALQKKKKKHESVLEAL 365
Query: 143 KKLVGEMKKSP 153
+K EMK P
Sbjct: 366 RKTYEEMKFKP 398
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,959,444
Number of Sequences: 28460
Number of extensions: 194138
Number of successful extensions: 937
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 924
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 933
length of query: 967
length of database: 4,897,600
effective HSP length: 99
effective length of query: 868
effective length of database: 2,080,060
effective search space: 1805492080
effective search space used: 1805492080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0340.18


