

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
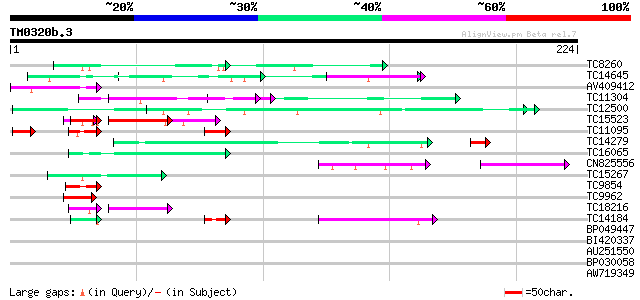
Query= TM0320b.3
(224 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8260 similar to UP|Q94ES8 (Q94ES8) Nodule extensin (Fragment),... 50 2e-07
TC14645 similar to UP|Q41122 (Q41122) Proline-rich protein precu... 50 3e-07
AV409412 48 2e-06
TC11304 similar to PIR|T04669|T04669 serine O-acetyltransferase ... 47 3e-06
TC12500 similar to GB|AAH45354.1|28279536|BC045354 FNBP4 protein... 47 3e-06
TC15523 similar to UP|GRPA_MAIZE (P10979) Glycine-rich RNA-bindi... 35 4e-06
TC11095 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 35 8e-06
TC14279 similar to UP|Q09085 (Q09085) Hydroxyproline-rich glycop... 42 1e-05
TC16065 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich... 44 2e-05
CN825556 42 2e-05
TC15267 similar to UP|EXTN_TOBAC (P13983) Extensin precursor (Ce... 44 3e-05
TC9854 weakly similar to UP|Q9LSN7 (Q9LSN7) Gb|AAC24081.1 (AT3g1... 44 3e-05
TC9962 similar to UP|Q8L7Z6 (Q8L7Z6) AT3g54680/T5N23_40, partial... 41 1e-04
TC18216 similar to GB|CAA28991.1|34041|HSKER65A keratin type II ... 35 2e-04
TC14184 similar to UP|Q93WZ6 (Q93WZ6) Abscisic stress ripening-l... 37 8e-04
BP049447 39 0.001
BI420337 38 0.001
AU251550 37 0.003
BP030058 36 0.006
AW719349 33 0.052
>TC8260 similar to UP|Q94ES8 (Q94ES8) Nodule extensin (Fragment), partial
(79%)
Length = 1017
Score = 50.4 bits (119), Expect = 2e-07
Identities = 42/149 (28%), Positives = 46/149 (30%), Gaps = 17/149 (11%)
Frame = -2
Query: 18 VTVEIGWWWW------WQRWWLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVV 71
V V + WWWW W W WWW V N G D V V V
Sbjct: 584 VRVSVNWWWWRRCIFVWLLMWWWWW-------RRMVNWVWVRVSVNWWWGRVVDWVRVRV 426
Query: 72 SVVAIEWWRR-------LWWWWR*GGGCGGGRAAVVGVAATSVVAVV----AAMVVVVLA 120
SV WWRR WWWWR VV V +V V+V L
Sbjct: 425 SV--NWWWRR*VLVMLLRWWWWR----------RVVNWVRVGVRVLVWWGWR*WVLVRLM 282
Query: 121 TIGCGGGSKAVAA**GGGDGGGWWWWWWQ 149
WWWWWW+
Sbjct: 281 ----------------------WWWWWWR 261
Score = 38.9 bits (89), Expect = 7e-04
Identities = 24/79 (30%), Positives = 26/79 (32%), Gaps = 9/79 (11%)
Frame = -2
Query: 18 VTVEIGWWWWWQR-----WWLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVS 72
V V + WWWWW R W L WW R VV
Sbjct: 299 VLVRLMWWWWWWRVVHWVWLLVWW-----------------------------RWRVVHW 207
Query: 73 VVAIEWWRRLW----WWWR 87
V + WWR W WWWR
Sbjct: 206 VRLLVWWR--W*VNRWWWR 156
Score = 34.3 bits (77), Expect = 0.018
Identities = 24/65 (36%), Positives = 29/65 (43%), Gaps = 22/65 (33%)
Frame = -3
Query: 125 GGGSKAVAA**GGGDGGGW-----WWWWWQ*GGG---------CGVGGGGD--------Y 162
GGG + + GGG GG W W + GGG CG GGGG+ Y
Sbjct: 412 GGGDEYL*CFLGGGGGGEW*TG*GWG*GYLCGGGGGDGYL*GLCGGGGGGEWYTGCGFSY 233
Query: 163 GGGGD 167
GGGG+
Sbjct: 232 GGGGE 218
Score = 33.1 bits (74), Expect = 0.040
Identities = 21/53 (39%), Positives = 25/53 (46%), Gaps = 8/53 (15%)
Frame = -3
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGD--------YGGGGD 167
G GGG + GGG GG W+ G G GGGG+ YGGGG+
Sbjct: 319 GGGGGDGYL*GLCGGGGGGEWYT-----GCGFSYGGGGEWYTG*GFSYGGGGE 176
Score = 30.4 bits (67), Expect = 0.26
Identities = 12/45 (26%), Positives = 15/45 (32%), Gaps = 25/45 (55%)
Frame = -2
Query: 17 VVTVEIGWWWW-----WQR--------------------WWLWWW 36
V+ + + WWWW W R WW WWW
Sbjct: 398 VLVMLLRWWWWRRVVNWVRVGVRVLVWWGWR*WVLVRLMWWWWWW 264
Score = 28.5 bits (62), Expect = 0.98
Identities = 12/31 (38%), Positives = 16/31 (50%), Gaps = 4/31 (12%)
Frame = -2
Query: 10 RLVLVAVVVTVEIGWWWWWQRW----WLWWW 36
R V+ V V V + WW W+ W +WWW
Sbjct: 365 RRVVNWVRVGVRVLVWWGWR*WVLVRLMWWW 273
Score = 27.7 bits (60), Expect = 1.7
Identities = 30/108 (27%), Positives = 37/108 (33%), Gaps = 16/108 (14%)
Frame = -3
Query: 121 TIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCG-----VGGGGDY-----------GG 164
T G GG + GGG GG W GCG GGGG++ GG
Sbjct: 568 TGGGGGDAYLYGFSCGGGGGGEW-------*TGCG*G*V*TGGGGEW*TG*G*G*V*TGG 410
Query: 165 GGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGVSW 212
GGD+ + + GGGGGG +G W
Sbjct: 409 GGDEYL*CFL------------------------GGGGGGEW*TG*GW 338
Score = 26.2 bits (56), Expect = 4.9
Identities = 27/87 (31%), Positives = 33/87 (37%), Gaps = 6/87 (6%)
Frame = -1
Query: 43 GGGKAVAATVEGGGNGGGGGGGDRVVVVVS------VVAIEWWRRLWWWWR*GGGCGGGR 96
G GK E G G G G + VV +S VV +E +L G GG
Sbjct: 486 GEGKCELVVGESGRLGEGEGKCELVVEEMSTCNAS*VVVVEESGKL-------GEGGGEG 328
Query: 97 AAVVGVAATSVVAVVAAMVVVVLATIG 123
VVGV +VVV T+G
Sbjct: 327 TCVVGVEVMGTCKAYVVVVVVESGTLG 247
>TC14645 similar to UP|Q41122 (Q41122) Proline-rich protein precursor,
partial (52%)
Length = 1060
Score = 50.1 bits (118), Expect = 3e-07
Identities = 34/120 (28%), Positives = 37/120 (30%)
Frame = -2
Query: 44 GGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVA 103
GG+ + G G G GG WW WWWW GG G G
Sbjct: 453 GGRVMVVYRAGAGAGEGG----------------WW---WWWWPVGGLTGTEVGG*TG-- 337
Query: 104 ATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYG 163
IG G G+ GGWWWWWW CG GG G
Sbjct: 336 ----------------GLIGAGAGA------------GGWWWWWW-----CGA*AGG*IG 256
Score = 45.1 bits (105), Expect = 1e-05
Identities = 21/40 (52%), Positives = 21/40 (52%), Gaps = 1/40 (2%)
Frame = -2
Query: 126 GGSKAVAA**GGGDG-GGWWWWWWQ*GGGCGVGGGGDYGG 164
GG V G G G GGWWWWWW GG G GG GG
Sbjct: 453 GGRVMVVYRAGAGAGEGGWWWWWWPVGGLTGTEVGG*TGG 334
Score = 43.5 bits (101), Expect = 3e-05
Identities = 37/104 (35%), Positives = 42/104 (39%), Gaps = 10/104 (9%)
Frame = -2
Query: 8 GGRLVLV--AVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGG--GGGG 63
GGR+++V A E GWWWWW W + GG + T GG GG G G
Sbjct: 453 GGRVMVVYRAGAGAGEGGWWWWW--WPV--------GG----LTGTEVGG*TGGLIGAGA 316
Query: 64 GDRVVVVVSVVAIEWWRRLWWWW--R*GGG----CGGGRAAVVG 101
G A WW WWWW * GG C GG G
Sbjct: 315 G----------AGGWW---WWWWCGA*AGG*IGECVGG*TGATG 223
Score = 25.0 bits (53), Expect(2) = 3.1
Identities = 11/29 (37%), Positives = 14/29 (47%), Gaps = 2/29 (6%)
Frame = -1
Query: 18 VTVEIGWWW--WWQRWWLWWW***GSGGG 44
V V + WW WW +W + W G GG
Sbjct: 223 VVVIVRWWSCNWWVKWNVVWKCYKGGNGG 137
Score = 20.0 bits (40), Expect(2) = 3.1
Identities = 7/8 (87%), Positives = 7/8 (87%)
Frame = -3
Query: 1 VGGGGGNG 8
VGGGGG G
Sbjct: 308 VGGGGGGG 285
>AV409412
Length = 426
Score = 47.8 bits (112), Expect = 2e-06
Identities = 19/41 (46%), Positives = 22/41 (53%), Gaps = 5/41 (12%)
Frame = -1
Query: 1 VGGGGGN-----GGRLVLVAVVVTVEIGWWWWWQRWWLWWW 36
+GG G + G R L V EIGWWWWW WW+ WW
Sbjct: 183 IGGSGRSLGR*WGRRRCLEQPVTAPEIGWWWWW--WWMTWW 67
Score = 33.1 bits (74), Expect = 0.040
Identities = 9/15 (60%), Positives = 9/15 (60%)
Frame = -1
Query: 141 GGWWWWWWQ*GGGCG 155
G WWWWWW CG
Sbjct: 102 GWWWWWWWMTWWCCG 58
Score = 29.6 bits (65), Expect = 0.44
Identities = 17/46 (36%), Positives = 19/46 (40%), Gaps = 3/46 (6%)
Frame = -1
Query: 52 VEGGGNGGGGGGGDRVVVVVSVVAIE---WWRRLWWWWR*GGGCGG 94
+ G G G G R + V A E WW WWWW CGG
Sbjct: 183 IGGSGRSLGR*WGRRRCLEQPVTAPEIGWWW---WWWWMTWWCCGG 55
Score = 27.3 bits (59), Expect = 2.2
Identities = 9/16 (56%), Positives = 9/16 (56%), Gaps = 3/16 (18%)
Frame = -1
Query: 140 GGGWWWW---WWQ*GG 152
G WWWW WW GG
Sbjct: 102 GWWWWWWWMTWWCCGG 55
Score = 26.6 bits (57), Expect = 3.7
Identities = 18/46 (39%), Positives = 21/46 (45%), Gaps = 4/46 (8%)
Frame = -2
Query: 42 GGGGKAVAATVE----GGGNGGGGGGGDRVVVVVSVVAIEWWRRLW 83
GGGG A + GGG GGGG G + I +WR LW
Sbjct: 149 GGGGDA*NNQ*QHQK*GGGGGGGG*HGG------AAAEIGFWRDLW 30
Score = 25.8 bits (55), Expect = 6.3
Identities = 10/14 (71%), Positives = 11/14 (78%)
Frame = -2
Query: 52 VEGGGNGGGGGGGD 65
VEGGG GGG GG+
Sbjct: 395 VEGGGGGGGEEGGE 354
Score = 22.3 bits (46), Expect(2) = 4.3
Identities = 7/13 (53%), Positives = 9/13 (68%)
Frame = -1
Query: 148 WQ*GGGCGVGGGG 160
W+ G CG+GG G
Sbjct: 207 WRREGRCGIGGSG 169
Score = 22.3 bits (46), Expect(2) = 4.3
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = -2
Query: 199 GGGGGGGANSGVS 211
GGGGGGG + G +
Sbjct: 98 GGGGGGG*HGGAA 60
>TC11304 similar to PIR|T04669|T04669 serine O-acetyltransferase F8D20.150
- Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (24%)
Length = 775
Score = 47.0 bits (110), Expect = 3e-06
Identities = 28/65 (43%), Positives = 33/65 (50%), Gaps = 5/65 (7%)
Frame = -1
Query: 40 GSGGGGKAVAA-----TVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGG 94
G GGGG+ A GGG+GGGGGG + V ++ R LWWW GGG GG
Sbjct: 427 GGGGGGRRRRACWGERASYGGGSGGGGGGSHK----PCEVGLK*GRELWWW---GGGGGG 269
Query: 95 GRAAV 99
G V
Sbjct: 268 GVVVV 254
Score = 43.9 bits (102), Expect = 2e-05
Identities = 25/78 (32%), Positives = 32/78 (40%)
Frame = -3
Query: 28 WQRWWLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR 87
W+ WW WWW + + GG G+R+VVV WW WWWW
Sbjct: 368 WR*WWWWWW-----------LTQAMRGGVE-----IGERIVVV------GWW---WWWW- 267
Query: 88 *GGGCGGGRAAVVGVAAT 105
G GG + V+ AT
Sbjct: 266 ---GGGGSKVVVLFALAT 222
Score = 42.4 bits (98), Expect = 7e-05
Identities = 27/100 (27%), Positives = 30/100 (30%)
Frame = -3
Query: 79 WRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGG 138
WR WWWW GG + +VVV
Sbjct: 368 WR*WWWWWWLTQAMRGG-------------VEIGERIVVV-------------------- 288
Query: 139 DGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVV 178
GWWWWWW GGGG KVVV + V
Sbjct: 287 ---GWWWWWW--------------GGGGSKVVVLFALATV 219
Score = 33.9 bits (76), Expect = 0.023
Identities = 26/71 (36%), Positives = 28/71 (38%), Gaps = 22/71 (30%)
Frame = -1
Query: 125 GGGSKAVAA**GGGDGGG-----WWWWWWQ*GGGCGVGGGGDY----------------- 162
GGG + GGG GGG W GGG G GGGG +
Sbjct: 463 GGGGRVQERRFGGGGGGGRRRRACWGERASYGGGSGGGGGGSHKPCEVGLK*GRELWWWG 284
Query: 163 GGGGDKVVVAA 173
GGGG VVV A
Sbjct: 283 GGGGGGVVVVA 251
Score = 31.6 bits (70), Expect = 0.12
Identities = 37/117 (31%), Positives = 39/117 (32%), Gaps = 2/117 (1%)
Frame = -1
Query: 42 GGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*--GGGCGGGRAAV 99
GGGG+ V+ GGGGGGG R RR W R GGG GGG
Sbjct: 463 GGGGR-----VQERRFGGGGGGGRR-------------RRACWGERASYGGGSGGG---- 350
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGV 156
GGGS G G WWW GGG GV
Sbjct: 349 -------------------------GGGSHKPCE---VGLK*GRELWWWGGGGGGGV 263
Score = 29.6 bits (65), Expect = 0.44
Identities = 26/83 (31%), Positives = 30/83 (35%)
Frame = -1
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGG 61
GGGGG + V + E LWWW GGGG
Sbjct: 358 GGGGGGSHKPCEVGLK*GRE-----------LWWW---------------------GGGG 275
Query: 62 GGGDRVVVVVSVVAIEWWRRLWW 84
GGG VVVV+ +WW L W
Sbjct: 274 GGG---VVVVA----KWWFFLHW 227
Score = 29.3 bits (64), Expect = 0.57
Identities = 17/53 (32%), Positives = 23/53 (43%), Gaps = 19/53 (35%)
Frame = -2
Query: 156 VGGGGDYGGGGDKVVVAAIMVVV-------------------ATPVVTVGRWW 189
VGG G++ G ++V VA ++VVV VV VG WW
Sbjct: 414 VGGAGEHVGESERVTVAVVVVVVVAHTSHARWG*NRGENCGGGVVVVVVGWWW 256
Score = 28.1 bits (61), Expect = 1.3
Identities = 28/97 (28%), Positives = 32/97 (32%), Gaps = 30/97 (30%)
Frame = -2
Query: 93 GGGRAAVVGVAATSVV-------------------AVVAAMVVVVLATI----------- 122
GGG + V V SVV VA +VVVV+A
Sbjct: 480 GGGESRAVVVGFRSVVLEAAAVVGGAGEHVGESERVTVAVVVVVVVAHTSHARWG*NRGE 301
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
CGGG V WWW Q GG +G G
Sbjct: 300 NCGGGVVVVVV----------GWWW*QSGGSFCIGHG 220
>TC12500 similar to GB|AAH45354.1|28279536|BC045354 FNBP4 protein {Danio
rerio;} , partial (5%)
Length = 678
Score = 47.0 bits (110), Expect = 3e-06
Identities = 59/184 (32%), Positives = 72/184 (39%), Gaps = 29/184 (15%)
Frame = -2
Query: 55 GGNGGGGGGGDRVVV-----------------VVSVVAIEWWRRLWWWWR*GGG------ 91
GG GGGGGGGDRV V V+S+ I R GGG
Sbjct: 674 GGVGGGGGGGDRVGVGGGASSDVSGVDSRGSGVLSIEDI---------GRGGGGMVATPY 522
Query: 92 CGGGRAAVVGVAATSVVAVVA-----AMVVVVLATIGCGGGSKAVAA**GGGDGGGWWW- 145
GGG G + V VV +V V G G G+ +VA GG+GG
Sbjct: 521 LGGGDGGCGGEGSGVWVIVVRELGGDGVVSDVRCVEGGGSGALSVADMSKGGEGGMVVTP 342
Query: 146 WWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGG 205
++ GGGCG G G + G ++A + A V GR DGGGG GG
Sbjct: 341 YFGGGGGGCG-GDGREQGKEASPDIIALPAITEAGGGVVGGR----------DGGGGCGG 195
Query: 206 ANSG 209
G
Sbjct: 194 GGEG 183
Score = 46.2 bits (108), Expect = 5e-06
Identities = 58/211 (27%), Positives = 76/211 (35%), Gaps = 7/211 (3%)
Frame = -2
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGG-- 59
GG GG GG V V+V E+G G G + VEGGG+G
Sbjct: 515 GGDGGCGGEGSGVWVIVVRELG------------------GDGVVSDVRCVEGGGSGALS 390
Query: 60 -----GGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAM 114
GG G VV ++ GGGCGG A+ ++A+ A
Sbjct: 389 VADMSKGGEGGMVVTP-------------YFGGGGGGCGGDGREQGKEASPDIIALPAIT 249
Query: 115 VVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAI 174
GGG GG DGGG GCG GG G+ G +
Sbjct: 248 EA--------GGGVV------GGRDGGG----------GCGGGGEGEQGRETSPTDLETF 141
Query: 175 MVVVATPVVTVGRWWRRRRRR*SDGGGGGGG 205
++ A P + + ++GGGGGGG
Sbjct: 140 EII-ALPAI-----------KGAEGGGGGGG 84
Score = 35.0 bits (79), Expect = 0.010
Identities = 36/123 (29%), Positives = 49/123 (39%), Gaps = 20/123 (16%)
Frame = -2
Query: 40 GSGGGGKAVAATVEGG----------GNGGGGGGGD-------RVVVVVSVVAI-EWWRR 81
G G G +VA +GG G GGGG GGD ++++ AI E
Sbjct: 413 GGGSGALSVADMSKGGEGGMVVTPYFGGGGGGCGGDGREQGKEASPDIIALPAITEAGGG 234
Query: 82 LWWWWR*GGGCGGGRAAVVG--VAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGD 139
+ GGGCGGG G + T + + + G GGG + GG +
Sbjct: 233 VVGGRDGGGGCGGGGEGEQGRETSPTDLETFEIIALPAIKGAEGGGGGGGRALSDFGGVE 54
Query: 140 GGG 142
GGG
Sbjct: 53 GGG 45
Score = 26.6 bits (57), Expect = 3.7
Identities = 7/14 (50%), Positives = 8/14 (57%)
Frame = -1
Query: 23 GWWWWWQRWWLWWW 36
GW W W + W W W
Sbjct: 663 GWGWGWGQSWGWRW 622
Score = 25.4 bits (54), Expect = 8.3
Identities = 12/24 (50%), Positives = 16/24 (66%)
Frame = -2
Query: 40 GSGGGGKAVAATVEGGGNGGGGGG 63
G GGGG+A++ + GG GGG G
Sbjct: 101 GGGGGGRALS---DFGGVEGGGSG 39
>TC15523 similar to UP|GRPA_MAIZE (P10979) Glycine-rich RNA-binding,
abscisic acid-inducible protein, complete
Length = 972
Score = 34.7 bits (78), Expect(2) = 4e-06
Identities = 11/19 (57%), Positives = 11/19 (57%), Gaps = 5/19 (26%)
Frame = +1
Query: 22 IGWWWW-----WQRWWLWW 35
I WWWW W RW LWW
Sbjct: 652 IRWWWW*PL*RWWRWRLWW 708
Score = 33.9 bits (76), Expect = 0.023
Identities = 37/120 (30%), Positives = 40/120 (32%), Gaps = 5/120 (4%)
Frame = +2
Query: 54 GGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAA 113
GGG GGGG GG R R GG GGG + G
Sbjct: 602 GGGGGGGGYGGGR--------------------REGGYGGGGGSRYSG------------ 685
Query: 114 MVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYG-----GGGDK 168
G GGG GG GGG+ GG GGGG YG GGG +
Sbjct: 686 ---------GGGGGY-------GGSGGGGY--------GGRREGGGGGYGGSREGGGGSR 793
Score = 33.9 bits (76), Expect = 0.023
Identities = 28/91 (30%), Positives = 37/91 (39%), Gaps = 6/91 (6%)
Frame = +2
Query: 137 GGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR* 196
GG GGG + + GG G GGG Y GGG + G + RR
Sbjct: 602 GGGGGGGGYGGGRREGGYGGGGGSRYSGGGGGGYGG-----------SGGGGYGGRREGG 748
Query: 197 SDG------GGGGGGANSGVSWQQ*RVVVMV 221
G GGGG +SG SW+ * ++ +
Sbjct: 749 GGGYGGSREGGGGSRYSSGGSWRD*SFMIFL 841
Score = 31.2 bits (69), Expect(2) = 4e-06
Identities = 20/55 (36%), Positives = 26/55 (46%), Gaps = 11/55 (20%)
Frame = +2
Query: 40 GSGGGGKAVAATVEGGGNGGG--GGGGDRV---------VVVVSVVAIEWWRRLW 83
GSGGGG GGG GG GGGG R ++ +V+I +W +W
Sbjct: 707 GSGGGGYGGRREGGGGGYGGSREGGGGSRYSSGGSWRD*SFMIFLVSIMYWF*VW 871
Score = 30.8 bits (68), Expect(2) = 1e-04
Identities = 14/25 (56%), Positives = 16/25 (64%)
Frame = +2
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGG 64
G GGGG + + GGG GG GGGG
Sbjct: 650 GYGGGGGSRYSGGGGGGYGGSGGGG 724
Score = 30.0 bits (66), Expect(2) = 1e-04
Identities = 9/17 (52%), Positives = 11/17 (63%), Gaps = 5/17 (29%)
Frame = +1
Query: 25 WWWWQRW----WL-WWW 36
WWW +RW W+ WWW
Sbjct: 616 WWWIRRWSS*RWIRWWW 666
Score = 29.6 bits (65), Expect = 0.44
Identities = 10/18 (55%), Positives = 10/18 (55%), Gaps = 5/18 (27%)
Frame = +1
Query: 25 WWWWQR---WWLW--WW* 37
WWWW WW W WW*
Sbjct: 658 WWWW*PL*RWWRWRLWW* 711
Score = 29.6 bits (65), Expect(2) = 0.003
Identities = 11/25 (44%), Positives = 11/25 (44%), Gaps = 12/25 (48%)
Frame = +1
Query: 24 WWW---W----WQRWWLW-----WW 36
WWW W W RWW W WW
Sbjct: 616 WWWIRRWSS*RWIRWWWW*PL*RWW 690
Score = 26.9 bits (58), Expect = 2.8
Identities = 15/31 (48%), Positives = 15/31 (48%), Gaps = 6/31 (19%)
Frame = +2
Query: 40 GSGGGGKAVAATVEGGGNGG------GGGGG 64
G GGGG EGG GG GGGGG
Sbjct: 605 GGGGGGGYGGGRREGGYGGGGGSRYSGGGGG 697
Score = 26.2 bits (56), Expect(2) = 0.003
Identities = 15/27 (55%), Positives = 16/27 (58%), Gaps = 3/27 (11%)
Frame = +2
Query: 41 SGGGGKAVAATVEGGGNGG---GGGGG 64
SGGGG + GGG GG GGGGG
Sbjct: 680 SGGGGGGYGGS-GGGGYGGRREGGGGG 757
Score = 25.8 bits (55), Expect = 6.3
Identities = 12/25 (48%), Positives = 13/25 (52%)
Frame = +2
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGG 64
G GGG + GGG GG GGG
Sbjct: 647 GGYGGGGGSRYSGGGGGGYGGSGGG 721
Score = 25.8 bits (55), Expect = 6.3
Identities = 7/11 (63%), Positives = 7/11 (63%)
Frame = +1
Query: 25 WWWWQRWWLWW 35
WWWW LWW
Sbjct: 745 WWWW----LWW 765
>TC11095 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (89%)
Length = 912
Score = 35.0 bits (79), Expect(2) = 8e-04
Identities = 9/14 (64%), Positives = 9/14 (64%), Gaps = 1/14 (7%)
Frame = +3
Query: 24 WW-WWWQRWWLWWW 36
WW WWW WW W W
Sbjct: 324 WWSWWWTLWWWWRW 365
Score = 33.1 bits (74), Expect = 0.040
Identities = 9/13 (69%), Positives = 9/13 (69%), Gaps = 1/13 (7%)
Frame = +3
Query: 25 WWWWQRWW-LWWW 36
W WW WW LWWW
Sbjct: 318 WRWWSWWWTLWWW 356
Score = 32.7 bits (73), Expect(2) = 8e-06
Identities = 9/13 (69%), Positives = 9/13 (69%)
Frame = +3
Query: 24 WWWWWQRWWLWWW 36
W W W RWW WWW
Sbjct: 306 WRWRW-RWWSWWW 341
Score = 32.0 bits (71), Expect(2) = 8e-06
Identities = 8/10 (80%), Positives = 8/10 (80%)
Frame = +3
Query: 78 WWRRLWWWWR 87
WW LWWWWR
Sbjct: 333 WWWTLWWWWR 362
Score = 30.8 bits (68), Expect = 0.20
Identities = 14/26 (53%), Positives = 16/26 (60%)
Frame = +1
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGD 65
G GGGG+ GG +GGGGGG D
Sbjct: 313 GGGGGGRG------GGRSGGGGGGSD 372
Score = 26.9 bits (58), Expect = 2.8
Identities = 15/31 (48%), Positives = 15/31 (48%)
Frame = +1
Query: 137 GGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGD 167
GG GGG GGG G G G GGG D
Sbjct: 304 GGGGGG--------GGGRGGGRSGGGGGGSD 372
Score = 26.2 bits (56), Expect = 4.9
Identities = 10/13 (76%), Positives = 10/13 (76%)
Frame = +1
Query: 54 GGGNGGGGGGGDR 66
GGG GGGG GG R
Sbjct: 307 GGGGGGGGRGGGR 345
Score = 25.8 bits (55), Expect = 6.3
Identities = 11/25 (44%), Positives = 13/25 (52%)
Frame = +1
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGG 64
G G ++ GGG GGGGG G
Sbjct: 262 GKNGWRVELSHNSRGGGGGGGGGRG 336
Score = 25.8 bits (55), Expect = 6.3
Identities = 11/21 (52%), Positives = 11/21 (52%)
Frame = +1
Query: 189 WRRRRRR*SDGGGGGGGANSG 209
WR S GGGGGGG G
Sbjct: 274 WRVELSHNSRGGGGGGGGGRG 336
Score = 22.7 bits (47), Expect(2) = 8e-04
Identities = 8/9 (88%), Positives = 8/9 (88%)
Frame = +1
Query: 2 GGGGGNGGR 10
GGGGG GGR
Sbjct: 307 GGGGGGGGR 333
>TC14279 similar to UP|Q09085 (Q09085) Hydroxyproline-rich glycoprotein
(HRGP) (Fragment), partial (57%)
Length = 941
Score = 42.0 bits (97), Expect(2) = 1e-05
Identities = 50/146 (34%), Positives = 56/146 (38%), Gaps = 20/146 (13%)
Frame = -1
Query: 42 GGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVG 101
GGGG+A T GGG GGGGD + V + + W+ G GGG +
Sbjct: 638 GGGGEA*M*T--GGGGDFVGGGGDL*IYVGAGEGEGGGGDV*WYGGGGNFTGGGGDL*MY 465
Query: 102 VAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDG-GGWWWWWWQ*GGGCGVGGGG 160
V A G GGG A GGGDG GG W GGG VGGGG
Sbjct: 464 VGAGD----------------GEGGGGDA*WYG-GGGDGEGGGGLT*W*GGGGDDVGGGG 336
Query: 161 D-------------------YGGGGD 167
D YGGGGD
Sbjct: 335 DL***GG*GMGDGGGGLL**YGGGGD 258
Score = 36.6 bits (83), Expect = 0.004
Identities = 27/50 (54%), Positives = 28/50 (56%), Gaps = 6/50 (12%)
Frame = -1
Query: 123 GCGGGSKAVAA**GGG---DGGGWWWWWWQ*GGGCGVGGGGD---YGGGG 166
G G G **GGG DGGG W + GGG G GGGGD YGGGG
Sbjct: 215 GDGDGGGGDL***GGGGECDGGGGDW**YG-GGGEGDGGGGDL**YGGGG 69
Score = 34.3 bits (77), Expect = 0.018
Identities = 22/71 (30%), Positives = 27/71 (37%), Gaps = 6/71 (8%)
Frame = -3
Query: 24 WWW------WWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIE 77
WWW WW R + WW G+ G G GG R VV +
Sbjct: 636 WWWGSINVNWWGRRFCWW---------GR*FVNIC-----GSG*RGGRRW*CVV----VW 511
Query: 78 WWRRLWWWWR* 88
WW + +WW R*
Sbjct: 510 WWWQFYWWGR* 478
Score = 33.5 bits (75), Expect = 0.030
Identities = 26/79 (32%), Positives = 29/79 (35%), Gaps = 7/79 (8%)
Frame = -3
Query: 16 VVVTVEIGWWWW------WQRW-WLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVV 68
VV + G WW W+RW W W W G+ V G G R
Sbjct: 327 VVGRMRNGRWWRRTFVVVWRRWRWRWRW--------GRFVIVR-----RGR*RGWWWR*F 187
Query: 69 VVVSVVAIEWWRRLWWWWR 87
VVV WWR WWW R
Sbjct: 186 VVVG----RWWRM*WWWGR 142
Score = 33.5 bits (75), Expect = 0.030
Identities = 27/94 (28%), Positives = 30/94 (31%), Gaps = 9/94 (9%)
Frame = -3
Query: 3 GGGGNGGRLVLVAVVVTVEIGWWWWWQRWW---------LWWW***GSGGGGKAVAATVE 53
G G GGR VV WWWW WW W W G + V
Sbjct: 558 GSG*RGGRRW*CVVV------WWWWQFYWWGR*FVNVCGSWRW-------GRRRW*CIVV 418
Query: 54 GGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR 87
G G ++V WRR WWWR
Sbjct: 417 WWGRRW*GWRRTHIMV-------RRWRR*RWWWR 337
Score = 32.7 bits (73), Expect = 0.052
Identities = 25/78 (32%), Positives = 27/78 (34%), Gaps = 16/78 (20%)
Frame = -3
Query: 26 WWWQ------RWWL--WWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIE 77
WWW+ RWW WWW G+ V V GG G R VV
Sbjct: 204 WWWR*FVVVGRWWRM*WWW--------GRLVV--VWGGRRG-------RWWWR*FVVVWR 76
Query: 78 WWRRL--------WWWWR 87
WW R WWW R
Sbjct: 75 WWGRFIIVWFWYGWWWRR 22
Score = 28.9 bits (63), Expect = 0.75
Identities = 20/36 (55%), Positives = 21/36 (57%), Gaps = 4/36 (11%)
Frame = -1
Query: 136 GGGDGGGWWWWWWQ*GGGC-GVGGGGDY---GGGGD 167
GGGDG G *GGG G GGGGD GGGG+
Sbjct: 269 GGGDGDGGGGDL***GGGGDGDGGGGDL***GGGGE 162
Score = 26.9 bits (58), Expect = 2.8
Identities = 18/41 (43%), Positives = 19/41 (45%), Gaps = 11/41 (26%)
Frame = -1
Query: 36 W***GSGGGGKAVAATV--EGGGNG---------GGGGGGD 65
W** G GG G + GGG G GGGGGGD
Sbjct: 140 W**YGGGGEGDGGGGDL**YGGGGGDL**YGFGTGGGGGGD 18
Score = 26.6 bits (57), Expect = 3.7
Identities = 20/74 (27%), Positives = 22/74 (29%), Gaps = 10/74 (13%)
Frame = -3
Query: 24 WW---WWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWR 80
WW WWW R + W GG R VV WW
Sbjct: 171 WWRM*WWWGRLVVVW-------------------------GGRRGRWWWR*FVVVWRWWG 67
Query: 81 R---LW----WWWR 87
R +W WWWR
Sbjct: 66 RFIIVWFWYGWWWR 25
Score = 22.3 bits (46), Expect(2) = 1e-05
Identities = 7/8 (87%), Positives = 7/8 (87%)
Frame = -3
Query: 183 VTVGRWWR 190
V VGRWWR
Sbjct: 186 VVVGRWWR 163
>TC16065 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich
glycoprotein DZ-HRGP precursor, partial (20%)
Length = 608
Score = 43.9 bits (102), Expect = 2e-05
Identities = 23/64 (35%), Positives = 24/64 (36%)
Frame = -3
Query: 24 WWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLW 83
WWWWW LWWW G A T + G R I WWRR
Sbjct: 297 WWWWWA--GLWWW-----GARWPWGATTAVWRPS*VSR*GEGRPFSARRARWI*WWRRRI 139
Query: 84 WWWR 87
WWWR
Sbjct: 138 WWWR 127
Score = 34.7 bits (78), Expect = 0.014
Identities = 40/131 (30%), Positives = 43/131 (32%), Gaps = 4/131 (3%)
Frame = -1
Query: 40 GSGG----GGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGG 95
G GG GG+ +GG GG GGGG G G GGG
Sbjct: 371 GGGGPGPQGGRGYGGPPQGGRGGGYGGGG------------------------GPGYGGG 264
Query: 96 RAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCG 155
GV G GG S+ GG GG GGG G
Sbjct: 263 GRGGHGVPQQQYGGPPEYQ------GRGRGGPSQQ------GGRGG-------YSGGGGG 141
Query: 156 VGGGGDYGGGG 166
GGGG GGGG
Sbjct: 140 YGGGGGRGGGG 108
Score = 31.2 bits (69), Expect = 0.15
Identities = 8/8 (100%), Positives = 8/8 (100%)
Frame = -3
Query: 79 WRRLWWWW 86
WRRLWWWW
Sbjct: 309 WRRLWWWW 286
Score = 28.5 bits (62), Expect = 0.98
Identities = 8/17 (47%), Positives = 10/17 (58%), Gaps = 4/17 (23%)
Frame = -3
Query: 24 WW----WWWQRWWLWWW 36
WW WWW+R WW+
Sbjct: 156 WWRRRIWWWRRPGRWWY 106
Score = 27.7 bits (60), Expect = 1.7
Identities = 26/75 (34%), Positives = 30/75 (39%), Gaps = 2/75 (2%)
Frame = -1
Query: 137 GGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR* 196
GG GGG+ GGG G GGGG G G + P GR ++
Sbjct: 317 GGRGGGYGG-----GGGPGYGGGGRGGHGVPQQQYGG-------PPEYQGRGRGGPSQQG 174
Query: 197 SDGG--GGGGGANSG 209
GG GGGGG G
Sbjct: 173 GRGGYSGGGGGYGGG 129
Score = 26.2 bits (56), Expect = 4.9
Identities = 15/28 (53%), Positives = 15/28 (53%)
Frame = -1
Query: 41 SGGGGKAVAATVEGGGNGGGGGGGDRVV 68
SGGGG GGG GGGG G R V
Sbjct: 158 SGGGGGYGG----GGGRGGGGMGSGRGV 87
Score = 25.4 bits (54), Expect = 8.3
Identities = 20/60 (33%), Positives = 21/60 (34%), Gaps = 3/60 (5%)
Frame = -1
Query: 40 GSGGGGKAVAAT---VEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGR 96
G G GG + GGG G GGGGG R GGG G GR
Sbjct: 206 GRGRGGPSQQGGRGGYSGGGGGYGGGGG----------------------RGGGGMGSGR 93
>CN825556
Length = 655
Score = 41.6 bits (96), Expect(2) = 2e-05
Identities = 29/65 (44%), Positives = 30/65 (45%), Gaps = 21/65 (32%)
Frame = -3
Query: 123 GCGG--------GSKAVAA**---GGGDGGGWWWWW---------WQ*GGGCGVG-GGGD 161
GCGG GS A + * GGG G GW W W W *G GCG G G G
Sbjct: 638 GCGGFCSGSPGEGSGASSG*GRDAGGGTGWGWGWGWGWG*GWG*GWG*GCGCGCGCGCGS 459
Query: 162 YGGGG 166
GGGG
Sbjct: 458 EGGGG 444
Score = 30.0 bits (66), Expect = 0.34
Identities = 17/40 (42%), Positives = 18/40 (44%)
Frame = -3
Query: 23 GWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGGG 62
GW W W W W *G G G + GGG G GGG
Sbjct: 542 GWGWGWG*GWG*GW-G*GCGCGCGCGCGSEGGGGTGCGGG 426
Score = 28.1 bits (61), Expect = 1.3
Identities = 33/112 (29%), Positives = 37/112 (32%), Gaps = 12/112 (10%)
Frame = -3
Query: 34 WWW***G---SGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEW-WRRLWWW---- 85
WW+* G SG G+ A+ G + GGG G W W W W
Sbjct: 650 WWF*GCGGFCSGSPGEGSGASSG*GRDAGGGTG--------------WGWGWGWGWG*GW 513
Query: 86 ---WR*GGGCGGG-RAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA 133
W *G GCG G G T GCGGGS A
Sbjct: 512 G*GWG*GCGCGCGCGCGSEGGGGT-----------------GCGGGSTGAKA 408
Score = 27.7 bits (60), Expect = 1.7
Identities = 17/42 (40%), Positives = 17/42 (40%)
Frame = -3
Query: 23 GWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGGGGG 64
GW W W W W * G G G GG G G GGG
Sbjct: 548 GWGWGWG-WG*GWG*GWG*GCGCGCGCGCGSEGGGGTGCGGG 426
Score = 21.6 bits (44), Expect(2) = 2e-05
Identities = 13/35 (37%), Positives = 15/35 (42%)
Frame = -2
Query: 187 RWWRRRRRR*SDGGGGGGGANSGVSWQQ*RVVVMV 221
RW RR+ + GANS S *R V V
Sbjct: 432 RWQHRRKSTHAKNAQPSNGANSPTSTTV*RAQVCV 328
>TC15267 similar to UP|EXTN_TOBAC (P13983) Extensin precursor (Cell wall
hydroxyproline-rich glycoprotein), partial (6%)
Length = 1003
Score = 43.5 bits (101), Expect = 3e-05
Identities = 23/66 (34%), Positives = 26/66 (38%), Gaps = 19/66 (28%)
Frame = -2
Query: 16 VVVTVEIGWWWW-------------------WQRWWLWWW***GSGGGGKAVAATVEGGG 56
VVV + WWWW W+ W LW W GGG V + G G
Sbjct: 258 VVVVCAVAWWWWWCGGGGWRG*KPRRE*RRGWRGWRLWRW--KLYGGGVDCVVVGLRGCG 85
Query: 57 NGGGGG 62
GGGGG
Sbjct: 84 GGGGGG 67
Score = 30.8 bits (68), Expect = 0.20
Identities = 9/33 (27%), Positives = 15/33 (45%), Gaps = 6/33 (18%)
Frame = -1
Query: 10 RLVLVAVVVTVEIGWW------WWWQRWWLWWW 36
R+ + ++ + WW W + WW WWW
Sbjct: 169 RMARMEIMAVEALRWWC*LRCCWSSRLWWWWWW 71
Score = 30.8 bits (68), Expect = 0.20
Identities = 10/25 (40%), Positives = 14/25 (56%), Gaps = 8/25 (32%)
Frame = -1
Query: 70 VVSVVAIEWW--------RRLWWWW 86
+++V A+ WW RLWWWW
Sbjct: 151 IMAVEALRWWC*LRCCWSSRLWWWW 77
Score = 30.8 bits (68), Expect = 0.20
Identities = 13/25 (52%), Positives = 14/25 (56%)
Frame = -2
Query: 70 VVSVVAIEWWRRLWWWWR*GGGCGG 94
VV V A+ WW WWW GGG G
Sbjct: 258 VVVVCAVAWW----WWWCGGGGWRG 196
Score = 30.8 bits (68), Expect = 0.20
Identities = 8/10 (80%), Positives = 8/10 (80%)
Frame = -1
Query: 24 WWWWWQRWWL 33
WWWWW WWL
Sbjct: 88 WWWWW--WWL 65
Score = 30.4 bits (67), Expect = 0.26
Identities = 48/148 (32%), Positives = 55/148 (36%), Gaps = 4/148 (2%)
Frame = -3
Query: 42 GGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVG 101
G GG+ V EG G G GG VVV GGG GG G
Sbjct: 419 GDGGELVHGGAEGEGEDGLWVGG----VVV-----------------GGGDGGD----AG 315
Query: 102 VAATSVVAVVAAMVV----VVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVG 157
V S VA A V ++L GGG V GG+G Q G
Sbjct: 314 VREASDVADGADGVG*VERLLLFVRWLGGGGGVVEE---GGEGES------QGESEEEDG 162
Query: 158 GGGDYGGGGDKVVVAAIMVVVATPVVTV 185
GDYGGG VVV +++V VV V
Sbjct: 161 EDGDYGGGSFTVVVLIALLLVFEVVVVV 78
Score = 30.0 bits (66), Expect = 0.34
Identities = 6/6 (100%), Positives = 6/6 (100%)
Frame = -1
Query: 143 WWWWWW 148
WWWWWW
Sbjct: 85 WWWWWW 68
Score = 30.0 bits (66), Expect = 0.34
Identities = 6/6 (100%), Positives = 6/6 (100%)
Frame = -1
Query: 143 WWWWWW 148
WWWWWW
Sbjct: 88 WWWWWW 71
Score = 29.6 bits (65), Expect = 0.44
Identities = 24/83 (28%), Positives = 24/83 (28%), Gaps = 21/83 (25%)
Frame = -2
Query: 144 WWWWWQ*GGGCGVGGGGD---------------------YGGGGDKVVVAAIMVVVATPV 182
WWWWW CG GG YGGG D VVV
Sbjct: 234 WWWWW-----CGGGGWRG*KPRRE*RRGWRGWRLWRWKLYGGGVDCVVVGLR-------- 94
Query: 183 VTVGRWWRRRRRR*SDGGGGGGG 205
GGGGGGG
Sbjct: 93 --------------GCGGGGGGG 67
Score = 28.9 bits (63), Expect = 0.75
Identities = 23/77 (29%), Positives = 24/77 (30%), Gaps = 15/77 (19%)
Frame = -2
Query: 143 WWWWWWQ*GG---------------GCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGR 187
WWWWW GG G + YGGG D VVV
Sbjct: 234 WWWWWCGGGGWRG*KPRRE*RRGWRGWRLWRWKLYGGGVDCVVVGL-------------- 97
Query: 188 WWRRRRRR*SDGGGGGG 204
R GGGGGG
Sbjct: 96 -------RGCGGGGGGG 67
Score = 26.9 bits (58), Expect = 2.8
Identities = 30/106 (28%), Positives = 38/106 (35%), Gaps = 26/106 (24%)
Frame = -3
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWW--WR*GGG------ 91
G G G V V GGG+GG G + V + RL + W GGG
Sbjct: 383 GEGEDGLWVGGVVVGGGDGGDAGVREASDVADGADGVG*VERLLLFVRWLGGGGGVVEEG 204
Query: 92 ------------------CGGGRAAVVGVAATSVVAVVAAMVVVVL 119
GGG VV + A +V V +VVVV+
Sbjct: 203 GEGESQGESEEEDGEDGDYGGGSFTVVVLIALLLVFEVVVVVVVVV 66
Score = 25.8 bits (55), Expect = 6.3
Identities = 18/39 (46%), Positives = 19/39 (48%), Gaps = 3/39 (7%)
Frame = -2
Query: 185 VGRW---WRRRRRR*SDGGGGGGGANSGVSWQQ*RVVVM 220
VGRW WRRRRRR G G S * VVV+
Sbjct: 363 VGRWGGRWRRRRRRCRSPGSE*RCRRCGWSRLS*EVVVV 247
>TC9854 weakly similar to UP|Q9LSN7 (Q9LSN7) Gb|AAC24081.1
(AT3g17100/K14A17_22_), partial (43%)
Length = 1108
Score = 43.5 bits (101), Expect = 3e-05
Identities = 11/14 (78%), Positives = 11/14 (78%)
Frame = -2
Query: 23 GWWWWWQRWWLWWW 36
GWWWWW WW WWW
Sbjct: 48 GWWWWW--WWWWWW 13
Score = 33.9 bits (76), Expect = 0.023
Identities = 8/11 (72%), Positives = 9/11 (81%)
Frame = -2
Query: 138 GDGGGWWWWWW 148
G+G WWWWWW
Sbjct: 54 GEGWWWWWWWW 22
Score = 32.7 bits (73), Expect = 0.052
Identities = 7/10 (70%), Positives = 8/10 (80%)
Frame = -2
Query: 24 WWWWWQRWWL 33
WWWWW WW+
Sbjct: 36 WWWWWWWWWV 7
Score = 30.4 bits (67), Expect = 0.26
Identities = 10/20 (50%), Positives = 10/20 (50%)
Frame = -2
Query: 129 KAVAA**GGGDGGGWWWWWW 148
K V G G WWWWWW
Sbjct: 75 KVVTGIRGEGWWWWWWWWWW 16
Score = 30.0 bits (66), Expect = 0.34
Identities = 6/6 (100%), Positives = 6/6 (100%)
Frame = -2
Query: 143 WWWWWW 148
WWWWWW
Sbjct: 30 WWWWWW 13
Score = 30.0 bits (66), Expect = 0.34
Identities = 6/6 (100%), Positives = 6/6 (100%)
Frame = -2
Query: 143 WWWWWW 148
WWWWWW
Sbjct: 27 WWWWWW 10
Score = 28.1 bits (61), Expect = 1.3
Identities = 10/12 (83%), Positives = 11/12 (91%)
Frame = -3
Query: 53 EGGGNGGGGGGG 64
+GGG GGGGGGG
Sbjct: 50 KGGGGGGGGGGG 15
Score = 28.1 bits (61), Expect = 1.3
Identities = 8/18 (44%), Positives = 9/18 (49%)
Frame = -2
Query: 69 VVVSVVAIEWWRRLWWWW 86
VV + WW WWWW
Sbjct: 72 VVTGIRGEGWWWWWWWWW 19
Score = 27.7 bits (60), Expect = 1.7
Identities = 10/11 (90%), Positives = 10/11 (90%)
Frame = -3
Query: 54 GGGNGGGGGGG 64
GGG GGGGGGG
Sbjct: 44 GGGGGGGGGGG 12
Score = 27.7 bits (60), Expect = 1.7
Identities = 10/11 (90%), Positives = 10/11 (90%)
Frame = -3
Query: 54 GGGNGGGGGGG 64
GGG GGGGGGG
Sbjct: 41 GGGGGGGGGGG 9
Score = 26.9 bits (58), Expect = 2.8
Identities = 6/9 (66%), Positives = 6/9 (66%)
Frame = -2
Query: 78 WWRRLWWWW 86
WW WWWW
Sbjct: 39 WWWWWWWWW 13
Score = 26.9 bits (58), Expect = 2.8
Identities = 6/9 (66%), Positives = 6/9 (66%)
Frame = -2
Query: 78 WWRRLWWWW 86
WW WWWW
Sbjct: 42 WWWWWWWWW 16
Score = 26.9 bits (58), Expect(2) = 0.33
Identities = 6/9 (66%), Positives = 6/9 (66%)
Frame = -2
Query: 78 WWRRLWWWW 86
WW WWWW
Sbjct: 36 WWWWWWWWW 10
Score = 26.6 bits (57), Expect = 3.7
Identities = 10/12 (83%), Positives = 10/12 (83%)
Frame = -3
Query: 53 EGGGNGGGGGGG 64
E GG GGGGGGG
Sbjct: 53 EKGGGGGGGGGG 18
Score = 26.2 bits (56), Expect = 4.9
Identities = 9/14 (64%), Positives = 9/14 (64%)
Frame = -3
Query: 199 GGGGGGGANSGVSW 212
GGGGGGG G W
Sbjct: 47 GGGGGGGGGGGGGW 6
Score = 21.6 bits (44), Expect(2) = 0.33
Identities = 8/13 (61%), Positives = 9/13 (68%)
Frame = -3
Query: 52 VEGGGNGGGGGGG 64
+E GGGGGGG
Sbjct: 65 LESEEKGGGGGGG 27
>TC9962 similar to UP|Q8L7Z6 (Q8L7Z6) AT3g54680/T5N23_40, partial (21%)
Length = 585
Score = 41.2 bits (95), Expect = 1e-04
Identities = 10/13 (76%), Positives = 11/13 (83%)
Frame = -1
Query: 22 IGWWWWWQRWWLW 34
+GWWWWW WWLW
Sbjct: 162 MGWWWWWCSWWLW 124
Score = 36.2 bits (82), Expect = 0.005
Identities = 31/107 (28%), Positives = 37/107 (33%), Gaps = 2/107 (1%)
Frame = -1
Query: 43 GGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWR--RLWWWWR*GGGCGGGRAAVV 100
G ++ V GGG+ G G G VVV+V WW R+W W G G G G
Sbjct: 402 GEARSRPIPVRGGGDPGHVGSGWVGKVVVAVAVQAWWGA*RVWEWVGSGRGSGHG----- 238
Query: 101 GVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWW 147
V V + K G GWWWWW
Sbjct: 237 -------VCWVRKQQNRKRKRMKKKEQEK--------GSWMGWWWWW 142
Score = 30.8 bits (68), Expect = 0.20
Identities = 25/85 (29%), Positives = 30/85 (34%)
Frame = -1
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGG 61
GG G+ G + VVV V + WW R W W GSG G V N
Sbjct: 366 GGDPGHVGSGWVGKVVVAVAVQAWWGA*RVWEWV----GSGRGSGHGVCWVRKQQNRKRK 199
Query: 62 GGGDRVVVVVSVVAIEWWRRLWWWW 86
+ S + WW WW W
Sbjct: 198 RMKKKEQEKGSWMGWWWWWCSWWLW 124
Score = 25.8 bits (55), Expect = 6.3
Identities = 23/69 (33%), Positives = 25/69 (35%)
Frame = -2
Query: 141 GGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGG 200
G W W W GG G G G G A V + G RRR +R G
Sbjct: 329 GRWLWLWRCRRGGVRDGFG---SGSGQDEDPAMGFVGCGNSRIGSGSV*RRRSKRRVLGW 159
Query: 201 GGGGGANSG 209
GGGG G
Sbjct: 158 VGGGGGVLG 132
>TC18216 similar to GB|CAA28991.1|34041|HSKER65A keratin type II (AA1-215)
{Homo sapiens;} , partial (16%)
Length = 501
Score = 34.7 bits (78), Expect(2) = 2e-04
Identities = 9/18 (50%), Positives = 10/18 (55%), Gaps = 5/18 (27%)
Frame = +1
Query: 24 WWWWWQR-----WWLWWW 36
WWW W WW+WWW
Sbjct: 46 WWWVWCSCCSH*WWIWWW 99
Score = 31.2 bits (69), Expect = 0.15
Identities = 10/22 (45%), Positives = 11/22 (49%), Gaps = 9/22 (40%)
Frame = +1
Query: 24 WWWW--WQRWWLW-------WW 36
WW W W RWW+W WW
Sbjct: 121 WWIWRSWLRWWVWGI*QPRKWW 186
Score = 30.8 bits (68), Expect = 0.20
Identities = 9/21 (42%), Positives = 11/21 (51%), Gaps = 8/21 (38%)
Frame = +1
Query: 24 WWWWWQ---RWWLW-----WW 36
WWW+W WW+W WW
Sbjct: 91 WWWFWCCSFSWWIWRSWLRWW 153
Score = 27.3 bits (59), Expect(2) = 0.13
Identities = 23/52 (44%), Positives = 24/52 (45%), Gaps = 8/52 (15%)
Frame = +2
Query: 125 GGGSKAVAA**GGGDGGGWWWWWWQ*G-GGCGVGGG-------GDYGGGGDK 168
GGG A AA GG GGG+ G GG G GGG G G GG K
Sbjct: 47 GGGFGAAAAPTSGGFGGGFGAVPSAGGFGGAGSGGGFGAFSNPGSGGFGGSK 202
Score = 25.4 bits (54), Expect(2) = 2e-04
Identities = 13/25 (52%), Positives = 14/25 (56%)
Frame = +2
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGG 64
G GGG AV + GG G GGG G
Sbjct: 86 GFGGGFGAVPSAGGFGGAGSGGGFG 160
Score = 22.7 bits (47), Expect(2) = 0.13
Identities = 5/9 (55%), Positives = 5/9 (55%)
Frame = +1
Query: 78 WWRRLWWWW 86
WW WWW
Sbjct: 28 WWCCFHWWW 54
>TC14184 similar to UP|Q93WZ6 (Q93WZ6) Abscisic stress ripening-like
protein, partial (57%)
Length = 1141
Score = 36.6 bits (83), Expect = 0.004
Identities = 19/71 (26%), Positives = 19/71 (26%), Gaps = 7/71 (9%)
Frame = +2
Query: 24 WW-WWW--QRW--WLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEW 78
WW WWW RW W WWW W
Sbjct: 383 WWRWWWILHRWRLWHWWW----------------------------------------RW 442
Query: 79 WRRL--WWWWR 87
WR WWWWR
Sbjct: 443 WRIFYRWWWWR 475
Score = 33.9 bits (76), Expect = 0.023
Identities = 11/26 (42%), Positives = 11/26 (42%), Gaps = 13/26 (50%)
Frame = +2
Query: 24 WWW---------WWQRWW----LWWW 36
WWW WW RWW WWW
Sbjct: 392 WWWILHRWRLWHWWWRWWRIFYRWWW 469
Score = 32.7 bits (73), Expect = 0.052
Identities = 11/21 (52%), Positives = 12/21 (56%), Gaps = 8/21 (38%)
Frame = +2
Query: 24 WWW-WWQ---RWWLW----WW 36
WWW WW+ RWW W WW
Sbjct: 428 WWWRWWRIFYRWWWWRILHWW 490
Score = 32.3 bits (72), Expect = 0.068
Identities = 8/14 (57%), Positives = 9/14 (64%), Gaps = 2/14 (14%)
Frame = +2
Query: 25 WWWWQ--RWWLWWW 36
WWWW+ WW W W
Sbjct: 461 WWWWRILHWWWWLW 502
Score = 30.0 bits (66), Expect = 0.34
Identities = 8/13 (61%), Positives = 8/13 (61%)
Frame = +2
Query: 24 WWWWWQRWWLWWW 36
WWWW W WWW
Sbjct: 461 WWWWRILHW-WWW 496
Score = 29.6 bits (65), Expect = 0.44
Identities = 17/35 (48%), Positives = 18/35 (50%), Gaps = 4/35 (11%)
Frame = +3
Query: 136 GGGDGGGWWWWWWQ*GGGCGVGGGG----DYGGGG 166
GGG G G + GGG G GGGG GGGG
Sbjct: 366 GGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGG 470
Score = 28.9 bits (63), Expect = 0.75
Identities = 15/29 (51%), Positives = 17/29 (57%), Gaps = 3/29 (10%)
Frame = +3
Query: 40 GSGGGGKAVAATVEGGGN---GGGGGGGD 65
G+GGGG +T GGG GGGG G D
Sbjct: 423 GTGGGGGGEYSTGGGGGGYSTGGGGYGND 509
Score = 27.3 bits (59), Expect(3) = 8e-04
Identities = 19/51 (37%), Positives = 22/51 (42%), Gaps = 4/51 (7%)
Frame = +3
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGG----DYGGGGDKV 169
G GG + G G GGG + GGG GGGG D G D+V
Sbjct: 384 GGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNRRDEV 536
Score = 27.3 bits (59), Expect = 2.2
Identities = 13/25 (52%), Positives = 15/25 (60%)
Frame = +3
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGG 64
G+GG G + GGG G GGGGG
Sbjct: 378 GTGGAGGGYST---GGGYGTGGGGG 443
Score = 26.9 bits (58), Expect = 2.8
Identities = 12/26 (46%), Positives = 14/26 (53%)
Frame = +3
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGD 65
G G GG + GG GGGGGG+
Sbjct: 372 GYGTGGAGGGYSTGGGYGTGGGGGGE 449
Score = 25.8 bits (55), Expect(3) = 8e-04
Identities = 8/10 (80%), Positives = 8/10 (80%)
Frame = +2
Query: 78 WWRRLWWWWR 87
WWR LW WWR
Sbjct: 365 WWR-LWHWWR 391
Score = 23.1 bits (48), Expect(3) = 8e-04
Identities = 8/25 (32%), Positives = 9/25 (36%), Gaps = 13/25 (52%)
Frame = +2
Query: 25 WWWWQRWWL-------------WWW 36
+WWW WL WWW
Sbjct: 239 FWWWL*NWLQQDIIILY**A*FWWW 313
>BP049447
Length = 478
Score = 38.5 bits (88), Expect = 0.001
Identities = 17/34 (50%), Positives = 18/34 (52%)
Frame = -1
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWW 35
GGGGG GG + E G WWWW WW WW
Sbjct: 247 GGGGGGGG------LEGRFEKGCWWWW--WW-WW 173
Score = 37.4 bits (85), Expect = 0.002
Identities = 14/26 (53%), Positives = 14/26 (53%), Gaps = 10/26 (38%)
Frame = -1
Query: 136 GGGDGGG----------WWWWWWQ*G 151
GGG GGG WWWWWW *G
Sbjct: 244 GGGGGGGLEGRFEKGCWWWWWWWW*G 167
Score = 36.2 bits (82), Expect = 0.005
Identities = 15/32 (46%), Positives = 15/32 (46%), Gaps = 7/32 (21%)
Frame = -1
Query: 124 CGGGSKAVAA**GGGDG-------GGWWWWWW 148
CGGG GGG G G WWWWWW
Sbjct: 250 CGGG--------GGGGGLEGRFEKGCWWWWWW 179
Score = 35.8 bits (81), Expect = 0.006
Identities = 19/63 (30%), Positives = 21/63 (33%)
Frame = -1
Query: 27 WWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWW 86
WW + WW G GGGG + E G WW WWWW
Sbjct: 286 WWVNFGSGWWWCCGGGGGGGGLEGRFEKG--------------------CWWW---WWWW 176
Query: 87 R*G 89
*G
Sbjct: 175 W*G 167
Score = 32.3 bits (72), Expect = 0.068
Identities = 12/34 (35%), Positives = 12/34 (35%), Gaps = 19/34 (55%)
Frame = -1
Query: 23 GWWW-------------------WWQRWWLWWW* 37
GWWW WW WW WWW*
Sbjct: 265 GWWWCCGGGGGGGGLEGRFEKGCWW--WWWWWW* 170
Score = 30.4 bits (67), Expect = 0.26
Identities = 6/7 (85%), Positives = 7/7 (99%)
Frame = -3
Query: 143 WWWWWWQ 149
WWWWWW+
Sbjct: 245 WWWWWWR 225
Score = 30.0 bits (66), Expect = 0.34
Identities = 14/28 (50%), Positives = 14/28 (50%), Gaps = 6/28 (21%)
Frame = -1
Query: 139 DGGGWW------WWWWQ*GGGCGVGGGG 160
DG WW WWW GGG GGGG
Sbjct: 298 DGCSWWVNFGSGWWWCCGGGG---GGGG 224
Score = 29.3 bits (64), Expect = 0.57
Identities = 8/13 (61%), Positives = 9/13 (68%)
Frame = -3
Query: 18 VTVEIGWWWWWQR 30
V V + WWWWW R
Sbjct: 263 VVVVLWWWWWWWR 225
Score = 28.5 bits (62), Expect = 0.98
Identities = 19/50 (38%), Positives = 21/50 (42%)
Frame = -2
Query: 161 DYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGV 210
D GGGG VVV + V+ VG GGGGGG GV
Sbjct: 270 DLGGGGVVVVVVVVEVLKGDLRRVVG-------------GGGGGGGEEGV 160
Score = 28.5 bits (62), Expect = 0.98
Identities = 8/19 (42%), Positives = 10/19 (52%)
Frame = -3
Query: 18 VTVEIGWWWWWQRWWLWWW 36
+ V + WWWW WWW
Sbjct: 269 IWVVVVLWWWW-----WWW 228
Score = 28.1 bits (61), Expect = 1.3
Identities = 8/18 (44%), Positives = 10/18 (55%), Gaps = 3/18 (16%)
Frame = -3
Query: 22 IGWWWWWQ---RWWLWWW 36
+G +W W WW WWW
Sbjct: 284 VGEFWIWVVVVLWWWWWW 231
Score = 27.7 bits (60), Expect = 1.7
Identities = 11/24 (45%), Positives = 13/24 (53%)
Frame = -1
Query: 140 GGGWWWWWWQ*GGGCGVGGGGDYG 163
G GWWW GGG G+ G + G
Sbjct: 271 GSGWWWCCGGGGGGGGLEGRFEKG 200
Score = 27.7 bits (60), Expect = 1.7
Identities = 8/13 (61%), Positives = 9/13 (68%)
Frame = -3
Query: 17 VVTVEIGWWWWWQ 29
VV V WWWWW+
Sbjct: 263 VVVVLWWWWWWWR 225
>BI420337
Length = 397
Score = 38.1 bits (87), Expect = 0.001
Identities = 25/87 (28%), Positives = 29/87 (32%), Gaps = 8/87 (9%)
Frame = -2
Query: 9 GRLVLVAVVVTVEIGWWWWWQRWWL--------WWW***GSGGGGKAVAATVEGGGNGGG 60
GR VLV ++ WWWWW R + WWW
Sbjct: 228 GRRVLVWLL------WWWWW*RVMVRRRR*VHWWWW------------------------ 139
Query: 61 GGGGDRVVVVVSVVAIEWWRRLWWWWR 87
RVV+V RLWWWWR
Sbjct: 138 -----RVVIV----------RLWWWWR 103
Score = 37.7 bits (86), Expect = 0.002
Identities = 23/88 (26%), Positives = 30/88 (33%), Gaps = 24/88 (27%)
Frame = -2
Query: 23 GWWW-------WWQRW---WLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVS 72
GWWW WW+RW W+ WW R +++VS
Sbjct: 372 GWWWRLVLVNWWWRRWVLVWVLWW---------------------------RRRRLIIVS 274
Query: 73 -------VVAIEW-WRR------LWWWW 86
+V + W W R LWWWW
Sbjct: 273 WCTWWWRLVLVHWRWGRRVLVWLLWWWW 190
Score = 30.4 bits (67), Expect(2) = 0.006
Identities = 20/48 (41%), Positives = 24/48 (49%), Gaps = 5/48 (10%)
Frame = -3
Query: 125 GGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGD-----YGGGGD 167
GGG + GGG G W GGG +GGGG+ +GGGGD
Sbjct: 233 GGGEGYLYGFFGGGGGEE*W*G----GGGECIGGGGEW***GFGGGGD 102
Score = 26.2 bits (56), Expect = 4.9
Identities = 7/14 (50%), Positives = 8/14 (57%), Gaps = 6/14 (42%)
Frame = -2
Query: 142 GWWW------WWWQ 149
GWWW WWW+
Sbjct: 372 GWWWRLVLVNWWWR 331
Score = 24.3 bits (51), Expect(2) = 0.006
Identities = 9/20 (45%), Positives = 11/20 (55%), Gaps = 5/20 (25%)
Frame = -2
Query: 73 VVAIEWWRRLW---W--WWR 87
+V + WW R W W WWR
Sbjct: 357 LVLVNWWWRRWVLVWVLWWR 298
>AU251550
Length = 329
Score = 37.0 bits (84), Expect = 0.003
Identities = 19/74 (25%), Positives = 25/74 (33%), Gaps = 7/74 (9%)
Frame = -2
Query: 20 VEIGWWW--WWQRWWLW-----WW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVS 72
++ GW W WW WW W WW G ++V R
Sbjct: 256 IKHGWRWGRWWAMWWWWMVMPEWW-----WWGSRSVRI--------------PRWTWRWW 134
Query: 73 VVAIEWWRRLWWWW 86
++ W R WWWW
Sbjct: 133 IIVTRWLWRTWWWW 92
Score = 31.2 bits (69), Expect = 0.15
Identities = 42/135 (31%), Positives = 44/135 (32%), Gaps = 8/135 (5%)
Frame = -3
Query: 40 GSGGGGKAVAATVEGGGNGGG-----GGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGG 94
G G GG G G GGG GGGGD GG CGG
Sbjct: 321 GPGPGGCGGCGGPGGPGGGGG**NMAGGGGD-----------------------GGPCGG 211
Query: 95 GRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGC 154
G G GGG+ A + GG GGG * GGC
Sbjct: 210 GGW*CPN---------------------GGGGGAGA*GS--RGGPGGGGL*----*PGGC 112
Query: 155 GVGGGGDY---GGGG 166
G GGG GGGG
Sbjct: 111 GGPGGGGLW*PGGGG 67
Score = 31.2 bits (69), Expect = 0.15
Identities = 22/46 (47%), Positives = 23/46 (49%), Gaps = 2/46 (4%)
Frame = -3
Query: 123 GCGGGSKAVAA**GGGDGG--GWWWWWWQ*GGGCGVGGGGDYGGGG 166
G GGG +A GGGDGG G W GGG G G G GG G
Sbjct: 273 GGGGG**NMAG--GGGDGGPCGGGGW*CPNGGGGGAGA*GSRGGPG 142
Score = 27.3 bits (59), Expect = 2.2
Identities = 9/27 (33%), Positives = 10/27 (36%), Gaps = 14/27 (51%)
Frame = -2
Query: 24 WW-----WWWQRWWLW---------WW 36
WW W W+ WW W WW
Sbjct: 139 WWIIVTRWLWRTWWWWIMVTRWRWTWW 59
>BP030058
Length = 455
Score = 35.8 bits (81), Expect = 0.006
Identities = 9/14 (64%), Positives = 10/14 (71%), Gaps = 1/14 (7%)
Frame = +3
Query: 24 WWWWW-QRWWLWWW 36
WWWWW + WW WW
Sbjct: 369 WWWWW*RGWWRRWW 410
Score = 34.7 bits (78), Expect = 0.014
Identities = 10/21 (47%), Positives = 10/21 (47%), Gaps = 8/21 (38%)
Frame = +3
Query: 24 WWWWWQRW--------WLWWW 36
WW WW RW W WWW
Sbjct: 315 WWRWWGRWRWWRWRWAWQWWW 377
Score = 33.9 bits (76), Expect = 0.023
Identities = 26/73 (35%), Positives = 28/73 (37%), Gaps = 4/73 (5%)
Frame = +1
Query: 136 GGGDGGGWWWWWWQ*GGGCGVGGGGDY----GGGGDKVVVAAIMVVVATPVVTVGRWWRR 191
GGG+GGG GGG G GGGGD GGGG+
Sbjct: 301 GGGEGGGG-------GGGDGGGGGGDGHGSGGGGGE------------------------ 387
Query: 192 RRRR*SDGGGGGG 204
DGGGGGG
Sbjct: 388 -----GDGGGGGG 411
Score = 33.1 bits (74), Expect = 0.040
Identities = 14/26 (53%), Positives = 16/26 (60%)
Frame = +1
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGD 65
G GGGG +G G+GGGGG GD
Sbjct: 316 GGGGGGDGGGGGGDGHGSGGGGGEGD 393
Score = 31.6 bits (70), Expect = 0.12
Identities = 8/13 (61%), Positives = 10/13 (76%), Gaps = 3/13 (23%)
Frame = +3
Query: 24 WWW---WWQRWWL 33
WWW WW+RWW+
Sbjct: 375 WWW*RGWWRRWWV 413
Score = 30.8 bits (68), Expect = 0.20
Identities = 20/42 (47%), Positives = 21/42 (49%)
Frame = +1
Query: 125 GGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGG 166
GGG + GGGDGGG G G G GGG GGGG
Sbjct: 298 GGGGEGGGG--GGGDGGGGGGD----GHGSGGGGGEGDGGGG 405
Score = 30.8 bits (68), Expect(2) = 0.079
Identities = 15/29 (51%), Positives = 17/29 (57%), Gaps = 5/29 (17%)
Frame = +1
Query: 42 GGGGKAVAATVE-----GGGNGGGGGGGD 65
GGG K + + GGG GGGGGGGD
Sbjct: 250 GGGDKGLQFCGQEGYFGGGGEGGGGGGGD 336
Score = 30.4 bits (67), Expect = 0.26
Identities = 15/29 (51%), Positives = 18/29 (61%), Gaps = 1/29 (3%)
Frame = +1
Query: 40 GSGGGGKAVAATVEGG-GNGGGGGGGDRV 67
G GGGG + GG G+GGGGGG +V
Sbjct: 337 GGGGGGDGHGSGGGGGEGDGGGGGGSPQV 423
Score = 30.0 bits (66), Expect = 0.34
Identities = 18/86 (20%), Positives = 19/86 (21%), Gaps = 23/86 (26%)
Frame = +3
Query: 24 WWWWWQ----------------RWWLWW--W***GSGGGGKAVAATVEGGGNGGGGGGGD 65
WWWW Q RWW WW W
Sbjct: 246 WWWW*QGVAILWAGGVLRRRR*RWWRWWGRW----------------------------- 338
Query: 66 RVVVVVSVVAIEWWR-----RLWWWW 86
WWR + WWWW
Sbjct: 339 -----------RWWRWRWAWQWWWWW 383
Score = 30.0 bits (66), Expect = 0.34
Identities = 9/15 (60%), Positives = 9/15 (60%), Gaps = 1/15 (6%)
Frame = +3
Query: 24 WWWWWQRW-WLWWW* 37
WW W W W WWW*
Sbjct: 342 WWRWRWAWQWWWWW* 386
Score = 29.6 bits (65), Expect = 0.44
Identities = 20/48 (41%), Positives = 25/48 (51%), Gaps = 3/48 (6%)
Frame = +1
Query: 122 IGCGGGSKAVAA**GGGDGGGWWWW---WWQ*GGGCGVGGGGDYGGGG 166
+GC G + GGGD G + ++ GG G GGGGD GGGG
Sbjct: 217 LGCPGTYMILG---GGGDKGLQFCGQEGYFGGGGEGGGGGGGDGGGGG 351
Score = 28.5 bits (62), Expect = 0.98
Identities = 13/25 (52%), Positives = 13/25 (52%)
Frame = +1
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGG 64
G GGGG GGG G GGGG
Sbjct: 331 GDGGGGGGDGHGSGGGGGEGDGGGG 405
Score = 28.1 bits (61), Expect = 1.3
Identities = 20/63 (31%), Positives = 21/63 (32%)
Frame = +1
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGG 61
GGGGG+GG G GG G GGG G GG
Sbjct: 319 GGGGGDGG------------------------------GGGGDGHGSGG---GGGEGDGG 399
Query: 62 GGG 64
GGG
Sbjct: 400 GGG 408
Score = 26.6 bits (57), Expect = 3.7
Identities = 8/14 (57%), Positives = 8/14 (57%), Gaps = 1/14 (7%)
Frame = +3
Query: 24 WWWWWQRWW-LWWW 36
W WWW WW WW
Sbjct: 363 WQWWW--WW*RGWW 398
Score = 20.0 bits (40), Expect(2) = 0.079
Identities = 11/21 (52%), Positives = 14/21 (66%)
Frame = +2
Query: 99 VVGVAATSVVAVVAAMVVVVL 119
VVG A VA+ A+VVVV+
Sbjct: 323 VVGEMAVVAVAMGMAVVVVVV 385
>AW719349
Length = 143
Score = 32.7 bits (73), Expect = 0.052
Identities = 17/80 (21%), Positives = 19/80 (23%), Gaps = 13/80 (16%)
Frame = +2
Query: 24 WWWWWQ-------------RWWLWWW***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVV 70
WW WW WWLWW
Sbjct: 5 WWLWWWLRRLWWRLWKYDEEWWLWW----------------------------------- 79
Query: 71 VSVVAIEWWRRLWWWWR*GG 90
WRR+WW W+ GG
Sbjct: 80 ------RLWRRVWWLWKYGG 121
Score = 31.2 bits (69), Expect = 0.15
Identities = 10/22 (45%), Positives = 11/22 (49%), Gaps = 9/22 (40%)
Frame = +2
Query: 24 WWWWWQR----WWLW-----WW 36
WW WW+ WWLW WW
Sbjct: 65 WWLWWRLWRRVWWLWKYGGKWW 130
Score = 28.9 bits (63), Expect = 0.75
Identities = 26/80 (32%), Positives = 28/80 (34%), Gaps = 4/80 (5%)
Frame = +3
Query: 90 GGCGGGRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ 149
GGCGGG GCGGG ++ GG GGG
Sbjct: 6 GGCGGGCG-------------------------GCGGGCGSMMK--SGGCGGGC------ 86
Query: 150 *GGGCGVGG----GGDYGGG 165
GGGCG G G GGG
Sbjct: 87 -GGGCGGCGNMVESGGCGGG 143
Score = 26.9 bits (58), Expect = 2.8
Identities = 20/57 (35%), Positives = 23/57 (40%), Gaps = 1/57 (1%)
Frame = +3
Query: 40 GSGGG-GKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGG 95
G GGG G + + GGG GGG GG +V GGCGGG
Sbjct: 30 GCGGGCGSMMKSGGCGGGCGGGCGGCGNMVE-------------------SGGCGGG 143
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.338 0.154 0.636
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,971,372
Number of Sequences: 28460
Number of extensions: 124236
Number of successful extensions: 16850
Number of sequences better than 10.0: 854
Number of HSP's better than 10.0 without gapping: 4466
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 12085
length of query: 224
length of database: 4,897,600
effective HSP length: 87
effective length of query: 137
effective length of database: 2,421,580
effective search space: 331756460
effective search space used: 331756460
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 53 (25.0 bits)
Lotus: description of TM0320b.3


