

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
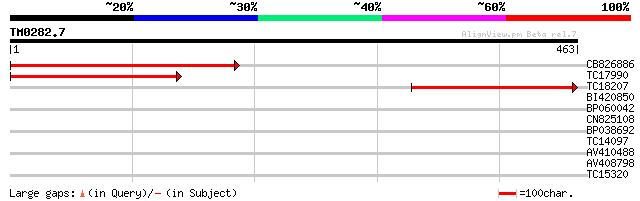
Query= TM0282.7
(463 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB826886 393 e-110
TC17990 similar to GB|AAM65958.1|21594040|AY088421 homogentisate... 299 6e-82
TC18207 similar to UP|Q9M6U1 (Q9M6U1) Homogentisate 1,2-dioxygen... 291 1e-79
BI420850 30 0.84
BP060042 29 1.9
CN825108 28 2.5
BP038692 28 2.5
TC14097 homologue to UP|GCSP_PEA (P26969) Glycine dehydrogenase ... 28 3.2
AV410488 27 5.5
AV408798 27 7.1
TC15320 weakly similar to GB|AAN28760.1|23505801|AY143821 At3g51... 27 9.3
>CB826886
Length = 631
Score = 393 bits (1009), Expect = e-110
Identities = 186/187 (99%), Positives = 186/187 (99%)
Frame = +3
Query: 1 MEHSTAGDDFNYLSGFGNHFSSEALAGALPEAQNSPLICPYGLYAEQISGTSFTSPRNLN 60
MEHSTAGDDFNYLSGFGNHFSSEALAGALPEAQNSPLICPYGLYAEQISGTSFTSP NLN
Sbjct: 69 MEHSTAGDDFNYLSGFGNHFSSEALAGALPEAQNSPLICPYGLYAEQISGTSFTSPWNLN 248
Query: 61 LFSWFYRVKPSVTHEPFKPRVPCNGRILSEFNHSNSSTNPTQLRWKPMDSPDSPTDFIDG 120
LFSWFYRVKPSVTHEPFKPRVPCNGRILSEFNHSNSSTNPTQLRWKPMDSPDSPTDFIDG
Sbjct: 249 LFSWFYRVKPSVTHEPFKPRVPCNGRILSEFNHSNSSTNPTQLRWKPMDSPDSPTDFIDG 428
Query: 121 LSTVCGSGSSFMRHGYAIHMYTANKSMNNCAFCNADGDFLIVPQQGRLFITTECGRLKVS 180
LSTVCGSGSSFMRHGYAIHMYTANKSMNNCAFCNADGDFLIVPQQGRLFITTECGRLKVS
Sbjct: 429 LSTVCGSGSSFMRHGYAIHMYTANKSMNNCAFCNADGDFLIVPQQGRLFITTECGRLKVS 608
Query: 181 PGEIAML 187
PGEIAML
Sbjct: 609 PGEIAML 629
>TC17990 similar to GB|AAM65958.1|21594040|AY088421 homogentisate
1,2-dioxygenase {Arabidopsis thaliana;} , partial (28%)
Length = 561
Score = 299 bits (766), Expect = 6e-82
Identities = 140/140 (100%), Positives = 140/140 (100%)
Frame = +2
Query: 1 MEHSTAGDDFNYLSGFGNHFSSEALAGALPEAQNSPLICPYGLYAEQISGTSFTSPRNLN 60
MEHSTAGDDFNYLSGFGNHFSSEALAGALPEAQNSPLICPYGLYAEQISGTSFTSPRNLN
Sbjct: 83 MEHSTAGDDFNYLSGFGNHFSSEALAGALPEAQNSPLICPYGLYAEQISGTSFTSPRNLN 262
Query: 61 LFSWFYRVKPSVTHEPFKPRVPCNGRILSEFNHSNSSTNPTQLRWKPMDSPDSPTDFIDG 120
LFSWFYRVKPSVTHEPFKPRVPCNGRILSEFNHSNSSTNPTQLRWKPMDSPDSPTDFIDG
Sbjct: 263 LFSWFYRVKPSVTHEPFKPRVPCNGRILSEFNHSNSSTNPTQLRWKPMDSPDSPTDFIDG 442
Query: 121 LSTVCGSGSSFMRHGYAIHM 140
LSTVCGSGSSFMRHGYAIHM
Sbjct: 443 LSTVCGSGSSFMRHGYAIHM 502
>TC18207 similar to UP|Q9M6U1 (Q9M6U1) Homogentisate 1,2-dioxygenase
(Fragment) , partial (23%)
Length = 583
Score = 291 bits (746), Expect = 1e-79
Identities = 135/135 (100%), Positives = 135/135 (100%)
Frame = +3
Query: 329 VAEHTFRPPYYHRNCMSEFMGLIHGGYEAKVDGFLPGGASLHSCMTPHGPDTKSYEATIA 388
VAEHTFRPPYYHRNCMSEFMGLIHGGYEAKVDGFLPGGASLHSCMTPHGPDTKSYEATIA
Sbjct: 3 VAEHTFRPPYYHRNCMSEFMGLIHGGYEAKVDGFLPGGASLHSCMTPHGPDTKSYEATIA 182
Query: 389 RGNDVGPNKITDTMAFMFESCLIPRISRWALESPFLDHDYYQCWIGLRSHFNVTEISPRT 448
RGNDVGPNKITDTMAFMFESCLIPRISRWALESPFLDHDYYQCWIGLRSHFNVTEISPRT
Sbjct: 183 RGNDVGPNKITDTMAFMFESCLIPRISRWALESPFLDHDYYQCWIGLRSHFNVTEISPRT 362
Query: 449 PTLDSETVAGDTKSS 463
PTLDSETVAGDTKSS
Sbjct: 363 PTLDSETVAGDTKSS 407
>BI420850
Length = 419
Score = 30.0 bits (66), Expect = 0.84
Identities = 17/57 (29%), Positives = 27/57 (46%), Gaps = 5/57 (8%)
Frame = +3
Query: 402 MAFMFESCLIPRISRWALESPFLDHDYYQ-----CWIGLRSHFNVTEISPRTPTLDS 453
+ F F++ + R W++ + + YQ CWI RSHF+V + R L S
Sbjct: 219 LGFKFKA*ICSRT*IWSINANSIRLHEYQGTSTRCWIQQRSHFHVRGVEARHQALSS 389
>BP060042
Length = 353
Score = 28.9 bits (63), Expect = 1.9
Identities = 16/38 (42%), Positives = 19/38 (49%)
Frame = +2
Query: 337 PYYHRNCMSEFMGLIHGGYEAKVDGFLPGGASLHSCMT 374
P R SEF+GL GG +AK GFL + C T
Sbjct: 149 PMNVRCIRSEFLGLNGGGSKAKQRGFLSCDVNTRKCWT 262
>CN825108
Length = 655
Score = 28.5 bits (62), Expect = 2.5
Identities = 21/63 (33%), Positives = 32/63 (50%), Gaps = 1/63 (1%)
Frame = -2
Query: 267 PFNVVAWRGNYVPYKYDLSKFCPYN-TVLFDHSDPSINTVLTAPTDKPGVALLDFVIFPP 325
P NVVA YVP + ++ P + T L H SI T+L+ + + LL ++ P
Sbjct: 195 PVNVVAKLKPYVPIRALFTRIIPTDVTTLNIHDATSIATLLSPLVSEDMLNLLPYL---P 25
Query: 326 RWL 328
+WL
Sbjct: 24 KWL 16
>BP038692
Length = 405
Score = 28.5 bits (62), Expect = 2.5
Identities = 13/33 (39%), Positives = 19/33 (57%), Gaps = 3/33 (9%)
Frame = +1
Query: 308 APTDKPGVALL---DFVIFPPRWLVAEHTFRPP 337
+P+D P LL D+ +F P WL+ H +PP
Sbjct: 280 SPSDFPSDPLLQMGDYQMFEPSWLMKYHRVQPP 378
>TC14097 homologue to UP|GCSP_PEA (P26969) Glycine dehydrogenase
[decarboxylating], mitochondrial precursor (Glycine
decarboxylase) (Glycine cleavage system P-protein) ,
partial (97%)
Length = 3660
Score = 28.1 bits (61), Expect = 3.2
Identities = 16/54 (29%), Positives = 22/54 (40%)
Frame = +3
Query: 92 NHSNSSTNPTQLRWKPMDSPDSPTDFIDGLSTVCGSGSSFMRHGYAIHMYTANK 145
NH+ S P+QL + S P D + S + F +H HM NK
Sbjct: 3117 NHTPGSAQPSQLHGSGLPSSGLPQDVLTTCMVTATSSAPFSQH----HMLLKNK 3266
>AV410488
Length = 428
Score = 27.3 bits (59), Expect = 5.5
Identities = 7/12 (58%), Positives = 12/12 (99%)
Frame = +1
Query: 332 HTFRPPYYHRNC 343
H+FRPP++H++C
Sbjct: 259 HSFRPPHFHKHC 294
>AV408798
Length = 324
Score = 26.9 bits (58), Expect = 7.1
Identities = 15/45 (33%), Positives = 21/45 (46%)
Frame = +3
Query: 71 SVTHEPFKPRVPCNGRILSEFNHSNSSTNPTQLRWKPMDSPDSPT 115
+ + PF P + RI + N + S NP L P + P SPT
Sbjct: 66 TTNNNPFNPFSKVSPRIPTPTNGFDFSRNPFSLSGLPKNHPISPT 200
>TC15320 weakly similar to GB|AAN28760.1|23505801|AY143821
At3g51730/T18N14_110 {Arabidopsis thaliana;}, partial
(52%)
Length = 1195
Score = 26.6 bits (57), Expect = 9.3
Identities = 14/45 (31%), Positives = 25/45 (55%)
Frame = -1
Query: 4 STAGDDFNYLSGFGNHFSSEALAGALPEAQNSPLICPYGLYAEQI 48
S DF + SGF + +S+A A NSP++ P+ +++ +I
Sbjct: 184 SAQTSDFLFNSGFASSLASQAQAAPSTIRNNSPIL-PFMVHSHEI 53
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.139 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,443,689
Number of Sequences: 28460
Number of extensions: 151226
Number of successful extensions: 634
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 633
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 634
length of query: 463
length of database: 4,897,600
effective HSP length: 93
effective length of query: 370
effective length of database: 2,250,820
effective search space: 832803400
effective search space used: 832803400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0282.7


