

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
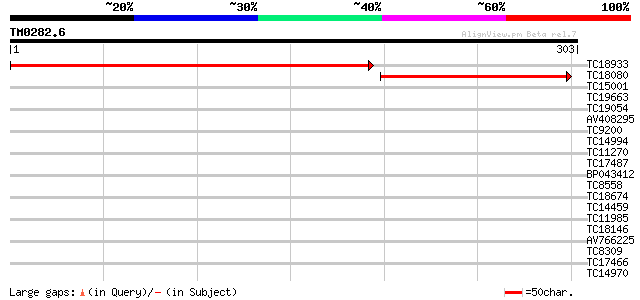
Query= TM0282.6
(303 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC18933 similar to GB|AAL90935.1|19699138|AY090274 AT3g18790/MVE... 396 e-111
TC18080 similar to GB|AAL90935.1|19699138|AY090274 AT3g18790/MVE... 197 2e-51
TC15001 similar to UP|GLE1_YEAST (Q12315) RNA export factor GLE1... 37 0.003
TC19663 similar to UP|Q8H6S8 (Q8H6S8) Translation initiation fac... 31 0.23
TC19054 similar to UP|Q8L7T5 (Q8L7T5) At1g07990/T6D22_5, partial... 30 0.39
AV408295 30 0.39
TC9200 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1, ... 30 0.50
TC14994 similar to UP|O82469 (O82469) Protein phosphatase-2C , p... 29 0.86
TC11270 similar to UP|Q9LM88 (Q9LM88) F2D10.15, partial (9%) 28 1.9
TC17487 UP|CAE45597 (CAE45597) SAR DNA-binding protein-like prot... 27 3.3
BP043412 27 3.3
TC8558 weakly similar to UP|Q9NDI0 (Q9NDI0) 200 kDa antigen p200... 27 3.3
TC18674 similar to UP|Q7Y0V3 (Q7Y0V3) Actin filament bundling pr... 27 4.3
TC14459 homologue to UP|Q93YH3 (Q93YH3) ATP citrate lyase b-subu... 27 4.3
TC11985 similar to GB|AAF40452.1|7211981|F13M7 ESTs gb|N65605, g... 27 4.3
TC18146 similar to UP|Q9FFK7 (Q9FFK7) Signal recognition particl... 27 5.6
AV766225 27 5.6
TC8309 UP|O22306 (O22306) Nlj21, complete 27 5.6
TC17466 similar to UP|Q9M427 (Q9M427) Oligouridylate binding pro... 26 7.3
TC14970 similar to UP|O24294 (O24294) Legumin (Minor small) prec... 26 7.3
>TC18933 similar to GB|AAL90935.1|19699138|AY090274 AT3g18790/MVE11_17
{Arabidopsis thaliana;}, partial (63%)
Length = 609
Score = 396 bits (1017), Expect = e-111
Identities = 194/194 (100%), Positives = 194/194 (100%)
Frame = +2
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE
Sbjct: 26 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 205
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP
Sbjct: 206 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 385
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL
Sbjct: 386 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 565
Query: 181 EPSAEAAMRREAAE 194
EPSAEAAMRREAAE
Sbjct: 566 EPSAEAAMRREAAE 607
>TC18080 similar to GB|AAL90935.1|19699138|AY090274 AT3g18790/MVE11_17
{Arabidopsis thaliana;}, partial (24%)
Length = 585
Score = 197 bits (500), Expect = 2e-51
Identities = 102/102 (100%), Positives = 102/102 (100%)
Frame = +3
Query: 199 LDRIRQEARKGVKSGEVAEVSAAVREILREEEEDVVEEERMRGREMKERLEENEREFVVH 258
LDRIRQEARKGVKSGEVAEVSAAVREILREEEEDVVEEERMRGREMKERLEENEREFVVH
Sbjct: 3 LDRIRQEARKGVKSGEVAEVSAAVREILREEEEDVVEEERMRGREMKERLEENEREFVVH 182
Query: 259 VPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAKDMLN 300
VPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAKDMLN
Sbjct: 183 VPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAKDMLN 308
>TC15001 similar to UP|GLE1_YEAST (Q12315) RNA export factor GLE1, partial
(4%)
Length = 988
Score = 37.4 bits (85), Expect = 0.003
Identities = 22/72 (30%), Positives = 39/72 (53%)
Frame = +2
Query: 217 EVSAAVREILREEEEDVVEEERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKK 276
E + V E +R+ E+ + E ++ E++ RLEE + +V V + EKE E ++E ++
Sbjct: 395 ETAKRVEEAIRKRVEESLNSEEVQV-EIQRRLEEGRKRLIVEVAVQLEKEKEAALIEARE 571
Query: 277 MELLNKYVSEDL 288
E + EDL
Sbjct: 572 KEEQARKEKEDL 607
>TC19663 similar to UP|Q8H6S8 (Q8H6S8) Translation initiation factor,
partial (10%)
Length = 485
Score = 31.2 bits (69), Expect = 0.23
Identities = 27/74 (36%), Positives = 36/74 (48%), Gaps = 2/74 (2%)
Frame = +3
Query: 227 REEEEDV--VEEERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYV 284
REEEE + EEER R E++ + EE R +KE EK L KKK E K +
Sbjct: 258 REEEEKLRKEEEERRRQEELERQAEEARRR---------KKEREKEKLLKKKQE--GKLL 404
Query: 285 SEDLMDEQSEAKDM 298
+ +EQ + M
Sbjct: 405 TGKQKEEQRRLEAM 446
>TC19054 similar to UP|Q8L7T5 (Q8L7T5) At1g07990/T6D22_5, partial (22%)
Length = 746
Score = 30.4 bits (67), Expect = 0.39
Identities = 16/37 (43%), Positives = 26/37 (70%)
Frame = -1
Query: 212 SGEVAEVSAAVREILREEEEDVVEEERMRGREMKERL 248
+GE AE+ A+ + + EEE +V EEER R ++ K++L
Sbjct: 245 TGEEAEIEASFQNMWLEEEGEV-EEERGRWKKQKQKL 138
>AV408295
Length = 426
Score = 30.4 bits (67), Expect = 0.39
Identities = 23/59 (38%), Positives = 31/59 (51%), Gaps = 1/59 (1%)
Frame = +3
Query: 211 KSGEVAEVSAAVREILREEEEDVVEEERMRG-REMKERLEENEREFVVHVPLPDEKEIE 268
K EVA +AA E + EEEE EEE+ E KE + E E++ P +EK+ E
Sbjct: 150 KKDEVA--AAATEEKVEEEEEKKTEEEKKPAEEEKKEEVVEEEKK-----PAEEEKKAE 305
>TC9200 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1, partial
(31%)
Length = 727
Score = 30.0 bits (66), Expect = 0.50
Identities = 22/69 (31%), Positives = 34/69 (48%), Gaps = 1/69 (1%)
Frame = +1
Query: 214 EVAEVSAAVRE-ILREEEEDVVEEERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVL 272
E EV V E + EEEE+ EEE E +E EE E E D++ I+K++
Sbjct: 193 EYEEVEEEVEEEVEEEEEEEEEEEE----GEEEEEEEEEEEEVEAEAEEEDDEPIQKLLE 360
Query: 273 EKKKMELLN 281
K ++++
Sbjct: 361 PMGKEQIIS 387
>TC14994 similar to UP|O82469 (O82469) Protein phosphatase-2C , partial
(89%)
Length = 1672
Score = 29.3 bits (64), Expect = 0.86
Identities = 15/29 (51%), Positives = 18/29 (61%)
Frame = +2
Query: 225 ILREEEEDVVEEERMRGREMKERLEENER 253
I REEEE+V EEE E +E EE E+
Sbjct: 152 IAREEEEEVEEEEEEEEEEEEEEEEEAEK 238
>TC11270 similar to UP|Q9LM88 (Q9LM88) F2D10.15, partial (9%)
Length = 735
Score = 28.1 bits (61), Expect = 1.9
Identities = 36/140 (25%), Positives = 53/140 (37%), Gaps = 24/140 (17%)
Frame = +3
Query: 168 GYRDDEDGILERLEPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILR 227
G R + ++E+L E E EE R +R E E E
Sbjct: 288 GGRKKKLKLVEKLNQGVEEEEEDEEDEEEQTPHPTRGRSRVEHAQEEDEEEENGEEE--E 461
Query: 228 EEEEDVVEEERMRGREMKERLEENEREFVVHV------------------------PLPD 263
EEEE VVE+E +G E ++ EE E VV V PLPD
Sbjct: 462 EEEEVVVEDE--QGNEDEDEDEEEEGVVVVEVKGRKVDSKGLHSGSGTPSKVAYVIPLPD 635
Query: 264 EKEIEKMVLEKKKMELLNKY 283
++ +E ++ + +K + Y
Sbjct: 636 KRTLELILDKLQKKDTYGVY 695
>TC17487 UP|CAE45597 (CAE45597) SAR DNA-binding protein-like protein, complete
Length = 1794
Score = 27.3 bits (59), Expect = 3.3
Identities = 17/55 (30%), Positives = 28/55 (50%), Gaps = 6/55 (10%)
Frame = +1
Query: 230 EEDVVEEERMRGREMKERLEENEREFVVHVPL------PDEKEIEKMVLEKKKME 278
+ED E + +E KE+ ++ E+ VPL +EKE+ K +KK+ E
Sbjct: 1492 DEDTHEPSADKKKEKKEKKKKKEKNEEKDVPLLADEDGDEEKEVVKKEKKKKRKE 1656
>BP043412
Length = 467
Score = 27.3 bits (59), Expect = 3.3
Identities = 13/33 (39%), Positives = 17/33 (51%)
Frame = -3
Query: 223 REILREEEEDVVEEERMRGREMKERLEENEREF 255
RE L E+D E + RGR K ++ NE F
Sbjct: 372 REFLLAAEDDECEGNQPRGRRSKGKIPRNEANF 274
>TC8558 weakly similar to UP|Q9NDI0 (Q9NDI0) 200 kDa antigen p200
(Fragment), partial (4%)
Length = 585
Score = 27.3 bits (59), Expect = 3.3
Identities = 23/74 (31%), Positives = 30/74 (40%)
Frame = +1
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEEDVVEEERMR 240
E S R AEE R+R+E ++ E A REE+E + E
Sbjct: 4 ESSPTLVAARLDAEERRNNIRLREEQDAAYRAA--LEADQARERQRREEQERLAREAAEA 177
Query: 241 GREMKERLEENERE 254
R+ KE E ERE
Sbjct: 178 ERKRKEEEEARERE 219
>TC18674 similar to UP|Q7Y0V3 (Q7Y0V3) Actin filament bundling protein
P-115-ABP, partial (17%)
Length = 952
Score = 26.9 bits (58), Expect = 4.3
Identities = 16/54 (29%), Positives = 26/54 (47%)
Frame = +2
Query: 98 PNYTRHSAKMTDLDGNIVDVPNPSGRGPGYRYFGAAKKLPGVRELFDKPPELRK 151
P+ ++ S++ + + V V GR P + A + PG R L PP +RK
Sbjct: 29 PDKSQRSSRSMSVSPDRVRV---RGRSPAFNALAANFENPGARNLSTPPPVVRK 181
>TC14459 homologue to UP|Q93YH3 (Q93YH3) ATP citrate lyase b-subunit ,
partial (48%)
Length = 777
Score = 26.9 bits (58), Expect = 4.3
Identities = 18/48 (37%), Positives = 24/48 (49%)
Frame = +2
Query: 71 LRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVP 118
+R L ++ KL K H V GGPNY + AKM L G + +P
Sbjct: 401 IRALKEKEQKLKEAKMHL---YVRRGGPNYQKGLAKMRAL-GEEIGIP 532
>TC11985 similar to GB|AAF40452.1|7211981|F13M7 ESTs gb|N65605, gb|N38087,
gb|T20485, gb|T13726, gb|N38339, gb|F15440 and gb|N97201
come from this gene. {Arabidopsis thaliana;}, partial
(88%)
Length = 1334
Score = 26.9 bits (58), Expect = 4.3
Identities = 23/88 (26%), Positives = 40/88 (45%), Gaps = 11/88 (12%)
Frame = +2
Query: 189 RREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEEDV-----------VEEE 237
R++ EE R++R R++ R + + E R EE+ EEE
Sbjct: 500 RKKKEEEEKRMEREREKER--------IRIGKELLEAKRNEEDKERKRWLA*RKAEKEEE 655
Query: 238 RMRGREMKERLEENEREFVVHVPLPDEK 265
R ++K++LEE++ E + LP E+
Sbjct: 656 RRAREKIKQKLEEDKAERRRKLGLPPEE 739
>TC18146 similar to UP|Q9FFK7 (Q9FFK7) Signal recognition particle
receptor beta subunit-like protein, partial (28%)
Length = 463
Score = 26.6 bits (57), Expect = 5.6
Identities = 13/49 (26%), Positives = 22/49 (44%)
Frame = +2
Query: 47 RQQIMREIGRKVAEIQNEGLGEHRLRDLNDEINKLIREKSHWERRIVEL 95
RQ +R+ R E EHR N+ ++ ++E HW+ + L
Sbjct: 23 RQICIRQPPRTAEETSRRRNSEHRTPHHNETLDFTMQELEHWKEELSRL 169
>AV766225
Length = 334
Score = 26.6 bits (57), Expect = 5.6
Identities = 26/84 (30%), Positives = 32/84 (37%), Gaps = 17/84 (20%)
Frame = +3
Query: 208 KGVKSGEVAEVSAAVREILREEEEDVVEEER------------MRGREMKERLEENER-- 253
K K + V V E EEEE VV R +R RE EN
Sbjct: 18 KRKKKNNIKTVVVVVPE--EEEEESVVFNSRSHLRLRFRSCSVVRERERMADHSENSNPK 191
Query: 254 ---EFVVHVPLPDEKEIEKMVLEK 274
+ VVH PLP ++KM + K
Sbjct: 192 HRIDCVVHAPLPYSSVVDKMGVAK 263
>TC8309 UP|O22306 (O22306) Nlj21, complete
Length = 1063
Score = 26.6 bits (57), Expect = 5.6
Identities = 25/97 (25%), Positives = 42/97 (42%), Gaps = 4/97 (4%)
Frame = +3
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEAR----KGVKSGEVAEVSAAVREILREEEEDVVEE 236
EP E EA +E ++ A + ++GE E + ++E +EEE V E
Sbjct: 450 EPKVEEKTDGEAKKEATETKETKESADPVEVQAPEAGEEPE-TVTLKEDASKEEEKVAEP 626
Query: 237 ERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLE 273
E E +E + E + DEK+ E+ V++
Sbjct: 627 EAKEEVVNTEAPDEKKEEEKLDTEGSDEKKEEEEVIK 737
>TC17466 similar to UP|Q9M427 (Q9M427) Oligouridylate binding protein,
partial (20%)
Length = 389
Score = 26.2 bits (56), Expect = 7.3
Identities = 12/18 (66%), Positives = 13/18 (71%)
Frame = -2
Query: 228 EEEEDVVEEERMRGREMK 245
EEEE+ EEE RG EMK
Sbjct: 58 EEEEEEEEEEEERGTEMK 5
>TC14970 similar to UP|O24294 (O24294) Legumin (Minor small) precursor,
partial (51%)
Length = 1566
Score = 26.2 bits (56), Expect = 7.3
Identities = 28/98 (28%), Positives = 39/98 (39%), Gaps = 2/98 (2%)
Frame = +1
Query: 171 DDEDGILERLEPSAEAAMR--REAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILRE 228
+D++ PS R RE E+ + RQ R+G + R RE
Sbjct: 535 EDQEQHGRHSRPSGRRTRRPSREEDEDEDEDEEERQGGRRGPGERQ--------RHFERE 690
Query: 229 EEEDVVEEERMRGREMKERLEENEREFVVHVPLPDEKE 266
EEE+ EE RGR + EE E E +P +E
Sbjct: 691 EEEER-EERGPRGRGKHWQREEEEEEEESRRTIPSRRE 801
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,757,652
Number of Sequences: 28460
Number of extensions: 42420
Number of successful extensions: 274
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 236
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 253
length of query: 303
length of database: 4,897,600
effective HSP length: 90
effective length of query: 213
effective length of database: 2,336,200
effective search space: 497610600
effective search space used: 497610600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0282.6


