

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0258c.2
(986 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
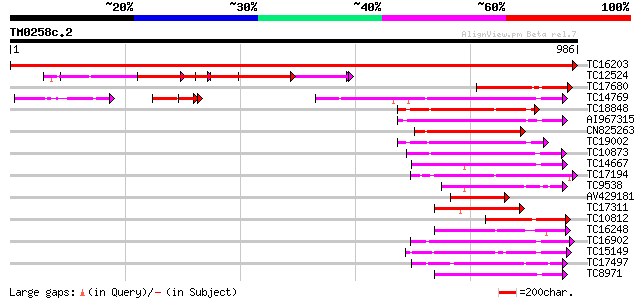
Score E
Sequences producing significant alignments: (bits) Value
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 1966 0.0
TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kina... 285 3e-77
TC17680 similar to UP|O65440 (O65440) CLV1 receptor kinase like ... 204 7e-53
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 201 4e-52
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 197 9e-51
AI967315 191 4e-49
CN825263 187 5e-48
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 187 7e-48
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 182 3e-46
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 178 4e-45
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 177 6e-45
TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein k... 176 1e-44
AV429181 176 2e-44
TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serin... 172 3e-43
TC10812 similar to UP|O65440 (O65440) CLV1 receptor kinase like ... 171 5e-43
TC16248 similar to UP|BAC87845 (BAC87845) Leucine-rich repeat re... 167 6e-42
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 166 1e-41
TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like pro... 158 5e-39
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 151 6e-37
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 150 1e-36
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 1966 bits (5093), Expect = 0.0
Identities = 986/986 (100%), Positives = 986/986 (100%)
Frame = +2
Query: 1 MRIRVSYLLVLCFTLIWFRWTVVYSSFSDLDALLKLKESMKGAKAKHHALEDWKFSTSLS 60
MRIRVSYLLVLCFTLIWFRWTVVYSSFSDLDALLKLKESMKGAKAKHHALEDWKFSTSLS
Sbjct: 110 MRIRVSYLLVLCFTLIWFRWTVVYSSFSDLDALLKLKESMKGAKAKHHALEDWKFSTSLS 289
Query: 61 AHCSFSGVTCDQNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLAS 120
AHCSFSGVTCDQNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLAS
Sbjct: 290 AHCSFSGVTCDQNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLAS 469
Query: 121 LTSLKVLNISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAG 180
LTSLKVLNISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAG
Sbjct: 470 LTSLKVLNISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAG 649
Query: 181 NYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAF 240
NYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAF
Sbjct: 650 NYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAF 829
Query: 241 GSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLS 300
GSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLS
Sbjct: 830 GSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLS 1009
Query: 301 INDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNL 360
INDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNL
Sbjct: 1010 INDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNL 1189
Query: 361 GGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVA 420
GGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVA
Sbjct: 1190 GGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVA 1369
Query: 421 NNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVISGESLGTLTLSNNLFTGKIPAAMK 480
NNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVISGESLGTLTLSNNLFTGKIPAAMK
Sbjct: 1370 NNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVISGESLGTLTLSNNLFTGKIPAAMK 1549
Query: 481 NLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSR 540
NLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSR
Sbjct: 1550 NLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSR 1729
Query: 541 NNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVPTGGQ 600
NNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVPTGGQ
Sbjct: 1730 NNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVPTGGQ 1909
Query: 601 FLVFNYDKTFAGNPNLCFPHRASCPSVLYDSLRKTRAKTARVRAIVIGIALATAVLLVAV 660
FLVFNYDKTFAGNPNLCFPHRASCPSVLYDSLRKTRAKTARVRAIVIGIALATAVLLVAV
Sbjct: 1910 FLVFNYDKTFAGNPNLCFPHRASCPSVLYDSLRKTRAKTARVRAIVIGIALATAVLLVAV 2089
Query: 661 TVHVVRKRRLHRAQAWKLTAFQRLEIKAEDVVECLKEENIIGKGGAGIVYRGSMPNGTDV 720
TVHVVRKRRLHRAQAWKLTAFQRLEIKAEDVVECLKEENIIGKGGAGIVYRGSMPNGTDV
Sbjct: 2090 TVHVVRKRRLHRAQAWKLTAFQRLEIKAEDVVECLKEENIIGKGGAGIVYRGSMPNGTDV 2269
Query: 721 AIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEW 780
AIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEW
Sbjct: 2270 AIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEW 2449
Query: 781 LHGAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFG 840
LHGAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFG
Sbjct: 2450 LHGAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFG 2629
Query: 841 LAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEF 900
LAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEF
Sbjct: 2630 LAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEF 2809
Query: 901 GDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRLSGYPLTSVIHMFNIAMMCVKEMGPAR 960
GDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRLSGYPLTSVIHMFNIAMMCVKEMGPAR
Sbjct: 2810 GDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRLSGYPLTSVIHMFNIAMMCVKEMGPAR 2989
Query: 961 PTMREVVHMLTNPPQSNTSTQDLINL 986
PTMREVVHMLTNPPQSNTSTQDLINL
Sbjct: 2990 PTMREVVHMLTNPPQSNTSTQDLINL 3067
>TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1,
partial (27%)
Length = 834
Score = 285 bits (728), Expect = 3e-77
Identities = 133/276 (48%), Positives = 191/276 (69%), Gaps = 1/276 (0%)
Frame = +2
Query: 223 ELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTG 282
EL+LGY N Y+GGIPP G++ L + A C L+GEIP LG L KL +LF+Q+N L+G
Sbjct: 2 ELYLGYYNNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSG 181
Query: 283 TIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNL 342
++ PEL + SL S+DLS N L+G++P SF++LKNLTL+N F+N+ G++P F+G++P L
Sbjct: 182 SLTPELGHLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPAL 361
Query: 343 ETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRG 402
E LQ+WENNF+ +P +LG NG+ D++ N LTG +PP +C RL+T I NF G
Sbjct: 362 EVLQLWENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLFG 541
Query: 403 PIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVIS-GESL 461
PIP+ +G+C SLT+IR+ NFL+G +P G+F LP +T E +N L+GE P S ++
Sbjct: 542 PIPESLGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVSHNI 721
Query: 462 GTLTLSNNLFTGKIPAAMKNLRALQSLSLDANEFIG 497
G +TLSNN +G +P+ + N ++Q L LD N+F G
Sbjct: 722 GQITLSNNKLSGPLPSTIGNFTSMQKLLLDGNKFSG 829
Score = 132 bits (331), Expect = 3e-31
Identities = 89/266 (33%), Positives = 131/266 (48%)
Frame = +2
Query: 88 GHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLFSGQFPGNITVGMT 147
G +PPEIG L +L + L+ ++P++L L L L + N+ SG + +
Sbjct: 35 GGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPELG-HLK 211
Query: 148 ELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSL 207
L+++D +N SG +P +L+ L L+L N G IPE E +LE L L N+
Sbjct: 212 SLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQLWENNF 391
Query: 208 TGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNL 267
TG +P+SL K L + L SN G +PP S L+ L L G IP SLG
Sbjct: 392 TGSIPQSLGKNGKLTLVDLS-SNKLTGTLPPHMCSGNRLQTLIALGNFLFGPIPESLGKC 568
Query: 268 TKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNK 327
L + + N L G+IP L + L ++ N L+GE PE+ S N+ + NK
Sbjct: 569 ESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQITLSNNK 748
Query: 328 FRGSLPSFIGDLPNLETLQVWENNFS 353
G LPS IG+ +++ L + N FS
Sbjct: 749 LSGPLPSTIGNFTSMQKLLLDGNKFS 826
Score = 124 bits (311), Expect = 7e-29
Identities = 80/271 (29%), Positives = 130/271 (47%), Gaps = 1/271 (0%)
Frame = +2
Query: 324 FQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPD 383
+ N ++G +P IG+L L S +P LG + + N L+G + P+
Sbjct: 17 YYNNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPE 196
Query: 384 LCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITEL 443
L LK+ +++N G +P E ++LT + + N L G +P V ++P++ + +L
Sbjct: 197 LGHLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQL 376
Query: 444 SNNRLNGELPSVISGESLGTLT-LSNNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGG 502
N G +P + TL LS+N TG +P M + LQ+L N G IP
Sbjct: 377 WENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLFGPIPES 556
Query: 503 VFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNL 562
+ + LT++ + N L G IP + LT V+ N L+GE P+ ++ + L
Sbjct: 557 LGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQITL 736
Query: 563 SRNEISGPVPDEIRFMTSLTTLDLSSNNFTG 593
S N++SGP+P I TS+ L L N F+G
Sbjct: 737 SNNKLSGPLPSTIGNFTSMQKLLLDGNKFSG 829
Score = 123 bits (308), Expect = 2e-28
Identities = 78/250 (31%), Positives = 126/250 (50%), Gaps = 2/250 (0%)
Frame = +2
Query: 350 NNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIG 409
NN+ +P +G + L FD L+G IP +L K +L T + N G + +G
Sbjct: 23 NNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPELG 202
Query: 410 ECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVISGE--SLGTLTLS 467
+SL + ++NN L G VP +L ++T+ L NRL+G +P + GE +L L L
Sbjct: 203 HLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFV-GEMPALEVLQLW 379
Query: 468 NNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTI 527
N FTG IP ++ L + L +N+ G +P + L + GN L GPIP ++
Sbjct: 380 ENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLFGPIPESL 559
Query: 528 THRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLS 587
SLT + + +N L G +PKG+ L L+ + N +SG P+ ++ + LS
Sbjct: 560 GKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQITLS 739
Query: 588 SNNFTGTVPT 597
+N +G +P+
Sbjct: 740 NNKLSGPLPS 769
Score = 112 bits (279), Expect = 4e-25
Identities = 76/252 (30%), Positives = 125/252 (49%), Gaps = 5/252 (1%)
Frame = +2
Query: 60 SAHCSFSG-VTCD----QNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQL 114
+A+C SG + + Q L + L V + L G L PE+G L+ L+++ +S N L+ Q+
Sbjct: 86 AAYCGLSGEIPAELGKLQKLDTLFLQVNV--LSGSLTPELGHLKSLKSMDLSNNMLSGQV 259
Query: 115 PSDLASLTSLKVLNISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLK 174
P+ A L +L +LN+ N G P M LE L ++N+F+G +P+ + K KL
Sbjct: 260 PASFAELKNLTLLNLFRNRLHGAIP-EFVGEMPALEVLQLWENNFTGSIPQSLGKNGKLT 436
Query: 175 YLHLAGNYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEG 234
+ L+ N +GT+P L+ L N L G +PESL K ++L + +G N G
Sbjct: 437 LVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLFGPIPESLGKCESLTRIRMG-QNFLNG 613
Query: 235 GIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSL 294
IP + L +E + L+GE P + + + + N L+G +P + + S+
Sbjct: 614 SIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQITLSNNKLSGPLPSTIGNFTSM 793
Query: 295 MSLDLSINDLTG 306
L L N +G
Sbjct: 794 QKLLLDGNKFSG 829
Score = 107 bits (267), Expect = 9e-24
Identities = 69/202 (34%), Positives = 103/202 (50%), Gaps = 1/202 (0%)
Frame = +2
Query: 398 NFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGEL-PSVI 456
N ++G IP IG L + A L G +P + +L + L N L+G L P +
Sbjct: 23 NNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPELG 202
Query: 457 SGESLGTLTLSNNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISG 516
+SL ++ LSNN+ +G++PA+ L+ L L+L N G IP V E+P L + +
Sbjct: 203 HLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQLWE 382
Query: 517 NNLTGPIPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIR 576
NN TG IP ++ LT VDLS N L G +P M + L L N + GP+P+ +
Sbjct: 383 NNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLFGPIPESLG 562
Query: 577 FMTSLTTLDLSSNNFTGTVPTG 598
SLT + + N G++P G
Sbjct: 563 KCESLTRIRMGQNFLNGSIPKG 628
Score = 34.3 bits (77), Expect = 0.099
Identities = 17/52 (32%), Positives = 28/52 (53%)
Frame = +2
Query: 86 LFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLFSGQ 137
L G P + + +T+S N L+ LPS + + TS++ L + N FSG+
Sbjct: 677 LSGEFPETGSVSHNIGQITLSNNKLSGPLPSTIGNFTSMQKLLLDGNKFSGR 832
>TC17680 similar to UP|O65440 (O65440) CLV1 receptor kinase like protein,
partial (16%)
Length = 685
Score = 204 bits (518), Expect = 7e-53
Identities = 108/167 (64%), Positives = 129/167 (76%), Gaps = 1/167 (0%)
Frame = +1
Query: 813 SPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTL 872
SPLIIHRDVKSNNILL+++FEAHVADFGLAKFL+D G SQ MSSIAGSYGYIAPEYAYTL
Sbjct: 13 SPLIIHRDVKSNNILLNSEFEAHVADFGLAKFLHDTGTSQCMSSIAGSYGYIAPEYAYTL 192
Query: 873 KVDEKSDVYSFGVVLLELIIGRKPVGEFG-DGVDIVGWVNKTMSELSQPSDTALVLAVVD 931
KVDEKSDVYSFGVVLLEL+ GR+PVG+FG +G++IV W +K ++ +Q V+ ++D
Sbjct: 193 KVDEKSDVYSFGVVLLELLTGRRPVGDFGEEGLNIVQW-SKVQTDWNQER----VVKILD 357
Query: 932 PRLSGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHMLTNPPQSNT 978
RL PL +F +AM+CV+E RP MREVV ML Q NT
Sbjct: 358 GRLCHIPLEEAKQVFFVAMLCVQEPSVERPNMREVVEMLAQAKQPNT 498
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 201 bits (512), Expect = 4e-52
Identities = 146/470 (31%), Positives = 233/470 (49%), Gaps = 32/470 (6%)
Frame = +3
Query: 533 LTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFT 592
+T +DLS +NL G +P + + +L LN+S N G VP + L ++DLS N+
Sbjct: 1599 ITKLDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPS-FPLSSLLISVDLSYNDLM 1775
Query: 593 GTVPTGGQFLVFNYDKTFAGNPNLCFPHRASCPSVLYDS-LRKTRAKTARVRAIVIGIAL 651
G +P L F N ++ A+ S L ++ + + K +R +++ A+
Sbjct: 1776 GKLPESIVKLPHLKSLYFGCNEHMSPEDPANMNSSLINTDYGRCKGKESRFGQVIVIGAI 1955
Query: 652 ATAVLLVAVTVHVV-----------------RKRRLHRAQAWKLTAFQRLEIKAEDV--- 691
LL+ + V+ +K + + L + IK+ +
Sbjct: 1956 TCGSLLITLAFGVLFVCRYRQKLIPWEGFAGKKYPMETNIIFSLPSKDDFFIKSVSIQAF 2135
Query: 692 ------VECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLG 745
V + + +IG+GG G VYRG++ +G +VA+K + S + F E+ L
Sbjct: 2136 TLEYIEVATERYKTLIGEGGFGSVYRGTLNDGQEVAVK-VRSSTSTQGTREFDNELNLLS 2312
Query: 746 KIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHG--AKGGHLRWEMRYKIAVEAAR 803
I+H N++ LLGY + D +L+Y +M NGSL + L+G AK L W R IA+ AAR
Sbjct: 2313 AIQHENLVPLLGYCNESDQQILVYPFMSNGSLQDRLYGEPAKRKILDWPTRLSIALGAAR 2492
Query: 804 GLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGY 863
GL Y+H +IHRD+KS+NILLD A VADFG +K+ G S + G+ GY
Sbjct: 2493 GLAYLHTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVSLEVRGTAGY 2672
Query: 864 IAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVD--IVGWVNKTMSELSQPS 921
+ PEY T ++ EKSDV+SFGVVLLE++ GR+P+ + +V W +
Sbjct: 2673 LDPEYYKTQQLSEKSDVFSFGVVLLEIVSGREPLNIKRPRTEWSLVEWATPYIR------ 2834
Query: 922 DTALVLAVVDPRL-SGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 970
+ V +VDP + GY ++ + +A+ C++ RP+M +V L
Sbjct: 2835 -GSKVDEIVDPGIKGGYHAEAMWRVVEVALQCLEPFSTYRPSMVAIVREL 2981
Score = 60.5 bits (145), Expect = 1e-09
Identities = 47/173 (27%), Positives = 79/173 (45%)
Frame = +3
Query: 9 LVLCFTLIWFRWTVVYSSFSDLDALLKLKESMKGAKAKHHALEDWKFSTSLSAHCSFSGV 68
L+ + ++ R + ++ +D+ + K++E + + + ALE W + + G+
Sbjct: 1395 LLNAYEILQVRPWIEETNQTDVGVIQKMREELLLQNSGNRALESWSGDPCILL--PWKGI 1568
Query: 69 TCDQNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLN 128
CD + N + V + L +S +NL +PS +A +T+L+ LN
Sbjct: 1569 ACDGS------NGSSV---------------ITKLDLSSSNLKGLIPSSIAEMTNLETLN 1685
Query: 129 ISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGN 181
ISHN F G P + L ++D N G LPE IVKL LK L+ N
Sbjct: 1686 ISHNSFDGSVPSFPLSSL--LISVDLSYNDLMGKLPESIVKLPHLKSLYFGCN 1838
Score = 58.2 bits (139), Expect = 6e-09
Identities = 33/80 (41%), Positives = 52/80 (64%), Gaps = 1/80 (1%)
Frame = +3
Query: 249 LEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPP-ELSSMMSLMSLDLSINDLTGE 307
L++++ NL G IP S+ +T L +L + N+ G++P LSS+ L+S+DLS NDL G+
Sbjct: 1608 LDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPSFPLSSL--LISVDLSYNDLMGK 1781
Query: 308 IPESFSKLKNLTLMNFFQNK 327
+PES KL +L + F N+
Sbjct: 1782 LPESIVKLPHLKSLYFGCNE 1841
Score = 41.2 bits (95), Expect = 8e-04
Identities = 19/42 (45%), Positives = 27/42 (64%)
Frame = +3
Query: 294 LMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSF 335
+ LDLS ++L G IP S +++ NL +N N F GS+PSF
Sbjct: 1599 ITKLDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPSF 1724
Score = 40.0 bits (92), Expect = 0.002
Identities = 23/86 (26%), Positives = 42/86 (48%)
Frame = +3
Query: 318 LTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLT 377
+T ++ + +G +PS I ++ NLETL + N+F +P + + + D++ N L
Sbjct: 1599 ITKLDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVP-SFPLSSLLISVDLSYNDLM 1775
Query: 378 GLIPPDLCKSGRLKTFIITDNFFRGP 403
G +P + K LK+ N P
Sbjct: 1776 GKLPESIVKLPHLKSLYFGCNEHMSP 1853
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 197 bits (500), Expect = 9e-51
Identities = 103/248 (41%), Positives = 155/248 (61%), Gaps = 2/248 (0%)
Frame = +2
Query: 675 AWKLTAFQRLEIKAEDVVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRND 734
+W++ ++ L ++N +G+GG G VY G +G +A+K+L S + +
Sbjct: 254 SWRIFTYKELHAATGG----FSDDNKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNS-KAE 418
Query: 735 YGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHG--AKGGHLRWE 792
F E+E LG++RH+N++ L GY D L++Y+YMPN SL LHG A L W+
Sbjct: 419 MEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQLNWQ 598
Query: 793 MRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQ 852
R KIA+ +A G+ Y+HH+ +P IIHRD+K++N+LL++DFE VADFG AK L G S
Sbjct: 599 KRMKIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAK-LIPEGVSH 775
Query: 853 SMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNK 912
+ + G+ GY+APEYA KV E DVYSFG++LLEL+ GRKP+ + + G V +
Sbjct: 776 MTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEK------LPGGVKR 937
Query: 913 TMSELSQP 920
T++E ++P
Sbjct: 938 TITEWAEP 961
>AI967315
Length = 1308
Score = 191 bits (486), Expect = 4e-49
Identities = 113/298 (37%), Positives = 166/298 (54%), Gaps = 2/298 (0%)
Frame = +1
Query: 675 AWKLTAFQRLEIKAEDVVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQ-GSGRN 733
+WK +++ L EN++GKGG VY+G + +G ++A+KRL R
Sbjct: 100 SWKCFSYEELF----HATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDERK 267
Query: 734 DYGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEM 793
+ F EI T+G + H N+M LLG + L++E GS+ +H K L W+
Sbjct: 268 EKEFLTEIGTIGHVCHSNVMPLLGCCIDNGL-YLVFELSTVGSVASLIHDEKMAPLDWKT 444
Query: 794 RYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQS 853
RYKI + ARGL Y+H C IIHRD+K++NILL DFE ++DFGLAK+L S
Sbjct: 445 RYKIVLGTARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHS 624
Query: 854 MSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNKT 913
++ I G++G++APEY VDEK+DV++FGV LLE+I GRKPV G + W
Sbjct: 625 IAPIEGTFGHLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKPVD--GSHQSLHTWAKPI 798
Query: 914 MSELSQPSDTALVLAVVDPRLSG-YPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 970
+S+ + +VDPRL G Y +T + A +C++ RPTM EV+ ++
Sbjct: 799 LSKWE-------IEKLVDPRLEGCYDVTQFNRVAFAASLCIRASSTWRPTMSEVLEVM 951
>CN825263
Length = 663
Score = 187 bits (476), Expect = 5e-48
Identities = 96/195 (49%), Positives = 131/195 (66%), Gaps = 2/195 (1%)
Frame = +1
Query: 705 GAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGYVSNKDT 764
G G+VY+G + +G DVA+K ++ + R F AE+E L ++ HRN+++L+G K T
Sbjct: 1 GFGLVYKGILNDGRDVAVK-ILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQT 177
Query: 765 NLLLYEYMPNGSLGEWLHGA--KGGHLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVK 822
L+YE +PNGS+ LHGA + G L W R KIA+ AARGL Y+H D +P +IHRD K
Sbjct: 178 RCLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFK 357
Query: 823 SNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYS 882
S+NILL+ DF V+DFGLA+ D G + + G++GY+APEYA T + KSDVYS
Sbjct: 358 SSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDVYS 537
Query: 883 FGVVLLELIIGRKPV 897
+GVVLLEL+ G KPV
Sbjct: 538 YGVVLLELLTGTKPV 582
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 187 bits (475), Expect = 7e-48
Identities = 103/265 (38%), Positives = 159/265 (59%), Gaps = 3/265 (1%)
Frame = +1
Query: 675 AWKLTAFQRLEIKAEDVVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRND 734
AW++ + + L + +N +G+GG G VY G + +G+ +A+KRL S + D
Sbjct: 16 AWRVFSLKELHSATNN----FNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLK-VWSNKAD 180
Query: 735 YGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGH--LRWE 792
F E+E L ++RH+N++ L GY + L++Y+YMPN SL LHG L W
Sbjct: 181 MEFAVEVEILARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWN 360
Query: 793 MRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQ 852
R IA+ +A G+ Y+HH +P IIHRD+K++N+LLD+DF+A VADFG AK + D GA+
Sbjct: 361 RRMNIAIGSAEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPD-GATH 537
Query: 853 SMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNK 912
+ + G+ GY+APEYA K +E DV+SFG++LLEL G+KP+ + + V +
Sbjct: 538 VTTRVKGTLGYLAPEYAMLGKANECCDVFSFGILLLELASGKKPLEK------LSSTVKR 699
Query: 913 TMSELSQPSDTALVLA-VVDPRLSG 936
++++ + P A DPRL+G
Sbjct: 700 SINDWALPLACAKKFTEFADPRLNG 774
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 182 bits (461), Expect = 3e-46
Identities = 112/283 (39%), Positives = 156/283 (54%), Gaps = 5/283 (1%)
Frame = +1
Query: 690 DVVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRH 749
D KE N+IG+GG G VY+G + G VA+K+L G + F E+ L + H
Sbjct: 436 DATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGR-QGFQEFVMEVLMLSLLHH 612
Query: 750 RNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGH--LRWEMRYKIAVEAARGLCY 807
N++RL+GY ++ D LL+YEYMP GSL + L L W R K+AV AARGL Y
Sbjct: 613 TNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEY 792
Query: 808 MHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPE 867
+H P +I+RD+KS NILLD +F ++DFGLAK + + + G+YGY APE
Sbjct: 793 LHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPE 972
Query: 868 YAYTLKVDEKSDVYSFGVVLLELIIGRKPV--GEFGDGVDIVGWVNKTMSELSQPSDTAL 925
YA + K+ KSD+YSFGVVLLEL+ GR+ + ++V W SD
Sbjct: 973 YAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYF------SDRRR 1134
Query: 926 VLAVVDPRLSG-YPLTSVIHMFNIAMMCVKEMGPARPTMREVV 967
+VDP L G +P + I MC++E RP + ++V
Sbjct: 1135FGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIV 1263
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 178 bits (451), Expect = 4e-45
Identities = 107/281 (38%), Positives = 154/281 (54%), Gaps = 10/281 (3%)
Frame = +2
Query: 700 IIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGYV 759
+IG+G G VY ++ +G VA+K+L + F ++ + ++++ N + L GY
Sbjct: 374 LIGEGSYGRVYYATLNDGNAVAVKKLDVSSEPETNNEFLTQVSMVSRLKNDNFVELHGYC 553
Query: 760 SNKDTNLLLYEYMPNGSLGEWLHGAKGGH-------LRWEMRYKIAVEAARGLCYMHHDC 812
+ +L YE+ GSL + LHG KG L W R +IAV+AARGL Y+H
Sbjct: 554 VEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQRVRIAVDAARGLEYLHEKV 733
Query: 813 SPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTL 872
P IIHRD++S+N+L+ D++A +ADF L+ D A + + G++GY APEYA T
Sbjct: 734 QPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTG 913
Query: 873 KVDEKSDVYSFGVVLLELIIGRKPVGEF--GDGVDIVGWVNKTMSELSQPSDTALVLAVV 930
++ +KSDVYSFGVVLLEL+ GRKPV +V W +SE V V
Sbjct: 914 QLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTWATPRLSE-------DKVKQCV 1072
Query: 931 DPRLSG-YPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 970
DP+L G YP V + +A +CV+ RP M VV L
Sbjct: 1073DPKLKGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKAL 1195
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 177 bits (450), Expect = 6e-45
Identities = 114/299 (38%), Positives = 168/299 (56%), Gaps = 10/299 (3%)
Frame = +1
Query: 698 ENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLG 757
+N IG+GG G VY + G AIK++ Q S F E++ L + H N++RL+G
Sbjct: 1096 DNKIGQGGFGAVYYAEL-RGKKTAIKKMDVQASTE----FLCELKVLTHVHHLNLVRLIG 1260
Query: 758 YVSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLII 817
Y + + L+YE++ NG+LG++LHG+ L W R +IA++AARGL Y+H P+ I
Sbjct: 1261 YCV-EGSLFLVYEHIDNGNLGQYLHGSGKEPLPWSSRVQIALDAARGLEYIHEHTVPVYI 1437
Query: 818 HRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEK 877
HRDVKS NIL+D + VADFGL K L + G S + + G++GY+ PEYA + K
Sbjct: 1438 HRDVKSANILIDKNLRGKVADFGLTK-LIEVGNSTLQTRLVGTFGYMPPEYAQYGDISPK 1614
Query: 878 SDVYSFGVVLLELIIGRKPVGEFGDGV-DIVGWVNKTMSELSQPSDTALVLAVVDPRL-S 935
DVY+FGVVL ELI + V + G+ V + G V L++ + +VDPRL
Sbjct: 1615 IDVYAFGVVLFELISAKNAVLKTGELVAESKGLVALFEEALNKSDPCDALRKLVDPRLGE 1794
Query: 936 GYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHMLTN--------PPQSNTSTQDLINL 986
YP+ SV+ + + C ++ RP+MR +V L +S+ +Q LINL
Sbjct: 1795 NYPIDSVLKIAQLGRACTRDNPLLRPSMRSLVVALMTLSSLTEDCDDESSYESQTLINL 1971
>TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein kinase 5
precursor , partial (10%)
Length = 1067
Score = 176 bits (447), Expect = 1e-44
Identities = 102/232 (43%), Positives = 138/232 (58%), Gaps = 12/232 (5%)
Frame = +2
Query: 751 NIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGH-----------LRWEMRYKIAV 799
NI+RLL +SN+ + LL+YEY+ N SL +WLH L W R KIA+
Sbjct: 5 NIVRLLCCISNEASMLLVYEYLENHSLDKWLHLKPKSSSVSGVVQQYTVLDWPKRLKIAI 184
Query: 800 EAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAG 859
AA+GL YMHHDCSP I+HRDVK++NILLD F A VADFGLA+ L PG MS++ G
Sbjct: 185 GAAQGLSYMHHDCSPPIVHRDVKTSNILLDKQFNAKVADFGLARMLIKPGELNIMSTVIG 364
Query: 860 SYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGD-GVDIVGWVNKTMSELS 918
++GYIAPEY T ++ EK DVYSFGVVLLEL G++ +GD + W + + S
Sbjct: 365 TFGYIAPEYVQTTRISEKVDVYSFGVVLLELTTGKE--ANYGDQHSSLAEWAWRHILIGS 538
Query: 919 QPSDTALVLAVVDPRLSGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 970
D L D + Y + + +F + +MC + RP+M+EV+ +L
Sbjct: 539 NVXD----LLXKDVMEASY-IDEMCSVFKLGVMCTATLPATRPSMKEVLPIL 679
>AV429181
Length = 313
Score = 176 bits (446), Expect = 2e-44
Identities = 81/103 (78%), Positives = 93/103 (89%)
Frame = +3
Query: 767 LLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNI 826
L+YEYM NGSLGE LHG KG L W MRYKI++++A+GLCY+HHDCSPLI+HRDVKSNNI
Sbjct: 3 LVYEYMRNGSLGEALHGKKGAFLSWNMRYKISIDSAKGLCYLHHDCSPLILHRDVKSNNI 182
Query: 827 LLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYA 869
LL+++FEAHVADFGLAKFL D GAS+ MSSIAGSYGYIAPEYA
Sbjct: 183 LLNSNFEAHVADFGLAKFLVDAGASEYMSSIAGSYGYIAPEYA 311
>TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serine-threonine
kinase 3, partial (7%)
Length = 606
Score = 172 bits (435), Expect = 3e-43
Identities = 83/166 (50%), Positives = 114/166 (68%), Gaps = 10/166 (6%)
Frame = +3
Query: 740 EIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLH----------GAKGGHL 789
E+E L IRH NI++L +S++++ LL+Y+Y+ N SL WLH + +
Sbjct: 3 EVEILSNIRHNNIVKLQCCISSEESLLLVYDYLENLSLDRWLHKKSKPPTVSGSVQNNII 182
Query: 790 RWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPG 849
W R IA+ A+GLCYMHHDCSP ++HRD+K +NILLD+ F A VADFGLA P
Sbjct: 183 DWPRRLHIAIGVAQGLCYMHHDCSPPVVHRDLKPSNILLDSQFNAKVADFGLAMMSVKPE 362
Query: 850 ASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRK 895
+MS++AG++GYIAPEYA T++V+EK DVYSFGV+LLEL G++
Sbjct: 363 ELATMSAVAGTFGYIAPEYALTIRVNEKIDVYSFGVILLELTTGKE 500
>TC10812 similar to UP|O65440 (O65440) CLV1 receptor kinase like protein,
partial (14%)
Length = 671
Score = 171 bits (433), Expect = 5e-43
Identities = 90/149 (60%), Positives = 109/149 (72%), Gaps = 1/149 (0%)
Frame = +3
Query: 828 LDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVL 887
L++DFEAH ADFGLAKF+ D G S+ MSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVL
Sbjct: 3 LNSDFEAHFADFGLAKFMQDTGTSECMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVL 182
Query: 888 LELIIGRKPVGEF-GDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRLSGYPLTSVIHMF 946
LELI GR+PVG+F +G+DIV W +++ + +V+ ++D RLS PL +F
Sbjct: 183 LELITGRRPVGDFEEEGLDIVQW-----TKMQTNWNKEMVMKILDERLSHIPLDEAEQVF 347
Query: 947 NIAMMCVKEMGPARPTMREVVHMLTNPPQ 975
+AM+CV E RPTMREVV ML Q
Sbjct: 348 FVAMLCVNEHSVERPTMREVVEMLAQAKQ 434
>TC16248 similar to UP|BAC87845 (BAC87845) Leucine-rich repeat receptor-like
protein kinase 1, partial (17%)
Length = 866
Score = 167 bits (424), Expect = 6e-42
Identities = 102/247 (41%), Positives = 147/247 (59%), Gaps = 11/247 (4%)
Frame = +2
Query: 739 AEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHG-AKGGHLRWEMRYKI 797
+EI+TL +IRHRN+++L G+ + + L+YE++ GSL + L+ + WE R +
Sbjct: 2 SEIQTLTEIRHRNVIKLHGFCLHSKFSFLVYEFLEGGSLDQVLNSDTQAAAFHWEKRVNV 181
Query: 798 AVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSI 857
A L YMHHDCSP IIHRD+ S N+LLD +EAHV+DFG AKFL PG S + ++
Sbjct: 182 VKGVANALSYMHHDCSPPIIHRDISSKNVLLDLQYEAHVSDFGTAKFL-KPG-SHTWTAF 355
Query: 858 AGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNKTMSEL 917
AG++GY APE A T++V+EK DVYSFGV LE+I+G P GD +S L
Sbjct: 356 AGTFGYAAPELAQTMQVNEKCDVYSFGVFALEIIMGMHP----GD----------LISSL 493
Query: 918 SQPSDTALV--LAVVD-----PRLSGYPL-TSVIHMFNIAMMCVKEMGPARPTMREV--V 967
PS +V L ++D P P+ VI + ++A+ C+++ +RPTM +V
Sbjct: 494 MSPSTVPMVNDLLLIDILDQRPHQVEKPIDEEVILIASLALACLRKNPHSRPTMDQVSKA 673
Query: 968 HMLTNPP 974
+L PP
Sbjct: 674 LVLGKPP 694
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 166 bits (421), Expect = 1e-41
Identities = 105/292 (35%), Positives = 158/292 (53%), Gaps = 6/292 (2%)
Frame = +3
Query: 697 EENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYG---FRAEIETLGKIRHRNIM 753
E ++G GG G VY G + GT VAIKR G+ ++ G F+ EIE L K+RHR+++
Sbjct: 3 EALLLGVGGFGKVYYGEVDGGTKVAIKR----GNPLSEQGVHEFQTEIEMLSKLRHRHLV 170
Query: 754 RLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVEAARGLCYMHHDCS 813
L+GY +L+Y++M G+L E L+ + L W+ R +I + AARGL Y+H
Sbjct: 171 SLIGYCEENTEMILVYDHMAYGTLREHLYKTQKPPLPWKQRLEICIGAARGLHYLHTGAK 350
Query: 814 PLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLK 873
IIHRDVK+ NILLD + A V+DFGL+K + + + GS+GY+ PEY +
Sbjct: 351 YTIIHRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPEYFRRQQ 530
Query: 874 VDEKSDVYSFGVVLLELIIGRKPVGE--FGDGVDIVGWVNKTMSELSQPSDTALVLAVVD 931
+ +KSDVYSFGVVL E++ R + + V + W + ++ ++ ++D
Sbjct: 531 LTDKSDVYSFGVVLFEILCARPALNPSLAKEQVSLAEWASHCYNK-------GILDQILD 689
Query: 932 PRLSGYPLTSVIHMF-NIAMMCVKEMGPARPTMREVVHMLTNPPQSNTSTQD 982
P L G F AM CV + G RP+M +V+ L Q S ++
Sbjct: 690 PYLKGKIAPECFKKFAETAMKCVSDQGIERPSMGDVLWNLEFALQLQESAEE 845
>TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like protein kinase
protein (AT3g17840/MEB5_6), partial (45%)
Length = 1489
Score = 158 bits (399), Expect = 5e-39
Identities = 106/296 (35%), Positives = 158/296 (52%), Gaps = 7/296 (2%)
Frame = +3
Query: 689 EDVVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIR 748
ED++ E ++GKG G Y+ + G VA+KRL + F+ +IET+G +
Sbjct: 264 EDLLRASAE--VLGKGTFGTAYKAVLETGLVVAVKRLKDVTISEKE--FKDKIETVGAMD 431
Query: 749 HRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGG---HLRWEMRYKIAVEAARGL 805
H++++ L Y ++D LL+Y+YMP GSL LHG KG L WE+R IA+ AARG+
Sbjct: 432 HQSLVPLRAYYFSRDEKLLVYDYMPMGSLSALLHGNKGAGRTPLNWEIRSGIALGAARGI 611
Query: 806 CYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIA 865
Y+H P + H ++K++NILL +EA V+DFGLA + G S + + +A GY A
Sbjct: 612 EYLHSQ-GPNVSHGNIKASNILLTKSYEAKVSDFGLAHLV---GPSSTPNRVA---GYRA 770
Query: 866 PEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEF--GDGVDIVGWVNKTMSELSQPSDT 923
PE KV +K+DVYSFGV+LLEL+ G+ P +GVD+ WV + E
Sbjct: 771 PEVTDPRKVSQKADVYSFGVLLLELLTGKAPTHALLNEEGVDLPRWVQSLVRE------- 929
Query: 924 ALVLAVVDPRLSGYPLT--SVIHMFNIAMMCVKEMGPARPTMREVVHMLTNPPQSN 977
V D L Y ++ + +A+ C RP++ EV + +SN
Sbjct: 930 EWTSEVFDLELLRYQNVEEEMVQLLQLAVDCAAPYPDRRPSIAEVTRSIEELCRSN 1097
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 151 bits (381), Expect = 6e-37
Identities = 91/272 (33%), Positives = 146/272 (53%)
Frame = +1
Query: 699 NIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGY 758
N +G+GG G VY+G + NG ++A+KRL SG+ F+ EI+ + +++HRN+++L G
Sbjct: 1684 NKLGEGGFGPVYKGLLANGQEIAVKRL-SNTSGQGMEEFKNEIKLIARLQHRNLVKLFGC 1860
Query: 759 VSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLIIH 818
++D N N + L + + W R +I ARGL Y+H D IIH
Sbjct: 1861 SVHQDENS-----HANKKMKILLDSTRSKLVDWNKRLQIIDGIARGLLYLHQDSRLRIIH 2025
Query: 819 RDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKS 878
RD+K++NILLD + ++DFGLA+ + G+YGY+ PEYA KS
Sbjct: 2026 RDLKTSNILLDDEMNPKISDFGLARIFIGDQVEARTKRVMGTYGYMPPEYAVHGSFSIKS 2205
Query: 879 DVYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRLSGYP 938
DV+SFGV++LE+I G+K +G F D + ++ + LV ++D +
Sbjct: 2206 DVFSFGVIVLEIISGKK-IGRFYDPHHHLNLLSHAWRLWIEERPLELVDELLDDPVIP-- 2376
Query: 939 LTSVIHMFNIAMMCVKEMGPARPTMREVVHML 970
T ++ ++A++CV+ RP M +V ML
Sbjct: 2377 -TEILRYIHVALLCVQRRPENRPDMLSIVLML 2469
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 150 bits (378), Expect = 1e-36
Identities = 89/235 (37%), Positives = 134/235 (56%), Gaps = 4/235 (1%)
Frame = +3
Query: 740 EIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAV 799
E+E L KI HRN+++LLG++ + +L+ EY+PNG+L E L G +G L + R +IA+
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHLDGLRGKILDFNQRLEIAI 185
Query: 800 EAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSS-IA 858
+ A GL Y+H IIHRDVKS+NILL A VADFG A+ G +S+ +
Sbjct: 186 DVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNGDQTHISTKVK 365
Query: 859 GSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPV--GEFGDGVDIVGWVNKTMSE 916
G+ GY+ PEY T ++ KSDVYSFG++LLE++ GR+PV + D + W + +E
Sbjct: 366 GTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAADERVTLRWAFRKYNE 545
Query: 917 LSQPSDTALVLAVVDPRLSGYPLTSVI-HMFNIAMMCVKEMGPARPTMREVVHML 970
S V+ ++DP + V+ M +++ C + RP M+ V L
Sbjct: 546 GS-------VVELMDPLMEEAVNADVLMKMLDLSFQCAAPIRTDRPNMKSVGEQL 689
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.138 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,264,427
Number of Sequences: 28460
Number of extensions: 226465
Number of successful extensions: 2360
Number of sequences better than 10.0: 383
Number of HSP's better than 10.0 without gapping: 1450
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1893
length of query: 986
length of database: 4,897,600
effective HSP length: 99
effective length of query: 887
effective length of database: 2,080,060
effective search space: 1845013220
effective search space used: 1845013220
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0258c.2


